Abstract
Aim
Multigene panel testing for Hereditary Breast and Ovarian Cancer (HBOC) using next generation sequencing is becoming more common in medical care.
We report our experience regarding deleterious mutations of high and moderate-risk breast cancer genes (BRCA1/2, TP53, STK11, CDH1, PTEN, PALB2, CHEK2, ATM), as well as more recently identified cancer genes, many of which have increased risk but less well-defined penetrance.
Methods
Genetic testing was performed in 130 consecutive cases with breast cancer referred to our clinic for surgical evaluation and who met the 2016 National Comprehensive Cancer Network (NCCN) criteria for genetic testing.
Results
82 patients had pathogenic/likely pathogenic mutations and VUS mutations, and 48 were negative; 36 of the pathogenic mutations were in the high-risk genes and 16 were in the moderate risk genes and only 5 cases in the intermediary risk group.
From the VUS mutation group 21 cases were in the intermediary risk group, 9 cases were in the moderate risk group and only 7 cases in high risk group.
The most frequent BRCA1 variant was c.3607C>T (7 cases) followed by c.5266dupC and c.4035delA (each in 4 cases). Regarding BRCA-2 mutations we identified c.9371A>T and c.8755-1G>A in 6 cases and we diagnosed VUS mutations in 3 cases.
Conclusion
Our study identified 2 mutations in the BRCA1 gene that are less common in the Romanian population, c.3607C>T and c.4035delA. Both variants had particular molecular phenotypes, c.3607C>T variant respecting the triple negative pattern of BRCA1 breast cancer while c.4035delA were Luminal B HER positive.
Keywords: BRCA1/2, hereditary breast and ovarian cancer, next generation sequencing hereditary cancer panels
Introduction
In the last decade, we witnessed an accelerated progress in understanding and also in the diagnosis and treatment of patients with hereditary cancer, studies demonstrating the importance and usefulness of genetic testing.
About 30% of breast cancers are caused by a family predisposition, but only 10% of the cases are due to the inheritance of a mutation in a major gene involved in the occurrence of this pathology [1].
Until recently, genetic diagnosis of hereditary breast and ovarian cancer (HBOC) was based mainly on determining mutations in the BRCA1-/-2 genes. Along with the US Supreme Court’s decision which held that human genetic material cannot be patented, and after introduction of Next Generation Sequencing (NGS) [2] this has changed. Furthermore genetic testing for HBOC has seen an even greater publicity in May 2013, when Angelina Jolie announced that she had undergone prophylactic bilateral mastectomy because she was a BRCA1 mutation carrier [3].
The aim of these genetic tests is to create the opportunity for multidisciplinary and individualized treatment for each patient, depending on the mutation they carried [4]. It also creates the possibility of genetic counseling to the relatives of these patients about the risk of developing breast cancer or another type of cancer caused by the inheritance of certain mutations [1,4].
Along with the technological progress and the use of NGS on a larger scale, based on the clonal amplification and multiple parallel sequencing, it was possible to investigate more genes in a shorter time and with lower costs [5,6]. Moreover, this new method increased the sensitivity for detecting individual mutations that occur less frequently or have a lower penetrance [2,7].
Another advantage of NGS technology is that it made possible to identify some mutation carriers originating from families with hereditary cancer, but with atypical or particular phenotypes [7].
Unfortunately, a major deficiency of using multigene test panels is the deterioration of the clinical genetic conduct [2] and also the lack of interpretation of some mutations in clinical context [7]. Furthermore, for some mutations we don’t have specific guidelines regarding genetic counseling or prophylactic surgery in mutation carriers or for their relatives [7].
Mutations in genes with high penetrance such as PTEN, CDH1, STK11 and TP53 [8–11] or in genes with moderate penetrance like PALB2, CHEK2, ATM [8,12–14] are known to be responsible for familial forms of breast cancer. Less attention has been paid to mutations in genes with a lower contribution in the occurrence of breast cancer, such as those responsible for Lynch syndrome (MLH1, MSH2, MSH6 and PMS2), or other rare genes like BARD 1, MUTYH, RAD50 etc. [15,16], but this has changed during the past years.
The objective of our study is to report our experience from the first 130 consecutive patients with breast cancer referred to our service for multipanel genetic testing, providing data on the frequency of pathogenic genetic variants by gene.
Material and method
Ethics
This study was conducted with the approval of the ethics committee of Iuliu Haţieganu University of Medicine and Pharmacy “, Cluj-Napoca (no.369 / 14.10.2016). We obtained the informed consent of all patients at the first medical evaluation.
Patients
Were enrolled in the study 130 consecutive patients diagnosed with breast cancer who presented for oncological examination to Oncosurg Surgical Oncology Clinic of Cluj-Napoca between January 2015 and June 2017 and met the 2016 National Comprehensive Cancer Network (NCCN) criteria for genetic testing [4]. 48 patients were excluded due to the negative result on genetic testing.
