Abstract
Abnormal apolipoprotein E (APOE) methylation has been demonstrated to be associated with Alzheimer's disease, which may have overlapping mechanisms with autism spectrum disorder (ASD). Thus, the purpose of the present study was to assess the possible link between APOE methylation and ASD. Genomic DNA was extracted from peripheral blood and subjected to a methylation assay. SYBR green-based quantitative methylation-specific polymerase chain reaction analysis was used to measure APOE methylation in 62 pediatric patients with ASD and 73 age-matched healthy subjects. The APOE methylation in each sample was expressed as a percentage of methylation of a reference (PMR). The results indicated that APOE methylation in pediatric patients with ASD was significantly higher than that in the healthy controls (median PMR, 33 vs. 11%; P=2.36×10−10). Receiver operating characteristic curve demonstrated that PMR of 15.4% was the optimal cut-off for predicting ASD (area under curve, 0.817; sensitivity, 93.5%; specificity, 72.6%; P=2.36×10−10). In summary, the present results indicated that APOE hypermethylation in peripheral blood DNA may be used as a diagnostic biomarker for ASD.
Keywords: apolipoprotein E, autism spectrum disorder, DNA methylation, peripheral blood, male
Introduction
Autism spectrum disorder (ASD) comprises a range complex neurological diseases with impairments in social skills, communication and repetitive behaviors (1). The incidence of ASD in males is four times of that in females (2). ASD causes great obstacles in interpersonal relationships (3), and it has become a heavy burden on society and the families of affected individuals (4,5). Early intervention may change the long-term prognosis of ASD patients (6).
The pathogenesis of ASD involves a complex interaction between heredity and environment (7). Epigenetics may provide the best bridge between genetic and environmental factors (8). DNA methylation is an important epigenetic modification that regulates the expression of numerous functional genes in the human nervous system (6,9). Aberrant DNA methylation has been reported in diseases or disorders of the central nervous system (10,11). These include hypermethylation of oxytocin R (12), Reelin (13), and SH3 and multiple ankyrin repeat domains 3 (14) in ASD.
Apolipoprotein E (APOE) encodes a glycoprotein associated with lipoproteins in the periphery and the brain (15,16). Abnormal methylation of APOE was reported to be associated with Alzheimer's disease (AD) (17), which may have overlapping mechanisms with ASD (18). A genetic study indicated that a variant of APOE is associated with ASD (19); however, it has remained elusive whether APOE methylation is linked to ASD.
In the light of those previous results, the goal of the present study was to explore whether APOE methylation is associated with ASD.
Materials and methods
Patients and samples
A total of 135 pediatric subjects, including 62 ASD patients (with no other disorders or diseases) and 73 healthy age-matched individuals, volunteered for the study and were recruited at Ningbo Kangning Hospital (Ningbo, China) between September 2015 and September 2017. The ASD patients were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, the Autism Diagnostic Observation Schedule and the Childhood Autism Rating Scales (CARS) (5). Blood samples were drawn from healthy controls and ASD cases prior to treatment. The clinical data acquired from patients mainly included maternal pregnancy history, family history, and disease duration. The study was approved by the Ethics Committees in Ningbo Kangning Hospital and Ningbo University (Ningbo, China), and written informed consent was obtained from the parents of all patients.
DNA isolation and bisulphite conversion
DNA was extracted from blood with the E.Z.N.A.™ Blood DNA Kit (Omega Bio-Tek, Norcross, GA, USA), according to the manufacturer's instructions. The Nanodrop2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to measure the DNA concentration. The EZ DNA Methylation-Gold kit™ (Zymo Research, Irvine, CA, USA) was used to convert unmethylated cytosines into the corresponding uracils, while the methylated cytosines remained in their positions.
