Abstract
Understanding how evolution of microbial resistance towards a given antibiotic influences susceptibility to other drugs is a challenge of profound importance. By combining laboratory evolution, genome sequencing, and functional analyses, recent works have charted the map of evolutionary trade-offs between antibiotics and have explored the underlying molecular mechanisms. Strikingly, mutations that caused multidrug resistance in bacteria simultaneously enhanced sensitivity to many other unrelated drugs (collateral sensitivity). Here, we explore how this emerging research sheds new light on resistance mechanisms and the way it could be exploited for the development of alternative antimicrobial strategies.
Keywords: collateral sensitivity, antibiotic resistance, experimental evolution, cross-resistance, multidrug resistance
Trade-offs are all around
The Apollo Lunar Module was the lander portion of the Apollo spacecraft built for the U.S. Apollo program. Its task was to carry a crew from the lunar orbit to the surface and back. While designing the module, engineers faced a formidable difficulty regarding the number of landing legs. Five legs assured safe landing, but were far too heavy; three legs substantially reduced the weight of the vehicle, but were unsafe. Finally, engineers reached a compromise of four landing legs. This example is by no means special: trade-off (see Glossary) is a serious problem in engineering, economy, and nature. Indeed, a wealth of comparative and experimental data have confirmed that, when organisms evolve to a given environment, the beneficial changes accumulated in one trait are generally linked to detrimental changes in other traits [1,2].
Glossary.
AcrAB efflux system: a proton-motive-force-dependent multidrug efflux system which confers resistance to multiple antimicrobial agents.
Collateral sensitivity: the phenomenon in which an organism that has developed resistance to one drug displays increased sensitivity to a second drug.
Cross-resistance: the phenomenon in which an organism that has developed resistance to one drug displays decreased sensitivity to a second drug.
Dihydrofolate reductase: an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as electron donor.
DNA supercoiling: DNA supercoiling is the under or over winding of a DNA strand. This process is important for DNA packaging and transcription within all cells.
Fluoroquinolones: broad-spectrum antibiotics which act by inhibiting the activity of both the DNA gyrase and the topoisomerase IV enzymes.
Hsp90: a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation.
Mutant-prevention concentration: an antibiotic concentration threshold above which the selective proliferation of resistant mutants is not expected to occur.
Pleiotropy: occurs when one gene affects multiple, seemingly unrelated, phenotypic traits.
Proton motive force (PMF): a form of energy stored as a proton gradient across a cellular membrane.
Tetracycline-resistance (tet) genes: genes that are usually acquired via transferable plasmids and/or transposons.
Topoisomerase genes: genes that are influential in regulating transcriptional DNA supercoiling.
Trade-off: a situation that involves losing one quality or aspect of something in return for gaining another quality or aspect.
In this opinion article we examine how the evolutionary principle of trade-offs could be applied to studying and manipulating the evolution of antibiotic resistance in the clinic. As the development of novel drugs substantially lags behind the rate at which pathogenic microbes evolve resistance to them, there is an increasing demand for new strategies which could extend the usage of existing antimicrobial agents or direct research for novel drugs. Indeed, manipulating drug resistance is a formidable task, not least because it requires detailed knowledge of the microbial genetic toolkits and relevant ecological conditions.
It has long been known that evolution of resistance towards a given antimicrobial agent frequently increases resistance to several other drugs. In sharp contrast, it has remained unclear how frequently evolution of resistance increases sensitivity to other drugs, a phenomenon usually termed ‘collateral sensitivity’. Except for a pioneering, but largely phenomenological, work more than 60 years ago [3], there was a long hiatus in follow-up microbial studies. The shortage of relevant publications is surprising for at least two reasons. First, there is an increasing realization that resistance reduces fitness in nonstressed conditions, and therefore could leave organisms vulnerable to specific drugs as well [4]. Second, collateral sensitivity has been extensively studied in cancer chemotherapy [5].
