Abstract
In the present study, hereditary non-polyposis colorectal cancer (HNPCC) susceptibility genes were screened for using whole exome sequencing in 3 HNPCC patients from 1 family and using single nucleotide polymorphism (SNP) genotyping assays in 96 other colorectal cancer and control samples. Peripheral blood was obtained from 3 HNPCC patients from 1 family; the proband and the proband's brother and cousin. High-throughput sequencing was performed using whole exome capture technology. Sequences were aligned against the HAPMAP, dbSNP130 and 1,000 Genome Project databases. Reported common variations and synonymous mutations were filtered out. Non-synonymous single nucleotide variants in the 3 HNPCC patients were integrated and the candidate genes were identified. Finally, SNP genotyping was performed for the genes in 96 peripheral blood samples. In total, 60.4 Gb of data was retrieved from the 3 HNPCC patients using whole exome capture technology. Subsequently, according to certain screening criteria, 15 candidate genes were identified. Among the 96 samples that had been SNP genotyped, 92 were successfully genotyped for 15 gene loci, while genotyping for HTRA1 failed in 4 sporadic colorectal cancer patient samples. In 12 control subjects and 81 sporadic colorectal cancer patients, genotypes at 13 loci were wild-type, namely DDX20, ZFYVE26, PIK3R3, SLC26A8, ZEB2, TP53INP1, SLC11A1, LRBA, CEBPZ, ETAA1, SEMA3G, IFRD2 and FAT1. The CEP290 genotype was mutant in 1 sporadic colorectal cancer patient and was wild-type in all other subjects. A total of 5 of the 12 control subjects and 30 of the 81 sporadic colorectal cancer patients had a mutant HTRA1 genotype. In all 3 HNPCC patients, the same mutant genotypes were identified at all 15 gene loci. Overall, 13 potential susceptibility genes for HNPCC were identified, namely DDX20, ZFYVE26, PIK3R3, SLC26A8, ZEB2, TP53INP1, SLC11A1, LRBA, CEBPZ, ETAA1, SEMA3G, IFRD2 and FAT1.
Keywords: hereditary non-polyposis colorectal cancer, susceptibility gene, whole exome sequencing, single nucleotide polymorphism, DDX20
Introduction
Hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch syndrome, is inherited as an autosomal dominant disease and is the most common hereditary colorectal cancer, accounting for ~50% of familial colorectal cancer and 3% of all colorectal cancer cases (1). Unlike with sporadic colorectal cancer, HNPCC is associated with specific genetic factors and significant clinicopathological features. These features are often associated with synchronous and metachronous colorectal cancer and cause a high incidence of extraintestinal malignant tumors, including endometrial, gastric, renal, pancreatic and ovarian cancer types (2). Inactivation of DNA mismatch repair (MMR) genes, including MLH1, MSH2, MSH6 and PMS2, is the molecular genetic basis of HNPCC pathogenesis. Mutation of MMR genes can result in loss of DNA MMR function, leading to aberrant DNA replication, increased spontaneous mutation frequency and microsatellite instability. This ultimately leads to the transformation of normal cells into malignant cells (3–5).
However, a previous study observed that certain HNPCC patients, diagnosed by the presence of MMR gene mutations, did not meet some of the clinical diagnostic criteria for HNPCC (6). Furthermore, in certain patients meeting the clinical diagnostic criteria for HNPCC, MMR gene mutations could not be detected (7,8). Bashyam et al (8) demonstrated that, among 48 patients with Lynch syndrome, only 58% had MMR gene expression defects, which indicated that other, as yet unidentified, causative genes may be involved in the pathogenesis of HNPCC.
Based upon this assumption, in the present study, whole exome sequencing was performed in 3 HNPCC patients from 1 family and unreported mutations were observed in 15 gene loci. Subsequently, peripheral blood was collected from control subjects, sporadic colorectal cancer patients and the aforementioned 3 HNPCC patients. Single nucleotide polymorphism (SNP) genotyping assays were also performed on the aforementioned 15 genes using the DNA MassARRAY Genetic Analysis system to further verify whether these genes were associated with HNPCC pathogenesis.
Materials and methods
Blood sample collection
All procedures in studies involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committee and the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. All patients signed informed consent forms prior to participation in the study and the study was approved by the Third Xiangya Hospital Ethics Committee (Changsha, China).