SYBR green-based quantitative methylation-specific polymerase chain reaction (qMSP)
qMSP was used for to assess the methylation of APOE. The reaction system contained 10 µl SYBR Green I Master Mix (Roche Diagnostics, Basel, Switzerland), 0.5 µl forward primer (10 µm), 0.5 µl reverse primer (10 m), 1.0 µl templates and 8 µl double-distilled (dd)H2O. The thermocycling conditions comprised an initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 30 sec, 58°C for 30 sec and 72°C for 30 sec. Subsequently, melting curve analysis was performed at 95°C for 15 sec and 60°C for 1 min, followed by increases in the temperature by 0.11°C per sec to 95°C. Human sperm DNA (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) was methylated as the positive control by excess SssI methyltransferase (Thermo Fisher Scientific, Inc.), and ddH2O as a negative control. ACTB was used as the internal reference to correct the differences in the quality and quantity between samples. The sequences of the qMSP primers were APOE forward, 5′-CGAGGTGTAGGTTATGTTC-3′ and reverse, 5′-TACGCAACTTACGCAAAT-3′; and β-actin (ACTB) forward, 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse, 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′. The percentage of methylation of a reference (PMR) of APOE in each sample was calculated by the following approach: ΔΔCq=sample DNA (CqAPOE gene-CqACTB control)-fully methylated DNA (CqAPOE gene-CqACTB control) (20,21).
Statistical analysis
All statistical analyses were performed using SPSS software version 18.0 (SPSS, Inc., Chicago, IL, USA). The independent-samples t-test was applied for the comparisons of differences in APOE methylation between ASD cases and controls. Receiver operating characteristic curves (ROC) were drawn to evaluate the diagnostic value of APOE methylation for ASD. The Spearman rank test was applied to assess the associations between APOE methylation and clinical indicators in ASD. P<0.05 was considered to indicate a statistically significant difference.
Results
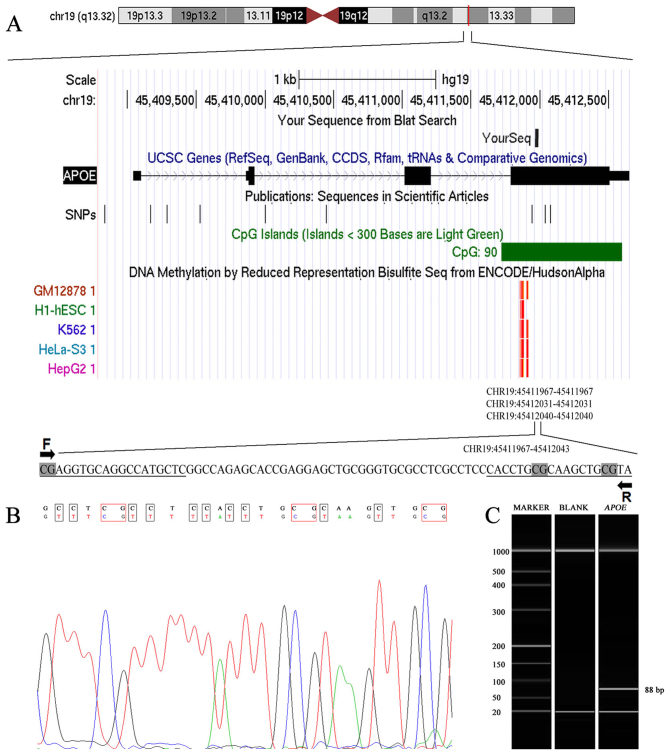
As presented in Fig. 1A, a methylation assay was performed on an 88-bp fragment of the CpG island of APOE. A successful bisulphite transformation was evidenced by detection of transformed thymine by Sanger sequencing (Fig. 1B). The target fragment length of 88 bp was confirmed by capillary electrophoresis (Fig. 1C).
Figure 1.
Characteristics of target sequences in the APOE gene. (A) APOE is located on chr19 (q13.32) and the 80-bp target fragment is located on the CpG island of APOE. (B) Sanger sequencing verified a successful bisulfite conversion. The upper row represents the original sequence and the lower row represents the converted sequence. (C) Results of capillary electrophoresis. MARKER denotes the DNA ladder marker and BLANK denotes an experimental negative control. The numbers on the left indicate the number of bps of the DNA ladder markers. The target fragment length was 88 bp. Chr, chromosome; APOE, apolipoprotein E; F, forward; R, Reverse; Seq, sequencing; UCSC, University of California, Santa Cruz; SNP, single nucleotide polymorphism.
