Abstract
Microrchidia 2 (MORC2) is important in DNA damage repair and lipogenesis, however, the clinical and functional role of MORC2 in liver cancer remains to be fully elucidated. The aim the present study was to clarify the role of MORC2 in liver cancer. Expression profile analysis, immunohistochemical staining, reverse transcription-quantitative polymerase chain reaction analysis and western blot analysis were performed to evaluate the levels of MORC2 in liver cancer patient specimens and cell lines; subsequently the expression of MORC2 was suppressed or increased in liver cancer cells and the effects of MORC2 on the cancerous transformation of liver cancer cells were examined in vitro and in vivo. MORC2 was upregulated in liver cancer tissues, and the upregulation was associated with certain clinicopathologic features of patients with liver cancer. MORC2 knockdown caused marked inhibition of liver cancer cell proliferation and clonogenicity, whereas the overexpression of MORC2 substantially promoted liver cancer cell proliferation. In addition, the knockdown of MORC2 inhibited the migratory and invasive ability of liver cancer cells, whereas increased migration and invasion rates were observed in cells with ectopic expression of MORC2. In a model of nude mice, the overexpression of MORC2 promoted tumorigenicity and markedly enhanced pulmonary metastasis of liver cancer. Furthermore, MORC2 regulated apoptosis and its expression level had an effect on the sensitivity of liver cancer cells to doxorubicin, 5-fluorouracil and cisplatin. Mechanically, MORC2 modulated the mitochondrial apoptotic pathway, possibly in a p53-dependent manner, and its dysregulation also resulted in the abnormal activation of the Hippo pathway. For the first time, to the best of our knowledge, the present study confirmed that MORC2 was a novel oncogene in liver cancer. These results provide useful insight into the mechanism underlying the tumorigenesis and progression of liver cancer, and offers clues into potential novel liver cancer therapies.
Keywords: microrchidia 2, oncogene, liver cancer, progression
Introduction
Liver cancer is a prevalent malignancy worldwide and ranks as one of the leading causes of cancer-associated mortality (1). The mechanism of liver cancer is complicated and heterogeneous, and is accompanied by various molecular abnormalities. It is necessary to identify novel oncogenes and tumor suppressors for further investigation and potential clinical application.
The microrchidia (MORC) family proteins are conserved proteins with important roles in multiple biological processes. MORC2, also known as ZCWCC1, ZCW3, KIAA0852 and AC004542.C22.1, is a member of this family (2,3). It contains an ATPase domain, a zinc finger type CW domain, and nuclear localization signal and coiled-coil domains (2–4). It has been reported that MORC2 can bind with histone deacetylase (HDAC)4 and functions as a transcriptional repressor by mediating the deacetylation of histone H3 (5). A limited number of studies have shown that MORC2 functions in chromatin remodeling, facilitating DNA damage repair and promoting lipogenesis (2,6,7), however, its function in cancer remains to be fully elucidated. As abnormal chromatin dynamics, enhancing DNA damage repair ability and de novo lipogenesis are crucial events in cancer cells, MORC2 may function as an oncogene by promoting the malignant phenotype of cancer cells. MORC2 can promote the migration and invasion of breast cancer cells, and is involved in a prognostic prediction model for breast cancer containing six genes (8,9). Its oncogenic role in gastric cancer has also been demonstrated (10–12). For example, it has been reported that MORC2 downregulates p21 by recruiting HDAC1 to the p21 promoter, in a p53-independent manner in gastric cancer; the phosphorylation of MORC2 increases the expression of cyclin D1-cyclin-dependent kinase (CDK)4 and cyclin D3-CDK6 complexes, promotes gastric cell cycle transition from the G1 to S stage, and indicates a poorer prognosis in patients with gastric cancer (11,12).
However, to date, no studies have reported on the clinicopathologic significance and functions of MORC2 in liver cancer. The present study presented the first evidence, to the best of our knowledge, of the expression pattern of MORC2 in human liver cancer and its clinical significance. The roles of MORC2 in the progression of liver cancer and its underlying mechanisms were investigated. The data demonstrated that MORC2 was upregulated in liver cancer, and contributed to the proliferation, metastasis and chemoresistance of liver cancer cells via the p53 and Hippo pathways.
Materials and methods
Cell culture, culture conditions and antibodies
The HepG2, Bel-7402, Huh7, PLC/PRF-5, SMMC7721 and LM3 liver cancer cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences Committee Type Culture Collection (Shanghai, China), and the normal L02 liver cell line was conserved at the Central Laboratory of Renmin Hospital of Wuhan University (Wuhan, China). The cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Zhejiang Tianhang Biotechnology Co., Ltd., Hangzhou, China) and 100 units penicillin/streptomycin. The cells were cultured at 37°C and 5% CO2 in a humidified chamber. Rabbit polyclonal anti-MORC2 antibody was purchased from Abcam (Cambridge, UK). Mouse monoclonal anti-β-actin antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Anti-rabbit and anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Promega Corporation (Madison, WI, USA).
Patients and histological and immunohistochemical (IHC) staining
The GSE14520 and GSE22058 mRNA expression profile were downloaded from the Gene Expression Omnibus (GEO) database (13–15). The Cancer Genome Atlas (TCGA) copy number-altered genome data for each patient was directly downloaded from cBioPortal for Cancer Genomics (16,17). All liver cancer samples and paired adjacent tissues were retrieved from patients receiving surgery between December 1 and December 31, 2014, from the Department of Pathology, Zhongnan Hospital of Wuhan University (Wuhan, China). All patients provided informed written consent prior to the investigation. The inclusion of human samples was approved by the Ethics Review Board of the Second People's Hospital of Guangdong Province (Guangdong, China; approval no. 2015-KYLL-023). The tissues were first stained with hematoxylin and eosin for histological examination. The deparaffinized sections were treated with 3% H2O2 and subjected to antigen retrieval by citric acid (pH 6.0). Following overnight incubation with primary antibody (anti-MORC2 antibody; 1:200) at 4°C, the sections were incubated for 30 min at room temperature with HRP-labeled polymer conjugated with secondary antibody (MaxVision™ kits) and incubated for 1 min with diaminobenzidine. The sections were then lightly counterstained with hematoxylin. Sections without primary antibody served as negative controls. The expression level of MORC2 was ascertained according to the average score of two pathologists' evaluations using a CKX41 microscope (Olympus Corporation, Tokyo, Japan). As MORC2 is mainly expressed in the nucleus, the positive nuclear staining of MORC2 was used to elucidate its expression level according to the following formula: Immunostaining score = percentage score × intensity score, where the percentage score represented the percentage of immunopositive cells, and was graded as 0 (<6%), 1 (6–33%), 2 (34–66%) and 3 (>66%). The intensity score represented the intensity of immunostaining, and was determined as 0 (absent), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). All cases were diagnosed by two certificated pathologists without discrepancy.
Small interfering RNA (siRNA) transfection and establishment of stable expressing cells
The siRNAs were designed and purchased commercially (Genepharma, Shanghai, China) as shown in Table I. The cells were transfected with 50 ng/µl targeting siRNA using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 48 h, following which the depletion efficiency was analyzed by western blot analysis. siRNA-homo-1760 was selected for the following experiments, as it exhibited the optimal efficiency. For the establishment of stable MORC2-overexpressing cells, the overexpression plasmid was constructed using the pLV-EGFP (2A) puro plasmid; the plasmid was packed using a lentivirus system (both from Inovogen Biotechnology Co., Ltd., Beijing, China). Following infection, the cells were selected for 4 weeks using DMEM with 10 µg/ml puromycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The expression of MORC2 was lowest in the PLC cell line, however, the overexpression efficiency was not satisfactory in this cell line, thus the SMMC7721 cell line was used to construct the stable MORC2-overexpressing cells in the present study.
Table I.
siRNA sequences used for the overexpression of MORC2.
| siRNA | Sense (5′-3′) | Antisense (5′-3′) |
|---|---|---|
| MORC2-homo-1760 | GCGGAACAUUGGUGAUCAUTT | AUGAUCACCAAUGUUCCGCTT |
| MORC2-homo-2439 | GGAGCCUACACACAACAAATT | UUUGUUGUGUGUAGGCUCCTT |
| MORC2-homo-3972 | GCAGCUGAGUGCUAUGAAUTT | AUUCAUAGCACUCAGCUGCTT |
| Control siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
siRNA, small interfering RNA; MORC2, microrchidia 2.