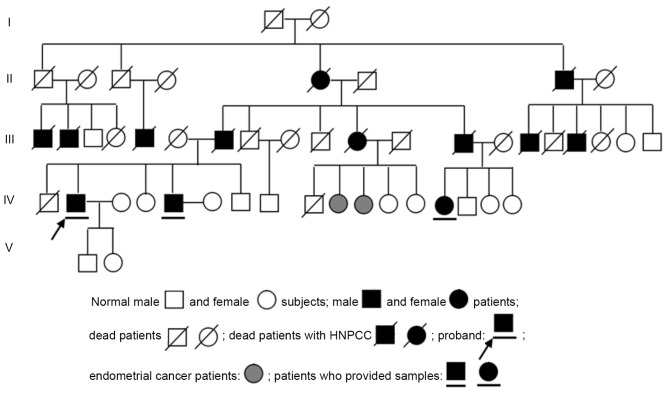
Blood samples were collected from 96 subjects, including 12 control subjects, 81 sporadic colorectal cancer patients who were diagnosed by histopathology from January 2014 to December 2016 at the Third Xiangya Hospital of Central South University and 3 HNPCC patients from the aforementioned hospital who met the Amsterdam Criteria (9), which is outlined as follows: i) ≥3 colorectal cancer cases in the same family diagnosed by histopathology, one case being a first-degree relative (parent or sibling) of the other two cases; ii) ≥2 successive generations affected; iii) ≥1 case with onset prior to the age of 50 years; and iv) familial adenomatous polyposis in HNPCC patients should be excluded. In the HNPCC family investigated in the present study, the proband's father had colorectal cancer that was diagnosed by histopathology and the other 2 cases who provided samples were a sibling and a cousin of the proband. The 3 patients experienced changes in their stools and abdominal bloating prior to being hospitalized. Colorectal cancer was diagnosed by histopathology (all pathology diagnoses were confirmed by two deputy or chief director pathologists) following radical surgery (Table I). The pedigree of the HNPCC family is presented in Fig. 1.
Table I.
Clinical characteristics of 3 hereditary non-polyposis colorectal cancer patients.
| Sample name | Sex | Age, years | Main symptoms | Pathological types |
|---|---|---|---|---|
| Lah | Male | 47 | Stool changes for 1 year, abdominal bloating for 2 months | Moderately-differentiated adenocarcinoma |
| Lyh | Male | 45 | Blood in stools, abdominal bloating, weight loss and stool changes for 2 years | Moderately-differentiated adenocarcinoma |
| Lyl | Female | 42 | Abdominal bloating and pain, hypodynamia and stool changes for 3 months. | Well-differentiated adenocarcinoma |
Figure 1.
Pedigree of the HNPCC family. HNPCC, hereditary non-polyposis colorectal cancer.
Whole exome sequencing
DNA was extracted from the peripheral blood of 3 HNPCC cases and purified using a DNeasy Blood and Tissue kit (cat. no. 69506; Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocols. Exome sequences were subjected to DNA sequencing on the Illumina platform using Illumina PE Flow Cell v3-HS (Illumina, Inc., San Diego, CA, USA). In accordance with the manufacturer's protocols, genomic DNA fragments were processed by end repair, addition of adenosine (A) to 3′ ends, ligation, DNA enrichment and hybridization. DNA libraries from samples were constructed. The concentration, purity and size of the libraries were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA) and a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The hybridization of sequencing primers and the generation of clusters were performed using cBot (HiSeq 2500; Illumina, Inc.) following the cBot User Guide (Part #15006165; Rev. F; lllumina, Inc.). A paired-end sequencing was then performed on a cluster-containing flow cell following the manufacturer's protocols (HiSeq 2500; Illumina, Inc.). Data acquisition software (Illumina, Inc.) was used for quality control and data analysis. The quality control standards for sequencing results were as follows: The average coverage for an exon region was ~100 times; if the average coverage was <90 times, it was resequenced; and at 100 times coverage, ≥85% of exon regions were covered by ≥1 sequence (Table II). The Burrows-Wheeler Alignment software package (version 0.5.9; Shanghai Biotechnology, China) was used to map sequences using human hg19 as the reference genome. Potential PCR duplicates were removed using rmdup of Samtools-0.1.18 (Shanghai Biotechnology, China), and mapping statistics were generated using Samtools flagstat (Shanghai Biotechnology, China) (Table III). Capture-enrichment methods were used to determine the amount of fragment from the captured target region and the coverage and depth of the target region.
Table II.
