Abstract
Background and aim
Breast cancer, a heterogeneous disease, is the most common cause of cancer-related death in women worldwide. Despite considerable developments in treatment modalities, a subset of patients with advanced-stage breast carcinoma display poor prognosis. Breast cancer heterogeneity and risk of recurrence could be explained with the help of cancer stem cell hypothesis. Stem cells have the capacity to self-renew and differentiate into multiple cell types. Aldehyde dehydrogenase-1 (ALDH1), an enzyme responsible for the oxidation of intracellular aldehydes, contributes to normal and tumor stem cell differentiation. Invasion and metastasis in breast cancer are found to be mediated by a subpopulation of tumor cells which exhibit stem cell-like features and express ALDH1.
The aim was to document ALDH1 expression in breast carcinoma and find its association with other clinico-pathologic prognostic parameters.
Study design
This was a cross-sectional observational study.
Methods
A total of 62 patients with breast carcinoma undergoing mastectomy were included in this study. The tumors were classified into molecular subtypes by assessing immunohistochemical (IHC) expression of ER, PgR, HER2 and Ki-67 according to St. Gallen Consensus Conference 2013. ALDH1 expression was studied by IHC and correlated with clinicoathological parameters.
Statistical analysis
Statistical analysis was done using Graph Pad software (Prism 5 version) for Windows 7. A p-value <0.05 was considered statistically significant.
Results and analysis
Out of 62 tumors, 35 tumors (56.4%) showed ALDH1 positivity. ALDH1 expression was significantly associated with larger size, lymph node involvement, higher grade, higher stage and HER2+ or triple negative tumors.
Conclusion
This study suggests that ALDH1 expression is associated with poor prognostic parameters and aggressive tumor behavior. Larger population-based prospective trials on Indian patients are required to validate these results.
Keywords: breast carcinoma, aldehyde dehydrogenase 1 (ALDH-1), stem cell marker
Introduction
Breast cancer is a global disease. Worldwide, invasive breast cancer is the most common carcinoma in women [1] accounting for 23% of all cancers in women. India is experiencing an unprecedented rise in the number of breast cancer cases across all sections of society and it is now the most common cancer in India as well as in the largest metropolitan city in eastern part of India where the present study was conducted [2,3]. Breast cancer is not a single entity but a heterogeneous disease which can be concluded from the fact that phenotypically similar breast tumors exhibit divergent clinical presentations and disease aggressiveness in addition to systemic investigations of gene expression pattern. Breast cancers are divided into at least five main molecular subtypes which show marked variation in terms of distinct races/ethnicities, risk factors distribution, prognosis, therapeutic treatment responsiveness, clinical outcomes and both overall survival and relapse-free survival. These are: luminal cell-like tumors, which are further subdivided into luminal A and B [both express Oestrogen receptor (ER) and show similar profiles to those of normal luminal cells of breast glands], basal cell like (BCL) phenotype [ER and progesterone receptor (PgR)-negative tumors with genes usually expressed by basal/ myoepithelial cells], HER2-enriched (Human epidermal growth factor receptor 2, amplification of the HER2/neu gene) and normal breast-like group [4]. Pathologists have observed for decades that not only the histologic features of tumors may vary between different patients, but a single tumor may exhibit varying morphologies and growth patterns. However, it was not until recently that the biological basis of histologic heterogeneity within breast cancer began to unravel. Currently prevailing models explaining intra-tumor heterogeneity include the clonal evolution and cancer stem cell hypothesis. According to the cancer stem cell (CSC) hypothesis, cancer stem cells are defined as a subset of tumor cells which have the capacity to self-renew and differentiate forming a heterogeneous tumor cell population. An example of the rapid progress made in this field is the use of a biomarker for CSC identification, aldehyde dehydrogenase-1 (ALDH1) [4]. ALDH1 is predominantly expressed in the epithelium of testis, brain, eye, liver, kidney, and is also found in high levels in hematopoietic and neural stem cells [5,6,7]. This enzyme is thought to play a role in the differentiation of hematopoietic and neural stem cells via the oxidation of retinal to retinoic acid [8]. Retinoic acid activates nuclear retinoic acid receptors (RARs) and RARs subsequently regulate the transcription of genes with retinoic acid response elements [9]. Numerous preclinical studies have shown that expression of ALDH1 in tumor cells has been associated with stem-like characteristics, including innate chemoresistance and clonal capacity. Ginestier et al. [10] used ALDH1 to isolate and identify stem cells from breast carcinoma tissues. They found that ALDH1-positive cells accounted for only 5% of all breast cancer cells and merely 500 ALDH1-positive cells were sufficient to generate a new tumor suggesting high tumorigenicity as well as self-renewal capacity of the positive cells. Moreover, expression of ALDH1 is associated with high histologic grade, HER2/neu overexpression, negative hormone receptors and overall poor survival [4].