Western blot analysis
In brief, cells were lysed in lysis buffer containing 50 mmol/l Tris (pH 8.0), 150 mmol/l NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mmol/l NaF, 1 mmol/l Na3VO4. Protein concentration was analyzed and adjusted using a bicinchoninic acid protein assay. Subsequently, equal quantities (50 µg) of samples were separated by 10% SDS-PAGE (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and transferred onto nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk in Tris-buffered saline containing 0.05% Tween-20, the membranes were incubated with primary antibodies against MORC2 (1:400, bs-0354R; BIOSS, Beijing, China), cytochrome c (1:200, ab53056), caspase-3 (1:500, ab47131) (both from Abcam), caspase-9 (1:150, sc-56076; Santa Cruz Biotechnology, Inc.), poly (ADP-ribose) polymerase (PARP) (1:150, ab4830; Abcam), p53 (1:500, sc-6243), B-cell lymphoma-2 (Bcl-2) (1:500, sc-783), Bcl-2-associated X protein (Bax) (1:300, sc-493) (all from Santa Cruz Biotechnology, Inc.), p53 upregulated modulator of apoptosis (PUMA)α (1:5000, ab33906; Abcam), Yes-associated protein 1 (YAP1) (1:500, orb89757; Biorbyt Ltd., Cambridge, UK), phosphorylated (p-)S127-YAP1 (1:10,000, ab76252; Abcam), Transcriptional co-activator with PDZ-binding motif (TAZ) (1:500, #23306-1-AP; ProteinTech Group, Inc., Chicago, IL, USA), p-S89-TAZ (1:500, sc-17610), and β-actin (1:1,000, sc-47778) (both from Santa Cruz Biotechnology, Inc.) at 4°C overnight. The primary antibodies coupled to sample proteins were then visualized by incubation for 1 h at 37°C with HRP-conjugated secondary antibodies (1:1,000, 074-1506 and 074-1806; KPL, Inc., Gaithersburg, MD, USA) using a chemiluminescence detection system (EMD Millipore) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from the tissues and cells was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 2 µg of total RNA from each sample was used for cDNA synthesis. RT-qPCR analysis was performed in triplicate with the SYBR® Green PCR Master mix (Takara Bio, Inc., Otsu, Japan) and β-actin was used as an internal control according to the manufacturer's protocol. Each sample included: 4 µl cDNA, 2 µl primers, 12.5 µl 2X SYBR Green master mix and 6.5 µl ddH2O. The PCR thermo-cycling conditions were as follows: 5 min at 94°C, followed by 40 cycles at 94°C for 20 sec, 60°C for 20 sec and at 72°C for 20 sec, and 5 min at 72°C. Relative expression levels of target genes were determined according to the 2−ΔΔCq method (18), where Cq represents the quantification cycle for each transcript. The primer pairs for human MORC2 and β-actin were designed as shown in Table II.
Table II.
Primers used for human MORC2 and β-actin.
| Primer | Sequence (5′-3′) |
|---|---|
| Homo-MORC2-F | GAAAGCCTGCCAACACTCTC |
| Homo-MORC2-R | CTCATCAGAAACTGCGACA |
| Homo-β-actin-F | CATTAAGGAGAAGCTGTGCT |
| Homo-β-actin-R | GTTGAAGGTAGTTTCGTGGA |
MORC2, microrchidia 2; F, forward; R, reverse.
Cell counting kit-8 (CCK-8) assay
In the cell proliferation assay, the cells (1×105/well) were plated onto 96-well plates on the first day and allowed to attach overnight. The following day, 10 µl of CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well and incubated at 37°C and 5% CO2 for 1 h between days 1 and 5. The absorbance was then detected with the multifunctional microplate reader at 490 nm. For the chemoresistance assay, following seeding of the cells (1×105/well) in the plates, the cells were treated with the chemotherapeutics, doxorubicin, 5-fluorouracil and cisplatin, at different concentrations (0, 1, 5, 10, 20, 40, 80 and 100 µM) for 72 h at 37°C, following which CCK-8 was added. Following incubation at 37°C and 5% CO2 for 1 h, the absorbance was detected and the half maximal inhibitory concentration (IC50) parameter was calculated.
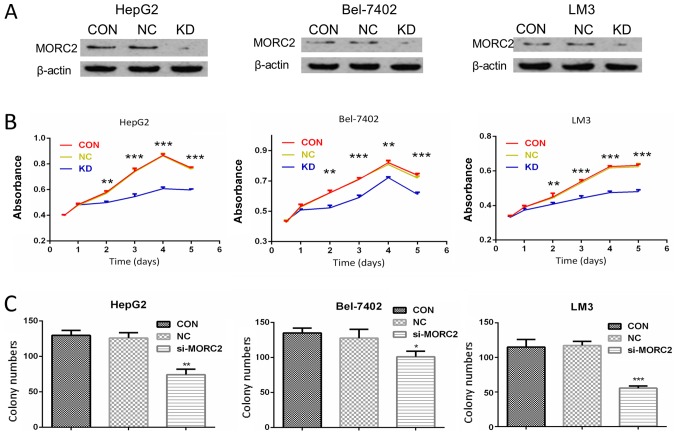
Plate colony formation assay
The cells were digested with trypsin, centrifuged at 110 × g for 5 min at 37°C, resuspended in DMEM supplemented with 10% FBS and seeded in 6-well plates (1,000 cells/well), following which the cells in each group were cultured for 2 weeks. The cells were then washed twice with PBS, fixed with 4% paraformaldehyde for 20 min, and then stained with 0.1% crystal violet for 30 min. The dishes were then carefully washed with PBS until the background was clear. Finally, the number of colonies was counted under a microscope (CKX41; Olympus Corporation).
Cell-cycle analysis
In brief, the cells were digested with trypsin, centrifuged at 110 × g for 5 min at 37°C, resuspended in PBS, washed twice with PBS and then fixed in 100% ice-cold methanol overnight at -20°C. The cells were then incubated with 50 mg/ml propidium iodide (PI) and 1 mg/ml RNAase in PBS for 20 min, following which the samples were analyzed with BD FACSAria (BD Biosciences, Franklin Lakes, NJ, USA).
In vitro migration and invasion assays
In the scratch wound healing assay, cells were cultured in serum-free medium for 24 h and wounded with pipette tips. Subsequently, the medium was replaced with fresh medium. The wound healing procedure was observed after 48 h, and images of the cells were captured under a microscope (CKX41; Olympus Corporation). Cell migration and invasion assays were performed using Transwell chambers, as previously described (19). The cells were harvested and resuspended in serum-free medium, and then added to the upper chamber. Following incubation for 48 h, cells remaining on the upper side of membrane were removed with a cotton swab. The cells migrated to the lower membrane surface were fixed and stained with 0.1% crystal violet for 30 min, and the number of cells was counted under a microscope. To assess invasion ability, the membranes were pre-coated with diluted Matrigel, whereas the membranes in migration experiments were not pre-coated with Matrigel.
Nude mice experiments
All animal experiments were approved by the Ethics Review Board of the Second People's Hospital of Guangdong Province (approval no. 2015-KYLL-063). For the experiments, 4-week-old male BALB/C nude mice (Hangzhou Hibio Technology Co., Ltd., Hangzhou, China) were used. The mice were maintained under the following pathogen-free conditions: 60% humidity; room temperature; 12-h light/dark cycle; ad libitum access to food and water. In a subcutaneous xenograft procedure, 1×106 cells (SMMC7721MORC2 or SMMC7721Vector) were resuspended in PBS solution and then injected subcutaneously into the left and right side of each of the mice (n=6). The tumor size was measured every 3 days, and tumor volume was calculated using the following formula: 1/2 length × width2. For the assessment of pulmonary metastasis, 10 mice were included in each group, and 1×106 cells (SMMC7721MORC2 or SMMC7721Vector) were injected into the caudal vena. After 2 weeks, the mice were sacrificed and lung colonization was quantified by pathological examination.
Analysis of apoptosis
Annexin V-FITC/PI staining was used to investigate whether MORC2 regulates the apoptosis of liver cancer cells. The cells were seeded into 6-well plates. When the cells in each group were at a log phase of growth, the cells were digested with trypsin, centrifuged at 110 × g for 5 min at 37°C and resuspended in PBS. The cells were then incubated with an ApoScreen Annexin V Apoptosis kit and PI. Every sample containing 10,000 cells was analyzed using BD FACSAria (BD Biosciences). The experiments were performed in triplicate.