Sequence quality control results for 3 hereditary non-polyposis colorectal cancer patients.
| Sample name | Orientation | Total reads, n | Total bases, n | Q20, % | Depth | 1×, % | Quality of sequencing results |
|---|---|---|---|---|---|---|---|
| Lah-1 | Forward | 48,356,713 | 4,835,671,300 | 98 | 105.69 | 99.95 | Good |
| Lah-2 | Reverse | 48,356,713 | 4,835,671,300 | 97 | Good | ||
| Lyl-1 | Forward | 73,411,952 | 7,341,195,200 | 97 | 149.87 | 99.86 | Good |
| Lyl-2 | Reverse | 73,411,952 | 7,341,195,200 | 95 | Good | ||
| Lyh-1 | Forward | 57,778,031 | 5,777,803,100 | 97 | 120.25 | 99.96 | Good |
| Lyh-2 | Reverse | 57,778,031 | 5,777,803,100 | 95 | Good |
The Q20 value refers to the probability of error given to the identified base in the base calling process. If the mass value is Q20, the probability of error recognition is 1%, that is, the error rate is 1% or the correct rate is 99%. The 1× value refers to the likelihood that there is at least one read coverage in the genome sequence.
Table III.
Sequence mapping information for 3 hereditary non-polyposis colorectal cancer patients.
| Sample name | Filtered reads, n | Mapped reads, n | Map ratio, % | Unique mapped reads, n | Unique mapped ratio, % |
|---|---|---|---|---|---|
| Lyh | 107,719,930 | 105,656,069 | 98.08 | 95,653,932 | 88.80 |
| Lyl | 136,284,036 | 133,688,255 | 98.10 | 120,411,348 | 88.35 |
| Lah | 92,361,758 | 91,224,395 | 98.77 | 83,257,847 | 90.14 |
Filtered reads, number of reads that pass filtering with sequenator; mapped reads, number of reads that map to each reference sequence; map ratio, ratio of mapped reads to filtered reads; unique mapped reads, number of reads that can map to each reference sequence after removing potential polymerase chain reaction duplicates using the Samtools rmdup tool; Unique Mapped Ratio, ratio of unique mapped reads to mapped reads.
SNP genotyping
DNA extraction
DNA was extracted from peripheral blood lymphocytes of the 96 samples using a DNeasy Blood and Tissue kit (Qiagen, Inc.), according to the manufacturer's protocols. DNA was quantified and assessed using a NanoDrop®ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and 0.8% agarose gel electrophoresis.
PCR amplification
PCR primer mixes were obtained from Invitrogen (Invitrogen, Thermo Fisher Scientific, Inc.) (Table IV). The Q-PCR Detection Kit was purchased from GeneCopoeia (Rockville, MD, USA). The First Strand cDNA Synthesis kit was purchased from Fermentas (Thermo Fisher Scientific, Inc.). Total PCR volume was 5 µl, including 1 µl template DNA, 1.8 µl ddH2O, 0.5 µl 10X PCR Buffer, 0.1 µl 25 mmol/l dNTPs, 0.4 µl 25 mmol/l MgCl2, 1 µl PCR Primer (0.5 mmol/l) and 0.2 µl Gold Tag PCR enzyme (Advanced Biotechnologies Inc., Eldersburg, MD, USA). PCR conditions were 95°C for 2 min, then 45 cycles of 95°C for 30 sec, 50°C for 30 sec, 72°C for 1 min and 72°C for 5 min.
Table IV.
Names and sequences of polymerase chain reaction primers.