Here we performed an institution-based study on mastectomy specimens from patients of breast carcinoma to detect breast cancer stem cells by immunohistochemistry (IHC), particularly the expression of stem cell marker ALDH1; test its association with traditional prognostic parameters as well as molecular subtypes, defined by IHC expression of ER, PgR, HER2/neu and Ki-67 and also discuss its possible utility as tissue based biomarker.
Materials and methods
Specimens and general information
A total of 62 specimens of breast carcinomas were studied, which were collected from patients undergoing modified radical mastectomy in the department of General Surgery of a tertiary care institute in eastern Indian from January 2014 to June 2015. Relevant information like age of the patient, menopausal status, family history and treatment history were collected. Patients were selected in such a manner that none of them received neo-adjuvant chemotherapy or preoperative endocrine or Her2 targeted therapy.
Histopathological examination
Grossing and reporting of mastectomy specimens with invasive breast carcinoma were done according to CAP (College of American Pathologists) protocol [11] which is based on AJCC/UICC TNM, 7th edition [12] which was followed during our study period. Specimens were fixed in 10% neutral buffered formalin. Representative areas were sampled and histopathological examination was done following proper tissue processing, paraffin embedding and staining with haematoxylin-eosin (H&E) respectively. Specific histologic subtype was assigned according to WHO 2012 classification of breast tumors [1]. Histologic grading of tumors was done according to the Nottingham combined histologic grade [Elston-Ellis modification of Scarff-Bloom-Richardson grading system] [13] and Nottingham Prognostic Index [NPI] was also calculated. Instead of commonly used six tier grouping system for NPI as advocated by Blamey et al. [14], a two tier system was adopted in the present study for analytic convenience. Patients were divided into two prognostic groups. Those having tumors with NPI ≤ 5.4 were allotted into good to moderate prognostic group while those with NPI ≥ 5.4 were categorized into poor outcome group. Presence and number of lymph node metastasis was also noted and pathologic (pTNM) staging was done according to 7th edition of AJCC Cancer Staging manual [12].
In this regard clarifications pertaining to certain parameters of our study deserve special mention since the 8th edition of AJCC Cancer Staging Manual [15] has been published recently and made some changes to the existing staging system, particularly by applying strict criteria for primary tumor (pT); defining tumors with prior neo-adjuvant therapy and including multigene panels as stage modifiers. However, no significant change has to be made to our study because mastectomy specimens were collected from patients who had not received neo-adjuvant chemotherapy and genetic analysis was not done. Moreover all the invasive tumors in our study were more than 10 mm in size and only measurement of maximum invasive tumor and largest contiguous tumor deposit were included for defining pT and pN categories respectively.
Immunohistochemistry (IHC) for ER, PgR, Her2/neu, Ki-67 and ALDH-1
Immunohistochemistry was performed on 3μ sections taken on poly-L-Lysine coated slides. Primary antibodies which were used for detection of ER, PgR, Her2/neu, Ki-67 and ALDH1 are as follows- ER: Mouse monoclonal Anti-Human, RTU, 6F11, Novocastra, Leica, PgR: Monoclonal Mouse Anti-Human, RTU, Novocastra, Leica, HER2: Polyclonal Rabbit Anti-Human, CB11, Cell Marque, Ki-67: Monoclonal Mouse Anti-Human, RTU, clone MIB-1, Novocastra, Leica, and ALDH1: Monoclonal Mouse Anti-Human, ALDH1/2 (H-8): sc 166362 (Santa Cruz Biotechnology, INC.). Di-amino benzidine (DAB) was used as chromogen. Positive controls which were used are as follows- (1) fibroadenoma for ER, PgR (2) previously known Her2/neu positive case of gastric adenocarcinoma for Her2/neu (3) tonsil for Ki-67 (4) previously known ALDH1 positive case of colorectal adenocarcinoma for ALDH1. Negative control was achieved by omitting primary antibody.