Recent technological advances in laboratory automation and genome sequencing have led to a renewed interest in collateral sensitivity. Systematic studies explored the extent to which microbial adaptive evolution in the presence of a single antibiotic increases the susceptibility to other drugs [6–8]. Unsurprisingly, these proof-of-concept laboratory studies provided no explicit guidelines for clinical practice. Despite this limitation, commentaries suggested that collateral sensitivities may inform, or even direct, future therapeutic interventions, leading to a better usage of existing antibiotics [9]. More generally, the results may be applicable for studying the rules governing evolutionary trade-offs in nature.
Here, we discuss the recent advances and implications of this emerging research with the aim of raising the interest of clinical microbiologists, systems biologists, and evolutionary biologists alike. We highlight the potentials and limitations of current approaches, review the underlying molecular mechanisms of collateral sensitivity, and suggest new directions for future research.
Single-drug treatment promotes the evolution of multidrug resistance
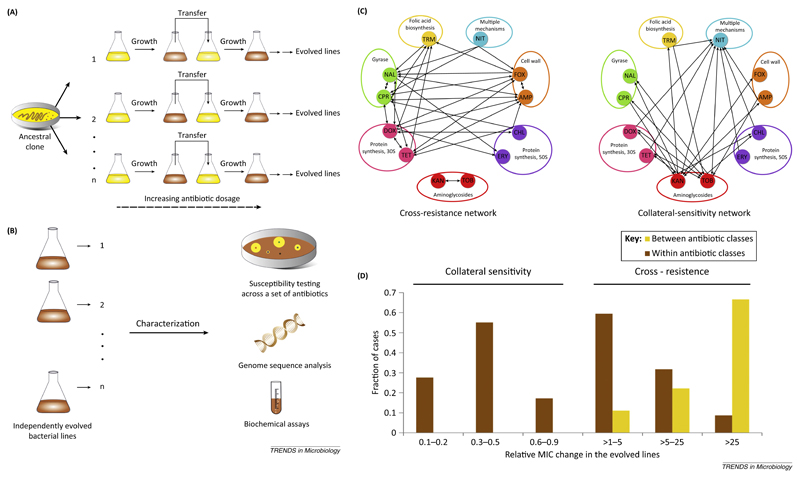
To chart the map of evolutionary trade-offs between antibiotics, recent studies initiated parallel laboratory evolutionary experiments [6–8,10]. Starting from a single clone of Escherichia coli, populations were allowed to evolve to increasing dosages of one of 20–25 antibiotics. Populations evolved to reach 20- to 300-fold increases in the minimum inhibitory concentrations (MICs) relative to their ancestor. As the next step, the corresponding changes in susceptibilities of the laboratory-evolved populations were measured against a panel of other antibiotics, allowing researchers to infer a network of cross-resistance and collateral-sensitivity interactions (Figure 1). Despite several limitations of laboratory evolution, these studies could recapitulate major patterns in the evolution of antibiotic resistance reported from the clinic. Notably, bacterial populations that reached high levels of resistance under strong selection pressure evolved resistance to multiple antibiotics, while mild antibiotic selection led to weaker cross-resistance [8]. Therefore, the use of high dosages of antibiotic may promote multidrug resistance in clinical settings.
Figure 1. Charting the maps of antibiotic cross-resistance and collateral sensitivity.
(A) Laboratory evolution experiments are conducted to evolve antibiotic-resistant bacteria in a controlled setting. They involve serial transfer of growing bacterial populations in parallel for hundreds of generations in the presence of an increasing dosage of one of 20–25 antibiotics. (B) Evolved lines are systematically profiled for changes in susceptibilities against a panel of antibiotics – and for genomic and biochemical alterations underlying these phenotypic changes. (C) Based on antibiotic susceptibility measurements, networks of cross-resistance (i.e., increased resistance to one or more agents) and collateral-sensitivity (increased sensitivity to one or more agents) interactions are inferred. The two networks shown here illustrate such evolutionary interactions between 12 antibiotics as determined previously [6,10]. An arrow from one antibiotic to another indicates that adaptation to one increases the resistance (sensitivity) to another. Aminoglycosides dominate the collateral-sensitivity network, with numerous links to other classes of antibiotics. Antibiotic abbreviations are ampicillin (AMP), cefoxitin (FOX), ciprofloxacin (CPR), nalidixic acid (NAL), nitrofurantoin (NIT), kanamycin (KAN), tobramycin (TOB), tetracycline (TET), doxycycline (DOX), chloramphenicol (CHL), erythromycin (ERY), and trimethoprim (TRM). ‘30S’ and ‘50S’ refer to the two main ribosomal subunits. (D) Distribution of the strength of cross-resistance and collateral-sensitivity interactions as determined previously [6,10]. Abbreviation: MIC, minimum inhibitory concentration.