All patients had a prior Tru-Cut biopsy to confirm the diagnosis and a complete histopathological report.
Genetic testing
Each patient was approached for genetic testing after the diagnosis. Genomic DNA was prepared and analyzed as described in our previous research [17].
The genes were grouped into three risk categories based on penetrance data:
High penetrance and high risk: BRCA1, BRCA2, TP53, CDH1, STK11, PTEN
Moderate penetrance and high risk: ATM, CHEK2, PALB2
Increased penetrance but less well-defined risk: BARD1, BLM, BRIP1, FAM175A, MEN1, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PMS2, RAD50, RAD51C, RAD51D, XRCC2.
Results
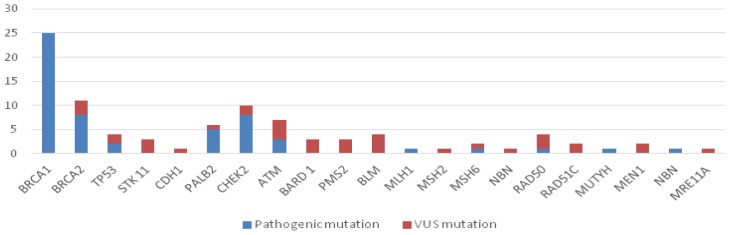
During the study, from the 130 patients with breast cancer which were eligible for genetic testing, 82 were included in the study. In the 82 patients tested we identified 93 mutations from the multigene panel (56 pathogenic variants and 37 VUS variants) – Table I, Figure 1.
Table I.
Distribution of mutations in the studied group.
| High penetrance breast genes | |||
|---|---|---|---|
| Gene | Cases | Pathogenic Mutation | VUS mutation |
| BRCA1 | 25 | c.3607C>T (7), c.5266dupC (4), c.181T>G (2), c.4035delA, c.3726 C>T, c.2241dupC, c.135-2A>G, c.1789G>A, c.737delT, c.3700_3704delGTAAA, c.843_846delCTCA, c.4035delA (3), c5329_5330insC | - |
| BRCA2 | 11 | c.9371A>T (3), c.8755-1G>A (3), c.1528G>T, c.8695 C>T | c.3547G>C, c.3562A>G (2) |
| TP53 | 4 | c.469G>T (2) | c.847C>T, c.480G>A |
| STK 11 | 3 | - | c.1225C>T (3) |
| CDH1 | 1 | - | c.1840A>G |
| Moderate penetrance breast genes | |||
| Gene | Cases | Pathogenic Mutation | VUS mutation |
| PALB2 | 6 | c.93dupA (2), c.3549C>G, c.509_510delGA, c.79G>T | c.2461A>T |
| CHEK2 | 10 | c.470T>C (5), c.1232G>A (2), 1283C>T | c.1521T>C, c.1313A>T |
| ATM | 7 | c.7630-2A>C, c.2250G>A, c.1564_1565delGA | c.9077T>G, c8734A>G, c.2735G>A, c.4768C>T |
| Additional genes | |||
| Gene | Cases | Pathogenic Mutation | VUS mutation |
| BARD 1 | 3 | - | c.1333G>A, c.2282G>A, c.2640del15 |
| PMS2 | 3 | - | c.620G>A, c.2012C>T, c.852A>G |
| BLM | 4 | - | c.3014T>C, c.3879A>G, c.4076+4T>G, c.1642C>T |
| MLH1 | 1 | c.2041G>A | - |
| MSH2 | 1 | - | c.1597C>G |
| MSH6 | 2 | c.2136delG | c.1068T>G |
| NBN | 1 | - | c.511A>G |
| RAD 50 | 4 | c.2165dupA | c.785T>G, c900G>A, c.1663A>G |
| Rad51C | 2 | - | c.790G>A (2) |
| MUTYH | 2 | c.1187G>A | c.158-3C>T |
| MEN1 | 1 | - | c.777G>A |
| NBN | 1 | c657_661delACAAA | - |
| MRE11A | 1 | - | c.1091G>A |
Figure 1.
Classification of mutations depending on the risk of pathogeny.
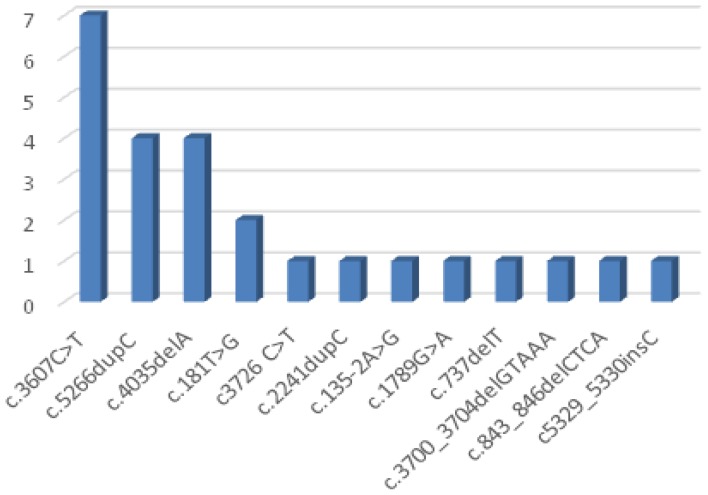
The most frequent BRCA1 mutation was c.3607C>T (28%), followed by c.5266dupC (16%) and c.4035delA (16%) and c.181T>G (8%) variant (Table I, Figure 2).