| Name of primer | Sequence of primer |
|---|---|
| BB14228-3chr1_112298707-F | ACGTTGGATGACCGCTCAGGATCTCAGCAG |
| BB14228-3chr14_68264412-F | ACGTTGGATGAGCAACCTTCCCGAAGATAC |
| BB14228-3chr1_46509382-F | ACGTTGGATGATCCTTGGTTCAGCACAACG |
| BB14228-3chr6_35911730-F | ACGTTGGATGCCCTCTGGTGAGTATGAATC |
| BB14228-3chr2_145156750-F | ACGTTGGATGAATTTTCAGCAGTTCATCGG |
| BB14228-3chr8_95952304-F | ACGTTGGATGCTGTTTACCGGCATCTCTTG |
| BB14228-3chr2_219249005-F | ACGTTGGATGCCTGAAGATCTGACTCGATG |
| BB14228-3chr4_151827481-F | ACGTTGGATGGAACTCAATTGCTATGCAGG |
| BB14228-3chr2_37439069-F | ACGTTGGATGACATCCATGAATGTTCCTCC |
| BB14228-3chr2_67630823-F | ACGTTGGATGGACAAATGCTTTGAAAGAGG |
| BB14228-3chr3_52474994-F | ACGTTGGATGTCACCTAGCTGGTCAAAGTG |
| BB14228-3chr3_50325883-F | ACGTTGGATGCAGAACAATGAGCTACTCCG |
| BB14228-3chr12_88514827-F | ACGTTGGATGTGAGAGAACAGCTGAACTGG |
| BB14228-3chr4_187541196-F | ACGTTGGATGAGTTATCGTTCCGATCACTG |
| BB14228-3chr10_124221227-F | ACGTTGGATGAGAGTCGCCATGCAGATCC |
| BB14228-3chr1_112298707-R | ACGTTGGATGAAAGCAGCAGTGACTCGAAG |
| BB14228-3chr14_68264412-R | ACGTTGGATGAGCTCTTCAGATTACCTGCC |
| BB14228-3chr1_46509382-R | ACGTTGGATGTATCTGCAAAGCGAGGGCAT |
| BB14228-3chr6_35911730-R | ACGTTGGATGATCATCCGTCTATGGCTTCC |
| BB14228-3chr2_145156750-R | ACGTTGGATGCCATCAACCCATACAAGGAC |
| BB14228-3chr8_95952304-R | ACGTTGGATGTCTCCTCCATTGGACATGAC |
| BB14228-3chr2_219249005-R | ACGTTGGATGTTCAGCCTGCGGAAGCTATG |
| BB14228-3chr4_151827481-R | ACGTTGGATGACCTTTTCAAGGCTATATCC |
| BB14228-3chr2_37439069-R | ACGTTGGATGACTTGGTAACCTGGATGACG |
| BB14228-3chr2_67630823-R | ACGTTGGATGGAGAAGAAAGTGATCGTGGG |
| BB14228-3chr3_52474994-R | ACGTTGGATGTGGAGCACTTTCCTCAAGGC |
| BB14228-3chr3_50325883-R | ACGTTGGATGTCTCAAAGCGTGGAACCTTG |
| BB14228-3chr12_88514827-R | ACGTTGGATGAACCTCTTCAGAGCCTCAAC |
| BB14228-3chr4_187541196-R | ACGTTGGATGTCTCTGCAATCTCTGCACTG |
| BB14228-3chr10_124221227-R | ACGTTGGATGAGCGGCCGGCCCGGGACAG |
| BB14228-3chr1_112298707-EX | CCGCCAACAGCACATCC |
| BB14228-3chr14_68264412-EX | GGCTCCTTCTTGTCCGA |
| BB14228-3chr1_46509382-EX | TCTGTGCATGAACAGGG |
| BB14228-3chr6_35911730-EX | TGGTCAGTGGAGAGGAGA |
| BB14228-3chr2_145156750-EX | ACTATGCTATGAACATGGA |
| BB14228-3chr8_95952304-EX | CCATTGGACATGACTCAAAC |
| BB14228-3chr2_219249005-EX | AAGCTATGGGCCTTCACGGGG |
| BB14228-3chr4_151827481-EX | CTATATCCTATTACCAAGAAGC |
| BB14228-3chr2_37439069-EX | GATGAAGTTTCTTTAGGAAGTA |
| BB14228-3chr2_67630823-EX | GAAAGTGATCGTGGGGTTTTAT |
| BB14228-3chr3_52474994-EX | TCCTCAAGGCCAGGCTGGTCTGCT |
| BB14228-3chr3_50325883-EX | TTGCAGGCCTTCAGGGCAGTGGCA |
| BB14228-3chr12_88514827-EX | AGCCTCAACTAATTCTTTATCCTTTT |
| BB14228-3chr4_187541196-EX | CTCTGCACTGTAGAAAGGTTTTTCAA |
| BB14228-3chr10_124221227-EX | GGCCGGCCCGGGACAGCTGCGCCGAG |
Shrimp alkaline phosphatase (SAP) purification
The total volume for the SAP purification reaction was 2 µl. This included 1.53 µl ddH2O, 0.17 µl SAP Buffer and 0.3 µl SAP enzyme (Sequenom, San Diego, CA, USA). Reaction conditions were 37°C for 40 min and 85°C for 5 min.