Statistical analysis
Statistical significances between values of different experimental groups were analyzed using Student's t-test or one-way analysis of variance. A χ2 test and Fisher's test were used in analyzing enumeration data and pathway analysis. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were conducted using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) or SPSS for Windows 17.0.1 software (SPSS, Inc., Chicago, IL, USA). Expression profile analysis was conducted on L02MORC2 and L02Vector data (Chen et al, unpublished data) using GCBI 1.0 software (Gminix Technology Co., Ltd., Shanghai, China) with the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/) as reference.
Results
MORC2 is overexpressed in liver cancer samples at the mRNA and protein levels
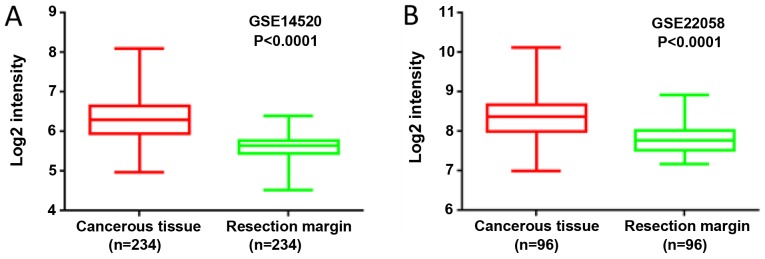
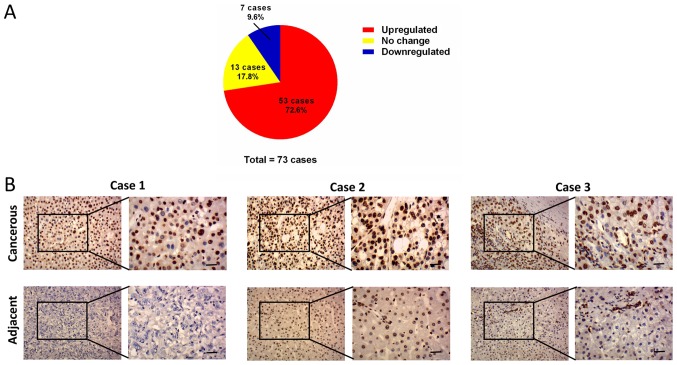
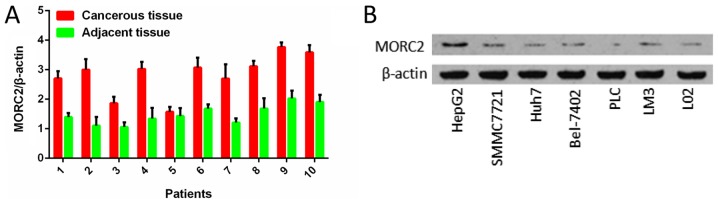
By analyzing the expression profile of liver cancer samples from GEO datasets GSE14520 and GSE22058, it was found that the mRNA levels of MORC2 were significantly upregulated in the liver cancer tissues, compared with the matched non-tumorous tissues (Fig. 1). The expression levels of MORC2 in 73 liver cancer and corresponding adjacent liver tissues specimens were then examined using IHC staining. Consistent with previous reports, MORC2 was expressed mainly in the nucleus and was relatively weak in the cytoplasm (Fig. 2) (3,10). The results showed that MORC2 was overexpressed in the majority (72.6%, 53/73) of the liver cancer samples (Fig. 2). RT-qPCR analysis was also used to examine the levels of MORC2 in 10 pairs of liver cancer and pair-adjacent liver tissue specimens; accordingly, the expression of MORC2 was upregulated in almost all the cancerous tissues (Fig. 3A). The expression level of MORC2 was then examined in HepG2, Bel-7402, Huh7, PLC/PRF-5, SMMC7721, LM3 and L02 cells. Higher expression of MORC2 was observed in the majority of the liver cancer cells, compared with the L02 cells (Fig. 3B). From these results, it was concluded that MORC2 was overexpressed in liver cancer at the mRNA and protein level, demonstrating the important role of MORC2 in the pathogenesis of liver cancer.
Figure 1.
Gene Expression Omnibus datasets show that the expression of MORC2 is increased in liver cancer tissue. In (A) GSE14520 and (B) GSE22058, the expression level of MORC2 in liver cancer tissues was upregulated, compared with that in adjacent tissues. MORC2, microrchidia 2.
Figure 2.
Protein expression level of MORC2 is increased in human liver cancer samples. (A) Expression of MORC2 was upregulated in 72.6% of liver cancer patient samples examined by immunohistochemistry. (B) Representative images of staining of MORC2 protein in three pairs of liver cancer and adjacent tissues (scale bar, 5 µm). MORC2, microrchidia 2.
Figure 3.
mRNA expression level of MORC2 is increased in human liver cancer samples and the protein expression level of MORC2 is increased in liver cancer cell lines. (A) Lysates from paired tissues of liver cancer and adjacent tissue were analyzed by reverse transcription-quantitative polymerase chain reaction analysis for the detection of MORC2. β-actin was used as a loading control. (B) Western blot analysis of the expression of MORC2 in the immortalized L02 normal liver cell line and six liver cancer cell lines. MORC2, microrchidia 2.
High expression of MORC2 leads to accumulation of copy number variations and unfavorable pathological characteristics
Dysfunctional DNA damage repair results in genetic alterations, including somatic copy number alteration, and this characteristic is increasingly recognized as common feature of human liver cancer (20). It has been reported that MORC2 is associated with chromatin remodeling, promotes the induction of γ-H2AX and modulates DNA damage repair (6). Utilizing the data of TCGA database (16,17), it was detected that the fraction of copy number alterations was higher in the genome of patients with liver cancer and higher MORC2 (Fig. 4). This result indicated that the upregulation of MORC2 may result in liver cancer progression via accumulating extra copies of DNA.
Figure 4.
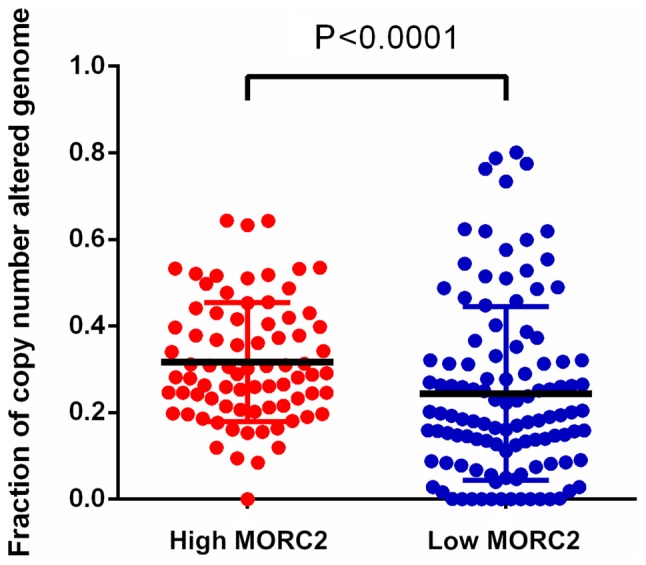
Higher expression of MORC2 is associated with increased somatic copy number variation in patients with liver cancer. Genome instability (DNA copy number alteration) was detected in patients with liver cancer in The Cancer Genome Atlas. Data are presented as the mean ± standard deviation. MORC2, microrchidia 2.
Using the clinical information of GSE14520, the correlation between the expression of MORC2 and clinicopathological features was assessed to determine its clinical significance. The samples were divided into two groups according to the expression level and were analyzed with χ2 test. As shown in Table III, a higher expression of MORC2 was associated with larger tumor volume (P=0.009) and higher American Joint Committee on Cancer T stage (P=0.007). These results indicated that MORC2 may be a potential prognostic biomarker in liver cancer.
Table III.
