Abstract
Ewing Sarcoma (ES) is a highly aggressive bone and soft tissue childhood cancer. The development of resistance to chemotherapy is common and remains the main cause of treatment failure. We herein evaluated the expression of genes associated with chemotherapy resistance in ES cell lines. A set of genes (CCAR1, TUBA1A, POLDIP2, SMARCA4 and SMARCB1) was data-mined for resistance against doxorubicin and vincristine, which are the standard drugs used in the treatment of patients with ES. The expression of each gene in SK-ES-1 ES cells was reported before and after exposure to a drug resistance-inducing protocol. There was a significant downregulation of CCAR1 and TUBA1A in doxorubicin-resistant cells, with low expression of TUBA1A in vincristine-resistant cells. By contrast, POLDIP2 was significantly upregulated in cells resistant to either drug, and the expression of the SMARCB1 and SMARCA4 genes was upregulated in doxorubicin-resistant cells. These findings indicate that resistance to specific chemotherapeutic agents was accompanied by differential changes in gene expression in ES tumors.
Keywords: doxorubicin, vincristine, chemoresistance, drug resistance, pediatric cancer
Introduction
Ewing Sarcoma (ES) is a bone and soft tissue tumor of possible neuroectodermal or mesenchymal origin that afflicts children and young adults. Approximately 20–25% of cases are metastatic at diagnosis, and the survival rates are poor in the advanced setting (1,2). ES is characterized by a frequent characteristic cytogenetic translocation of the EWSR1 (22q12) and FLI-1 (11q24) genes. The resulting fusion protein EWS-FLI-1 is responsible for oncogene activation, inhibition of tumor suppression, chromatin remodeling and epigenomic reprogramming (3–5).
Standard multimodality treatment consists of induction chemotherapy followed by local control with surgery and/or radiotherapy and then consolidation chemotherapy with multiple drugs. Active chemotherapy agents in the first-line setting include doxorubicin, vincristine, cyclophosphamide, etoposide and ifosfamide. Treatment efficacy appears to reach a plateau after dose intensification and interval compression of all active agents (6,7), and the development of resistance to chemotherapy remains as the main cause of treatment failure (8,9). Cooperative groups have examined different chemotherapy combinations, dose-intensifying regimens with bone marrow transplantation, metronomic therapy and other synergistic mechanisms associated with the EWS fusion protein in ES. However, these approaches have failed to improve survival rates in clinical trials thus far (10,11). Elucidating the molecular mechanisms underlying the development of chemotherapy resistance in ES may help develop new agents with improved synergy.
Doxorubicin belongs to the class of anthracyclines and acts by topoisomerase II poisoning, creation of double-strand DNA breaks (DSBs), and impairment of DNA repair and supercoiling, leading to changes in epigenetic processes (12,13). Mechanisms involved in resistance to doxorubicin include drug efflux transporters, alterations in the ability of doxorubicin to form DSBs, and alterations in downstream apoptosis signaling triggered by DNA damage (14).
Vincristine, a natural alkaloid extracted from Vinca rosea, interferes with microtubule formation and stability through depolymerization, resulting in cell cycle arrest and apoptosis (15). In addition to affecting chromatin stability (16) by interfering with DNA binding and histone eviction, vincristine also affects topoisomerase IIa levels (17). Resistance to vinca alkaloids involves overexpression of ATP-binding cassette (ABC) transporters such as P-glycoprotein, alterations of β-tubulin (βII, βIII and βIV) and multidrug-resistance proteins (18).
The aim of the present study was to investigate whether the expression of genes associated with resistance to chemotherapy in other tumor types is different in chemotherapy-resistant ES cells. The literature was first data-mined for genes associated with resistance to drugs used in ES treatment, focusing on doxorubicin and vincristine, and then SK-ES-1 cells resistant to these drugs were developed. Subsequently, the expression of the selected genes was evaluated with quantitative polymerase chain reaction (qPCR) analysis.