Extension reaction
The extension reaction was performed using a 9700 PCR instrument (Sequenom, Inc., San Diego, CA, USA) according to the manufacturer's protocol. The Complete iPLEX® Gold Genotyping Reagent Set was purchased from Sequenom. The total volume of the extension reaction was 2 µl and included 0.619 µl ddH2O, 0.2 µl iPLEX GOLD Buffer, 0.2 µl iPLEXTermination mix, 0.94 µl iPLEX Extension Primer mix and 0.041 µl iPLEX Enzyme. Extension reaction conditions were 40 cycles of 94°C for 30 sec and 94°C for 5 sec, and 5 cycles of 52°C for 5 sec and 80°C for 5 sec, followed by 1 cycle of 72°C for 3 min. The PCR products were purified using resin, were spotted onto a chip and were analyzed on the MassARRAY Platform SEQUENOM Analyzer 4 (Sequenom, Inc.).
Results
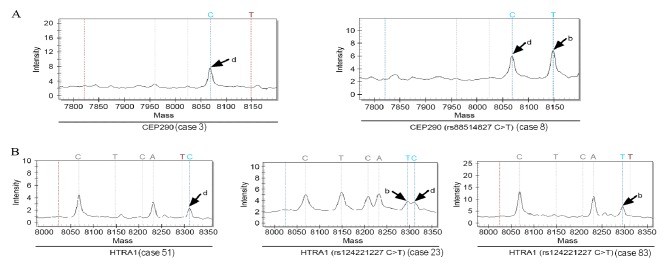
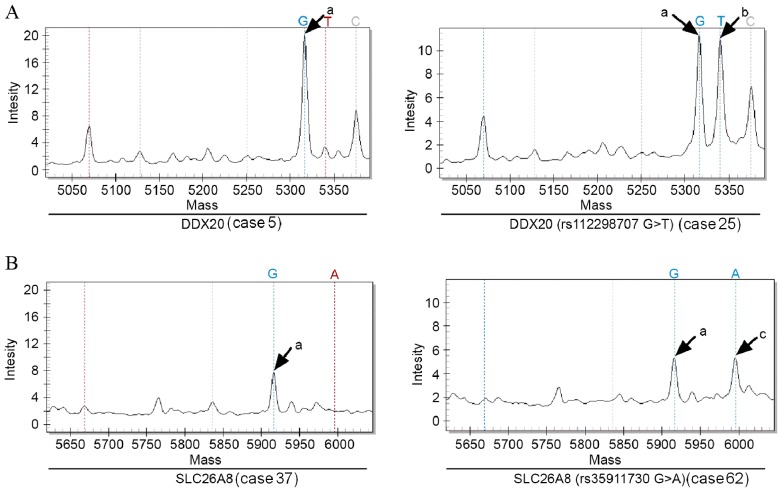
Whole exome sequencing generated 60.4 Gb of data from the 3 HNPCC patients. These data were screened as follows: i) Remove bases with low scores in accordance with the quality control specifications that require the sample coverage to be <5 and fraction variation in a single nucleotide to be <40; ii) filter and eliminate bases that do not fall in exonic regions; iii) filter and eliminate proven mutations in controls and common mutations carried by controls that are archived in public genetic mutation databases, namely HAPMAP (ftp://ftp.ncbi.nlm.nih.gov/hapmap/), 1,000 Genomes (http://www.internationalgenome.org/1000-genomes-project-publications) and dbSNP (https://www.ncbi.nlm.nih.gov/snp/); iv) reserve single nucleotide loci changes in non-synonymous mutations and filter out single nucleotide changes in synonymous mutations; and v) select the non-synonymous mutations that are common to the 3 cases. From this analysis the following mutations were identified in 15 genes (Table V): DDX20 (rs112298707), ZFYVE26 (rs68264412), PIK3R3 (rs46509382), SLC26A8 (rs35911730), ZEB2 (rs145156750), TP53INP1 (rs95952304), SLC11A1 (rs219249005), LRBA (rs151827481), CEBPZ (rs37439069), ETAA1 (rs67630823), SEMA3G (rs52474994), IFRD2 (rs50325883), FAT1 (rs18754119), CEP290 (rs88514827) and HTRA1 (rs124221227). SNP genotyping of these 15 genes was then performed in 96 subjects using the DNA MassARRAY Genetic Analysis system (Sequenom) (Table VI). Among the 96 samples, SNP genotyping was successful at all 15 loci in 92, but genotyping of HTRA1 (rs124221227C>T) failed in 4 of the sporadic colorectal cancer samples (Fig. 2). The genotype of CEP290 (rs88514827C>T) in all 12 control subjects was wild-type, while 1 of the 81 patients with sporadic colorectal cancer had a mutation in CEP290 (rs88514827C>T) (Fig. 2A). A total of 5/12 control subjects and 30/81 sporadic colorectal cancer patients had mutations in HTRA1 (rs124221227C>T). The genotypes of the other 13 genes in the 12 control subjects and 81 sporadic colorectal cancer patients were all wild-type, namely DDX20 (rs112298707G>T), ZFYVE26 (rs68264412C>T), PIK3R3 (rs46509382T>C), SLC26A8 (rs35911730G>A), ZEB2 (rs145156750C>A), TP53INP1 (rs95952304T>C), SLC11A1 (rs219249005C>G), LRBA (rs151827481C>T), CEBPZ (rs37439069A>G), ETAA1 (rs67630823A>G), SEMA3G (rs52474994G>A), IFRD2 (rs50325883 T>G) and FAT1 (rs187541196A>C) (two of these were selected as examples and are presented in Fig. 3). In all 3 HNPCC patients, all 15 genes carried the same mutations.