Evaluation of IHC staining
Reporting of ER, PgR and Her2/neu was done according to the CAP protocols [12]. For Ki-67, invasive edge of tumor was scored and at least 3 high-power fields (400X) were selected to represent spectrum of staining on initial overview of whole section. If there were clear-cut hot spots, data from these were included in overall score. Only nuclear staining was considered positive and staining intensity was not assessed. Scoring involved counting of 1000 malignant invasive cells. Ki-67 index was expressed as percentage of positive staining cells among total number of invasive cells in the area scored. We used our own laboratory cut-off value of 20% according to St. Gallen 2015 consensus conference guidelines [16]. Interpretation of ALDH1 immunostaining was done by using the criteria as described by Ginestier et al. [10] which is as follows:
Negative: < 5% positive tumor cells
1+: ≥ 5% but < 10% positive tumor cells
2+: ≥ 10% but < 50% positive tumor cells
3+: ≥ 50% positive tumor cells
Cytoplasmic expression in malignant epithelial component was evaluated, whereas nuclear staining alone was considered nonspecific. Stromal immunoreactivity in neoplastic growth and adjacent non-neoplastic breast tissue was observed as well but not incorporated in scoring. A minimum of five different areas for each tumor were evaluated, and the mean was assessed. A score of 2+ or 3+ was considered to be positive in our study.
Classification of breast cancer molecular subtypes according to IHC expression based on St. Gallen 2013 consensus conference guidelines [17]
The lesions were classified into 5 molecular subtypes: Luminal A-like ( ER and PgR positive, Her2 negative, low Ki-67 index); Luminal B-like (Her 2 negative subtype showing ER positivity and at least one: high Ki-67, negative or low PgR), Luminal B-like ( Her 2 positive subtype showing ER positivity, any Ki-67 value, any PgR staining ); Her2 positive (ER and PgR absent) and Triple-negative (Her2 negative, ER and PgR absent). A further sub classification of triple negative tumors into basal-like tumors was not done as basal cell markers were not employed in our study.
Statistical analysis
All statistical data analysis was done using Graph Pad software (Prism 5 version) for Windows 7. Chi-square test was carried out to see the correlation between ALDH1 expression and different prognostic parameters of breast carcinoma. Fisher’s exact test was used when expected cell counts were less than five. A p-value <0.05 was considered as statistically significant.
Results
Distribution of breast carcinomas according to molecular classification and expression of ALDH1 in various subtypes
A total of 62 cases of breast carcinoma were studied. Among them 26 cases were Luminal A-like (42%); Luminal B-like cases were 15 (24%), out of which 8 cases were Luminal B-like HER2+ and 7 cases were Luminal B-like HER2-.Total 13 (21%) cases belonged to HER2 enriched subgroup and 8 (13%) cases were of triple-negative category. Significant differences in the levels of ALDH1 expression were observed among various molecular subtypes (Table I). High expression levels were found in Her2 over expression type (Figure 1) and triple-negative subtype (Figure 2).
Table I.
Correlation of ALDH1 expression with molecular subtypes of breast carcinoma.
| Molecular subtypes | ALDH1+(n=35) | ALDH1-(n=27) | p value |
|---|---|---|---|
| Luminal A like(n=26) | 6 (17.1%) | 20 (74.1%) | 0.0001 |
| Luminal B,HER2+(n=8) | 5 (14.3%) | 3 (11.1%) | |
| Luminal B,HER2-(n=7) | 3 (8.6%) | 4 (14.8%) | |
| HER2+ (n=13) | 13 (37.1%) | 0 | |
| Triple Negative (n=8) | 8 (22.9%) | 0 |
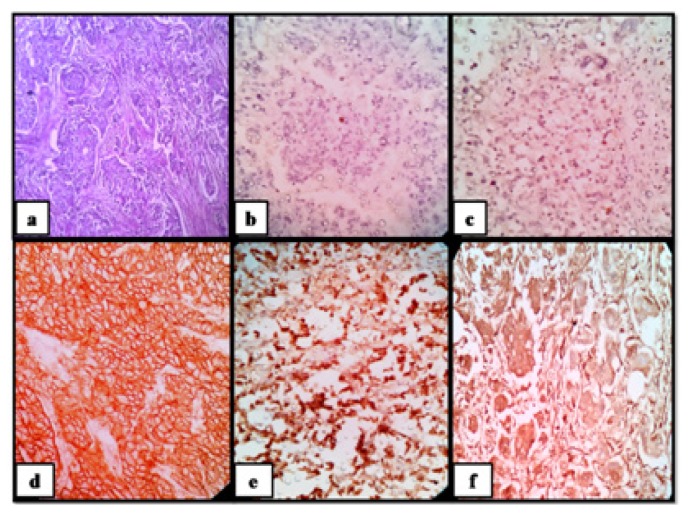
Figure 1.