Associations between the expression of MORC2 and the clinicopathologic features of liver cancer (GSE14520).
| Characteristic | Patients (n) | Expression ofmicrorchidia 2
|
P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | ||||
| ≤55 | 166 | 88 | 77 | 0.166 |
| >55 | 76 | 33 | 43 | |
| Sex | ||||
| Male | 211 | 108 | 103 | 0.336 |
| Female | 31 | 13 | 18 | |
| AFP (ng/ml) | ||||
| ≤200 | 128 | 58 | 70 | 0.119 |
| >200 | 110 | 61 | 49 | |
| ALT (U/l) | ||||
| ≤50 | 142 | 64 | 78 | 0.068 |
| >50 | 100 | 57 | 43 | |
| Cirrhosis | ||||
| Yes | 223 | 112 | 111 | 0.811 |
| No | 19 | 9 | 10 | |
| Tumor size (d/cm) | ||||
| <5 | 153 | 67 | 86 | 0.009 |
| ≥5 | 88 | 54 | 34 | |
| Tumor number | ||||
| Solitary | 190 | 91 | 99 | 0.211 |
| Multiple | 52 | 30 | 22 | |
| AJCC T stage | ||||
| T1 | 96 | 34 | 62 | 0.007 |
| T2 | 78 | 43 | 35 | |
| T3 | 51 | 30 | 21 | |
| BCLC stage | ||||
| 0 | 20 | 8 | 12 | 0.283 |
| A | 152 | 68 | 84 | |
| B | 24 | 13 | 11 | |
| C | 29 | 18 | 11 | |
| CLIP stage | ||||
| 0 | 98 | 41 | 57 | 0.297 |
| 1 | 79 | 40 | 39 | |
| 2,3,4,5 | 48 | 26 | 22 | |
| PRMS classification | ||||
| High | 121 | 74 | 47 | <0.001 |
| Low | 121 | 47 | 74 | |
Data are presented as numbers. AJCC, American Joint Committee on Cancer; BCLC, Barcelona Clinic Liver Cancer; CLIP, Cancer Liver Italian Program; PRMS, Predicted risk Metastasis Signature.
In liver cancer cells, knockdown of MORC2 inhibits proliferation in vitro
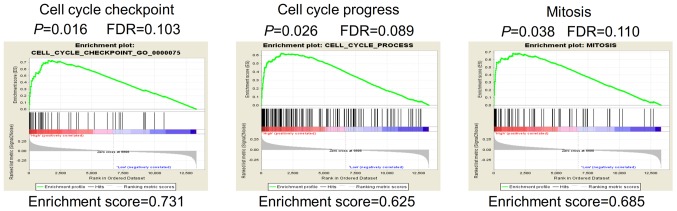
Gene set enrichment analysis (GSEA) was performed in order to identify potential genes modulated by MORC2. The mRNA expression profiling data in GSE14520 were used to outline MORC2-correlated genes. In the GSEA analysis of Gene Ontology terms, it showed that gene sets involved in 'cell cycle process' and 'mitosis' were enriched in the MORC2-high expression samples (Fig. 5). Enhanced mitogenic signaling and aberrant process of cell cycle are essential for cell proliferation and cancer progression (20). The bioinformatics data provided a possible explanation of why a high expression of MORC2 was associated with unfavorable clinicopathological features, including tumor size and T stage (Table III). The results suggested that MORC2 has an important function in the proliferation of liver cancer cells.
Figure 5.
GSEA results indicate that MORC2 modulates the proliferation of liver cancer cells. GSEA analysis of Gene Ontology terms showed MORC2 may regulate gene sets associated with cell cycle checkpoint (left), cell cycle progress (middle) and mitosis (right). GSEA, gene set enrichment analysis; MORC2, microrchidia 2.
To verify the biological role of MORC2 in the proliferation of liver cancer, MORC2 was knocked down using siRNAs in HepG2, Bel-7402 and LM3 cells, which had a higher expression of MORC2. Western blot analysis was used to determine the knockdown efficiency (Fig. 6A).
Figure 6.
MORC2 modulates proliferation of liver cancer cells in vitro. (A) Knockdown of MORC2 in HepG2 (left), Bel-7402 (middle) and LM3 (right) cell lines by siRNA targeting MORC2 was confirmed by western blot analysis; β-actin was used as a loading control. (B) Effect of MORC2-knockdown on the proliferation of HepG2 (left), Bel-7402 (middle) and LM3 (right) cells was determined using a cell counting kit-8 assay. (C) Effect of MORC2-knockdown on colony numbers was determined using a colony formation assay in HepG2 (left), Bel-7402 (middle) and LM3 (right) cells. *P<0.05; **P<0.01; ***P<0.001. MORC2, microrchidia 2; siRNA, small interfering RNA; CON, control; KD, knockdown; NC, negative control.
The effect of MORC2 on cell proliferation was then examined. As shown in Fig. 6B, compared with the control groups, following MORC2 knockdown, the HepG2, Bel-7402 and LM3 cells exhibited a significantly lower cell proliferation rate (Fig. 6B). Furthermore, cell proliferation was measured using a plate colony formation assay. Compared with the control cells, MORC2 knockdown in the HepG2, Bel-7402 and LM3 cells resulted in markedly decreased colony formation abilities (Fig. 6C).
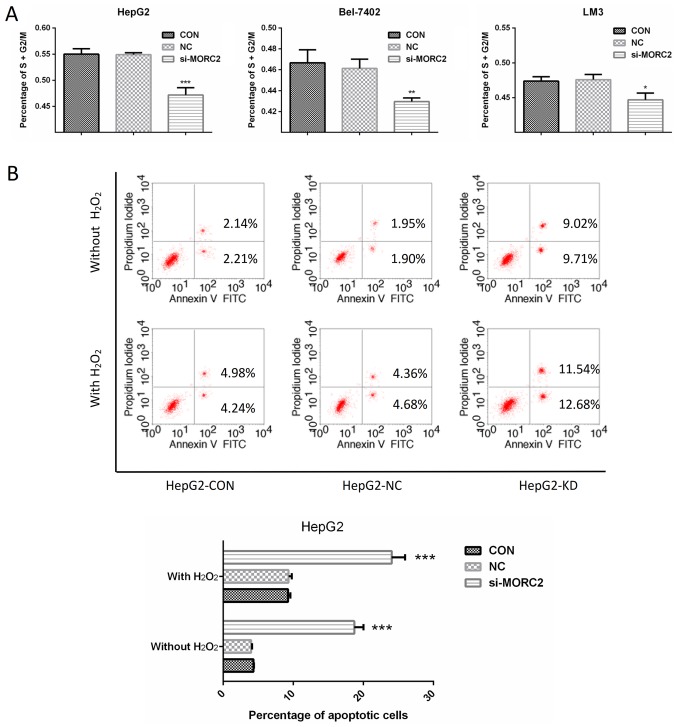
In liver cancer cells, inhibition of MORC2 promotes cell cycle arrest and induces apoptosis
Enhanced cell cycle progression and reduced apoptotic signaling are two important factors causing sustained proliferation. To examine the factors involved in MORC2 regulating the proliferation of liver cancer cells, the cell cycle was detected in liver cancer cells through flow cytometry. Following transfection with si-MORC2 or si-NC for 48 h, the inhibition of MORC2 led to a significant accumulation of cells at the G0/G1-phase and a marked decrease in cells at the S/G2/M-phase in HepG2, Bel-7402 and LM3 cells (Fig. 7A). Subsequently, the effects of MORC2 on apoptosis under normal condition or oxidative stress (treated with H2O2) were investigated. The knockdown of MORC2 markedly increased apoptosis of the HepG2 cells (Fig. 7B). These results indicated that the downregulation of MORC2 promoted cell cycle arrest and induced the apoptosis of liver cancer cells.
Figure 7.
MORC2 regulates liver cancer cell proliferation via cell cycle and apoptosis. (A) Effects of MORC2-knockdown on the percentage of cells in the S/G2/M-phase were determined by FACS analysis in HepG2 (left), Bel-7402 (middle) and LM3 (right) cells. (B) Following Annexin V-FITC/propidium iodide staining of the indicated cells following treatment without or with H2O2 (1 mmol) for 12 h, effects of MORC2-knockdown on HepG2 cell apoptosis were determined by FACS analysis (above) and quantitative analysis of apoptotic cell numbers (below). Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01; ***P<0.001. FACS, fluorescence-activated cell sorting; MORC2, microrchidia 2; si, small interfering RNA; CON, control; KD, knockdown; NC, negative control.