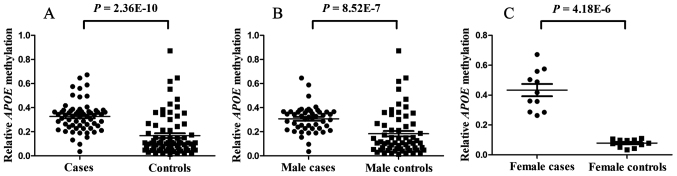
The present results indicated that APOE methylation in pediatric ASD patients was significantly higher than that in healthy age-matched individuals (median PMR, 33.36 vs. 10.61%; P=2.36×10−10; Fig. 2A). APOE hypermethylation was identified in 36 out of 62 subjects with ASD. Considering the higher prevalence of ASD in males, a breakdown analysis by gender was performed. The results indicated that APOE methylation levels were higher in male ASD cases than in healthy male controls (median PMR, 32.12 vs. 11.00%; P=8.51×10−7; Fig. 2B). Similar results were also obtained in females (mean PMR, 43.35 vs. 7.87%; P=4.18×10−6; Fig. 2C).
Figure 2.
Comparison of the relative APOE methylation between ASD cases and healthy controls. (A) Comparison between ASD cases and controls. The median PMR in cases and controls was 33% (interquartile range, 25 and 37%) and 11% (interquartile range, 6 and 23%), respectively. The P-value for the comparison between the cases and the controls was 2.36×10−10. (B) Comparison between male ASD cases and male controls. The median PMR values of cases and controls were 32% (23, 37%) and 11% (6, 27%), respectively. The P-value for the comparison between the male cases and the male controls was 8.52×10−10. (C) Comparison between female ASD cases and female controls. The median PMR values of cases and controls were 0.42 (0.29, 0.56) and 0.08 (0.05, 0.10), respectively. The P-value for the comparison between the female cases and the female controls was 4.18×10−6. APOE, apolipoprotein E; ASD, autism spectrum disorder; PMR, percentage of methylation of a reference.
In addition, ROC curve analysis was performed to quantitatively evaluate the diagnostic value. As presented in Fig. 3A, the area under the curve (AUC) was 0.817 [95% confidence interval (CI), 0.741–0.893]. The ROC curves demonstrated that PMR of 15.4% was the optimal cut-off for predicting ASD (sensitivity, 93.5%, specificity, 72.6%; P=2.36×10−10; Fig. 3A). Further subgroup analysis by gender provided a similar result in males (AUC=0.772; 95% CI, 0.682–0.865; sensitivity, 93.9%; specificity, 67.7%; P=8.52×10−7; Fig. 3B), PMR of 15.3% was the optimal cut-off for predicting ASD. Due to a limited number of female individuals, it was not possible to perform any ROC curve analysis in females.
Figure 3.
ROC curve of APOE methylation for the diagnosis of ASD. The ROC curve was generated to demonstrate the sensitivity (Y-axis) and the specificity (X-axis) of the continuous indicators. (A) A ROC curve was used to estimate the diagnostic value of APOE methylation for ASD. The AUC was 0.817 (95% CI: 0.741–0.893) with a sensitivity of 93.5% and a specificity of 72.6% when the cut-off was 15.4%. (B) ROC curve analysis in the male sub-population (AUC=0.773; 95% CI, 0.682–0.865; sensitivity, 93.9%; specificity, 67.7%). AUC, area under the curve; ROC, receiver operating characteristic; APOE, apolipoprotein E; ASD, autism spectrum disorder.
Discussion
APOE is a multifunctional protein in the brain, and it is known as a carrier of cholesterol and other lipids in the central nervous system (22–24). APOE methylation levels have been reported to be significantly elevated in AD patients (17). Previous studies have indicated that ASD and AD have various common mechanisms in the course of their development (25–27). Therefore, it was speculated that APOE is associated with the occurrence and the development of ASD. The present results demonstrated that APOE hypermethylation is significantly associated with ASD. Based on the ROC curve, APOE hypermethylation may be regarded as a potential biomarker for the diagnosis of ASD.
APOE polymorphisms (APOE ε2 and APOE ε4) are involved in the etiological complexity of the predisposition for ASD (19,28). A transmission distortion of the APOE ε2 allele has been reported in families with cases of ASD. Compared with APOE ε3 and APOE ε4, the APOE ε2 protein product displays the lowest receptor binding affinity (28). DNA methylation is an epigenetic modification and regulates gene expression in response to environmental stimuli (29). Gene methylation is generally inversely correlated with gene expression (30–32). APOE expression has been reported to be downregulated by APOE hypermethylation in malignant transformed TRL 1215 cells (33). Due to material restrictions, it was not possible to measure the correlation between APOE expression and APOE methylation in the present study. Thus, The Cancer Genome Atlas (TCGA) database was used to collect the methylation and the transcription data of APOE. This data analysis in TCGA indicated that APOE hypermethylation was indeed inversely correlated with lower expression (P=1.96×10−27, r=−0.604). In the present case-control study, APOE hypermethylation was detected in pediatric patients with ASD compared with healthy age-matched individuals. Therefore, it was speculated that APOE hypermethylation may reduce APOE expression, eventually leading to the onset of ASD.