Invasive carcinoma of breast of no special type ( grade 2), molecular subtype-Her 2+ve; (a) H&E stained section (100X); tumor cells show; (b) ER negativity (100X); (c) PgR negativity (100X); (d) 3+ Her2 positivity (100X); (e) Ki-67≥20% (100X); (f) diffuse cytoplasmic ALDH1 positivity (100X).
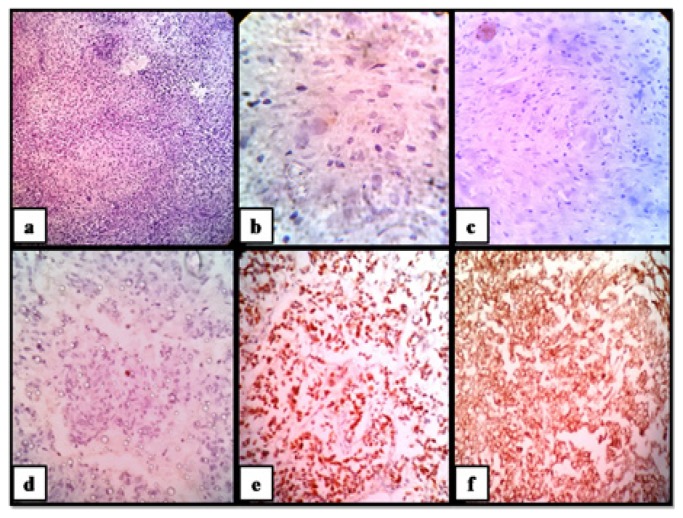
Figure 2.
Metaplastic carcinoma of breast, molecular subtype-Triple Negative/ Basal Cell Like; (a) H&E stained section (100X); malignant spindle cells show; (b) ER negativity (400X); (c) PgR negativity (100X); (d) Her2 negativity (100X); (e) Ki-67≥20% (100X); (f) diffuse cytoplasmic ALDH1 positivity (100X).
Relationship between ALDH1 and various clinico-pathological parameters of breast carcinomas
ALDH1 positivity was noted in 35 cases (56.4%) of breast carcinoma, while 27 cases (43.6%) were ALDH1 negative. Age of the patients showing ALDH1 positivity ranged from 37 to 61 years (mean age-47.4± SD 7.6years), while those who lacked ALDH1 expression were aged between 39 to 73 years (mean age-51.3 ± SD 8.9 years). There was no significant correlation of ALDH1 expression with the age of the patient (P=0.0655). All the cases were females except a single 45-year-old male, who presented with stage IIIA (T2N2M0) invasive carcinoma of no special type (luminal B-like HER2+ molecular subtype) and showed strong ALDH1 expression. Out of 61 female patients, 27 cases were pre-menopausal and 34 were post-menopausal. A total of 34 female patients showed ALDH1 positivity. Among them, 19 cases (51.9%) were pre-menopausal and 15 cases (44.1%) were post-menopausal. Rest of 27 ALDH1 negative cases included 8 pre-menopausal (29.6%) and 19 post-menopausal (70.4%) patients respectively. ALDH1 expression showed no significant correlation with menopausal status (P=0.0733). Total 2 out of 62 cases (3.2%) had positive family history of breast cancer in first degree relatives. Both of them presented with invasive carcinoma of no special type (Nottingham histologic grade 3), belonged to triple-negative subgroup and showed ALDH1 positivity. Invasive carcinoma of no special type was the most prevalent histologic subtype noted in our study (56 cases) and ALDH1 positivity was found in 31 cases (55.4%) while rest 25 cases (44.6%) were ALDH1−. There were 2 cases of invasive lobular carcinoma (both luminal A-like, one ALDH1+ and another ALDH1−) and one case each of mucinous carcinoma (luminal A-like, ALDH1−), invasive papillary carcinoma (Her2+, ALDH1+), carcinoma with medullary features (triple-negative, ALDH1+) and metaplastic carcinoma (triple-negative, ALDH1+) respectively. ALDH1 expression showed significant association with mean tumor size, higher histologic grade, lymphovascular invasion, lymph node involvement, higher tumor stage, negative hormone receptor status, Her2/neu over expression and high Ki-67 index (Table II).
