Abstract
The aetiology of oral squamous cell carcinoma (OSCC) remains unclear. Numerous single nucleotide polymorphisms (SNPs) associated with cancer have been identified using genome-wide association studies (GWAS). The present study was designed to identify common SNPs associated with cancer susceptibility and to evaluate their involvement in OSCC. Susceptible loci were identified by analysing a cancer GWAS catalogue. A multicentre case-control study using an OSCC and control population was performed for selected SNPs. The function of the selected locus and its associated gene was explored using a reverse transcription-polymerase chain reaction, enzyme linked immunosorbent assay and immunohistochemistry. The association between genotypes and clinical parameters was assessed in 76 patients with OSCC. Rs2294008 located in the prostate stem cell antigen gene (PSCA) was selected. It was identified that the rs2294008 polymorphism was associated with OSCC susceptibility and PSCA may be involved in the development, progression and prognosis of OSCC.
Keywords: oral squamous cell carcinoma, single nucleotide polymorphism, prostate stem cell antigen gene, rs2294008
Introduction
An estimated 300,400 novel cases and 145,400 deaths from oral cavity cancer occurred in 2012 globally (1). Oral cancer is the most common head and neck cancer. Among the different types of oral cancers, the incidence of oral squamous cell carcinoma (OSCC) is the highest (1). The prevention, early diagnosis, treatment, and prognosis of OSCC remains unsatisfactory. The occurrence, development, and prognosis of OSCC results from between environmental and genetic factors interactions (2). The mechanisms underlying oral mucosal carcinogenesis remain unknown, and this hinders treatment. Researchers in the field have been focusing on exploring the mechanisms underlying oral mucosa carcinogenesis and identifying markers for OSCC diagnosis and treatment (3,4).
Genome-wide association studies (GWAS) use microarray technology and DNA samples from patients enrolled in large-scale case-control studies to identify genetic factors influencing the risk of complex diseases. GWAS does not depend on a specific gene selected based on a pathogenic hypothesis (5). Association studies of single nucleotide polymorphisms (SNPs) and complex disease phenotypes provide important genetic markers for the diagnosis and prevention of complex diseases (5–7).
The occurrence of tumours is caused by many factors influencing cell proliferation or apoptosis (8–10). Different types of tumours have some common underlying mechanisms in terms of the proteins and signalling pathways involved in the onset of the disease (11–13). For example, a type of tumour susceptibility loci may be associated with the onset of other tumour types, especially if the tumours are of the same origin (11,12,14). Thus, the susceptible loci reported in the Cancer GWAS catalogue [2012-03-09; (http://www.genome.gov/gwastudies)] (15) might be associated with the onset of OSCC (11,12).
In the present study, a meta-analysis was performed to screen common genetic susceptibility loci of malignant tumours using the GWAS catalogue, leading to the identification of the OSCC genetic susceptibility locus, rs2294008, which is located in the prostate stem cell antigen (PSCA) gene.
The influence of PSCA gene polymorphisms on the prognosis of patients with OSCC was explored through the analysis of clinical data and postoperative follow-up of patients enrolled in a case-control study. PSCA mRNA and protein expression was assessed in OSCC cell lines and the mechanisms underlying the role of PSCA in oral mucosa carcinogenesis were investigated.
Materials and methods
Screening for common SNPs associated with susceptibility for malignant tumour using GWAS data
Data retrieval
Seven cancers having the highest incidences of malignant tumour formation, including lung, breast, colorectal, upper aerodigestive tract, prostate, liver, and bladder cancer, and two cancers closely associated with oral cavity disease, skin cancer and nasopharyngeal carcinoma (NPC), were chosen for the screening of common genetic susceptibility loci, and these loci may be susceptible to OSCC as well. The official directory Catalogue of GWAS studies (2012-03-09) was downloaded, and relevant GWAS data on the selected nine cancers were retrieved, including details of authors, publication year, journal, tumour type, sample size, SNP ID, P-value, OR value, and 95% confidence interval.
Statistical analysis
Regarding the nine types of malignant tumours as a whole, different P-values of the same locus reported in different studies (same tumour type or different tumour type) were merged through Fisher's exact probability method using Stata 11.0 (StataCorp LP, College Station, USA). For the nine different types of malignant tumours, related sites for each tumour type were analysed, and different P-values of the same locus for the same tumour type from different publications were merged through the same method.
All susceptibility loci associated with malignant tumour morbidity were imported to the dbSNP database, and gene loci in the exon that may affect the corresponding encoding functions were screened.
Association study between the selected SNPs and OSCC
Study case inclusion
Twelve SNP loci located in the exons were selected from the 225 SNP loci associated with the nine types of malignant tumour in the GWAS Catalogue and utilized for the following experiments. To determine a possible association between these 12 SNPs and OSCC, a multicentre case-control study, including Chinese Han patients with OSCC and healthy volunteers, was performed, and the distribution of the 12 loci alleles and genotypes was explored to determine the genetic susceptibility loci associated with OSCC.
Two-hundred and seventy-nine patients with OSCC (162 males, 114 females; average age: 59 years) who were treated at the West China Hospital of Stomatology (Sichuan University), the Beijing Stomatological Hospital (Capital Medical University), and the Guangdong Provincial Stomatological Hospital from January 2003 to December 2011 (the case group) and 218 healthy volunteers (the control group; 126 males, 112 females; average age, 54 years) who were examined in the West China Hospital (Sichuan University) and Stomatological Hospital of the Fourth Military Medical University were enrolled in the present study. No patient in the case group had a history of other tumours. The control group consisted of healthy volunteers without a history of cancer.
Reagents
Primers for Polymerase Chain Reaction (PCR) amplification and single-base extension were designed with Genotyping Tools and MassARRAY Assay Design 3.1 software (Sequenom, San Diego, CA, USA), and synthesized by Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). QIAamp DNA Blood Midi kit (QIAGEN GmbH, Hilden, Germany) was used for DNA extraction, and iPLEX® Gold Reagent kit purchased from Sequenom was used for SNP genotyping.
DNA extraction
DNA in blood samples was extracted, and quantified by using a spectrophotometer, and quality tested by using agarose gel electrophoresis. The concentration of DNA samples that passed the quality inspection was adjusted to 50 ng/µl, and samples were stored at −20°C for later use.
PCR amplification and purification
Multiplex PCR amplification was performed. The total reaction volume was 5 µl, which included 10 ng of template DNA, 0.5 U of Hotstar Taq, 0.5 pmol amplification primer, and 0.1 µl of 25 mM dNTPs. The reaction conditions were as follows: pre-denaturation at 94°C for 4 min, followed by 45 cycles of 94°C for 20 sec, 56°C for 30 sec, and 72°C for 1 min. A final extension was performed at 72°C for 3 min, and the samples were stored at 4°C.