Table V.
15 single nucleotide mutations detected following integration.
| Gene name | Chromosome number | Position | Reference base | Sequencing base | Mutation type | Gene position |
|---|---|---|---|---|---|---|
| DDX20 | chr1 | 112298707 | G | T | Nonsense | Exon |
| ZFYVE26 | chr14 | 68264412 | C | T | Nonsense | Exon |
| PIK3R3 | chr1 | 46509382 | T | C | Nonsense | Exon |
| SLC26A8 | chr6 | 35911730 | G | A | Nonsense | Exon |
| ZEB2 | chr2 | 145156750 | C | A | Nonsense | Exon |
| TP53INP1 | chr8 | 95952304 | T | C | Nonsense | Exon |
| SLC11A1 | chr2 | 219249005 | C | G | Nonsense | Exon |
| LRBA | chr4 | 151827481 | C | T | Nonsense | Exon |
| CEBPZ | chr2 | 37439069 | A | G | Nonsense | Exon |
| ETAA1 | chr2 | 67630823 | A | G | Nonsense | Exon |
| SEMA3G | chr3 | 52474994 | G | A | Nonsense | Exon |
| IFRD2 | chr3 | 50325883 | T | G | Nonsense | Exon |
| FAT1 | chr4 | 187541196 | A | C | Nonsense | Exon |
| CEP290 | chr12 | 88514827 | C | T | Nonsense | Exon |
| HTRA1 | chr10 | 124221227 | C | T | Nonsense | Exon |
Table VI.
Single nucleotide polymorphism genotyping results at 15 gene loci.
| SNP genotype | |||
|---|---|---|---|
| Gene locus | Control subjects | Sporadic colorectal cancer patients | HNPCC family |
| DDX20 (rs112298707) | GG | GG | GT |
| ZFYVE26 (rs68264412) | CC | CC | CT |
| PIK3R3 (rs46509382) | TT | TT | CT |
| SLC26A8 (rs35911730) | GG | GG | GA |
| ZEB2 (rs145156750) | CC | CC | CA |
| TP53INP1 (rs95952304) | TT | TT | CT |
| SLC11A1 (rs219249005) | CC | CC | CG |
| LRBA (rs151827481) | CC | CC | CT |
| CEBPZ (rs37439069) | AA | AA | GA |
| ETAA1 (rs67630823) | AA | AA | GA |
| SEMA3G (rs52474994) | GG | GG | GA |
| IFRD2 (rs50325883) | TT | TT | GT |
| FAT1 (rs187541196) | AA | AA | CA |
| CEP290 (rs88514827) | CC | CC (80/81) CT (1/81) | CT |
| HTRA1 (rs124221227) | CC (7/12) TC (5/12) | CC (47/81) TC (29/81) TT (1/81) not detected (4/81) | TC |
SNP, single nucleotide polymorphism; HNPCC, hereditary non-polyposis colorectal cancer.
Figure 2.
DNA spectrums of (A) CEP290 and (B) HTRA1. b, T-base dissociation absorption peak; d, C-base dissociation absorption peak.
Figure 3.
DNA spectrums of (A) DDX20 and (B) SLC26A8. a, G-base dissociation absorption peak; b, T-base dissociation absorption peak; c, A-base dissociation absorption peak.