Diverse resistance mechanisms result in pleiotropic effects
According to textbook examples, there are four major resistance mechanisms: modification of cellular targets so that antibiotic binding is diminished, physical removal of an antibiotic from the cell through modification of efflux pumps, reduced cellular uptake, and enzymatic inactivation of the antibiotic [11]. While these canonical mechanisms can spontaneously evolve in the laboratory, whole-genome sequence analysis of the laboratory-evolved lines suggests that the picture is far more complex. First, more than half of the evolved populations reached high levels of resistance without mutations affecting the cellular target [10]. The absence of target-affecting mutations most likely reflects unusually high fitness costs but other factors may also play a role [12,13]. Second, adaptive mutations repeatedly occurred in cellular subsystems with no obvious link to the mechanisms mentioned above. For example, transcriptional regulatory genes were frequently mutated during evolution of antibiotic resistance [10,13]. This is consistent with the suggestion that enhanced tolerance can be achieved through transcriptional re-wiring of stress-response pathways [14]. The observed mutated genes mediate general defense in times of stress, including osmotic, acidic, nutrient, membrane, and oxidative stresses. The resistance-conferring mutations detected in these studies have a wide range of pleiotropic effects. Many caused substantial fitness loss in a stress-free medium, perturbed genomic expression, and altered susceptibilities to other antibiotics [10,15–17]. Pleiotropic effects were not restricted to mutations occurring in regulatory genes. Most surprisingly, even a canonical resistance mutation in the target protein of fluoroquinolone influenced resistance to multiple nonquinolone antibiotics (see below) [10,18].
Emergence of collateral sensitivity during antibiotic selection
Given the prevalence of resistance-conferring mutations with pleiotropic effects, multidrug resistance may yield hypersensitivity to other classes of antibiotics as a byproduct. Laboratory studies have demonstrated that this is indeed so. According to one study, 74% of the laboratory-evolved resistant lines showed enhanced sensitivities to one or more drugs [7]. Thus, not only cross-resistance but also collateral-sensitivity interactions frequently occur during the evolution of antibiotic resistance. Here, we argue that various mechanisms of resistance, including target mutations and those mutations affecting drug uptake and efflux, are prone to induce collateral sensitivity.
Understanding the mechanisms underlying the interactions of collateral sensitivity is still at an early stage. The best-described example concerns the costs and benefits of membrane-potential-altering mutations [6] (Figure 2A). Cellular uptake of one particular class of antibiotics, aminoglycosides, is unique as it demands an active proton motive force (PMF) [19]. Consequently, resistance to aminoglycosides, in both laboratory and clinic settings, is achieved partly by reduction of the PMF through membrane-potential-altering mutations [6,8,20]. Such mutations influence oxidative phosphorylation, heme biosynthesis, or proton/potassium ion symport, leading to a reduced cellular uptake of aminoglycosides. However, this resistance mechanism comes at a high cost, as removal of many other antibiotics from the cell relies on PMF-dependent efflux pumps. Indeed, reduction of PMF in aminoglycoside-resistant lines diminishes the activity of PMF-dependent major efflux pumps, leading to susceptibility to several unrelated classes of antibiotics. The activity of the AcrAB efflux system, a major multidrug resistance mechanism in E. coli, is especially strongly affected, and this makes a major contribution to the collateral sensitivity pattern observed [6]. In sum, altering the bacterial membrane potential is a double-edged sword as it modulates intracellular antibiotic concentrations in an antagonistic manner.
Figure 2. Molecular mechanisms of collateral sensitivity.