PCR products were processed with 0.5 U shrimp alkaline phosphatase (SAP), and excess dNTP was removed. The total reaction volume was 7 µl, which included the PCR product (5 µl) and SAP mixture (2 µl; 0.5 U of SAP, 0.17 µl of buffer). The reaction was performed at 37°C for 20 min and 85°C for 5 min, and the samples were stored at 4°C.
Single-base extension and purification
The total reaction volume for single base extension was 9 µl including 7 µl of PCR products following processing with SAP, primer extension reaction mixture (0.804 µl), iPLEX enzyme (0.041 µl). The reaction conditions were as follows: Pre-denaturation at 94°C for 30 sec and 94°C for 5 sec, followed by 5 cycles of 52°C for 5 sec and 80°C for 5 sec; and subsequently return to 94°C for 5 sec for a total 40 cycles, finally followed by extension at 72°C for 3 min and stored at 4°C. Each extension reaction product was purified with 6 mg Spectro CLEAN™ resin (Sequenom) (16).
Mass spectrometric detection
Purified products were placed in a 384-well SpectroCHIP (Sequenom) chip, and the signals were analysed via Matrix-assisted laser desorption/Ionization time of a flight (MALDI-TOF) mass spectrometry; the results were classified with TYPER 4.0 (Sequenom).
Statistical analysis
The case and control groups were tested using the Hardy Weinberg-genetic equilibrium (HWE) test. Between groups, the allele frequency at various points was calculated with the chi-squared test, and Fisher's exact probability test was used to assess differences in allele distribution between the OSCC and control groups. Bilateral probability was estimated for the statistical test (α=0.05). P<0.05 was considered to indicate a statistically significant difference.
Cell lines
Oral cancer cells, H314; tongue cancer cells, HSC-3, HSC-4, UM1, and UM2; buccal cancer cells, H413; pharyngeal cancer cells, HN30, HN31; and normal oral mucosa epithelial keratinocytes, NOK and HOK, were frozen at −150°C and cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% heat inactivated foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U of penicillin G and 100 U of streptomycin sulphate under 5% CO2 and a humidified air atmosphere at 37°C. UM1 and UM2 cells (17) were provided by Dr Xiaofeng Zhou (Centre for Molecular Biology of Oral Diseases, College of Dentistry, University of Illinois at Chicago, Chicago, USA), HSC-3 and HSC-4 cells were obtained from the Japan Cancer Research Resources Bank, H314 and H413 cells were obtained from the European Collection of Cell Cultures, and NOK, HOK, HN30 and HN31 cells were kindly provided by Dr JS Gutkind (National Institute of Dental and Craniofacial Research, Bethesda, MD, USA).
Sera and paraffin specimens
Sera from 32 patients with OSCC and 10 healthy volunteers were collected at the clinical laboratory of the West China Hospital of Stomatology, Sichuan University from 2009 to 2011. Nine paraffin specimens (normal oral epithelia and squamous cell carcinoma tissue) of OSCC were collected by the department of pathology of West China Hospital of Stomatology, Sichuan University from 2009 to 2011 (sera samples were obtained from all 32+10 patients and healthy controls, respectively, but paraffin specimens were obtained from only 9+8 patients and healthy controls, respectively). Clinical pathology data were obtained from medical records and histopathologic reports. Eight normal oral epithelia specimens were obtained from healthy patients who underwent oral and maxillofacial plastic surgery.
Reagents
Reagent kit (including 5 X PrimeScriptTM buffer, PrimeScriptTM RT Enzyme Mix I, Oligo dT Primer, Random 6 mers, RNase Free dH2O and EASY Dilution buffer) for reverse transcription (RT) and PCR was purchased from Takara Bio Inc. (Otsu, Japan). Rabbit polyclonal anti-PSCA antibody was purchased from Abcam (Cambridge, UK). Immunohistochemical kit was purchased from DAKO (Agilent Technologies, Inc., Santa Clara, CA, USA). Human Prostate Stem Cell Antigen (PSCA) ELISA kit (cat no. CSB-EL018840HU) was purchased from CUSABIO BIOTECH (Stratech, Scientific, Ltd., UK).
RNA extraction and RT-PCR
Cells were separately homogenized using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Following the complete dissociation of nucleoprotein complexes, phase separation was achieved using chloroform and centrifugation (12,000 × g at 4°C for 15 min). The precipitated RNA from the aqueous phase was washed using 75% ethanol. RNA was dried and dissolved in RNase-free water. Following RNA extraction, and the concentration was measured by using a spectrophotometer. The RNA samples were reverse-transcribed to cDNA. The obtained cDNA was used for PCR amplification. Primer sequences of PSCA for RT-PCR were designed as follows: PSCA forward, ATGGCAGGCTTGGCCCTGCAGCCAGGC and reverse, CTAGAGCTGGCCGGGTCCCCAGAGCAG; GAPDH forward, GAGTCAACGGATTTGGTCGT and reverse, TTGATTTTGGAGGGATCTCG was which was used as reference gene. PCR amplification of the cDNA was performed at an initial temperature of 94°C for 5 min followed by 30 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, followed by a final extension at 72°C for 10 min. The PCR products were loaded into a 2% agarose gel for electrophoresis.
Enzyme-linked immunosorbent assay (ELISA)
The expression of PSCA proteins in different samples was detected by ELISA kit. The main steps are as follows: a standard curve was drawn. The sample to be tested was diluted and 100 µl solution was added to each well. Test solution (50 µl) was added and incubated at room temperature for 1 h and chromogenic substrate was added. The absorbance values were measured at 450 nm, and the protein concentration in each sample was calculated according to the standard curve given previously.
Immunohistochemistry (IHC)
All specimens were detected using a two-step IHC method in the same conditions. Following deparaffinization, antigen retrieval was performed in a 10 mM citrate buffer (self-prepared as follows: Storage solution: Solution A: 0.1 M citric acid monohydrate (C6H8O7•H2O FW = 210.14); Solution B: 0.1 M trisodium citrate, dihydrate, (C6H5O7Na3•2H2O FW = 294.12). Working solution: 18 ml Solution A+ 82 ml Solution B+ 900 ml water) at pH 6.0 in a pressure cooker for 3 min. Endogenous peroxidase was blocked with 3% hydrogen peroxide in phosphate buffered saline for 10 min at room temperature. The sections were incubated with primary antibodies against PSCA (1:100; cat no. ab64919; Abcam) at 4°C overnight, and subjected to incubation with an goat anti-mouse and rabbit IgG antibody conjugated with horseradish peroxidase (HRP; stock solution; cat. no. GK600505; Gene Tech Biotechnology Co., Ltd., Shanghai, China) for 30 min at room temperature. Positive staining was observed as brown yellow granules. An immunoreactive score (IRS) was calculated for all samples. Five fields were randomly selected at high magnification (magnification, ×400), and the staining intensity (SI) and percentage of positive cells (PP) were scored (SI evaluation) as follows: Colourless - 0, buff - 1 point, tan - 2 points, brown - 3 points. PP evaluation scores were as follows: Negative - 0, positive cells 10% or less - 1 point, 11–50% - 2 points, 51–80% - 3 points, >80% - 4 points. IRS was calculated using the equation, IRS=SI × PP. IRS >3 points was considered positive. All of these were analyzed using AperioImageScope software (Version 11; Leica Microsystems, Inc., Buffalo Grove, IL, USA).