Early intervention of ASD may change the long-term prognosis of ASD patients (6). The current diagnostic methods for ASD comprise the ASD behavior checklist (ABC) (34), the CARS (5) and the Social Communication Questionnaire (SCQ) (35). The ABC scale has a specificity of 97% and a sensitivity of 38% with an idiomatic cutoff score at 67 (36). In addition, the CARS has a sensitivity of 83% and a specificity of 82% (37), and the SCQ scale has a sensitivity of 85% and a specificity of 75% (35). These tests are also substantially used in the clinic.
A large variety of molecular biomarkers have been studied for their diagnostic value in ASD, including paraoxonase-1 (PON-1), lipoprotein-associated phospholipase A2 (Lp-PLA2) (38), microRNAs (39,40), interleukin (IL)-6 and tumor necrosis factor (TNF)α (41). However, only few biomarkers are actually used as an auxiliary in the diagnosis of ASD in the clinic. The sensitivity of Lp-PLA2 is 70% and its specificity is 77%; furthermore, PON-1 has a relatively low AUC of 0.660 (sensitivity, 59%; specificity, 63%) (38). IL-6 and TNFα have a low sensitivity of 84.0 and 76.0%, respectively, for the diagnosis of ASD (41). In the present study, APOE methylation yielded a higher AUC of 0.817 (sensitivity, 93.5%; specificity, 72.6%). It is possible to accurately measure DNA methylation in a variety of materials, including plasma and serum, and it is considered an ideal marker for experimental determination (42,43). The results of the present study indicated that APOE methylation has a high diagnostic value for ASD, suggesting that APOE hypermethylation in peripheral blood may be a biomarker for the early diagnosis of ASD.
In conclusion, the present study indicated that APOE hypermethylation is associated with ASD. Future studies may be necessary to confirm the utility of APOE hypermethylation in peripheral blood DNA as a diagnostic marker for ASD.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Medical Science and Technology Project of Zhejiang Province (grant no. 2017207569) and the K.C. Wong Magna Fund of Ningbo University.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
ZH, YY and SD contributed to the conception, design and final approval of the submitted version. YZ, HY, XY, DZ, JZ, ZZ, JL, RP, WZ and FC contributed by performing the experiments, interpreting the data and designing the figures. All authors read and approved the final manuscript.
Ethical approval and consent to participate
The study was approved by the Ethics Committees in Ningbo Kangning Hospital and Ningbo University and written informed consent was obtained from the parents of all the children.
Consent for publication
Written informed consent was obtained for the publication of the patient's data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shpyleva S, Ivanovsky S, de Conti A, Melnyk S, Tryndyak V, Beland FA, James SJ, Pogribny IP. Cerebellar oxidative DNA damage and altered DNA methylation in the BTBR T+tf/J mouse model of autism and similarities with human post mortem cerebellum. PLoS One. 2014;9:e113712. doi: 10.1371/journal.pone.0113712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginsberg MR, Rubin RA, Falcone T, Ting AH, Natowicz MR. Brain transcriptional and epigenetic associations with autism. PLoS One. 2012;7:e44736. doi: 10.1371/journal.pone.0044736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird G, Cook R. Mixed emotions: The contribution of alexithymia to the emotional symptoms of autism. Transl Psychiatry. 2013;3:e285. doi: 10.1038/tp.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buescher AV, Cidav Z, Knapp M, Mandell DS. Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr. 2014;168:721–728. doi: 10.1001/jamapediatrics.2014.210. [DOI] [PubMed] [Google Scholar]