Figure 2.
Distribution of BRCA1 variants.
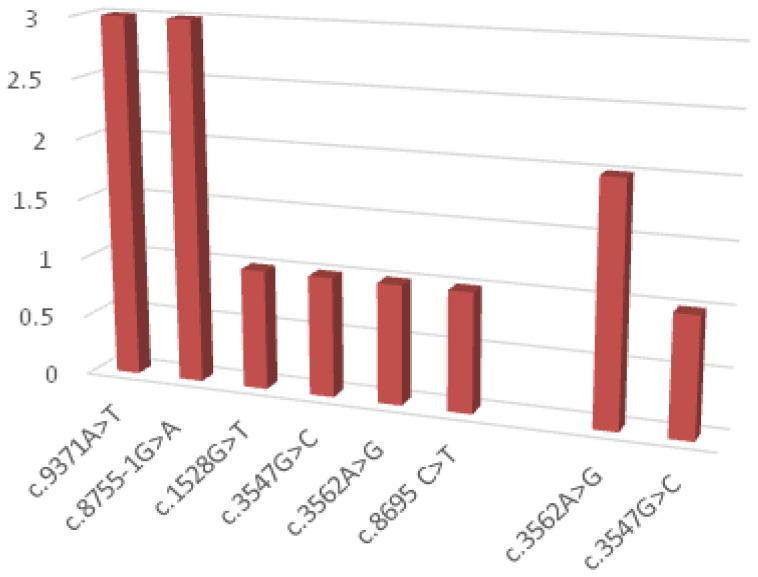
Regarding BRCA2 gene mutations c.9371A>T and c.8755-1G>A variants were found each in 27.27% of cases. 3 VUS mutations have been identified in the BRCA2 gene (27.27%)- Figure 3.
Figure 3.
Distribution of BRCA2 variants.
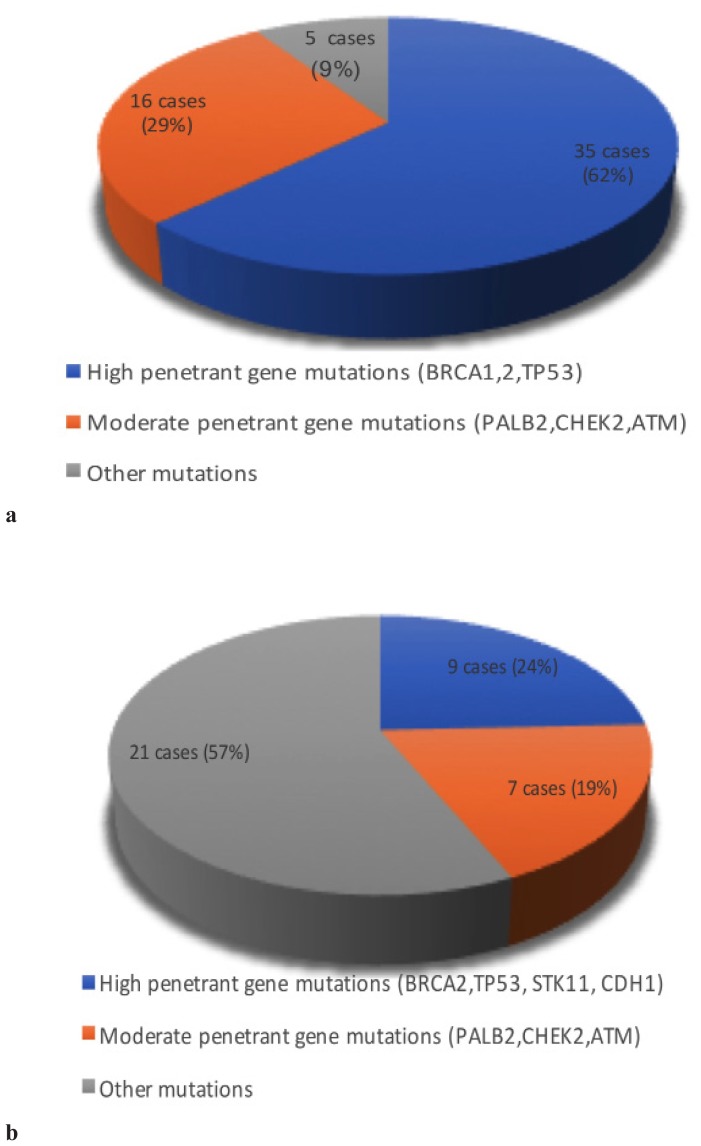
Mutations in the additional genes were more often VUS variants, in only 5 cases being pathogenic (Figure 4a and 4b).