Statistical analysis
PSCA expression in OSCC and normal tissues was compared via Fisher's exact probability test, and statistical analysis was performed by SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Association study of PSCA polymorphism and clinical prognosis of patients with OSCC
Previous experiments in the present study identified the PSCA gene polymorphism locus, rs2294008, as a genetic susceptibility locus of OSCC, and the CC genotype frequency was significantly higher in patients with OSCC compared with controls. OSCC patients clinical and postoperative follow-up data were analysed to explore the association between PSCA polymorphism and clinical prognosis of patients with OSCC.
Study case inclusion
Seventy-six OSCC cases (as aforementioned) were derived from patients diagnosed as OSCC and who underwent oral and maxillofacial surgery at West China Hospital of Stomatology, Sichuan University. Recurrence or mortality incidence was recorded based on a telephone follow-up. Rs2294008 genotype information of patients with OSCC was obtained from the foregoing OSCC case-control study results.
Statistical analysis
The association between rs2294008 locus gene type and the clinical indicators was analysed through the single factor chi-squared test, and Fisher's exact probability method was used to compare values between groups. Survival curves were obtained through the Kaplan-Meier method, and the independent impact of the genotype and clinical variables on the prognosis of OSCC risk were analysed by using the Cox proportional hazards regression model. P<0.05 was considered to indicate a statistically significant difference.
Results
Meta-analysis results
The GWAS Catalogue (2012-03-09) screening resulted in the identification of 91 articles published in 26 journals, and 225 SNP loci associated with nine types of malignant tumour were identified. Among these, 11 loci were associated with bladder cancer, 50 loci with breast cancer, 19 loci with colorectal cancer, 7 loci with liver cancer, 30 loci with lung cancer, 11 loci with nasopharyngeal carcinoma, 74 loci with prostate cancer, 2 loci with skin cancer, and 23 loci with digestive tract carcinoma. Two loci were associated with two tumour types: rs401681 was associated with lung and bladder cancer, and rs6983267 was associated with prostate and colorectal cancer. Twelve loci (Table I) were located in the exons of 11 genes, probably affecting genes functions. Finally, there were two loci associated with more than one malignant tumor.
Table I.
SNPs located in exons.
| SNP | Gene symbol | SNP | Gene symbol |
|---|---|---|---|
| rs671 | ALDH2 | rs2274223 | PLCE1 |
| rs8170 | C19orf62 | rs3765524 | PLCE1 |
| rs130067 | CCHCR1 | rs2294008 | PSCA |
| rs971074 | ADH7 | rs2292884 | MLPH |
| rs1051730 | CHRNA3 | rs4072037 | MUC1 |
| rs1229984 | ADH1B | rs10936599 | MYNN |
SNP, single nucleotide polymorphism.
Multicentre case-control study
The multicentre case-control study of a Chinese Han population led to the identification of 12 polymorphism loci whose distribution in patients with OSCC and healthy volunteers is demonstrated in Table II.
Table II.
Allele distribution of the 12 selected SNPs.
| SNP ID | Gene symbol | A/a | HWE | Risk allele | MAF in case | MAF in control | P (Chi-squared) |
|---|---|---|---|---|---|---|---|
| rs671 | ALDH2 | G/A | 0.2648 | A | 0.226 | 0.179 | 0.0674 |
| rs8170 | C19orf62 | C/T | 1b | T | 0.005 | 0.002 | 0.4569 |
| rs130067 | CCHCR1 | A/C | 0.1356 | C | 0.346 | 0.309 | 0.2178 |
| rs971074 | ADH7 | G/A | 0.6897 | G | 0.104 | 0.129 | 0.2115 |
| rs1051730 | CHRNA3 | C/T | 1b | T | 0.037 | 0.032 | 0.662 |
| rs1229984 | ADH1B | A/G | 0.0462b | A | 0.315 | 0.339 | 0.4224 |
| rs2274223 | PLCE1 | A/G | 0.549 | G | 0.228 | 0.198 | 0.2528 |
| rs2292884 | MLPH | A/G | 0.5995 | A | 0.279 | 0.29 | 0.6836 |
| rs2294008 | PSCA | C/T | 0.8509 | C | 0.258 | 0.323 | 0.0239a |
| rs3765524 | PLCE1 | C/T | 0.4102 | T | 0.227 | 0.195 | 0.2204 |
| rs4072037 | MUC1 | A/G | 2.29E-14b | A | 0.007 | 0.019 | 0.1023 |
| rs10936599 | MYNN | T/C | 0.0884 | T | 0.429 | 0.456 | 0.4001 |
P<0.05.
Allelic distribution was not in Hardy Weinberg-genetic equilibrium (HWE). MAF, minor allele frequency; SNP, single nucleotide polymorphism.
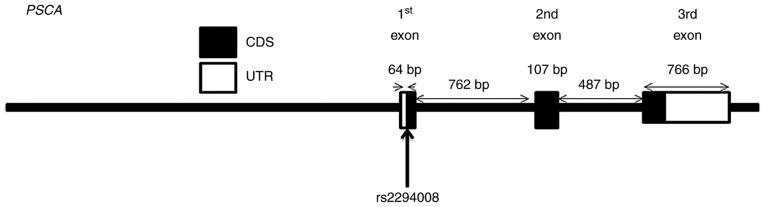
The allele distribution of rs4072037 in MUC1, rs1229984 in ADH1B, rs8170 in C19orf62, and rs1051730 in CHRNA3 did not conform HWE and were therefore excluded from further analysis. The allele distribution of rs2294008 (Fig. 1) was significantly different between patients with OSCC and healthy volunteers. The C allele seems to be a risk allele for OSCC as its frequency was higher in patients with OSCC compared with healthy volunteers [T vs. C; OR=0.728 (0.552, 0.959); P<0.05].
Figure 1.
The rs2294008 polymorphism in PSCA. OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
The genotype distribution of the eight remaining loci in patients with OSCC and normal controls is illustrated in Table III. Using the recessive inheritance model, rs2292884 and rs3765524 polymorphisms were identified to be associated with OSCC (P<0.05). Rs2294008 in PSCA was significantly different between patients with OSCC and healthy controls (P<0.01). The CC vs. CT + TT model demonstrated that the CC genotype frequency was significantly higher in patients with OSCC than in the normal group (P<0.01). The CC genotype was associated with OSCC risk [OR = 1.699 (1.188, 2.429); P<0.01]. No statistical difference was observed (P>0.05) in the CC + CT vs. TT model (Table III).
Table III.