Materials and methods
Data mining and refining
To select drug resistance genes, the literature was searched for studies investigating resistance to doxorubicin and vincristine in all cancer types. Approximately 270 genes were identified, but only 23 genes appropriately validated by experimental methodologies (mutational and knockout gene analysis) were selected. This database was further enriched with information from online tools, such as DrugBank, Gene Ontology and UniProt. Finally, a set of five genes (CCAR1, TUBA1A, POLDIP2, SMARCA4 and SMARCB1) was selected based on pathways that are important for ES development.
Cell culture
The standard Ewing sarcoma cell line SK-ES-1 (American Type Culture Collection, Manassas, VA, USA) was cultured in RPMI-1640 medium (Gibco®; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% bovine serum, 4 mg/ml gentamicin (Nova Farma, São Paulo, Brazil) and amphotericine B (Fungizone®, Invitrogen; Thermo Fisher Scientific, Inc.).
Chemoresistance induction
For chemotherapy resistance induction, the cells underwent treatment for 72 h, with increasing drug concentration every 2 weeks, for a period of ≥10 weeks. The concentrations were 10, 20, 30, 40 and 50 mM for doxorubicin (Libbs, São Paulo, Brazil) and 0.5, 1, 2, 3 and 4 nM for vincristine (Pfizer, Inc., New York, NY, USA). Untreated cells served as control (9).
Cell proliferation
SK-ES-1 cells were seeded at a density of 2.104 cells per well in 48-well plates. After 24 h, the cells were treated with vincristine and doxorubicin individually, including control. The concentrations for this treatment were 10, 30 and 50 nM for doxorubicin and 1, 3 and 5 nM for vincristine. The medium was removed after 72 h of treatment; the cells were washed with phosphate-buffered saline, detached with 0.25% trypsin solution (no EDTA; Invitrogen; Thermo Fisher Scientific) and then counted with the trypan blue exclusion method in a hemocytometer, as previously described (19). The mean of three experiments for each dose was utilized for calculations.
RNA extraction and cDNA synthesis
RNA was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific) according to the manufacturer's protocol. cDNA was synthesized using the reverse transcription (RT)-PCR kit SuperScriptTM III First-Strand Synthesis SuperMix (Invitrogen; Thermo Fisher Scientific).
qPCR
qPCR was performed using the AB 7500 system (Thermo Fisher Scientific Inc.), with positive and negative controls. Reactions were prepared with KiCqStart® qPCR Ready mix™ (Sigma-Aldrich; Merck KGaA, St. Louis, MO, USA), using a 0.5-µl sample cDNA. Expression levels were evaluated using the 2-ΔΔCq method, with GAPDH used as the housekeeping gene. The primers used for CCAR1, TUBA1A, POLDIP2, SMARCA4 and SMARCB1 are listed in Table I.
Table I.
Quantitative polymerase chain reaction primer sequences.
| Gene | Primer sequence | Product |
|---|---|---|
| SMARCB1 | F: 5′-TCCGTATGTTCCGAGGTTCT-3′ | 154 |
| R: 5′-CTGGTGGCTAGAGTCGTGTA-3′ | ||
| SMARCA4 | F: 5′-GCTCCGAGGTCTGATAGTGA-3′ | 133 |
| R: 5′-CGCTGTCTGGATCTGGAATC-3′ | ||
| CCAR1 | F: 5′-AGAGTTCGACGTGTTGTTCC-3′ | 90 |
| R: 5′-GCGCCTTAGTTCCATCATGT-3′ | ||
| TUBA1A | F: 5′-TTGTTCACTGGTACGTTGGG-3′ | 105 |
| R: 5′-AATCCACACCAACCTCCTCA-3′ | ||
| POLDIP2 | F: 5′-TTCCAGTATAGCAGCCACGT-3′ | 97 |
| R: 5′-GAACATCAAAGTGGGAGCCA-3′ | ||
| GAPDH | F: 5′-CAAGATCATCAGCAATGCCTCC-3′ | 103 |
| R: 5′-GACTGTGGTCATGAGTCCTTCC-3′ |
F, forward; R, reverse.