Figure 4.
Distribution of mutations according to penetrance: a- Pathogenic mutations; b- VUS mutations.
The majority of VUS variants were from the patients with rare mutations, while in the BRCA 1 mutation group we did not identify any VUS variants (Table I, Figure 1).
Nine patients had 2 mutations (BRCA1c.2241dupC + MEN1 c.777G>A; BRCA1 c.135-2A>G +MUTYH c.158-3C>T; BRCA1 c.843_846delCTCA + CHEK2 c.470T>C; BRCA2 c.9371A>T + ATM c.2735G>A; CHEK2 c.1232G>A + RAD50 c900G>A; c.470T>C + BLM c.4076+4T>G; MSH6 c.2136delG + MUTYH c.1187G>A; NBN 657_661delACAAA +MRE11A c.1091G>A; RAD50 c.1663A>G + RAD51 c.790G>A) and one patient had 3 mutations (CHEK2 1283C>T + BLM c.1642C>T + ATM c.4768C>T).
Discussion
BRCA1
The most common BRCA1 mutations in our study were c.3607C>T (7 cases), c.5266dupC (4 cases), c.4035delA (4 cases) and c.181T>G (2 cases).
BRCA1 c.3607C>T (p.Arg1203*), exon 11 - This sequence change creates a premature translational stop signal at codon 1203 (p.Arg1203*), causing the absence or disruption of protein synthesis [18]. BRCA1 c.3607C> T variant was diagnosed in 7 patients and was not reported until now in Romanian population, being the most frequent in the study group, all the patients originating from northwestern towns of Romania [17].
In 6 cases we identified the association of this variant with triple negative breast cancer (TNBC), which is in accordance with the general immunohistochemical phenotype of BRCA1 mutations [17].
BRCA1 c.5266dupC (p.Gln1756Profs*74), exon 20 - This mutation determines the insertion of one nucleotide (cytosine), creating a novel STOP codon after 74 amino acid residues and therefore will produce a truncated and a non-functional protein. This variant has been described as being the most prevalent mutation in the north-eastern Romania population, in the only two studies conducted so far in the BRCA1 mutation carriers from this region [19,20].
BRCA1 c.5266dupC is one of the BRCA1 founder mutations encountered in Hebrews and is most common in Polish population. This is due to the expulsion of Jews from countries like England, Spain, Portugal, Germany and Italy, around 1650, Poland totaling nowadays about 30% of Hebrew population worldwide [21]. In Polish population the c.5266dupC BRCA1 mutation is commonly associated with ovarian cancer [22], only one patient in our study from the four mutation carriers had a personal history of ovarian cancer, the breast cancer being diagnosed four years later.
C.5266dupC mutation is more commonly encountered in Russian population and in the Nordic countries near Russia (Estonia Lithuania, Latvia) [21], where it is considered to have originated, but an increased incidence of this variant is also found in countries bordering with Romania, like Hungary [23], Serbia and Montenegro [24].
BRCA1 c.4035delA variant is described as the third most common mutation in Polish people and alongside c.5266dupC and c.181T> G they’re representing 90% of pathogenic mutations in this population. Its origin seems to be more of Baltic origin, rather than Slavic, being described with an increased frequency among Estonian population [25].
In our study, we identified this mutation in 4 patients, of which 3 were relatives (mother and 2 daughters). What is more interesting is that all patients had as immunohistochemical characteristic the overexpression of HER2, being known that most BRCA1 mutations are triple negative.
BRCA1 c.181T>G (p.Cys61Gly), exon 5 – This mutation causes a sequence change that replaces cysteine with glycine at codon 61 of the BRCA1 protein causing a disruption in protein function.
This variant is the most common mutation in the BRCA 1 gene in the Austrian population [26] and 2nd after c.5266dupC variant in Czech [27], Slovenia [28], Hungary [29] and Polish population [30]. In our study, 2 patients were diagnosed with breast cancer and BRCA1 c.181T> G variant both of Hungarian ethnicity, mother and daughter.
For the rest of BRCA1 mutations found in one case each in our study (c.3726 C>T, c.2241dupC, c.135-2A>G, c.1789G>A, c.737delT, c.3700_3704delGTAAA, c.843_846delCTCA, c5329_5330insC) there is limited data available.
BRCA2
We found 11 BRCA 2 variants, eight pathogenic and likely pathogenic (c.9371A> T (3 cases), c.8755-1G> A (3 cases), c.1528G> T (1 case) and c.8695 C>T(1 case)) and 3 VUS mutations.