Genotype distribution of the 8 SNPs.
| SNP ID | Gene symbol | A/a | Population | AA | Aa | aa | P-value | P (AA+Aa vs. aa) | P (AA vs. Aa+aa) |
|---|---|---|---|---|---|---|---|---|---|
| rs671 | ALDH2 | G/A | OSCC | 163 | 101 | 12 | |||
| Control | 139 | 67 | 4 | 0.142 | 0.135 | 0.108 | |||
| rs130067 | CCHCR1 | A/C | OSCC | 125 | 119 | 38 | |||
| Control | 107 | 86 | 24 | 0.49 | 0.417 | 0.269 | |||
| rs971074 | ADH7 | C/A | OSCC | 223 | 56 | 1 | |||
| Control | 165 | 48 | 4 | 0.207 | 0.1 | 0.335 | |||
| rs2274223 | PLCE1 | A/G | OSCC | 169 | 96 | 16 | |||
| Control | 134 | 77 | 4 | 0.099 | 0.031a | 0.621 | |||
| rs2292884 | MLPH | A/G | OSCC | 141 | 122 | 17 | |||
| Control | 111 | 86 | 20 | 0.351 | 0.185 | 0.86 | |||
| rs2294008 | PSCA | C/T | OSCC | 161 | 98 | 24 | |||
| Control | 94 | 103 | 18 | 0.009a | 0.966 | 0.004a | |||
| rs3765524 | PLCE1 | C/T | OSCC | 169 | 98 | 15 | |||
| Control | 137 | 77 | 4 | 0.128 | 0.043a | 0.507 | |||
| rs10936599 | MYNN | T/C | OSCC | 85 | 153 | 45 | |||
| Control | 61 | 113 | 42 | 0.581 | 0.301 | 0.662 |
A. Major allele; a. Minor allele.
P<0.05.
PSCA transcription and expression in OSCC cell lines and patient serum and specimens
With universal screening of genetic susceptibility loci of malignant tumours and the multicentre case-control study, rs2294008 (located in exon 1 of PSCA gene) was identified as the OSCC genetic susceptibility locus. This result suggests that changes in PSCA function may affect the occurrence of OSCC. Thus, this locus was further investigated. As illustrated in Fig. 2, semi-quantitative PCR resulted in the amplification of a band of 345 bp corresponding to PSCA. PSCA mRNA expression was detected in OSCC cell lines, UM2, H314, HSC-3, and HSC-4, and in HN30 pharyngeal cancer cells, but not in HOK and NOK epithelial cells.
Figure 2.
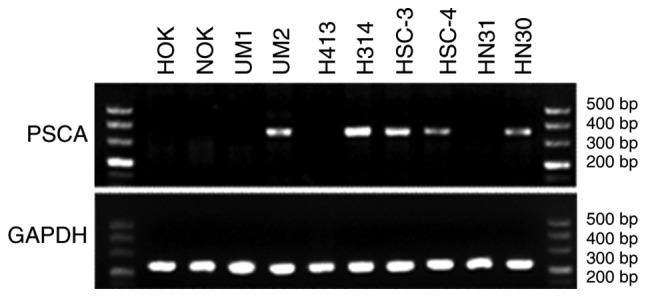
PSCA expression pattern in eight cancer and two normal cell lines. OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
PSCA was detected in the sera of 14 of 32 patients with OSCC. Six of 11 patients with OSCC presented with the CC genotype and were positive for PSCA, while eight of 21 patients with OSCC presented with the CT/TT genotype and were positive for PSCA. No PSCA expression was detected in the serum of the 10 healthy subjects (Table IV).
Table IV.
PSCA expression in OSCC and normal serum.
| Serum type | Number of patients | PSCA+ | PSCA- |
|---|---|---|---|
| Normal serum | 10 | 0 | 10 |
| Serum of OSCC | 32 | 14 | 18 |
| OSCC CC-genotype | 11 | 6 | 5 |
| OSCC CT/TT-genotype | 21 | 8 | 13 |
OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
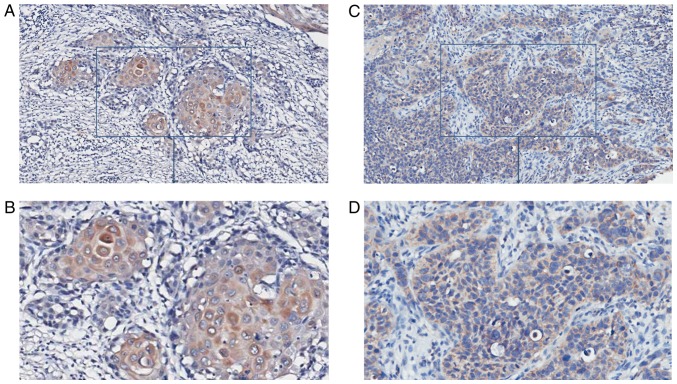
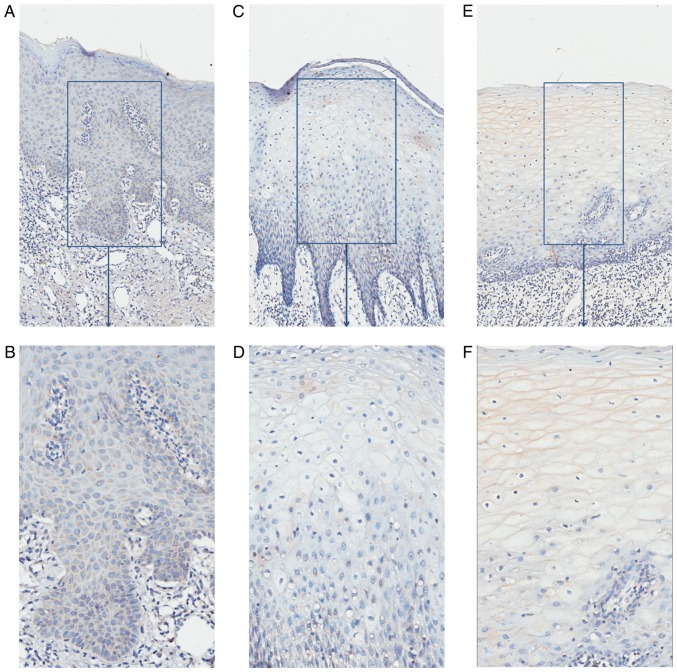
Paraffin specimens from nine patients with OSCC (five patients with serum that was PSCA-positive, four serum PSCA-negative; four CC genotypes, and five CT/TT genotypes) and eight controls were collected and immunochemically stained; the results are demonstrated in Table V. In specimens from the nine OSCC cases, epithelium and nests of eight cases were shaded, and the IRS scores of six cases were above 3 and were considered PSCA-positive (Fig. 3). It demonstrated that PSCA protein may be highly expressed in the OSCC tissues. In specimens from eight controls, one presented an IRS score of 3, while the IRS scores of the remaining were <3 and were considered PSCA-negative (Fig. 4). The percentage of PSCA-positive staining was significantly higher in the OSCC than in the normal control group (P<0.05). PSCA staining was mainly observed in the cytoplasm in OSCC and normal tissues. PSCA immunohistochemical staining was positive in specimens from five patients with OSCC who were positive for serum, while it was only positive in one of four patients with OSCC who were negative for serum PSCA; the difference was significant (P<0.05; Table V). The percentage of PSCA positive staining was not statistically different between OSCC patients with different genotypes (P>0.05). However, PSCA protein localisation was different in OSCC patients with different genotypes. The T allele of rs2294008 encodes a translation initiation codon upstream of the reported site and changes protein localization from the cytoplasm to the cell surface (18). In OSCC patients with the CC genotype, the cytoplasm of cells from the epithelium and nests was PSCA-positive. In OSCC patients with the CT/TT genotype, the membrane of the epithelial cornification layer cells was PSCA-positive, and the cytoplasm of the epithelial basal layer cells and nests was PSCA-positive (Fig. 5).