Statistical analysis
Statistical differences were analyzed using one-way analysis of variance, with the Sidak correction method for multiple comparison tests. Experiments were conducted three times and in triplicates. All statistical analyses were performed using SPSS 16.0 for Windows. The differences were considered statistically significant when P-values were <0.05.
Results
Protein network integration
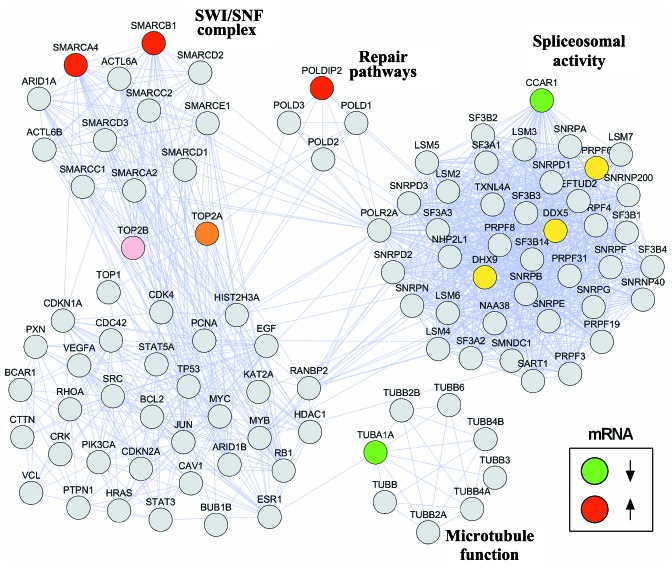
Protein interaction networks are crucial for understanding the biological cellular processes. We designed a protein network to visualize the interactions between the selected genes and molecular pathways associated with ES. A set of genes associated with resistance to doxorubicin and vincristine was manually curated. The open-source software programs Cytoscape version 3.6.0 and String version 10.5 were used to build the network. The interactions between the investigated genes (CCAR1, TUBA1A, POLDIP2, SMARCA4 and SMARCB1) and DNA-topoisomerase II (TOP2A), a target of both doxorubicin and vincristine, and genes directly binding to EWS-FLI1 fusion protein, including those associated with spliceosomal activity that is crucial to ES pathogenesis, are shown in Fig. 1.
Figure 1.
Protein interaction network of genes involved in resistance to chemotherapy. Genes selected for this study are represented in red or green according to higher or lower relative mRNA expression, respectively, between control and chemoresistant Ewing sarcoma cells. DNA-topoisomerase (TOP)2A and TOP2B (pink/orange) were included due to their role as doxorubicin targets. Genes involved in spliceosomal activity and EWS/FLI1 activity are shown in yellow. The network was constructed using the STRING database, version 10.5.
Induction of chemotherapy resistance
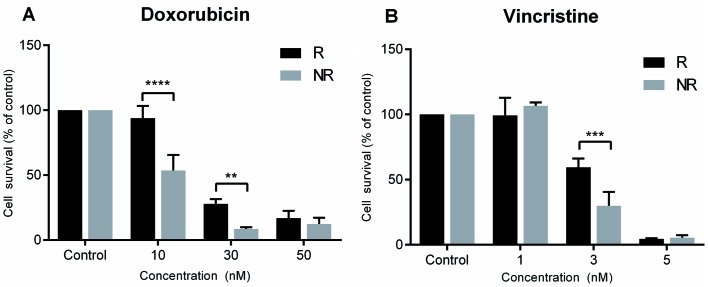
Chemotherapy resistance was successfully induced for both doxorubicin and vincristine in SK-ES-1 cells. Doxorubicin resistance was evidenced at concentrations of 10 nM (P<0.0001 compared with controls) and 30 nM (P<0.005 compared with controls), and vincristine resistance at a concentration of 3 nM (P<0.0005 compared with controls; Fig. 2).
Figure 2.