BRCA2 c.9371A>T, (p.Asn3124Ile), exon 25 - is a mutation caused by a single base substitution, asparagine with isoleucine at position 3124, this change leading to an alteration in the protein synthesis of BRCA2 gene [31]. This variant is a rare but deleterious mutation in BRCA2 protein synthesis, described as associated with an increased risk of breast and ovarian cancer, and is considered more likely to be pathogenic [30,32].
BRCA 2 c.8755-1G>A, intron 21- This mutation results in an incorrect mismatching of the mRNA causing a reading alteration and a non-functional protein synthesis after this point [33]. The pathogenesis of this variant is still a subject of debate, but most studies suggest that it is most likely pathogenic [27,33].
BRCA 2 c.8695C>T (p.Gln2899Ter) – is a rare variant described in literature as pathogenic and mentioned in Romanian population in a recent study [34].
BRCA2 c.1528G>T (p.Glu510Ter, p.Glu510Glu) is a rare pathogenic mutation in the BRCA2 gene described in the population of Slovenia and present in our study in one patient [35].
TP53
The two patients with TP53 mutation had both c.469G>T variants. This sequence change replaces Valine with Phenylalanine at codon 157 of the TP53 protein (p.Val157Phe).
The valine residue is highly conserved and there is little difference between the physicochemical properties of Valine and Phenylalanine, but this sequence change was reported in literature in breast and liver cancer patients [36].
PALB2
Among the patients with PALB2 mutations we identified five pathogenic variants: c.93dupA (2 cases), c.3549C>G, c.509_510delGA, c.79G>T.
PALB2 c.93dupA (p.Leu32Thrfs * 11) – This sequence change inserts 1 nucleotide in exon 2 of the PALB2 mRNA (c.93dupA), causing a frameshift at codon 32. This creates a premature translational stop signal 11 amino acid residues later and causes the production of a modified protein.
This variant has been described in literature in a patient with triple negative breast cancer [37]; in our study, only 1 patient was diagnosed with this molecular form, the 2nd being diagnosed with LUMINAL B HER negative type.
PALB2 c.3549C>G (p.Tyr1183*) – This sequence change results in the replacement of Tyrosine with a terminal codon at position 1183 of the PALB2 protein, resulting in a truncated PALB2 protein missing the last three amino acids [38]. This variant has been reported in literature for individuals people affected by breast cancer, pancreatic cancer and Fanconi anemia and it is considered pathogenic[39].
PALB2 c.509_510delGA (p.Arg170Ilefs*14) – This mutation is caused by the deletion of 2 base pairs at codon 509 of the PALB2 gene. It is a frame-shift mutation that will result in the creation of an unrecognized protein after amino acid 170 and a new stop codon after 14 amino acids, causing the absence or an abnormal protein synthesis [40]. This mutation is recognized as pathogenic, being identified primarily in the Polish population [30,40], where it is found at a higher frequency compared to the rest of Europe.
PALB2 c.79G>T (p.Glu29*) – This variant causes Glutamate to be replaced by a terminal codon at position 29 resulting in the modification of the PALB2 protein synthesis. For this reason, this variant it is considered to be more likely pathogenic and it is associated with breast and vulvar cancer and most international guidelines recommend testing this mutation [41].
CHEK2
We diagnosed 10 CHEK2 mutations, 8 pathogenic (c.470T>C (5 cases), c.1232G>A (2 cases), 1283C>T (1 case), and 2 VUS cases.
CHEK2 c.470T>C (p.Ile157Thr) – This sequence is due to the replacement of Isoleucine with Threonine at codon 157. This change leads to the synthesis of a modified CHEK 2 protein, which is associated with increased risk of lobular breast cancer (OR=1.48–1.58) [42], but our study has not confirmed this observation because the 3 patients were diagnosed with invasive ductal carcinoma (Table 2).
CHEK2 1283C>T (p.Ser428Phe) - This sequence is due to the replacement of Serine with Phenylalanine at codon 428, causing changes in protein kinase domain [43]. This mutation is known to be a pathogenic and associated with breast and prostate cancer, especially in the Ashkenazy Jews who have a twofold increased risk for developing breast cancer.
CHEK2 c.1232G>A (p.Trp411*) - This sequence change creates a premature translational stop signal at codon 411 of the CHEK2 protein causing absent or abnormal protein product.
ATM
ATM c.7630-2A>C – This sequence change occurs in 2 nucleotides before exon 52 of the ATM gene. This mutation determines the synthesis of a truncated protein, and literature data associates this variant with a higher breast cancer risk [44].
ATM c.2250G>A – This variant occurs at the last base of exon 14, a position that is highly conserved in the human genome. Although it does not result in an amino acid substitution, it has been demonstrated to affect splicing of the mRNA, specifically in frame skipping of the entire exon 14 [45]. This variation is cited in literature as pathogenic and the carriers of this mutation are at increased risk for breast cancer [46].