Table V.
PSCA protein expression in OSCC and normal tissues.
| Tissue type | Number | PSCA+ | PSCA- | P-valueb |
|---|---|---|---|---|
| OSCC | 9 | 6 | 3 | |
| Normal tissue | 8 | 1 | 7 | 0.022a |
| OSCC Elisa+ | 5 | 5 | 0 | |
| OSCC Elisa- | 4 | 1 | 3 | 0.012a |
| OSCC CC-genotype | 4 | 3 | 1 | |
| OSCC CT/TT-genotype | 5 | 3 | 2 | 0.685 |
P<0.05.
Calculated by Fisher's method. OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen; SNP, single nucleotide polymorphism.
Figure 3.
PSCA expression in OSCC tissues. (A and B) Representative images indicating PSCA expression in tissues from patients with OSCC and PSCA-positive serum (IRS=6; A, 100×; B, 200×). (C and D) PSCA expression in tissues from patients with OSCC and PSCA-negative serum (IRS=2; C, ×100; D, ×200). OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
Figure 4.
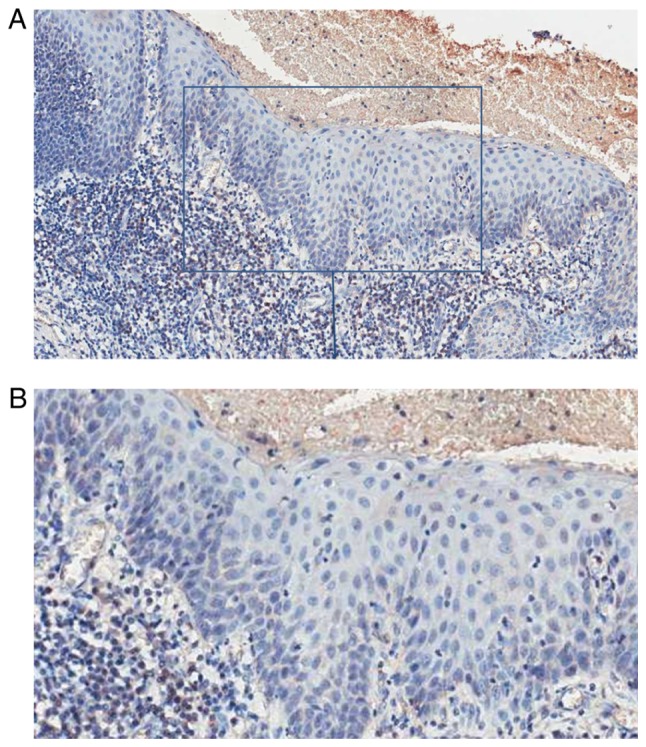
PSCA expression in normal epithelium. Representative images from normal tissue with an IRS=0 (A, ×100; B, ×200). OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
Figure 5.
PSCA expression in OSCC epithelium with different genotypes. (A and B) Representative images indicating PSCA expression in tissues from OSCC patients presenting the CC genotype (A, ×100; B, ×200). (C and D) Representative images indicating PSCA expression in OSCC patients presenting the CT genotype (C, ×100; D, ×200). Representative images indicating PSCA expression in tissues from OSCC patients presenting the TT genotype (E, ×100; F, ×200). OSCC, oral squamous cell carcinoma; PSCA, prostate stem cell antigen.
Association between rs229008 polymorphism and clinical and pathological characteristics of patients with OSCC
The association between rs2294008 polymorphism distribution and the clinical characteristics of 76 patients with OSCC is illustrated in Table VI. Patients with the CT/TT genotype drank less alcohol (P<0.05) compared with patients with the CC genotype. The CT/TT genotype was also associated with tumour size and T3/T4 stage when compared with the CC genotype (P<0.05). In addition, some weak associations (P<0.1) between the CT/TT genotype and smoking (P=0.085), lymph node metastasis (P=0.052), and higher clinical stage (P=0.075) were observed.
Table VI.
Rs2294008 polymorphism and clinical parameters.
| Total | CC | CT&TT | P-value | |
|---|---|---|---|---|
| Sex | ||||
| Female | 27 | 19 | 8 | |
| Male | 49 | 25 | 24 | 0.102 |
| Age, years | ||||
| ≤65 | 58 | 34 | 24 | |
| >65 | 18 | 10 | 8 | 0.818 |
| Smoking | ||||
| Yes | 42 | 28 | 14 | |
| No | 34 | 16 | 18 | 0.085 |
| Drinking | ||||
| Yes | 48 | 33 | 15 | |
| No | 28 | 11 | 17 | 0.012a |
| Grading | ||||
| 1 | 49 | 27 | 22 | |
| 2&3 | 27 | 17 | 10 | 0.507 |
| Nodal status | ||||
| Negative | 52 | 34 | 18 | |
| Positive | 24 | 10 | 14 | 0.052 |
| Tumor size | ||||
| T1 | 22 | 17 | 5 | |
| T2 | 35 | 20 | 15 | |
| T3 | 5 | 2 | 3 | |
| T4 | 13 | 4 | 9 | 0.045a |
| Tumor stage | ||||
| 1 | 20 | 15 | 5 | |
| 2 | 23 | 14 | 9 | |
| 3 | 14 | 8 | 6 | |
| 4 | 18 | 6 | 12 | 0.075 |
P<0.05.
Since the follow-up time was <3 years for many patients, the mortality percentage was <50%, and, therefore, the results were not suitable for survival analysis based on mortality. Therefore, the rate of survival was analysed based on cancer recurrence in the 76 patients with OSCC (Table VII). The resulting survival curve based on postoperative recurrence and the different genotypes is demonstrated in Fig. 6. The recurrence rate in OSCC patients presenting the CT/TT genotype was 62.5% and that in OSCC patients presenting the CC genotype was 47.7%; the difference was not statistically significant (P>0.05). Postoperative recurrence tended to increase in OSCC patients presenting the CT/TT genotype. As depicted in Table VIII, the COX proportional hazards regression model indicated that the different genotypes at the rs2294008 locus did not significantly influence postoperative recurrence in patients with OSCC [OR=0.9248 (0.4513, 1.8951), P>0.05]. Tumour size (P<0.05) and tumour clinical stage (P<0.05) were independent factors influencing postoperative recurrence in patients with OSCC.