Discussion
HNPCC is the most common hereditary colorectal cancer and exhibits familial aggregation; it is often accompanied by synchronous and metachronous colorectal cancer. The incidence of extraintestinal malignant tumors in HNPCC patients was previously revealed to be significantly higher than that in normal subjects (2). MMR gene defects are the molecular genetic basis of HNPCC pathogenesis, and ~90% of MMR gene mutations occur in the hMSH2 and hMLHl genes (10). However, in certain patients who meet the clinical diagnostic criteria for HNPCC, MMR gene defects cannot be detected (11,12).
In the present study, 3 HNPCC cases underwent whole exome sequencing. Mutations were newly identified at 15 gene loci. These 15 genes were investigated using an SNP genotyping assay in 96 subjects, including HNPCC patients, sporadic colorectal cancer patients and control subjects. The 15 loci carried the same mutations in all 3 HNPCC patients. However, in the 12 control subjects and 81 sporadic colorectal cancer patients, genotypes were wild-type at 13 of the 15 gene loci, indicating that mutations in these 13 genes may be associated with HNPCC pathogenesis. A number of these 13 genes have been revealed to be associated with the development and progression of malignant tumors (13–28), autoimmune diseases, tuberculosis and other infectious diseases (28), and sperm differentiation (29). However, the consequences of mutations in these 13 genes have not previously been reported in the pathology of colorectal cancer.
The results of the present study revealed that certain sporadic colorectal cancer patients and control subjects carry mutations in the HTRA1 gene. The expression level of the HTRA1 gene is associated with the prognosis of various types of malignant cancer, including liver cancer, breast cancer and mesothelioma (30,31). Additionally, 1 of the 81 sporadic colorectal cancer patients in the present study carried a mutation in the CEP290 gene that was also present in colorectal cancer patients from the HNPCC family. However, there have been no reports of a correlation between CEP290 mutations and the pathogenesis of malignant tumors. Future studies will further verify whether HTRA1 and CEP290 are susceptibility genes for HNPCC by expanding sample sizes.
In the present study, 13 genes that may be susceptibility genes for HNPCC were identified by whole exome sequencing and SNP genotyping experiments. In the future, studies will focus on large-scale genetic screening and in vivo and in vitro experiments in order to investigate the mechanisms of the confirmed mutations in the development and progression of colorectal cancer. It is anticipated that more pathogenic genes will be discovered and that our understanding of the molecular genetic basis of HNPCC will be improved, thereby providing theoretical guidance for the diagnosis and treatment of HNPCC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81572965) and the 125 Talent Project/New Xiangya Project of the Third Xiangya Hospital of Central South University.
References
- 1.Kastrinos F, Stoffel EM. History, genetics, and strategies for cancer prevention in Lynch syndrome. Clin Gastroenterol Hepatol. 2014;12:715–727. doi: 10.1016/j.cgh.2013.06.031. quiz e41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson P, Riley B. The tumor spectrum in the Lynch syndrome. Fam Cancer. 2005;4:245–248. doi: 10.1007/s10689-004-7994-z. [DOI] [PubMed] [Google Scholar]
- 3.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 4.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 5.Whitehouse A, Meredith DM, Markham AF. DNA mismatch repair genes and their association with colorectal cancer (Review) Int J Mol Med. 1998;1:469–474. doi: 10.3892/ijmm.1.2.469. [DOI] [PubMed] [Google Scholar]
- 6.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 8.Bashyam MD, Kotapalli V, Raman R, Chaudhary AK, Yadav BK, Gowrishankar S, Uppin SG, Kongara R, Sastry RA, Vamsy M, et al. Evidence for presence of mismatch repair gene expression positive Lynch syndrome cases in India. Mol Carcinog. 2015;54:1807–1814. doi: 10.1002/mc.22244. [DOI] [PubMed] [Google Scholar]