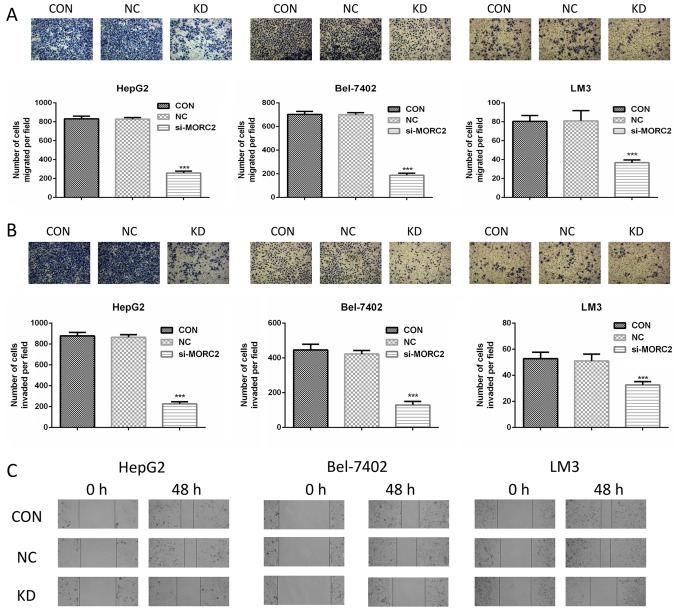
Knockdown of MORC2 inhibits cell migration and invasion of liver cancer cells in vitro
The clinicopathological analysis also revealed that a higher expression level of MORC2 was significantly associated with a gene expression signature of higher metastatic potential (P<0.001, Table III), however, the role of MORC2 in cancer cell metastasis has not been investigated previously. Cell migration and invasion are critical during the multistep process of cancer cell metastasis. The present study assessed whether MORC2 was a crucial molecule involved in cell migration and invasion using Transwell assays. MORC2-loss of function inhibited the migration and invasion rates of the HepG2, Bel-7402 and LM3 cells (Fig. 8A and B). To confirm this result, a scratch wound healing assay was also used to evaluate the effect of MORC2 on cell movement. Consistent with the previous observations, the inhibition of MORC2 attenuated the mobility of the liver cancer cells (Fig. 8C).
Figure 8.
MORC2 modulates migration, invasion and metastasis of liver cancer cells in vitro. (A) Cell migration was assessed using a Transwell assay in HepG2 (left), Bel-7402 (middle) and LM3 (right) cells following transfection of the cells with MORC2 siRNA for 48 h. The cells that migrated into the bottom surface of the filters were stained. Magnification, ×40. (B) Cell invasion was assessed using a Transwell assay with Matrigel in HepG2 (left), Bel-7402 (middle) and LM3 (right) cells following transfection of the cells with MORC2 siRNA for 48 h. The cells that invaded into the lower surface of the filters were stained. Magnification, ×40. (C) Movement ability was detected by scratch wound healing assays in HepG2 (left), Bel-7402 (middle) and LM3 (right) cells following transfection of the cells with MORC2 siRNA for 48 h. Magnification, ×40. Data are presented as the mean ± standard deviation of six independent experiments. ***P<0.001. MORC2, microrchidia 2; siRNA, small interfering RNA; CON, control; KD, knockdown; NC, negative control.
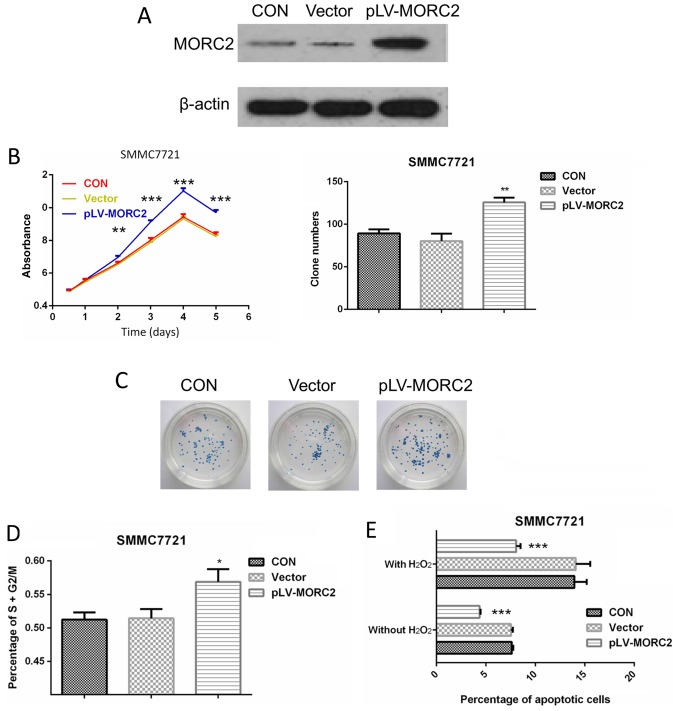
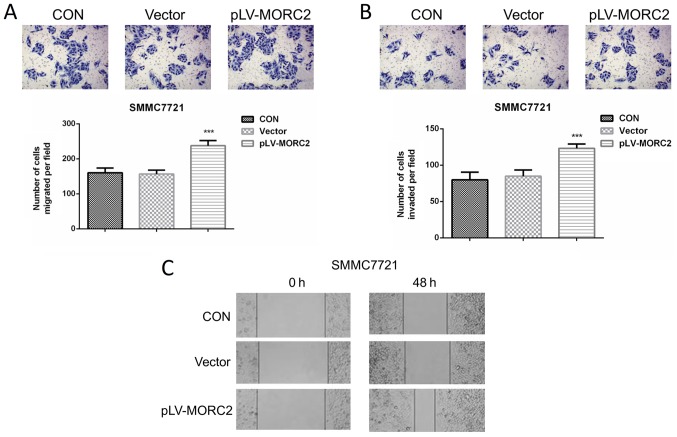
Overexpression of MORC2 promotes the malignant phenotypes of the SMMC7721 liver cancer cell line in vitro and in vivo
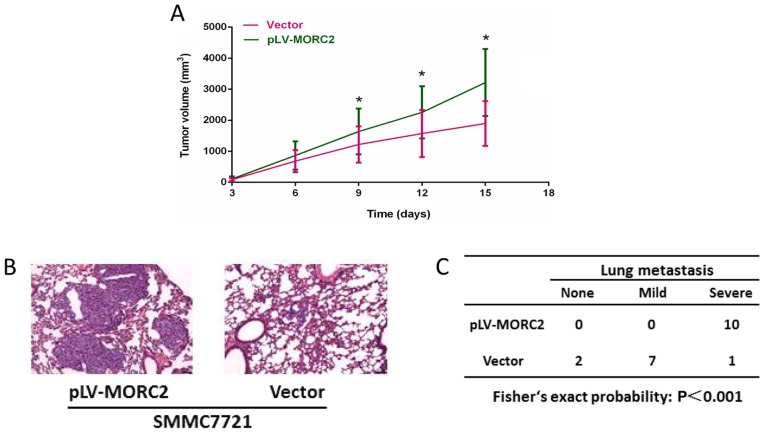
The SMMC7721 cell line, which had a relatively lower expression of MORC2, was used to construct cells overexpressing MORC2 using a lentivirus-mediated packed pLV-MORC2 vector. Western blot analysis was used to determine the knockdown and ectopic expression (Fig. 9A). Compared with the control group, SMMC7721 cells exhibited a significantly higher cell viability rate when MORC2 was overexpressed (Fig. 9B). Similarly, the MORC2-overexpressing SMMC7721 cells exhibited significantly increased colony formation (Fig. 9C). The overexpression of MORC2 increased the cell cycle progression and decreased the number of apoptotic cells (Fig. 9D and E). Furthermore, SMMC-7721 cells stably expressing MORC2 exhibited enhanced migration, invasion and movement, compared with the control cells (Fig. 10). The in vivo experiments showed that the overexpression of MORC2 significantly increased tumor volume in a subcutaneous xenograft model (Fig. 11A). Colonization at a distant site is the last key step in the metastatic cascade (21). The present study also used a tail vein injection model to imitate the pathophysiological process to determine whether MORC2 was involved in the distant colonization of liver cancer cells. At 2 weeks post-injection, the mice were sacrificed by cervical dislocation, and lung colonization was quantified by pathological examination. In line with the observation in vitro, all 10 mice exhibited severe lung metastasis in the MORC2-overexpression group, the incidence of which was significantly higher, compared that in the empty vector group (1/10; P<0.001) (Fig. 11B and C). These results revealed the promoting role of MORC2 in liver cancer metastasis.