Chemotherapy resistance in SK-ES-1 Ewing sarcoma cells. Evidence of resistance was observed at concentrations of 10 and 30 nM for doxorubicin and 3 nM for vincristine. Values are presented as percentage of survival of resistant (R) vs. non-resistant (NR) SK-ES-1 cells, for three independent experiments; **P<0.005; ***P<0.0005; and ****P<0.0001 compared with controls cells.
Gene expression
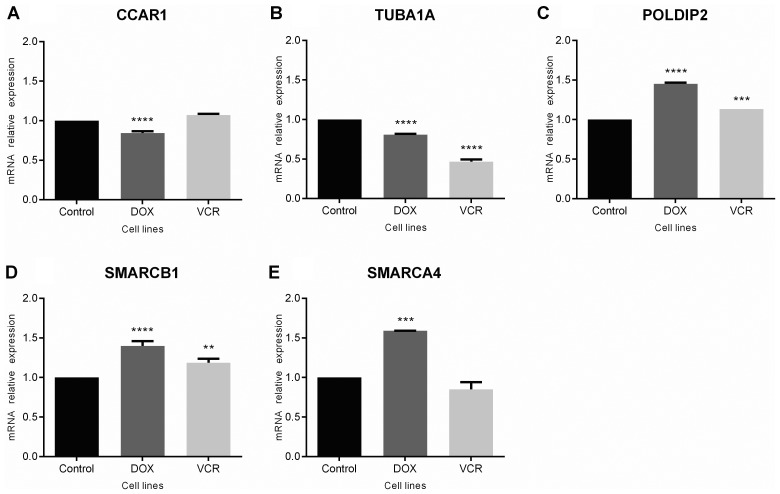
Changes in expression for the selected genes in chemoresistant ES cells were analyzed. The results revealed a modest, but statistically significant downregulation of CCAR1 (P<0.0001 compared with control cells; Fig. 3A) and TUBA1A (P<0.0001 compared with controls) in doxorubicin-resistant cells, with decreased expression of TUBA1A also observed in vincristine-resistant cells (P<0.0001 compared with controls; Fig. 3B). By contrast, POLDIP2 was upregulated in cells resistant to either drug (P<0.0001 for doxorubicin-resistant and P<0.0005 for vincristine-resistant cells; Fig. 3C). For SMARCB1 and SMARCA4, gene expression was upregulated in cells resistant to doxorubicin (P<0.0001 and P<0.0005, respectively), whereas changes were observed in vincristine-resistant cells for SMARCB1 only (P<0.0005; Fig. 3D and E).
Figure 3.
Gene expression analysis of (A) CCAR1, (B) TUBA1A, (C) POLDIP2, (D) SMARCB1 and (E) SMARCA4 in chemoresistant and non-resistant SK-ES-1 Ewing sarcoma cells. Values represent the relative expression of genes in doxorubicin (DOX)-resistant, vincristine (VCR)-resistant, and control SK-ES-1 cells (cell lines); **P<0.005; ***P<0.0005; and ****P<0.0001 compared with control cells.
Discussion
Resistance to chemotherapy remains the main reason for treatment failure in patients with ES. In the present study, a group of genes was selected (TUBA1A, POLDIP2, SMARCA4, SMARCB1 and CCAR1), known in other tumors to be associated with resistance to drugs commonly used in ES, and the gene expression in chemotherapy-resistant ES cells was evaluated. It was demonstrated that the TUBA1A expression levels were lower in cells resistant to either doxorubicin or vincristine, when compared with the non-resistant cells. The TUBA1A gene encodes the α-tubulin protein, which belongs to the tubulin family of proteins that form and organize microtubules that are required for cell division and movement. Given that vincristine is a microtubule-depolymerizing agent, resistance to this agent may develop from changes in tubulin levels (20). Doxorubicin may interfere in TUBA1A levels, with lower gene expression observed in resistant MCF-7 breast cancer cells (21).