Table VII.
Recurrence analysis of 76 patients with OSCC.
| Genotype | Total | No recurrence | Percentage (%) | P-value |
|---|---|---|---|---|
| CC | 44 | 23 | 52.30 | |
| CT+TT | 32 | 12 | 37.50 | 0.261 |
| Overall | 76 | 35 | 46.10 |
OSCC, oral squamous cell carcinoma.
Figure 6.
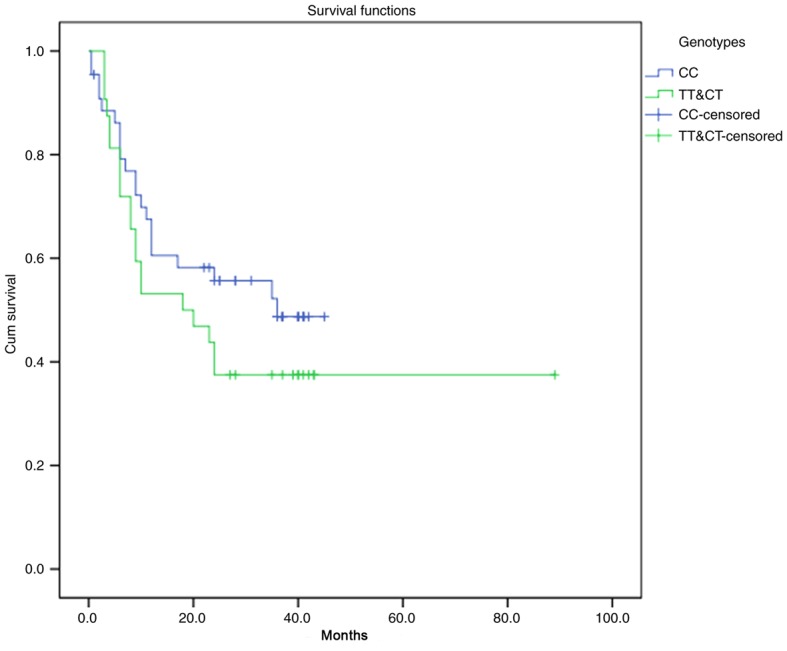
Survival analysis of 76 patients with OSCC with different genotypes.
Table VIII.
Result of COX proportional Hazards model.
| 95.0% CI for Exp(B) | ||||
|---|---|---|---|---|
| OR | Lower | Upper | P-value | |
| Genotype | 0.9248 | 0.4513 | 1.8951 | 0.8310 |
| Sex | 1.2570 | 0.4548 | 3.4739 | 0.6593 |
| Smoking | 0.7874 | 0.2831 | 2.1902 | 0.6470 |
| Drinking | 0.9204 | 0.3286 | 2.5784 | 0.8747 |
| Age | 1.4636 | 0.6350 | 3.3734 | 0.3713 |
| Grading | 1.4347 | 0.5838 | 3.5259 | 0.4315 |
| Tumor size | 0.0456a | |||
| T1 | 61.8182 | 3.2625 | 1171.3486 | 0.0060 |
| T2 | 10.8660 | 1.6237 | 72.7161 | 0.0139 |
| T3 | 17.3644 | 0.6132 | 491.7257 | 0.0943 |
| Nodal status | 0.7779 | |||
| Negative | 1.0067 | 0.1147 | 8.8342 | 0.9952 |
| Positive | 1.6820 | 0.3030 | 9.3381 | 0.5522 |
| Tumor stage | 0.0191a | |||
| 1 | 0.0049 | 0.0002 | 0.1463 | 0.0021 |
| 2 | 0.0740 | 0.0067 | 0.8171 | 0.0336 |
| 3 | 0.0449 | 0.0038 | 0.5359 | 0.0142 |
P<0.05. OSCC, OR, odds ratio; CI, confidence interval.
Discussion
Tumours are divided into many types according to their location and histological characteristics and genetic background may influence their development (13). In the current study, seven types of cancers with the highest incidence and two types of cancer, skin cancer and NPC, which have the same origin as OSCC, were used to identify common SNPs associated with cancer susceptibility and to evaluate their involvement in OSCC.
PSCA is located on chromosome 8 q24.3 and it belongs to the Thy-1/Ly-6 super family. PSCA is highly expressed in prostate cancer, and its expression is associated with classification, clinical stage, metastasis, and prognosis of patients with prostate cancer (19,20). It is a marker for prostate cancer diagnosis and prognosis and a novel target for the treatment of prostate cancer (19,21,22). PSCA is also highly expressed in bladder, pancreatic, and kidney cancer (23–25), while its expression level is low in other tumours such as gastric, oesophageal, and gallbladder cancer (26,27). As a type of membrane proteins with GPI anchoring, PSCA may serve a role in T cell activation (21). As a member of the Thy-1 family, is involved in the process of T cell activation, proliferation and survival of stem cells, and cell factor response; PSCA may also be involved in tumour formation, and activation and adhesion of tumour cells (28,29). In 2002, De Nooij-van Dalen et al (27) reported that PSCA was transcribed in cancer cells and normal head and neck squamous tissue. PSCA transcription was significantly different between head and neck squamous carcinoma and normal tissues, suggesting it may be involved in the development of head and neck squamous cell carcinomas.
Although little is known regarding the mechanisms underlying the regulation of PSCA expression, androgens are known to be involved in this process (30). Previous studies have detected androgen response elements in prostate epithelial PSCA promoter regions (30). Bahrenberg et al (31)demonstrated that growing the bladder cancer cell line RT112 in a hydrophobic medium resulted in cell clustering, while PSCA mRNA levels increased significantly, suggesting that PSCA expression may be involved in the regulation of epithelial cell adhesion. Furthermore, following addition of phorbol-12-myristate-13-acetate, PSCA mRNA levels increased significantly, suggesting that PSCA transcription may be associated with and indirectly regulated by protein kinase. In contrast, epidermal growth factor, platelet-derived growth factor (PDGF), tumour necrosis factor-alpha, and interferon gamma could not increase PSCA transcription level (31). The authors pointed out that PSCA expression was likely regulated by a strong induction type of promoter by specific extracellular signals (31). A previous study has demonstrated that PSCA expression is downregulated in telomerase-transduced urethral epithelial cells, suggesting that PSCA may be regulated by a mechanism involving telomerases (32).