Table II.
ALDH1 expression and its significant association with clinico-pathological parameters.
| Parameters | ALDH1+(n=35) | ALDH1-(n=27) | p value |
|---|---|---|---|
| Mean tumor size(cm) | 4.669 | 2.993 | 0.0001 |
| T1 (n=9) | 0 (0%) | 9 (33.3%) | |
| T2 (n=34) | 19 (54.3%) | 15 (55.6%) | |
| T3 (n=17) | 15 (42.8%) | 2 (7.4%) | |
| T4 (n=2) | 1 (2.9%) | 1 (3.7%) | |
| Grade 1 (n=4) | 0 (0%) | 4 (14.8%) | 0.0003 |
| Grade 2 (n=45) | 22 (62.9%) | 23 (85.2%) | |
| Grade 3 (n=13) | 13 (37.1%) | 0 (0%) | |
| LVI present (n=39) | 28 (80%) | 11 (40.8%) | 0.0036 |
| LVI absent (n=23) | 07 (20%) | 16 (59.2%) | |
| N0 (n=23) | 6 (17.2%) | 17 (63%) | 0.0003 |
| N1 (n=6) | 3 (8.5%) | 3 (11.1%) | |
| N2 (n=16) | 10 (28.6%) | 6 (22.2%) | |
| N3 (n=17) | 16 (45.7%) | 1 (3.7%) | |
| NPI ≥5.4 (n=32) | 26 (74.3%) | 6 (22.2%) | 0.0001 |
| NPI <5.4 (n=30) | 9 (25.7%) | 21 (77.8%) | |
| Stage I (n=9) | 0 (0%) | 9 (33.3%) | 0.0003 |
| Stage II (n=19) | 9 (25.7%) | 10 (37%) | |
| Stage III (n=30) | 22 (62.9%) | 8 (29.7%) | |
| Stage IV (n=4) | 4 (11.4%) | 0 (0%) | |
| ER− (n=22) | 22 (62.8%) | 0 (0%) | 0.0001 |
| ER+ (n=40) | 13 (37.2%) | 27 (100%) | |
| PR− (n=26) | 23 (65.7%) | 3 (11.1%) | 0.0001 |
| PR+ (n=36) | 12 (34.3%) | 24 (88.9%) | |
| HER2− (n=42) | 17 (48.6%) | 25 (92.6%) | 0.0007 |
| HER2+ (n=20) | 18 (51.4%) | 2 (7.4%) | |
| Ki-67 ≥20% (n=36) | 29 (82.9%) | 7 (26%) | 0.0001 |
| Ki-67 <20% (n=26) | 6 (17.1%) | 20 (74%) |
Discussion
Numerous markers have been recognized for the identification of cancer stem cells, among which ALDH1 is the most reported one in literatures. ALDH1 is a cytoplasmic detoxifying enzyme responsible for oxidation of aldehydes (retinal) into retinoids [18]. In 2003, Al-Hajj et al. [19] used cell surface markers CD44 and CD24 to distinguish tumorigenic and non- tumorigenic cells in breast cancers. Later on, Ginestier et al. [10] stated that ALDH1 is a better marker for identification of breast cancer stem cells because fewer ALDH1+ tumor cells than CD44+ and CD24- cells were able to form tumor in immunodeficient mice, establishing the high tumorigenic capacity of ALDH+ cells. On this background, the purpose of our study was to examine the expression of candidate stem cell marker ALDH1 in breast cancer and assess its association with various clinicopathological parameters and molecular subtypes.
In our study, ALDH1 expression was observed in 56.4% of cases, which corroborates with the findings of Zong Y et al. [20] where ALDH1 expression was noted in 63% of cases. Our findings indicate a higher frequency of ALDH1 expression in this series of breast cancer for an Indian population, compared with 19 and 30% in two different Caucasian populations described by Ginestier et al. [10]. These differences among the studies may be attributed to the diversity of detection techniques and samples under investigation, especially since more than 90% of the cases examined in our study were invasive carcinoma of no special type. This data provide evidence of enrichment of ALDH1+ cancer cells in invasive carcinoma tissue.