Upregulation of POLDIP2 in the chemotherapy-resistant ES cells was also observed. POLDIP2 encodes a protein implicated in the activity of translesional polymerases. The translesion synthesis polymerase and primer extension activities of PrimPol play a role in DNA damage tolerance (22); its involvement in repair processes may explain the increases in cell sensitivity to oxidative stress when POLDIP2 is silenced (23). In addition, POLDIP2 plays an important role in DNA replication/repair and regulation of reactive oxygen species and participates in cytoskeletal reorganization as well as key pathways in cancer, including those involved in autophagy and cell cycle regulation (24). POLDIP2 is also involved in vascular integrity, smooth cell migration and adhesion (25). These processes are important for tumor development and survival, and its upregulation in drug-resistant cells may contribute to increased tumor aggressiveness.
Both SMARCB1 and SMARCA4 are part of the SWI/SNF chromatin-remodeling complex, which recruits TOP2A to DNA and leads to the formation of DSBs and cell death. Loss of the SWI/SNF complex results in drug resistance, including DSBs and repair pathways (26). Knockdown of SMARCA4 and SMARCB1 leads to increased chemotherapy resistance (27). Our results revealed higher expression levels of SMARCA4 and SMARCB1 in doxorubicin-resistant ES cells, suggesting a different landscape in ES. The oncogenic EWS-FLI1 fusion induces chromatin-remodeling patterns, stimulating or repressing enhancers and establishing a modified oncogenic regulatory and interaction network that may explain divergences in drug resistance mechanisms among different tumors (4).
The results of the present study are consistent with previous evidence indicating that loss of SMARCB1 is found in a small percentage of ES patients, and this may interfere favorably with outcome. It is possible that the combination of EWSR1 translocation and SMARCB1 loss increases susceptibility of tumor cells to treatment (28).
CCAR1 is a biphasic regulator of cell growth and apoptosis, and plays an important role in tumorigenesis in gastric cancer (29) and hepatocellular carcinoma (30). CCAR1 can target gene activation by estrogen and glucocorticoid receptors in breast cancer cells (31), and androgen receptors in prostate cancer (32). EWS-FLI1 alters mRNA splicing in ES cells, giving rise to several protein isoforms implicated in oncogenesis (33), and CCAR1 is associated with spliceosomal activity. An unexpected decrease in CCAR1 expression levels was observed in doxorubicin-resistant cells. Further experiments are required to elucidate how this gene is associated with chemotherapy resistance and alternative splicing in ES. Although the genes selected in the present study have different functions and are involved in diverse pathways, our integration network reveals a possible connection among these different mechanisms. Therefore, repair pathways, SWI/SNF chromatin remodeling, microtubule rearrangements and spliceosomal activity may be interacting to maintain chemoresistance mechanisms in ES.
In summary, the present findings provide early evidence revealing novel changes in gene expression associated with chemotherapy resistance in ES cells. Gene knockout assays, characterization of resistance pathways and the use of tumor samples from patients are among the next steps required to confirm and extend these findings, and validate this set of genes as possible targets to counteract therapy resistance in ES.
Acknowledgements
Not applicable.
Funding
This research was supported by PRONON/Ministry of Health, Brazil (grant no. 25000.162.034/2014-21 to C.B.F); the Children's Cancer Institute (ICI); the National Council for Scientific and Technological Development (CNPq; grant no. 303276/2013-4 to R.R.) and the Clinical Hospital institutional research fund (FIPE/HCPA - 150419).
Availability of data and materials
The materials included in the manuscript will be made freely available to any researchers who wish to use them for non-commercial purposes, while preserving any necessary confidentiality and anonymity.