Some studies have revealed that PSCA is associated with the inhibition of tumour growth and metastasis and with prolonged survival (33,34). In contrast, in a gastric cancer cell line, PSCA inhibited cell growth, but had no effect on cell apoptosis (35). PSCA is upregulated or downregulated in various human tumours, suggesting that PSCA may be carcinogenic or act as tumour suppressor, depending on the cell type and the nature of the interaction between molecules and signalling pathways (36,37).
H314 and H413 cells are both mutated for P53, but the mutation position is different, suggesting that P53 mutation at different sites may affect PSCA expression through complex signalling pathways.
PSCA mRNA expression levels were higher in OSCC cell lines than that in normal keratin epithelial cells.
The multicentre case-control study demonstrated that the CC genotype frequency was higher in patients with OSCC than in healthy subjects and that the CC genotype was an OSCC susceptibility factor. However, in the present analysis of clinical and pathological indices in patients with OSCC, the CT/TT genotype was associated with tumour size, stage, and lymph node metastasis. Although the association between the CT/TT genotype and recurrence was not statistically significant, the CT/TT genotype may have an adverse impact for recurrence based on the survival curve. This seemingly contradictory phenomenon has also been reported by other groups (35,38–41). The T allele in the rs2294008 loci was confirmed as a risk allele of diffuse gastric cancer (35,38–40), while survival analysis by Wang et al (41) demonstrated that the CT/TT genotype had a positive protective effect on the prognosis of patients with gastric cancer. The study speculated that patients with the CC genotype may be more resistant to radiation and chemotherapy than patients with the CT/TT genotype, and hence lead to a poor prognosis. Nevertheless, the role of the rs2294008 polymorphism and PSCA in tumour pathogenesis remains unclear.
The present study has limitations given its small sample size and the short follow-up time. In further research, the influence of genotype should be verified at the mRNA and protein levels.
To conclude, rs2294008 polymorphism may be associated with OSCC development. The corresponding gene, PSCA, was expressed in OSCC but not in normal cell lines. No association was detected between the rs2294008 polymorphism and OSCC recurrence, but positive association was observed between the CT/TT genotype and smoking history, drinking history, lymph node metastasis, tumour size, and clinical stage. The present study demonstrated that PSCA may serve a role in OSCC and provided a novel perspective for future OSCC study.
Acknowledgements
Not applicable
Glossary
Abbreviations
- OSCC
oral squamous cell carcinoma
- GWAS
genome-wide association study
- SNPs
single nucleotide polymorphisms
- PSCA
prostate stem cell antigen
- UADT
upper aerodigestive tract
- HWE
Hardy Weinberg equilibrium
- PCR
polymerase chain reaction
- ELISA
enzyme-linked immunosorbent assay
- IHC
immunohistochemistry
- IRS
immunoreactive score
- PKC
protein kinase C
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81321002, 81472533 and 81500864), and the Nonprofit Industry Research Specific Fund of National Health and Family Planning Commission of China (grant no. 201502018).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KZ and XL conceived and designed the study. JL guided the experiments. KZ, XL, JL, JW and WG performed the experiments. LJ and HD collected and processed the clinical data. HX and JW analyzed and interpreted the patient data. KZ and XL wrote the paper, and JL, XZ and QC reviewed and edited the manuscript. XZ and QC contributed to the conception of the study and helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was conducted in accordance with the standards of the Declaration of Helsinki and approved by the Ethical Review Committee of West China Hospital of Stomatology, Sichuan University, China (approval no. 2010056). All patients provided written informed consent for their information to be stored and used in the hospital database.
Consent for publication
All patients provided informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Podlodowska J, Szumiło J, Podlodowski W, Starosławska E, Burdan F. Epidemiology and risk factors of the oral carcinoma. Pol Merkur Lekarski. 2012;32:135–137. (In Polish) [PubMed] [Google Scholar]
- 3.Wang Z, Jiang L, Huang C, Li Z, Chen L, Gou L, Chen P, Tong A, Tang M, Gao F, et al. Comparative proteomics approach to screening of potential diagnostic and therapeutic targets for oral squamous cell carcinoma. Mol Cell Proteomics. 2008;7:1639–1650. doi: 10.1074/mcp.M700520-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noguti J, De Moura CF, De Jesus GP, Da Silva VH, Hossaka TA, Oshima CT, Ribeiro DA. Metastasis from oral cancer: An overview. Cancer Genomics Proteomics. 2012;9:329–335. [PubMed] [Google Scholar]
- 5.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habibi I, Sfar I, Kort F, Aounallah-Skhiri H, Chebil A, Chouchene I, Bouraoui R, Limaiem R, Largheche L, Jendoubi-Ayed S, et al. Y402H polymorphism in complement factor H and age-related macular degeneration in the Tunisian population. Ophthalmic Res. 2013;49:177–184. doi: 10.1159/000345068. [DOI] [PubMed] [Google Scholar]
- 7.Varghese JS, Easton DF. Genome-wide association studies in common cancers-what have we learnt. Curr Opin Genet Dev. 2010;20:201–209. doi: 10.1016/j.gde.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Wang JY, Zhang XP, Lv ZW, Fu D, Lu YC, Hu GH, Luo C, Chen JX. RLIP76 is overexpressed in human glioblastomas and is required for proliferation, tumorigenesis and suppression of apoptosis. Carcinogenesis. 2013;34:916–926. doi: 10.1093/carcin/bgs401. [DOI] [PubMed] [Google Scholar]
- 9.Tan B, Anaka M, Deb S, Freyer C, Ebert LM, Chueh AC, Al-Obaidi S, Behren A, Jayachandran A, Cebon J, et al. FOXP3 over-expression inhibits melanoma tumorigenesis via effects on proliferation and apoptosis. Oncotarget. 2014;5:264–276. doi: 10.18632/oncotarget.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma GZ, Liu CH, Wei B, Qiao J, Lu T, Wei HC, Chen HD, He CD. Baicalein inhibits DMBA/TPA-induced skin tumorigenesis in mice by modulating proliferation, apoptosis, and inflammation. Inflammation. 2013;36:457–467. doi: 10.1007/s10753-012-9566-y. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Cao C, Luo H, Xiong S, Xu Y, Xiong W. Tumour necrosis factor alpha −308G/A polymorphism and risk of the four most frequent cancers: A meta-analysis. Int J Immunogenet. 2011;38:311–320. doi: 10.1111/j.1744-313X.2011.01014.x. [DOI] [PubMed] [Google Scholar]