(A) Altered membrane potential decreases the uptake of one class of antibiotics while it diminishes the efflux of others. Resistance to aminoglycoside (red dots) can be achieved partly through reduction in the proton motive force (PMF) across the inner membrane of Escherichia coli. As a side effect, the activity of PMF-dependent major efflux pumps is diminished, leading to hypersensitivity to numerous other antibiotics (blue dots) [6]. (B) Resistance mutation causes genome-wide reprogramming of expression with pleiotropic effects. For example, a specific mutation in a gyrase subunit causes resistance to quinolones (gyrase inhibitors). Simultaneously, the same mutation changes susceptibility to numerous other antibiotics by altering DNA supercoiling and thereby global genomic expression [18]. Expression reprogramming can affect various other cellular subsystems modulating antibiotic resistance, including cell-wall thickness and cell-surface charge in an RNA polymerase mutant [38]. (C) Two drugs inhibiting the same enzyme exhibit collateral sensitivity. A resistance mutation alters the structure of the enzyme in such a way that it becomes resistant to one inhibitor while exhibiting hypersensitivity to the other.
Another study focused on the extrachromosomal expression of tet genes, which are frequently the subjects of plasmid-mediated transfer between pathogenic strains [21]. The tet genes not only confer tetracycline resistance via increased efflux, they simultaneously increase the susceptibilities of Gram-negative bacteria to aminoglycosides by enhancing the uptake of these antibiotics. The expression of tet genes caused no change in membrane potential, suggesting that such expression increases aminoglycoside uptake by modulating the availability of specific carriers or by lowering the minimum membrane potential required for uptake.
More generally, changes in genomic expression associated with drug resistance may be another reason for collateral sensitivity (Figure 2B). For example, fluoroquinolone-resistant strains that evolved clinically or in the laboratory accumulate mutations in the target topoisomerase gene (gyrA) that controls bacterial DNA supercoiling [22,23]. Surprisingly, one of these clinically relevant mutations (Asp87Gly) also alters susceptibility to many nonquinolone drugs [10,18], including doxycycline and nitrofurantoin – to which sensitivity is increased [10]. The possible mechanism is that resistance to quinolones alters DNA supercoiling, and such changes fundamentally affect transcription across the genome [18]. Most likely, DNA supercoiling also modulates the expression of genes important in bacterial survival under stress (genes encoding RpoE, RpoS, and RecA).
Further examples support the view that even mutations in the molecular targets of an antibiotic can lead to hypersensitivity to unrelated drugs. Fusidic acid resistance is generally caused by mutations in the target gene, fusA, which encodes the elongation factor G [24]. However, such mutations have several pleiotropic side effects. They are associated with a slow growth rate, reduced levels of nutritional stress sensors (ppGpp), reduced heme levels, and increased sensitivity to oxidative stress and DNA damage [25,26]. Consequently, the corresponding mutants display increased susceptibility to antibiotics of several classes (β-lactam, fluoroquinolone, aminoglycoside, rifampicin, and chloramphenicol) [27].
Collateral sensitivity is not restricted to drugs with unrelated mechanisms of action: drugs acting on the same target protein can also have antagonistic effects (Figure 2C). For example, researchers investigated two compounds that both act on dihydrofolate reductase in the malaria parasite, Plasmodium vivax; they found that resistance to one of the compounds leads to hypersensitivity to the other [28].
Strong trade-offs between resistance and fitness could explain the rarity of resistance to certain drugs in the clinic. In recent work, Lindquist and colleagues studied laboratory evolution of resistance to amphotericin B (AmB) in the major fungal pathogen Candida albicans [29]. Resistance to AmB in the clinic is nondetectable despite prolonged and wide usage of the drug. This could be explained in two ways. Resistance mutations may either occur at extremely low rates or incur high fitness costs in the host environment – and are therefore selected against. The latter explanation turned out to be closer to the truth. AmB resistance mutations generated an internal cellular stress which demanded elevated expression of the molecular chaperone Hsp90 for survival. As a consequence, strains carrying these mutations were hypersensitive to oxidative stress, temperature, and killing by neutrophils, and they also had defects in filamentation and tissue invasion [29]. Thus, trade-offs associated with resistance can occur not only between susceptibilities to different drugs but also between drug tolerance and the ability to cause disease.