Authors' contributions
LH, MS, CAS, DBO and CBF made substantial contributions to the design of the present study; LH, MS, CAS and DBO performed the experiments; LH, MS, and CBF analysed the data; LH, MS, CAS, DBO, LJG, ALB, ATB, RR and CBF critically revised the manuscript for important intellectual content, wrote and reviewed the manuscript; LH, MS, ATB, RR and CBF wrote the manuscript. All authors critically revised the manuscript and all authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, Kovar H, Grimer R, Whelan J, Claude L, et al. Ewing sarcoma: Current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–3046. doi: 10.1200/JCO.2014.59.5256. [DOI] [PubMed] [Google Scholar]
- 3.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: Promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schönegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23:386–395. doi: 10.1038/nm.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE, Marcus K, Sailer S, Healey JH, Dormans JP, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: A report from the Children's Oncology Group. J Clin Oncol. 2012;30:4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJ, Pritchard DJ, Gebhardt MC, Dickman PS, Perlman EJ, Meyers PA, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing's sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348:694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed AA, Zia H, Wagner L. Therapy resistance mechanisms in Ewing's sarcoma family tumors. Cancer Chemother Pharmacol. 2014;73:657–663. doi: 10.1007/s00280-014-2392-1. [DOI] [PubMed] [Google Scholar]
- 9.Heinen TE, Dos Santos RP, da Rocha A, Dos Santos MP, Lopez PL, Filho Silva MA, Souza BK, Rivero LF, Becker RG, Gregianin LJ, et al. Trk inhibition reduces cell proliferation and potentiates the effects of chemotherapeutic agents in Ewing sarcoma. Oncotarget. 2016;7:34860–34880. doi: 10.18632/oncotarget.8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson PM, Bielack SS, Gorlick RG, Skubitz K, Daw NC, Herzog CE, Monge OR, Lassaletta A, Boldrini E, Pápai Z, et al. A phase II study of clinical activity of SCH 717454 (robatumumab) in patients with relapsed osteosarcoma and Ewing sarcoma. Pediatr Blood Cancer. 2016;63:1761–1770. doi: 10.1002/pbc.26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner LM, Fouladi M, Ahmed A, Krailo MD, Weigel B, DuBois SG, Doyle LA, Chen H, Blaney SM. Phase II study of cixutumumab in combination with temsirolimus in pediatric patients and young adults with recurrent or refractory sarcoma: A report from the Children's Oncology Group. Pediatr Blood Cancer. 2015;62:440–444. doi: 10.1002/pbc.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang B, Qiao X, Janssen L, Velds A, Groothuis T, Kerkhoven R, Nieuwland M, Ovaa H, Rottenberg S, van Tellingen O, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox J, Weinman S. Mechanisms of doxorubicin resistance in hepatocellular carcinoma. Hepat Oncol. 2016;3:57–59. doi: 10.2217/hep.15.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 16.Mohammadgholi A, Rabbani-Chadegani A, Fallah S. Mechanism of the interaction of plant alkaloid vincristine with DNA and chromatin: Spectroscopic study. DNA Cell Biol. 2013;32:228–235. doi: 10.1089/dna.2012.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skladanowski A, Côme MG, Sabisz M, Escargueil AE, Larsen AK. Down-regulation of DNA topoisomerase IIalpha leads to prolonged cell cycle transit in G2 and early M phases and increased survival to microtubule-interacting agents. Mol Pharmacol. 2005;68:625–634. doi: 10.1124/mol.105.013995. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Yang SH, Guo XL. New insights into Vinca alkaloids resistance mechanism and circumvention in lung cancer. Biomed Pharmacother. 2017;96:659–666. doi: 10.1016/j.biopha.2017.10.041. [DOI] [PubMed] [Google Scholar]
- 19.Flores DG, de Farias CB, Leites J, de Oliveira MS, Lima RC, Tamajusuku AS, Di Leone LP, Meurer L, Brunetto AL, Schwartsmann G, et al. Gastrin-releasing peptide receptors regulate proliferation of C6 Glioma cells through a phosphatidylinositol 3-kinase-dependent mechanism. Curr Neurovasc Res. 2008;5:99–105. doi: 10.2174/156720208784310240. [DOI] [PubMed] [Google Scholar]