- 5.Howsmon DP, Kruger U, Melnyk S, James SJ, Hahn J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput Biol. 2017;13:e1005385. doi: 10.1371/journal.pcbi.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernell E, Eriksson MA, Gillberg C. Early diagnosis of autism and impact on prognosis: A narrative review. Clin Epidemiol. 2013;5:33–43. doi: 10.2147/CLEP.S41714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Giri M, Xia Z, Subedi YN, Li Y. Genetic and epigenetic mechanisms of epilepsy: A review. Neuropsychiatr Dis Treat. 2017;13:1841–1859. doi: 10.2147/NDT.S142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: A view from the tip of the iceberg. Clin Immunol. 2003;109:80–88. doi: 10.1016/S1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 9.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J, Wang Y, Zhou K, Wang L, Li J, Zhuang Q, Xu X, Xu L, Zhang K, Dai D, et al. Male-specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS One. 2014;9:e89128. doi: 10.1371/journal.pone.0089128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai D, Cheng J, Zhou K, Lv Y, Zhuang Q, Zheng R, Zhang K, Jiang D, Gao S, Duan S. Significant association between DRD3 gene body methylation and schizophrenia. Psychiatry Res. 2014;220:772–777. doi: 10.1016/j.psychres.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 12.Yuksel Elagoz M, Yuceturk B, Karatas OF, Ozen M, Dogangun B. The altered promoter methylation of oxytocin receptor gene in autism. J Neurogenet. 2016;30:280–284. doi: 10.1080/01677063.2016.1202951. [DOI] [PubMed] [Google Scholar]
- 13.Lintas C, Sacco R, Persico AM. Differential methylation at the RELN gene promoter in temporal cortex from autistic and typically developing post-puberal subjects. J Neurodev Disord. 2016;8:18. doi: 10.1186/s11689-016-9151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, Bowman R, Yang H, Goldstein J, Li YJ, Jiang YH. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum Mol Genet. 2014;23:1563–1578. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamboli IY, Heo D, Rebeck GW. Extracellular proteolysis of apolipoprotein E (apoE) by secreted serine neuronal protease. PLoS One. 2014;9:e93120. doi: 10.1371/journal.pone.0093120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raiford KL, Shao Y, Allen IC, Martin ER, Menold MM, Wright HH, Abramson RK, Worley G, DeLong GR, Vance JM, et al. No association between the APOE gene and autism. Am J Med Genet B Neuropsychiatr Genet. 2004;125B:57–60. doi: 10.1002/ajmg.b.20104. [DOI] [PubMed] [Google Scholar]
- 17.Foraker J, Millard SP, Leong L, Thomson Z, Chen S, Keene CD, Bekris LM, Yu CE. The APOE gene is differentially methylated in Alzheimer's disease. J Alzheimers Dis. 2015;48:745–755. doi: 10.3233/JAD-143060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Napoli E, Ross-Inta C, Wong S, Hung C, Fujisawa Y, Sakaguchi D, Angelastro J, Omanska-Klusek A, Schoenfeld R, Giulivi C. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: Interplay between Pten and p53. PLoS One. 2012;7:e42504. doi: 10.1371/journal.pone.0042504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giunco CT, de Oliveira AB, Carvalho-Salles AB, Souza DS, Silva AE, da Rocha SS, Fett-Conte AC. Association between APOE polymorphisms and predisposition for autism. Psychiatr Genet. 2009;19:338. doi: 10.1097/YPG.0b013e3283328e41. [DOI] [PubMed] [Google Scholar]
- 20.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive melting analysis after real time-methylation specific PCR (SMART-MSP): High-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Hu H, Liu J, Yang Y, Liu G, Ying X, Chen Y, Li B, Ye C, Wu D, Duan S. FOXF2 promoter methylation is associated with prognosis in esophageal squamous cell carcinoma. Tumour Biol. 2017;39:1010428317692230. doi: 10.1177/1010428317692230. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Mahley RW. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer's diseases. Neurobiol Dis. 2014;72:3–12. doi: 10.1016/j.nbd.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Park S, Allington G, Prelli F, Sun Y, Sun Y, Martá-Ariza M, Scholtzova H, Biswas G2, Brown B, Verghese PB, et al. Targeting Apolipoprotein E/Amyloid β Binding by Peptoid CPO_Aβ17-21 P Ameliorates Alzheimer's disease related pathology and cognitive decline. Sci Rep. 2017;7:8009. doi: 10.1038/s41598-017-08604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao N, Liu CC, Qiao W, Bu G. Apolipoprotein E, Receptors, and Modulation of Alzheimer's Disease. Biol Psychiatry. 