ATM c.1564_1565delGA - This sequence change deletes 2 nucleotides in exon 10 of the ATM gene, causing a frameshift after codon 522 and the creation of a premature translational stop signal 43 amino acid residues later - p.(Glu522Ilefs*43) [47].
For high penetrance mutations but with less well-defined risk data on the association of breast cancer is reduced, RAD50 c.2165dupA (p.Glu723Glyfs*5) [48], MSH6 c.2136delG (p.Asp713Ilefs*23) [49], MUTYH c.1187G>A (p.Gly396Asp) [50], MLH1 c.2041G>A (p.Ala681Thr) [51] and NBN c657_661delACAAA [52], this association being described only in isolated cases.
Conclusion
Our study has successfully identified 2 mutations in the BRCA1 gene that are less common in the Romanian population, c.3607C>T and c.4035delA. Furthermore, the two pathogenic variants were associated with different histological and molecular phenotypes. The c.3607C> T variant was associated with triple negative breast cancer (negative ER, PR and HER2), respecting the general phenotype of patients with BRCA1 mutations while variant c.4035delA was associated with HER positive, hormone-negative forms, an atypical element for patients with BRCA1 mutations.
For the rest of the genes, the information could be useful for establishing specific variants of these mutations in the Romanian population and thus for their targeted identification and thus to a decrease of costs and a reduced time for obtaining results.
The absence of VUS mutations in patients in the BRCA1 group is most likely due to the fact that mutations in the BRCA1 gene were more extensively studied compared to those of other genes. This could be also the explanation for the numerous VUS mutations identified in the group of moderate penetrance and intermediate risk genes, where larger studies are missing.
Acknowledgements
The authors would like to thank all patients who participated in this study as well as GeneKor M.S.A. for providing the information regarding methodology.
References
- 1.Hoffman BL, Williams JW. Williams gynecology. New York: McGraw-Hill Medical; 2012. pp. 333–355. [Google Scholar]
- 2.Kurian AW, Kingham KE, Ford JM. Next-generation sequencing for hereditary breast and gynecologic cancer risk assessment. Curr Opin Obstet Gynecol. 2015;27(1):23–33. doi: 10.1097/GCO.0000000000000141. [DOI] [PubMed] [Google Scholar]
- 3.Desai S, Jena AB. Do celebrity endorsements matter? Observational study of BRCA gene testing and mastectomy rates after Angelina Jolie’s New York Times editorial. BMJ. 2016 Dec 14;355:i6357. doi: 10.1136/bmj.i6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly MB, Pilarski R, Axilbund JE, Berry M, Buys SS, Crawford B, et al. Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2015. J Natl Compr Canc Netw. 2016;14(2):153–162. doi: 10.6004/jnccn.2016.0018. [DOI] [PubMed] [Google Scholar]
- 5.Kwong A, Shin VY, Au CH, Law FB, Ho DN, Ip BK, et al. Detection of Germline Mutation in Hereditary Breast and/or Ovarian Cancers by Next-Generation Sequencing on a Four-Gene Panel. J Mol Diagn. 2016;18(4):580–594. doi: 10.1016/j.jmoldx.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Kwong A. Genetic testing for hereditary breast cancer in Asia-moving forward. Chin Clin Oncol. 2016 Jun;5(3):47. doi: 10.21037/cco.2016.05.11. [DOI] [PubMed] [Google Scholar]
- 7.LaDuca H, Stuenkel AJ, Dolinsky JS, Keiles S, Tandy S, Pesaran T, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med. 2014;16(11):830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295(12):1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 9.McCuaig JM, Armel SR, Novokmet A, Ginsburg OM, Demsky R, Narod SA, et al. Routine TP53 testing for breast cancer under age 30: ready for prime time? Fam Cancer. 2012;11(4):607–613. doi: 10.1007/s10689-012-9557-z. [DOI] [PubMed] [Google Scholar]
- 10.Pharoah PD, Guilford P, Caldas C International Gastric Cancer Linkage Consortium. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121(6):1348–1353. doi: 10.1053/gast.2001.29611. [DOI] [PubMed] [Google Scholar]
- 11.Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12(10):3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 12.Kwong A, Chen JW, Shin VY. A new paradigm of genetic testing for hereditary breast/ovarian cancers. Hong Kong Med J. 2016;22(2):171–177. doi: 10.12809/hkmj154634. [DOI] [PubMed] [Google Scholar]