Figure 9.
Overexpression of MORC2 promotes proliferation of SMMC7721 cells in vitro. (A) Overexpression of MORC2 in SMMC7721 cells by lentivirus-mediated packed pLV-MORC2 vector was confirmed by western blot analysis; β-actin was used as a loading control. (B) Effect of overexpression of MORC2 on the proliferation on SMMC7721 cells was determined using a cell counting kit-8 assay. (C) Effect of overexpression of MORC2 on colony numbers was determined by a colony formation assay in SMMC7721 cells. (D) Effect of overexpression of MORC2 on the percentage of cells in the S/G2/M-phase and on (E) SMMC7721 cell apoptosis were determined by fluorescence-activated cell sorting analysis. Data are presented as the mean ± standard deviation of three or six independent experiments. *P<0.05; **P<0.01; ***P<0.001. MORC2, microrchidia 2; CON, control.
Figure 10.
Overexpression of MORC2 promotes migration, invasion and motility of SMMC7721 cells in vitro. (A) Cell migration was assessed using a Transwell assay in SMMC7721MORC2 and SMMC7721vector cells; the cells that migrated to the bottom surface of the filters were stained. Magnification, ×100. (B) Cell invasion was assessed using a Transwell assay with Matrigel in SMMC7721MORC2 and SMMC7721vector cells; cells, which migrated to the bottom surface of the filters, were stained. Magnification, ×100. (C) Movement ability was detected using scratch wound healing assays in SMMC7721MORC2 and SMMC7721vector cells. Magnification, ×40. Date are presented as the mean ± standard deviation of three or six independent experiments. ***P<0.001. MORC2, microrchidia; CON, control.
Figure 11.
Overexpression of MORC2 promotes malignant phenotypes of SMMC7721 cells in vivo. (A) Representative data showing that the overexpression of MORC2 significantly promoted tumor growth in nude mice xenograft model (n=6). (B) Representative images showing the colonization of SMMC7721MORC2 and SMMC7721vector cells in the lung of recipient mice. Magnification, ×100. (C) Incidence and severity of lung metastasis in pulmonary metastasis model with SMMC7721MORC2 and SMMC7721vector cells. *P<0.05. MORC2, microrchidia.
Inhibition of MORC2 improves the sensitivity of liver cancer cells to chemotherapeutic drugs
The present study also examined whether MORC2 has the potential to be applied in clinical liver cancer treatment. Chemotherapy provides an optional strategy in the treatment of liver cancer, particularly for patients with advanced tumors. However, chemotherapy is unsatisfactory due to chemoresistance (22). The present study hypothesized that the negative effect of MORC2 on the apoptosis of liver cancer cells may also contribute to the drug resistance of liver cancer cells. To test this hypothesis, chemosensitivity to the three most common chemotherapeutic drugs, doxorubicin, cisplatin and 5-fluorouracil, was assayed in MORC2-knockdown groups and control groups. The results showed that the IC50 values of all the chemotherapeutic drugs were significantly decreased by the knockdown of MORC2 (Table IV). Therefore, MORC2 contributed to enhancing the chemotherapeutic sensitivity of liver cancer cells.
Table IV.
siRNA-mediated knockdown of MORC2 increases the sensitivity of liver cancer cells to chemotherapeutics.
| Cell group | IC50 values
|
||
|---|---|---|---|
| Doxorubicin (µM) | Cisplatin (µM) | 5-fluorouracil (µM) | |
| HepG2 | |||
| si-NC | 33.53 | 25.88 | 42.82 |
| si-MORC2 | 8.86 | 7.89 | 14.04 |
| Bel-7402 | |||
| si-NC | 34.24 | 39.78 | 50.54 |
| si-MORC2 | 15.27 | 27.84 | 31.12 |
| LM3 | |||
| si-NC | 29.72 | 43.23 | 23.68 |
| si-MORC2 | 19.89 | 35.77 | 12.06 |
MORC2, microrchidia 2; si, small interfering RNA; NC, negative control; IC50, half maximal inhibitory concentration.
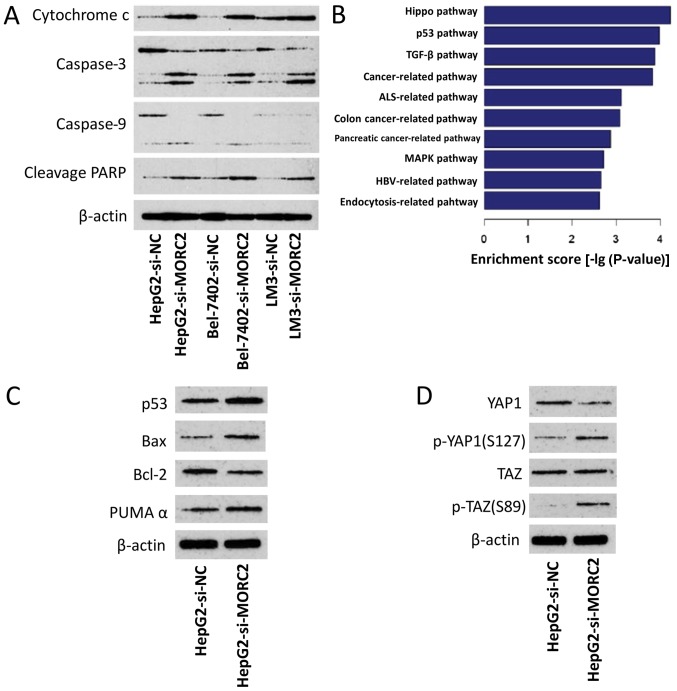
To determine the mechanisms by which MORC2 affects apoptosis and chemoresistance, the expression levels of apoptosis-related proteins were examined following the knockdown of MORC2 in the cells. As shown in Fig. 12A, in the HepG2, Bel-7402 and LM3 cells, si-MORC2 transfection induced the cleavage of caspase-9, caspase-3 and PARP, and the release of mitochondrial cytochrome c into the cytosol. These results demonstrated that MORC2 affected apoptosis and chemoresistance by modulating the mitochondrial apoptotic pathway.
Figure 12.
Dyregulation of MORC2 disrupts several crucial cancer-related pathways. (A) Knockdown of MORC2 activates apoptotic pathways. (B) Based on the Kyoto Encyclopedia of Genes and Genomes database, gene expression profile analysis identified significant different pathways modulated by MORC2. (C) In HepG2 cells, knockdown of MORC2 activated the p53 pathway. (D) In HepG2 cells, knockdown of MORC2 inhibited the Hippo pathway. MORC2, microrchidia; PARP, poly (ADP-ribose) polymerase; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X protein; PUMAα, p53 upregulated modulator of apoptosis α; p-, phosphorylated; si, small interfering RNA; NC, negative control; YAP1, Yes-associated protein 1; TAZ, transcriptional co-activator with PDZ-binding motif.
Dysregulation of MORC2 disrupts p53 and Hippo pathways
The p53 tumor-suppressor gene regulates apoptosis through the transcriptional activation of its target genes, and the mitochondrial apoptotic pathway is regulated by several p53-target genes, including Bax, Bcl-2, p53AIP1, Noxa and Puma (23–25). Therefore, the present study examined the possible association between p53 and MORC2 in liver cancer. By comparing the expression profile of L02MORC2 and L02Vector, we found that the overexpression of MORC2 significantly enriched genes involved in the p53 pathway (P<0.001) (Fig. 12B), which was consistent with our hypothesis. To confirm this result, the effect of MORC2-knockdown on the expression level of p53 and its target genes, including Bax, Bcl-2 and PUMAα, were examined. It was found that the knockdown of MORC2 increased the expression of p53, Bax and PUMAα, and reduced the expression of Bcl-2, compared with the control (Fig. 12C). The overexpression of MORC2 also notably enriched genes involved in the Hippo pathway (Fig. 12B). Further validating these results, the results of the western blot analysis showed that MORC2-knockdown increased phosphorylated Yes-associated protein (YAP) and TAZ (Fig. 12D). These results suggested that MORC2 may promote Hippo pathway activation in tumorigenesis. Collectively, the dysregulation of MORC2 in liver cancer disrupted the p53 and Hippo pathways during cancer progression.