We also found a significant association of tumor size with ALDH1 expression (p=0.0001) which corroborated with the findings of Khoury T et al. [21] who also found that ALDH1 expression was associated with larger tumor size (p=0.05) as well as Yoshioka T et al. [22] who noted that ALDH1 expression in node positive breast cancers significantly correlated with larger tumor size (p=0.007).
ALDH1 expression was also associated with high histological grade (p=0.0003) which was similar to the findings of H Nalwoga et al. [23] who noted that ALDH1 expression in breast cancers of an African population was associated with high histological grade (p value<0.0005).Our findings further corroborated with the findings of Ginestier C et al. [10] and Yoshioka T et al. [22] who also observed that high grade tumors were more likely to be ALDH1+. Ricardo S et al. [24] also documented the association of ALDH1 expression with high grade tumors where 78.8% of positive cases were grade 3 (p=0.003).
Furthermore, we noted that ALDH1+ tumors were associated with lymph node metastasis (p=0.0003), with 82.8% of ALDH1+ tumors showing evidence of nodal involvement. Similar finding was noted by Khoury T et al. [21] who also found that tumors with positive lymph nodes expressed ALDH1 more frequently than node-negative tumors (p=0.006), with 59% of ALDH1+ tumors showing evidence of lymph node metastasis.
We also observed that ALDH1 expression was intimately associated with tumor stage. ALDH1+ tumors were frequently found to be stage III/IV tumors (p=0.0003).Our finding corroborated with the study by Khoury T et al21 who noted that ALDH1 was expressed most commonly in stage III tumors followed by stage II tumors (p=0.0006). Stage IV tumors were excluded from their study.
In our study, we found that ALDH1 positivity was significantly associated with ER/PgR negativity, HER2 positivity and higher Ki-67 index. These were in keeping with findings of Khoury T et al. [21] and H Nalwoga et al. [23] who noted that ALDH1 expression was associated with ER/PgR negativity. Additional studies by Ginestier C et al. [10], Yoshioka T et al. [22] and Hosni HN et al. [18] showed that ALDH1 positivity in breast cancer was associated with HER2 overexpression along with ER/PgR negativity.
A study conducted by Morimoto K et al. [25] to investigate the clinico-pathological characteristics of breast cancer with ALDH1+ cancer stem cells, showed a significant correlation of ALDH1 positivity with ER/PgR negativity, HER2 overexpression and also high Ki-67 expression (p<0.001). In order to detect ALDH1+ tumor cells more accurately, they carried out immunohistochemical double staining of ALDH1 and Ki67 and were able to demonstrate that ALDH1+ tumor cells were mostly Ki67–, and, interestingly, ALDH1–tumor cells were more likely to be Ki67+. These results showed that breast cancer stem cells proliferate slowly than the surrounding tumor cells. This is consistent with the theory that cancer stem cells are relatively dormant but give rise to a rapidly dividing population of progenitor cells. Thus, this observation indicates that ALDH1+ cancer stem cells tumors are surrounded by a higher proportion of tumor cells which are ALDH1– and Ki67+. This was an interesting observation made by Morimoto K et al. [25], on the distribution of ALDH1+ tumor cells and Ki67+ tumor cells but it needs to be confirmed by future studies.
In our study we found a significant association (p=0.0001) between molecular subtypes and ALDH1 expression. Out of 35 tumors which showed ALDH1 positivity, 40% tumors belonged to Luminal subtypes while 37.1% and 22.9% tumors belonged to HER2+ and triple-negative subtype respectively. Our findings corroborated with findings of Morimoto K et al. [25] who noted that ALDH1 expression was highest in the HER2 overexpression type followed by triple-negative tumors. They also mentioned that triple-negative tumors, although heterogeneous in nature, are found to be basal-like in majority of cases. Bi et al. [4] who studied the correlations of ALDH1 expression with molecular subtypes in breast cancer documented that ALDH1 expression was found in 54.5% of HER2+ and 33.3% of basal-like tumors (p=0.003). Studies conducted by Khoury T et al. [21], Ricardo S et al. [24] and Park YS et al. [26] revealed similar findings with ALDH1+ cells being more frequent in basal-like and HER2+ subtypes than luminal subtypes.