- 13.Evans MK, Longo DL. PALB2 mutations and breast-cancer risk. N Engl J Med. 2014;371(6):566–568. doi: 10.1056/NEJMe1405784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldgar DE, Healey S, Dowty JG, Da Silva L, Chen X, Spurdle AB, et al. Rare variants in the ATM gene and risk of breast cancer. Breast Cancer Res. 2011 Jul 25;13(4):R73. doi: 10.1186/bcr2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahdi KM, Nassiri MR, Nasiri K. Hereditary genes and SNPs associated with breast cancer. Asian Pac J Cancer Prev. 2013;14(6):3403–3409. doi: 10.7314/apjcp.2013.14.6.3403. [DOI] [PubMed] [Google Scholar]
- 16.Lincoln SE, Kobayashi Y, Anderson MJ, Yang S, Desmond AJ, Mills MA, et al. A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. J Mol Diagn. 2015;17(5):533–544. doi: 10.1016/j.jmoldx.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Goidescu I, Eniu DT, Caracostea G, Cruciat Gh, Stamatian F. Associations of pathogenic mutations responsible for breast cancer risk with histology and immunohistochemistry in Romanian population. RJLabM. doi: 10.1515/rrlm-2017-0037. [DOI] [Google Scholar]
- 18.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108(44):18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Negura L, Uhrhammer N, Negura A, Artenie V, Carasevici E, Bignon YJ. Complete BRCA mutation screening in breast and ovarian cancer predisposition families from a North-Eastern Romanian population. Fam Cancer. 2010;9(4):519–523. doi: 10.1007/s10689-010-9361-6. [DOI] [PubMed] [Google Scholar]
- 20.Burcoş T, Cimponeriu D, Ion DA, Spandole S, Apostol P, Toma M, et al. Analysis of several BRCA1 and BRCA2 mutations in a hospital-based series of unselected breast cancer cases. Chirurgia (Bucur) 2013;108(4):468–472. [PubMed] [Google Scholar]
- 21.Hamel N, Feng BJ, Foretova L, Stoppa-Lyonnet D, Narod SA, Imyanitov E, et al. On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur J Hum Genet. 2011;19(3):300–306. doi: 10.1038/ejhg.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koczkowska M, Zuk M, Gorczynski A, Ratajska M, Lewandowska M, Biernat W, et al. Detection of somatic BRCA1/2 mutations in ovarian cancer - next-generation sequencing analysis of 100 cases. Cancer Med. 2016;5(7):1640–1646. doi: 10.1002/cam4.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Looij M, Cleton-Jansen AM, van Eijk R, Morreau H, van Vliet M, Kuipers-Dijkshoorn N, et al. A sporadic breast tumor with a somatically acquired complex genomic rearrangement in BRCA1. Genes Chromosomes Cancer. 2000;27(3):295–302. doi: 10.1002/(sici)1098-2264(200003)27:3<295::aid-gcc10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 24.Papp J, Raicevic L, Milasin J, Dimitrijevic B, Radulovic S, Olah E. Germline mutation analysis of BRCA1 and BRCA2 genes in Yugoslav breast/ovarian cancer families. Oncol Rep. 1999;6(6):1435–1438. doi: 10.3892/or.6.6.1435. [DOI] [PubMed] [Google Scholar]
- 25.Tamboom K, Kaasik K, Aršavskaja J, Tekkel M, Lilleorg A, Padrik P, et al. BRCA1 mutations in women with familial or early-onset breast cancer and BRCA2 mutations in familial cancer in Estonia. Hered Cancer Clin Pract. 2010 Apr 9;8(1):4. doi: 10.1186/1897-4287-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janavičius R. Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J. 2010;1(3):397–412. doi: 10.1007/s13167-010-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machackova E, Foretova L, Lukesova M, Vasickova P, Navratilova M, Coene I, et al. Spectrum and characterisation of BRCA1 and BRCA2 deleterious mutations in high-risk Czech patients with breast and/or ovarian cancer. BMC Cancer. 2008 May 20;8:140. doi: 10.1186/1471-2407-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krajc M, Teugels E, Zgajnar J, Goelen G, Besic N, Novakovic S, et al. Five recurrent BRCA1/2 mutations are responsible for cancer predisposition in the majority of Slovenian breast cancer families. BMC Med Genet. 2008 Sep 10;9:83. doi: 10.1186/1471-2350-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Looij M, Szabo C, Besznyak I, Liszka G, Csokay B, Pulay T, et al. Prevalence of founder BRCA1 and BRCA2 mutations among breast and ovarian cancer patients in Hungary. Int J Cancer. 2000;86(5):737–740. doi: 10.1002/(sici)1097-0215(20000601)86:5<737::aid-ijc21>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Wojcik P, Jasiowka M, Strycharz E, Sobol M, Hodorowicz-Zaniewska D, Skotnicki P, et al. Recurrent mutations of BRCA1, BRCA2 and PALB2 in the population of breast and ovarian cancer patients in Southern Poland. Hered Cancer Clin Pract. 2016 Feb 3;14:5. doi: 10.1186/s13053-016-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biswas K, Das R, Eggington JM, Qiao H, North SL, Stauffer S, et al. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum Mol Genet. 2012;21(18):3993–4006. doi: 10.1093/hmg/dds222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surowy HM, Sutter C, Mittnacht M, Klaes R, Schaefer D, Evers C, et al. Clinical and molecular characterization of the BRCA2 p.Asn3124Ile variant reveals substantial evidence for pathogenic significance. Breast Cancer Res Treat. 2014;145(2):451–460. doi: 10.1007/s10549-014-2943-5. [DOI] [PubMed] [Google Scholar]