Discussion
The DNA damage response (DDR), is associated with oncogenesis (26,27). The dysregulation of DNA damage repair-related genes results in the genomic instability, promoting the accumulation of DNA mutations and chromosomal aberrations of the cancer genome (20,26). Increasing evidence indicates that a high expression of certain DDR-related genes is crucial in causing chemoresistance, which eventually results in unsatisfactory treatment in cancer patients (28–31). It has been reported that p21 protein (Cdc42/Rac)-activated kinase 1 phosphorylation of MORC2 on serine 739 modulates ATPase-dependent chromatin remodeling following double-strand break damage, and facilitates efficient DNA damage repair (6). On finding that MORC2 was upregulated in liver cancer tissue, it was hypothesized that liver cancer cells with high expression levels of MORC2 are able to elicit more effective homologous recombination DNA repair, and may be less sensitive to apoptotic signals, leading to aberrant cell cycle progression, and higher survival ability and chemoresistance. Accordingly, in patients with liver cancer, the present study found that a higher fraction of copy number alterations in the genome was detected with higher expression of MORC2. It was also found that MORC2-knockdown induced cell cycle arrest and endogenous apoptotic pathways. The knockdown of MORC2 also sensitized liver cancer cells to doxorubicin, 5-fluorouracil and cisplatin, and markedly increased IC50 values, which suggested that MORC2 may be involved in the chemoresistance of liver cancer.
The p53 and Hippo pathways are two crucial pathways in cancer progression. p53 has been investigated intensively as a major tumor suppressor (23–25). p53 can be activated and transcriptionally induces several target genes in response to various stress signals. Its downstream genes include modulators controlling cell proliferation, apoptosis, DNA repair, autophagy, migration and metabolism (23). In the majority of types of human cancer, p53 is inactivated or missing through multiple mechanisms, resulting in tumorigenesis, cancer progression and metastasis (23–25). It has been demonstrated that the Hippo signaling pathway is pivotal in the regulation of tissue and organ size during development (32). The dysregulation of Hippo signaling leads to the inhibition of apoptosis and uncontrolled cellular proliferation (32,33). Despite diverse upstream mechanisms that regulate the Hippo pathway in cancer, the result is the common activation of YAP and TAZ (34). Previous studies have revealed that reorganization of the cell skeleton is a crucial factor affecting the phosphorylation levels of YAP and TAZ, and disrupting the structure of F-actin results in the activation of LATS1, which in turn inactivates YAP and TAZ (35–37). It is reported that ArgBP2 is a crucial protein involved in the formation of F-actin, and its upregulation inhibits the normal polymerization of actin (38,39). It has been demonstrated that MORC2 can inhibit the transcription of ArgBP2 via H2K27 trimethylation (40); therefore, MORC2 may be involved in maintaining the normal structure of F-actin and promoting the dephosphorylation of YAP/TAZ via the suppression of ArgBP2. This hypothesis will be tested in future investigations. When YAP and TAZ are activated, the proteins remain unphosphorylated, interacting with transcriptional factors TEA domain family member 1–4, translocating into the nucleus and activating the transcription of target genes, including connective tissue growth factor, insulin-like growth factor binding protein 3, integrin subunit β2, survivin, GLI family zinc finger 2 and AXL receptor tyrosine kinase, promoting cancer progression (34). In the present study, it was demonstrated that the dysregulation of MORC2 in liver cancer disrupted p53 and Hippo pathways, clarifying why p53 and Hippo pathways are aberrant in liver cancer.
The prognosis of patients with liver cancer is poor with high incidences of early metastasis and postoperative recurrence (22,41,42). Radical surgery, radiotherapy and liver transplantation are effective mainly for primary tumors, therefore, molecular-targeted therapy has been considered as a potential treatment tool (22,42). Clarifying the molecular mechanisms involved in the distant metastasis in liver cancer will enable promotion of the advancement of molecular-targeted therapies. In the present study, another novel finding was that MORC2 promoted the migration and invasion of cancer cells, which has not, to the best of our knowledge, been reported previously. It was also found that MORC2-knockdown sensitized liver cancer cells to chemotherapeutic drugs. These results indicated that MORC2-targeted therapy may be a potentially promising regimen to inhibit liver cancer metastasis and to improve the effect of chemotherapeutics for patients with advanced liver cancer and metastasis.
The present study had a number of limitations. It was found that MORC2-knockdown affected the proliferation and metastasis of liver cancer cells in vitro, however the present study did not investigate the effects of inhibiting MORC2 on the proliferation and lung metastasis of liver cancer cells in vivo. In addition, the study did not provide data on whether MORC2 modulated the p53, hippo and apoptotic pathways in vivo. The comprehensive underlying mechanism involved in MORC2 promoting liver cancer proliferation remains to be fully elucidated. Sánchez-Solana et al demonstrated that MORC2 promoted the activity of adenosine triphosphate citrate lyase (ACLY) by phosphorylation, and in turn activated acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN) (7). Of note, de novo lipogenesis is considered to be crucial in oncogenesis, and enhanced lipogenesis in cancer cells is reflected by improved activities of lipogenic enzymes, including ACLY, ACC and FASN (7,43–45). It is appropriate to suggest that MORC2 may promote cancer progression partly via this route. Additionally, by containing a CW zinc finger motif, which is predicted to be involved in DNA binding, MORC2 is considered to function as a transcriptional regulator (5). The high expression of MORC2 may alter the expression levels of certain crucial downstream tumor suppressors or oncogenes, in turn promoting the progression of cancer. These potential mechanisms require investigation and confirmation in the future. Additionally, it has been reported that the mutations of MORC2 may be involved in the development of Charcot-Marie-Tooth disease (46,47). The present study determined the mutation frequencies of MORC2 in tumor tissues using TCGA data, and it was demonstrated that the mutation frequencies of MORC2 in liver cancer and other types of cancer were low (data not shown). Therefore, it was hypothesized that mutation of MORC2 is not an important factor in tumor biology, which also requires validation in future investigations.
Acknowledgments
The authors would like to thank Professor Dongfeng Chen and Mr. Tao Wang (Daping Hospital, Chongqing, China) for providing access to the expression profile data of MORC2-overexpressing cells prior to publication.
Funding
This study was supported by the Natural Science Foundation of China (grant nos. 81172129, 81472798 and 81703030).
Availability of data and materials
The original expression profile or RNA sequencing data used in this study can be obtained from the following websites: TCGA database (http://www.cbioportal.org/) and GEO datasets (GSE14520 and GSE22058) (https://www.ncbi.nlm.nih.gov/gds). The authors also declare that the datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QD, HY, YG and FZ conceived and designed the experiments; QD, ZP and LW performed the experiments; QD, ZP,QG, LW and MT conducted the statistical analysis; ZP, QG, QD, LW and MT wrote the paper.