2018;83:347–357. doi: 10.1016/j.biopsych.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folmsbee SS, Wilcox DR, Tyberghein K, De Bleser P, Tourtellotte WG, van Hengel J, van Roy F, Gottardi CJ. αT-catenin in restricted brain cell types and its potential connection to autism. J Mol Psychiatry. 2016;4:2. doi: 10.1186/s40303-016-0017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blomqvist ME, Andreasen N, Bogdanovic N, Blennow K, Brookes AJ, Prince JA. Genetic variation in CTNNA3 encoding alpha-3 catenin and Alzheimer's disease. Neurosci Lett. 2004;358:220–222. doi: 10.1016/j.neulet.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Martin ER, Bronson PG, Li YJ, Wall N, Chung RH, Schmechel DE, Small G, Xu PT, Bartlett J, Schnetz-Boutaud N, et al. Interaction between the alpha-T catenin gene (VR22) and APOE in Alzheimer's disease. J Med Genet. 2005;42:787–792. doi: 10.1136/jmg.2004.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persico AM, D'Agruma L, Zelante L, Militerni R, Bravaccio C, Schneider C, Melmed R, Trillo S, Montecchi F, Elia M, et al. Enhanced APOE2 transmission rates in families with autistic probands. Psychiatr Genet. 2004;14:73–82. doi: 10.1097/01.ypg.0000128768.37838.17. [DOI] [PubMed] [Google Scholar]
- 29.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):S245–S254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 30.Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatry. 2007;12(519):593–600. doi: 10.1038/sj.mp.4001965. [DOI] [PubMed] [Google Scholar]
- 31.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M, Takeda S, Teraoka-Nishitani N, Yamagata A, Tanaka T, Sasaki M, Yasuda N, Oda M, Okano T, Yamahira K, et al. Cadmium-induced malignant transformation of rat liver cells: Potential key role and regulatory mechanism of altered apolipoprotein E expression in enhanced invasiveness. Toxicology. 2017;382:16–23. doi: 10.1016/j.tox.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Juneja M, Sharma S, Mukherjee SB. Sensitivity of the autism behavior checklist in Indian autistic children. J Dev Behav Pediatr. 2010;31:48–49. doi: 10.1097/DBP.0b013e3181c7241a. [DOI] [PubMed] [Google Scholar]
- 35.Dykens EM, Roof E, Hunt-Hawkins H, Dankner N, Lee EB, Shivers CM, Daniell C, Kim SJ. Diagnoses and characteristics of autism spectrum disorders in children with Prader-Willi syndrome. J Neurodev Disord. 2017;9:18. doi: 10.1186/s11689-017-9200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordin V, Gillberg C. Autism spectrum disorders in children with physical or mental disability or both. II: Screening aspects. Dev Med Child Neurol. 1996;38:314–324. doi: 10.1111/j.1469-8749.1996.tb12097.x. [DOI] [PubMed] [Google Scholar]
- 37.García-López C, Narbona J. Clinical usefulness of IDEA and CARS: Concordance with DSM-IV-TR in children and adolescents with suspicion of PDD. An Pediatr (Barc) 2014;80:71–76. doi: 10.1016/j.anpedi.2013.05.012. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 38.Hayek J, Cervellati C, Crivellari I, Pecorelli A, Valacchi G. Lactonase activity and lipoprotein-phospholipase A2 as possible novel serum biomarkers for the differential diagnosis of autism spectrum disorders and rett syndrome: Results from a pilot study. Oxid Med Cell Longev. 2017;2017:5694058. doi: 10.1155/2017/5694058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, Wakuda T, Iwata K, Tsujii M, Sugiyama T, Mori N. Serum microRNA profiles in children with autism. Mol Autism. 2014;5:40. doi: 10.1186/2040-2392-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016;16:52. doi: 10.1186/s12887-016-0586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Ansary AK, Ben Bacha AG, Al-Ayadhi LY. Proinflammatory and proapoptotic markers in relation to mono and di-cations in plasma of autistic patients from Saudi Arabia. J Neuroinflammation. 2011;8:142. doi: 10.1186/1742-2094-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu P, Cao Z, Wu S. New progress of epigenetic biomarkers in urological cancer. Dis Markers. 2016;2016:9864047. doi: 10.1155/2016/9864047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.