- 33.Acedo A, Hernandez-Moro C, Curiel-Garcia A, Diez-Gomez B, Velasco EA. Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Hum Mutat. 2015;36(2):210–221. doi: 10.1002/humu.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eniu AE, Antone NZ, Stoian A, Dronca E, Matei RD, Ligtenberg M, et al. BRCA 1/2 mutations by next-generation sequencing testing in 200 Romanian high-risk patients with breast cancer. Journal of Clinical Oncology. 2017;35(15_suppl):e13116–e16. [Google Scholar]
- 35.Stegel V, Krajc M, Zgajnar J, Teugels E, De Grève J, Hočevar M, et al. The occurrence of germline BRCA1 and BRCA2 sequence alterations in Slovenian population. BMC Med Genet. 2011 Jan 14;12:9. doi: 10.1186/1471-2350-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo Nigro C, Vivenza D, Monteverde M, Lattanzio L, Gojis O, Garrone O, et al. High frequency of complex TP53 mutations in CNS metastases from breast cancer. Br J Cancer. 2012;106(2):397–404. doi: 10.1038/bjc.2011.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33(4):304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 39.Tischkowitz M, Capanu M, Sabbaghian N, Li L, Liang X, Vallée MP, et al. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33(4):674–680. doi: 10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cybulski C, Kluźniak W, Huzarski T, Wokolorczyk D, Kashyap A, Jakubowska A, et al. Clinical outcomes in women with breast cancer and a PALB2 mutation: a prospective cohort analysis. Lancet Oncol. 2015;16(6):638–644. doi: 10.1016/S1470-2045(15)70142-7. [DOI] [PubMed] [Google Scholar]
- 41.Foley SB, Rios JJ, Mgbemena VE, Robinson LS, Hampel HL, Toland AE, et al. Use of Whole Genome Sequencing for Diagnosis and Discovery in the Cancer Genetics Clinic. EBioMedicine. 2015;2(1):74–81. doi: 10.1016/j.ebiom.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C, Wang Y, Wang QS, Wang YJ. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13(4):1355–1360. doi: 10.7314/apjcp.2012.13.4.1355. [DOI] [PubMed] [Google Scholar]
- 43.Roeb W, Higgins J, King MC. Response to DNA damage of CHEK2 missense mutations in familial breast cancer. Hum Mol Genet. 2012;21(12):2738–2744. doi: 10.1093/hmg/dds101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18(8):823–832. doi: 10.1038/gim.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandoval N, Platzer M, Rosenthal A, Dörk T, Bendix R, Skawran B, et al. Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum Mol Genet. 1999;8(1):69–79. doi: 10.1093/hmg/8.1.69. [DOI] [PubMed] [Google Scholar]
- 46.Castéra L, Krieger S, Rousselin A, Legros A, Baumann JJ, Bruet O, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet. 2014;22(11):1305–1313. doi: 10.1038/ejhg.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seifert BA, O’Daniel JM, Amin K, Marchuk DS, Patel NM, Parker JS, et al. Germline Analysis from Tumor-Germline Sequencing Dyads to Identify Clinically Actionable Secondary Findings. Clin Cancer Res. 2016;22(16):4087–4094. doi: 10.1158/1078-0432.CCR-16-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaDuca H, Farwell KD, Vuong H, Lu HM, Mu W, Shahmirzadi L, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One. 2017 Feb 2;12(2):e0170843. doi: 10.1371/journal.pone.0170843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46(2):107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Out AA, Wasielewski M, Huijts PE, van Minderhout IJ, Houwing-Duistermaat JJ, Tops CM, et al. MUTYH gene variants and breast cancer in a Dutch case-control study. Breast Cancer Res Treat. 2012;134(1):219–227. doi: 10.1007/s10549-012-1965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Win AK, Lindor NM, Jenkins MA. Risk of breast cancer in Lynch syndrome: a systematic review. Breast Cancer Res. 2013 Mar 19;15(2):R27. doi: 10.1186/bcr3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao P, Ma N, Li M, Tian QB, Liu DW. Functional variants in NBS1 and cancer risk: evidence from a meta-analysis of 60 publications with 111 individual studies. Mutagenesis. 2013;28(6):683–697. doi: 10.1093/mutage/get048. [DOI] [PubMed] [Google Scholar]