Ethics approval and consent to participate
The inclusion of human samples was approved by the Ethics Review Board of the Second People's Hospital of Guangdong Province (approval no. 2015-KYLL-023). All patients provided informed written consent prior to the investigation. All animal experiments were approved by the Ethics Review Board of the Second People's Hospital of Guangdong Province (approval no. 2015-KYLL-063).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Li DQ, Nair SS, Kumar R. The MORC family: New epigenetic regulators of transcription and DNA damage response. Epigenetics. 2013;8:685–693. doi: 10.4161/epi.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang GL, Wang CY, Cai XZ, Chen W, Wang XH, Li F. Identification and expression analysis of a novel CW-type zinc finger protein MORC2 in cancer cells. Anat Rec (Hoboken) 2010;293:1002–1009. doi: 10.1002/ar.21119. [DOI] [PubMed] [Google Scholar]
- 4.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Y, Li Y, Zhang J, Liu D, Liu F, Zhao Y, Shen T, Li F. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010;38:2813–2824. doi: 10.1093/nar/gkq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li DQ, Nair SS, Ohshiro K, Kumar A, Nair VS, Pakala SB, Reddy SD, Gajula RP, Eswaran J, Aravind L, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez-Solana B, Li DQ, Kumar R. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochimica et Biophysica Acta (BBA) Mol Cell Res. 2014;1843:316–326. doi: 10.1016/j.bbamcr.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LH, Kuo W-H, Tsai M-H, Chen P-C, Hsiao CK, Chuang EY, Chang LY, Hsieh FJ, Lai LC, Chang KJ. Identification of prognostic genes for recurrent risk prediction in triple negative breast cancer patients in Taiwan. PLoS One. 2011;6:e28222. doi: 10.1371/journal.pone.0028222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao XH, Zhang Y, Dong WJ, Shao ZM, Li DQ. Chromatin remodeling protein MORC2 promotes breast cancer invasion and metastasis through a PRD domain-mediated interaction with CTNND1. Oncotarget. 2017;8:97941–97954. doi: 10.18632/oncotarget.18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Y, Li Y, Gu H, Wang C, Liu F, Shao Y, Li F. HSF1, in association with MORC2, downregulates ArgBP2 via the RC2 family in gastric cancer cells. Biochim Biophys Acta. 2018;1864:1104–1114. doi: 10.1016/j.bbadis.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Song Y, Liu T, Wang C, Zhang Q, Liu F, Cai X, Miao Z, Xu H, Xu H, et al. PAK1-mediated MORC2 phosphorylation promotes gastric tumorigenesis. Oncotarget. 2015;6:9877–9886. doi: 10.18632/oncotarget.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Song Y, Chen W, Wang X, Miao Z, Cao L, Li F, Wang G. By recruiting HDAC1, MORC2 suppresses p21 Waf1/Cip1 in gastric cancer. Oncotarget. 2015;6:16461–16470. doi: 10.18632/oncotarget.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBio-Portal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Ding Q, He K, Luo T, Deng Y, Wang H, Liu H, Zhang J, Chen K, Xiao J, Duan X, et al. SSRP1 Contributes to the Malignancy of Hepatocellular Carcinoma and Is Negatively Regulated by miR-497. Mol Ther. 2016;24:903–914. doi: 10.1038/mt.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, Wang L, Zhou J, Ren ZG, Li YX, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Lin Y, Zhang Y, Zhu Z, Huo K. SSX2IP promotes metastasis and chemotherapeutic resistance of hepatocellular carcinoma. J Transl Med. 2013;11:52. doi: 10.1186/1479-5876-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arakawa H. p53, apoptosis and axon-guidance molecules. Cell Death Differ. 2005;12:1057–1065. doi: 10.1038/sj.cdd.4401601. [DOI] [PubMed] [Google Scholar]
- 24.Wu B, Chu X, Feng C, Hou J, Fan H, Liu N, Li C, Kong X, Ye X, Meng S. Heat shock protein gp96 decreases p53 stability by regulating Mdm2 E3 ligase activity in liver cancer. Cancer Lett. 2015;359:325–334. doi: 10.1016/j.canlet.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 25.Cui X, Choi HK, Choi YS, Park SY, Sung GJ, Lee YH, Lee J, Jun WJ, Kim K, Choi KC, et al. DNAJB1 destabilizes PDCD5 to suppress p53-mediated apoptosis. Cancer Lett. 2015;357:307–315. doi: 10.1016/j.canlet.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 26.Kumar Y, Yang J, Hu T, Chen L, Xu Z, Xu L, Hu XX, Tang G, Wang JM, Li Y, et al. Massive interstitial copy-neutral loss-of-heterozygosity as evidence for cancer being a disease of the DNA-damage response. BMC Med Genomics. 2015;8:42. doi: 10.1186/s12920-015-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuhldreier F, Kassel S, Schumacher L, Wesselborg S, Proksch P, Fritz G. Pleiotropic effects of spongean alkaloids on mechanisms of cell death, cell cycle progression and DNA damage response (DDR) of acute myeloid leukemia (AML) cells. Cancer Lett. 2015;361:39–48. doi: 10.1016/j.canlet.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 28.Ho H, Aruri J, Kapadia R, Mehr H, White MA, Ganesan AK. RhoJ regulates melanoma chemoresistance by suppressing pathways that sense DNA damage. Cancer Res. 2012;72:5516–5528. doi: 10.1158/0008-5472.CAN-12-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia H, Cong Q, Chua JF, Liu H, Xia X, Zhang X, Lin J, Habib SL, Ao J, Zuo Q, et al. p57Kip2 is an unrecognized DNA damage response effector molecule that functions in tumor suppression and chemoresistance. Oncogene. 2015;34:3568–3581. doi: 10.1038/onc.2014.287. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Mosel AJ, Oakley GG, Peng A. Deficient DNA damage signaling leads to chemoresistance to cisplatin in oral cancer. Mol Cancer Ther. 2012;11:2401–2409. doi: 10.1158/1535-7163.MCT-12-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SF, Chang CW, Wei RJ, Shiue YL, Wang SN, Yeh YT. Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed Res Int. 2014;2014:153867. doi: 10.1155/2014/153867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong L, Cai Y, Jiang M, Zhou D, Chen L. The Hippo signaling pathway in liver regeneration and tumorigenesis. Acta Biochim Biophys Sin (Shanghai) 2015;47:46–52. doi: 10.1093/abbs/gmu106. [DOI] [PubMed] [Google Scholar]
- 33.Zheng T, Wang J, Jiang H, Liu L. Hippo signaling in oval cells and hepatocarcinogenesis. Cancer Lett. 2011;302:91–99. doi: 10.1016/j.canlet.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 36.Bae JS, Kim SM, Lee H. The Hippo signaling pathway provides novel anti-cancer drug targets. Oncotarget. 2017;8:16084–16098. doi: 10.18632/oncotarget.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anekal PV, Yong J, Manser E. Arg kinase-binding protein 2 (ArgBP2) interaction with α-actinin and actin stress fibers inhibits cell migration. J Biol Chem. 2015;290:2112–2125. doi: 10.1074/jbc.M114.610725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cestra G, Toomre D, Chang S, De Camilli P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:1731–1736. doi: 10.1073/pnas.0409376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Y, Li Y, Gu H, Wang C, Liu F, Shao Y, Li J, Cao L, Li F. Microchidia protein 2, MORC2, downregulates the cytoskeleton adapter protein, ArgBP2, via histone methylation in gastric cancer cells. Biochem Biophys Res Commun. 2015;467:821–827. doi: 10.1016/j.bbrc.2015.10.059. [DOI] [PubMed] [Google Scholar]
- 41.Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z, Guo C, Liu Y, Chen Z, Wang X, et al. MicroRNA-362-5p promotes tumor growth and metastasis by targeting CYLD in hepatocellular carcinoma. Cancer Lett. 2015;356:809–818. doi: 10.1016/j.canlet.2014.10.041. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Han P, Li M, Yan W, Liu J, Liu J, He J, Tu W, Xia Y, Zhou Z, et al. The histidine-rich calcium binding protein (HRC) promotes tumor metastasis in hepatocellular carcinoma and is upregulated by SATB1. Oncotarget. 2015;6:6811–6824. doi: 10.18632/oncotarget.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, Destefanis G, Delogu S, Zimmermann A, Ericsson J, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–1083. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Rajput S, Watabe K, Liao DF, Cao D. Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Front Biosci (Schol Ed) 2010;2:515–526. doi: 10.2741/s82. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, He D, Yang L, Wen B, Dai J, Zhang Q, Kang J, He W, Ding Q, He D. TRIM26 functions as a novel tumor suppressor of hepatocellular carcinoma and its downregulation contributes to worse prognosis. Biochem Biophys Res Commun. 2015;463:458–465. doi: 10.1016/j.bbrc.2015.05.117. [DOI] [PubMed] [Google Scholar]
- 46.Laššuthová P, Šafka Brožková D, Krůtová M, Mazanec R, Züchner S, Gonzalez MA, Seeman P. Severe axonal Charcot-Marie-Tooth disease with proximal weakness caused by de novo mutation in the MORC2 gene. Brain. 2016;139:e26. doi: 10.1093/brain/awv411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao X, Li X, Hu Z, Liu L, Xie Y, Tian T, Man J, Wang J, Zi X, Xia K, et al. MORC2 mutations in a cohort of Chinese patients with Charcot-Marie-Tooth disease type 2. Brain. 2016;139:e56. doi: 10.1093/brain/aww156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original expression profile or RNA sequencing data used in this study can be obtained from the following websites: TCGA database (http://www.cbioportal.org/) and GEO datasets (GSE14520 and GSE22058) (https://www.ncbi.nlm.nih.gov/gds). The authors also declare that the datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.