- 9.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 10.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition-Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindor NM. Familial colorectal cancer type X: The other half of hereditary nonpolyposis colon cancer syndrome. Surg Oncol Clin N Am. 2009;18:637–645. doi: 10.1016/j.soc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieminen TT, O'Donohue MF, Wu Y, Lohi H, Scherer SW, Paterson AD, Ellonen P, Abdel-Rahman WM, Valo S, Mecklin JP, et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology. 2014;147:595–598.e5. doi: 10.1053/j.gastro.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-6426. [DOI] [PubMed] [Google Scholar]
- 14.Shin EM, Hay HS, Lee MH, Goh JN, Tan TZ, Sen YP, Lim SW, Yousef EM, Ong HT, Thike AA, et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J Clin Invest. 2014;124:3807–3824. doi: 10.1172/JCI73451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagona AP, Nezis IP, Bache KG, Haglund K, Bakken AC, Skotheim RI, Stenmark H. A tumor-associated mutation of FYVE-CENT prevents its interaction with Beclin 1 and interferes with cytokinesis. PLoS One. 2011;6:e17086. doi: 10.1371/journal.pone.0017086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen JF BB, Zhang XB, Jiang-Ping XU. Expression and significance of ZFYVE26 in hepatocellular carcinoma. J Prac Med. 2012;28:1939–1942. [Google Scholar]
- 17.Cao G, Dong W, Meng X, Liu H, Liao H, Liu S. MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol. 2015;36:4453–4459. doi: 10.1007/s13277-015-3085-z. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Yang X, Li C, Cao X, Luo X, Hu J. PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol Cancer Ther. 2014;13:1837–1847. doi: 10.1158/1535-7163.MCT-14-0049. [DOI] [PubMed] [Google Scholar]
- 19.Wong TS, Gao W, Chan JY. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. Biomed Res Int. 2014;2014:921564. doi: 10.1155/2014/921564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahbazi J, Lock R, Liu T. Tumor protein 53-induced nuclear protein 1 enhances p53 function and represses tumorigenesis. Front Genet. 2013;4:80. doi: 10.3389/fgene.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, Ma L, Li J, Gu J, Shi Q, Yu R. Effects of SEMA3G on migration and invasion of glioma cells. Oncol Rep. 2012;28:269–275. doi: 10.3892/or.2012.1796. [DOI] [PubMed] [Google Scholar]
- 22.Valletta D, Czech B, Spruss T, Ikenberg K, Wild P, Hartmann A, Weiss TS, Oefner PJ, Müller M, Bosserhoff AK, Hellerbrand C. Regulation and function of the atypical cadherin FAT1 in hepatocellular carcinoma. Carcinogenesis. 2014;35:1407–1415. doi: 10.1093/carcin/bgu054. [DOI] [PubMed] [Google Scholar]
- 23.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JW, Gamsby JJ, Highfill SL, Mora LB, Bloom GC, Yeatman TJ, Pan TC, Ramne AL, Chodosh LA, Cress WD, et al. Deregulated expression of LRBA facilitates cancer cell growth. Oncogene. 2004;23:4089–4097. doi: 10.1038/sj.onc.1207567. [DOI] [PubMed] [Google Scholar]
- 25.Herold T, Metzeler KH, Vosberg S, Hartmann L, Röllig C, Stölzel F, Schneider S, Hubmann M, Zellmeier E, Ksienzyk B, et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124:1304–1311. doi: 10.1182/blood-2013-12-540716. [DOI] [PubMed] [Google Scholar]
- 26.Wu DI, Liu L, Ren C, Kong D, Zhang P, Jin X, Wang T, Zhang G. Epithelial-mesenchymal interconversions and the regulatory function of the ZEB family during the development and progression of ovarian cancer. Oncol Lett. 2016;11:1463–1468. doi: 10.3892/ol.2016.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M, Li D, Neale RE, Olson SH, Scelo G, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911–916. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archer NS, Nassif NT, O'Brien BA. Genetic variants of SLC11A1 are associated with both autoimmune and infectious diseases: Systematic review and meta-analysis. Genes Immun. 2015;16:275–283. doi: 10.1038/gene.2015.8. [DOI] [PubMed] [Google Scholar]
- 29.Lohi H, Kujala M, Makela S, Lehtonen E, Kestila M, Saarialho-Kere U, Markovich D, Kere J. Functional characterization of three novel tissue-specific anion exchangers SLC26A7, -A8 and -A9. J Biol Chem. 2002;277:14246–14254. doi: 10.1074/jbc.M111802200. [DOI] [PubMed] [Google Scholar]
- 30.Zhu F, Jin L, Luo TP, Luo GH, Tan Y, Qin XH. Serine protease HtrA1 expression in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2010;9:508–512. [PubMed] [Google Scholar]
- 31.Lehner A, Magdolen V, Schuster T, Kotzsch M, Kiechle M, Meindl A, Sweep FC, Span PN, Gross E. Downregulation of serine protease HTRA1 is associated with poor survival in breast cancer. PLoS One. 2013;8:e60359. doi: 10.1371/journal.pone.0060359. [DOI] [PMC free article] [PubMed] [Google Scholar]