- 12.Kotnis A, Kannan S, Sarin R, Mulherkar R. Case-control study and meta-analysis of sult1a1 arg213his polymorphism for gene, ethnicity and environment interaction for cancer risk. Br J Cancer. 2008;99:1340–1347. doi: 10.1038/sj.bjc.6604683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: The role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C, Lao-Sirieix P, Gammon MD, Corley DA, Shaheen NJ, et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett's esophagus. Nat Genet. 2013;45:1487–1493. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindorff LA, MacArthur J, (European Bioinformatics Institute), Morales J (European Bioinformatics Institute) Junkins HA, Hall PN, Klemm AK, Manolio TA. A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies. [Sep 3;2012 ];A Catalog of Published Genome-Wide Association Studies. [Google Scholar]
- 16.Hartmer R, Storm N, Boecker S, Rodi CP, Hillenkamp F, Jurinke C, van den Boom D. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res. 2003;31:e47. doi: 10.1093/nar/gng047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama S, Sasaki A, Mese H, Alcalde RE, Matsumura T. Establishment of high and low metastasis cell lines derived from a human tongue squamous cell carcinoma. Invasion Metastasis. 1998;18:219–228. doi: 10.1159/000024515. [DOI] [PubMed] [Google Scholar]
- 18.Tanikawa C, Urabe Y, Matsuo K, Kubo M, Takahashi A, Ito H, Tajima K, Kamatani N, Nakamura Y, Matsuda K. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet. 2012;44:430–434. doi: 10.1038/ng.1109. [DOI] [PubMed] [Google Scholar]
- 19.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 20.Reiter RE, Sato I, Thomas G, Qian J, Gu Z, Watabe T, Loda M, Jenkins RB. Coamplification of prostate stem cell antigen (PSCA) and MYC in locally advanced prostate cancer. Genes Chromosomes Cancer. 2000;27:95–103. doi: 10.1002/(SICI)1098-2264(200001)27:1<95::AID-GCC12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, et al. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naz RK, Shiley B. Prophylactic vaccines for prevention of prostate cancer. Front Biosci (Schol Ed) 2012;4:932–940. doi: 10.2741/s309. [DOI] [PubMed] [Google Scholar]
- 23.Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, Malats N, Tang W, Liu L, Garcia-Closas M, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci USA. 2012;109:4974–4979. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao JY, Yang YL, Li S, Qian XL, Liu FF, Fu L. PSCA expression in invasive micropapillary carcinoma of breast. Zhonghua Bing Li Xue Za Zhi. 2011;40:382–386. (In Chinese) [PubMed] [Google Scholar]
- 25.Wang D, Wang Z, Tian J, He X, Chowdhury WH, Zhang X, Li S, Rodriguez R. Prostate stem cell antigen enhancer and uroplakin II promoter based bladder cancer targeted tissue-specific vector. Urol Oncol. 2010;28:164–169. doi: 10.1016/j.urolonc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Ono H, Hiraoka N, Lee YS, Woo SM, Lee WJ, Choi IJ, Saito A, Yanagihara K, Kanai Y, Ohnami S, et al. Prostate stem cell antigen, a presumable organ-dependent tumor suppressor gene, is down-regulated in gallbladder carcinogenesis. Genes Chromosomes Cancer. 2012;51:30–41. doi: 10.1002/gcc.20928. [DOI] [PubMed] [Google Scholar]
- 27.de Nooij-van Dalen AG, van Dongen GA, Smeets SJ, Nieuwenhuis EJ, Stigter-van Walsum M, Snow GB, Brakenhoff RH. Characterization of the human Ly-6 antigens, the newly annotated member Ly-6K included, as molecular markers for head-and-neck squamous cell carcinoma. Int J Cancer. 2003;103:768–774. doi: 10.1002/ijc.10903. [DOI] [PubMed] [Google Scholar]
- 28.Bamezai A. Mouse Ly-6 proteins and their extended family: Markers of cell differentiation and regulators of cell signaling. Arch Immunol Ther Exp (Warsz) 2004;52:255–266. [PubMed] [Google Scholar]
- 29.Hruska M, Keefe J, Wert D, Tekinay AB, Hulce JJ, Ibañez-Tallon I, Nishi R. Prostate stem cell antigen is an endogenous lynx1-like prototoxin that antagonizes alpha7-containing nicotinic receptors and prevents programmed cell death of parasympathetic neurons. J Neurosci. 2009;29:14847–14854. doi: 10.1523/JNEUROSCI.2271-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain A, Lam A, Vivanco I, Carey MF, Reiter RE. Identification of an androgen-dependent enhancer within the prostate stem cell antigen gene. Mol Endocrinol. 2002;16:2323–2337. doi: 10.1210/me.2002-0004. [DOI] [PubMed] [Google Scholar]
- 31.Bahrenberg G, Brauers A, Joost HG, Jakse G. PSCA expression is regulated by phorbol ester and cell adhesion in the bladder carcinoma cell line RT112. Cancer Lett. 2001;168:37–43. doi: 10.1016/S0304-3835(01)00497-9. [DOI] [PubMed] [Google Scholar]
- 32.Chapman EJ, Kelly G, Knowles MA. Genes involved in differentiation, stem cell renewal, and tumorigenesis are modulated in telomerase-immortalized human urothelial cells. Mol Cancer Res. 2008;6:1154–1168. doi: 10.1158/1541-7786.MCR-07-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saffran DC, Raitano AB, Hubert RS, Witte ON, Reiter RE, Jakobovits A. Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc Natl Acad Sci USA. 2001;98:2658–2663. doi: 10.1073/pnas.051624698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Z, Yamashiro J, Kono E, Reiter RE. Anti-prostate stem cell antigen monoclonal antibody 1G8 induces cell death in vitro and inhibits tumor growth in vivo via a Fc-independent mechanism. Cancer Res. 2005;65:9495–9500. doi: 10.1158/0008-5472.CAN-05-2086. [DOI] [PubMed] [Google Scholar]
- 35.Study Group of Millennium Genome Project for Cancer, corp-author. Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 36.Yang L, Han Y, Saiz Suarez F, Minden MD. A tumor suppressor and oncogene: The WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404770. [DOI] [PubMed] [Google Scholar]
- 37.Radtke F, Raj K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor. Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 38.Wu C, Wang G, Yang M, Huang L, Yu D, Tan W, Lin D. Two genetic variants in prostate stem cell antigen and gastric cancer susceptibility in a Chinese population. Mol Carcinog. 2009;48:1131–1138. doi: 10.1002/mc.20565. [DOI] [PubMed] [Google Scholar]
- 39.Ou J, Li K, Ren H, Bai H, Zeng D, Zhang C. Association and haplotype analysis of prostate stem cell antigen with gastric cancer in Tibetans. DNA Cell Biol. 2010;29:319–323. doi: 10.1089/dna.2009.0960. [DOI] [PubMed] [Google Scholar]
- 40.Matsuo K, Tajima K, Suzuki T, Kawase T, Watanabe M, Shitara K, Misawa K, Ito S, Sawaki A, Muro K, et al. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int J Cancer. 2009;125:1961–1964. doi: 10.1002/ijc.24519. [DOI] [PubMed] [Google Scholar]
- 41.Wang M, Bai J, Tan Y, Wang S, Tian Y, Gong W, Zhou Y, Gao Y, Zhou J, Zhang Z. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int J Cancer. 2011;129:1207–1213. doi: 10.1002/ijc.25740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.