Conclusion
We observed a high prevalence of ALDH1 positivity in invasive carcinoma of the breast and it was significantly associated with features of aggressive tumor. Assessment of ALDH1 expression might help recognize a high risk subgroup of breast cancers in this population. Available cancer therapeutic agents which emphasize on tumor regression, kill differentiated tumor cells while sparing the small cancer stem cell population. Thus, the development of more effective cancer therapies is essential to target this important cancer stem cell population. The success of these new approaches depends on the identification, isolation and characterization of cancer stem cells and ALDH1 may play a crucial role in such future ventures of developing targeted therapies with a difference.
References
- 1.Lakhani S. WHO classification of tumors of the breast. 4th ed. Lyon: International Agency for Research on Cancer; 2012. [Google Scholar]
- 2.Indian council of Medical Research. BULLETIN. 2010 Feb;40(2):11. [Google Scholar]
- 3.Statistics of breast cancer India. Available from: http:www.breastcancerindia.net/bc/statistics/stat_kolkata.htm.
- 4.Bi X, Wu C, Han M, Cai J. Correlations of ALDH1 expression with molecular subtypes and ABCG2 in breast cancer. Gland Surg. 2012;1(1):12–19. doi: 10.3978/j.issn.2227-684X.2012.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142–1151. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 7.Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins SJ. Retinoic acid receptors, hematopoiesis and leukemogenesis. Curr Opin Hematol. 2008;15:346–351. doi: 10.1097/MOH.0b013e3283007edf. [DOI] [PubMed] [Google Scholar]
- 9.Magni M, Shammah S, Schiró R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87:1097–1103. [PubMed] [Google Scholar]
- 10.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.College of American pathologists [certified] Protocol for the Examination of Specimens from Patients with Invasive Carcinoma of the Breast. [Protocol web posting date: December 2013]. Available from: http://www.cap.org/ShowProperty?nodePath=/UCMCon/Contribution%20Folders/WebContent/pdf/cp-breast-invasive-13protocol-3200.pdf.
- 12.Edge SB, Byrd DR, Carducci MA, Compton CC, editors. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer; 2009. [Google Scholar]
- 13.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 14.Blamey RW, Ellis IO, Pinder SE, Lee AH, Macmillan RD, Morgan DA, et al. Survival of invasive breast cancer according to the Nottingham Prognostic Index in cases diagnosed in 1990–1999. Eur J Cancer. 2007;43:1548–1555. doi: 10.1016/j.ejca.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 15.American Joint Committee on Cancer (AJCC) AJCC Cancer Staging Manual. 8th ed. New York: Springer; 2017. [Google Scholar]
- 16.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosni HN, Daoud SA, Bassam AM. Immunohistochemical Study of Stem Cell Marker ALDH1 and BRCA1 in Breast Cancer. Academic Journal of Cancer Research. 2014;7(1):01–07. [Google Scholar]
- 19.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong Y, Lin Y, Shen S, Zhou Y, Mao F, Guan J, et al. Expression of ALDH1 in breast invasive ductal carcinoma: an independent predictor of early tumor relapse. Cancer Cell Int. 2013 Jun 15;13:60. doi: 10.1186/1475-2867-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoury T, Ademuyiwa FO, Chandrasekhar R, Jabbour M, Deleo A, Ferrone S, et al. Aldehyde dehydrogenase 1A1 expression in breast cancer is associated with stage, triple negativity, and outcome to neoadjuvant chemotherapy. Mod Pathol. 2012;25:388–397. doi: 10.1038/modpathol.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshioka T, Umekita Y, Ohi Y, Souda M, Sagara Y, Sagara Y, et al. Aldehyde dehydrogenase 1 expression is a predictor of poor prognosis in node-positive breast cancers: a long-term follow-up study. Histopathology. 2011;58(4):608–616. doi: 10.1111/j.1365-2559.2011.03781.x. [DOI] [PubMed] [Google Scholar]
- 23.Nalwoga H, Arnes JB, Wabinga H, Akslen LA. Expression of aldehyde dehydrogenase 1 (ALDH1) is associated with basal-like markers and features of aggressive tumors in African breast cancer. Br J Cancer. 2010;102(2):369–375. doi: 10.1038/sj.bjc.6605488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricardo S, Vieira AF, Gerhard R, Leitão D, Pinto R, Cameselle-Teijeiro JF, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto K, Kim SJ, Tanei T, Shimazu K, Tanji Y, Taguchi T, et al. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009;100(6):1062–1068. doi: 10.1111/j.1349-7006.2009.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16(3):876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]