- 20.Huzil JT, Chen K, Kurgan L, Tuszynski JA. The roles of beta-tubulin mutations and isotype expression in acquired drug resistance. Cancer Inform. 2007;3:159–181. doi: 10.1177/117693510700300028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villeneuve DJ, Hembruff SL, Veitch Z, Cecchetto M, Dew WA, Parissenti AM. cDNA microarray analysis of isogenic paclitaxel- and doxorubicin-resistant breast tumor cell lines reveals distinct drug-specific genetic signatures of resistance. Breast Cancer Res Treat. 2006;96:17–39. doi: 10.1007/s10549-005-9026-6. [DOI] [PubMed] [Google Scholar]
- 22.Guilliam TA, Bailey LJ, Brissett NC, Doherty AJ. PolDIP2 interacts with human PrimPol and enhances its DNA polymerase activities. Nucleic Acids Res. 2016;44:3317–3329. doi: 10.1093/nar/gkw175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maga G, Crespan E, Markkanen E, Imhof R, Furrer A, Villani G, Hübscher U, van Loon B. DNA polymerase δ-interacting protein 2 is a processivity factor for DNA polymerase λ during 8-oxo-7,8-dihydroguanine bypass. Proc Natl Acad Sci USA. 2013;110:18850–18855. doi: 10.1073/pnas.1308760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown DI, Lassègue B, Lee M, Zafari R, Long JS, Saavedra HI, Griendling KK. Poldip2 knockout results in perinatal lethality, reduced cellular growth and increased autophagy of mouse embryonic fibroblasts. PLoS One. 2014;9:e96657. doi: 10.1371/journal.pone.0096657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutliff RL, Hilenski LL, Amanso AM, Parastatidis I, Dikalova AE, Hansen L, Datla SR, Long JS, El-Ali AM, Joseph G, et al. Polymerase delta interacting protein 2 sustains vascular structure and function. Arterioscler Thromb Vasc Biol. 2013;33:2154–2161. doi: 10.1161/ATVBAHA.113.301913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wijdeven RH, Pang B, van der Zanden SY, Qiao X, Blomen V, Hoogstraat M, Lips EH, Janssen L, Wessels L, Brummelkamp TR, et al. Genome-wide identification and characterization of novel factors conferring resistance to topoisomerase II poisons in cancer. Cancer Res. 2015;75:4176–4187. doi: 10.1158/0008-5472.CAN-15-0380. [DOI] [PubMed] [Google Scholar]
- 27.Dubey R, Lebensohn AM, Bahrami-Nejad Z, Marceau C, Champion M, Gevaert O, Sikic BI, Carette JE, Rohatgi R. Chromatin-remodeling complex SWI/SNF controls multidrug resistance by transcriptionally regulating the drug efflux pump ABCB1. Cancer Res. 2016;76:5810–5821. doi: 10.1158/0008-5472.CAN-16-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahromi MS, Putnam AR, Druzgal C, Wright J, Spraker-Perlman H, Kinsey M, Zhou H, Boucher KM, Randall RL, Jones KB, et al. Molecular inversion probe analysis detects novel copy number alterations in Ewing sarcoma. Cancer Genet. 2012;205:391–404. doi: 10.1016/j.cancergen.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang TS, Wei KL, Lu CK, Chen YH, Cheng YT, Tung SY, Wu CS, Chiang MK. Inhibition of CCAR1, a coactivator of beta-catenin, suppresses the proliferation and migration of gastric cancer cells. Int J Mol Sci. 2017;18:E460. doi: 10.3390/ijms18020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha SY, Kim JH, Yang JW, Kim J, Kim B, Park CK. The overexpression of CCAR1 in hepatocellular carcinoma associates with poor prognosis. Cancer Res Treat. 2016;48:1065–1073. doi: 10.4143/crt.2015.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo WY, Jeong BC, Yu EJ, Kim HJ, Kim SH, Lim JE, Kwon GY, Lee HM, Kim JH. CCAR1 promotes chromatin loading of androgen receptor (AR) transcription complex by stabilizing the association between AR and GATA2. Nucleic Acids Res. 2013;41:8526–8536. doi: 10.1093/nar/gkt644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, Yu S, Liu X, Paulsen MT, Ljungman ME, et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc Natl Acad Sci USA. 2015;112:E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The materials included in the manuscript will be made freely available to any researchers who wish to use them for non-commercial purposes, while preserving any necessary confidentiality and anonymity.