Clinical implications
How can the trade-offs between resistance and evolutionary fitness under clinical conditions be exploited to prevent the spread of resistant pathogens? Combination therapy – the concurrent application of two or more antimicrobials – is one possibility. In the treatment of infectious diseases such as HIV, malaria, and tuberculosis, combination therapy is the golden rule [30–32], but among common bacterial infections combinations of drugs are only rarely deployed.
There are several requirements for two drugs to be effective in combinations. First, it is generally believed that two drugs that show synergistic effects (that is, their combined effect exceeds the sum of their individual effects) maximize killing efficiency and therapeutic selectivity [33]. Second, certain drug combinations are expected to delay the evolution of resistance. Unless mutations that provide resistance to both drugs are exceptionally rare, combination therapy will specifically select for the emergence of multidrug-resistant bacteria. Therefore, both physiologic interactions (e.g., synergism) and evolutionary interactions (cross-resistance) between drugs are expected to affect the long-term efficiency of combinatorial antibiotic treatments.
A recent study on malaria parasites supported this notion. Simultaneous administration of two compounds that target wild-type and chloroquine-resistant Plasmodium falciparum, respectively, showed a strong synergy and successfully hindered the evolution of resistance in an in vitro evolutionary experiment [34]. How synergy and collateral sensitivity affect the evolution of resistance in combination therapies was addressed in a more systematic fashion by a laboratory evolution study of E. coli adapting to five different single antibiotics and their ten pairwise combinations [35]. Notably, the nature of antibiotic interactions (i.e., synergistic, additive or antagonistic) did not correlate with the degree of resistance evolution in the combination treatments, not least because the interactions themselves were subject to evolutionary change. In contrast, the presence of collateral sensitivity between two antibiotics was a robust predictor of decreased resistance evolution during simultaneous exposure to two drugs [35] (Figure 3A). These in vitro results indicate that – while synergy might help to achieve a stronger eradication of pathogens in the initial phase of combination therapy – the rate of resistance evolution can be understood mostly in terms of collateral sensitivity patterns.
Figure 3. Applying collateral sensitivity to combat resistance.
(A) Simultaneous administration or rapid cycling of a collateral-sensitive antibiotic pair decelerates resistance evolution by constraining the set of available mutational trajectories. (B) Two antibiotics (1,2), showing reciprocal collateral sensitivity, are administered in an alternating fashion. Prolonged treatment with antibiotic 1 selects for variants that have an increased resistance to 1 (blue) and an increased susceptibility to 2. Switching to antibiotic treatment 2 eradicates these variants and selects for mutations that increase susceptibility to 1 (red), hence enabling 1 to be reused. Abbreviation: WT, wild type.
Another possibility to slow the rate of resistance evolution is temporal cycling of different antibiotics. Recent works strongly indicate that the success of such a strategy depends on the choice of antibiotics: treatment with a single antibiotic and then switching to a collateral-sensitive partner could be a good strategy [7,36]. In one study, numerous promising drug pairs exhibiting reciprocal collateral sensitivity were identified, and it has been demonstrated that cyclical usage of one such pair, gentamicin and cefuroxime, selects against resistance to either drug. Here, levels of wild-type resistance were maintained by periodic eradication of resistant cells by exposure to the collateral-sensitive partner [7] (Figure 3B). Another study cycled collateral-sensitive antibiotics with a short switching time (i.e., 1 day) and showed that resistance evolution was decelerated [36]. This effect was not due to the accumulation of mutations inducing collateral sensitivity – rather, the set of mutational trajectories towards resistance became constrained (Figure 3A). Indeed, genome sequencing of laboratory-evolved lines revealed that, under antibiotic cycling, mutations occurred in genes that are distinct from those mutated under the corresponding single-drug treatments [31].
The experimental map of cross-resistance/collateral sensitivity could also serve as a unique resource in drug development. For example, information on cross-resistance across a large number of drugs could potentially advise researchers on the long-term efficacy of novel antimicrobial compounds. Indeed, by integrating available data on antibiotic properties, researchers unveiled some general principles governing the evolution of cross-resistance patterns [10]. Notably, similarity in the intrinsic resistome (i.e., set of genes that influence susceptibility) between antibiotics emerged as the strongest predictor of cross-resistance. By contrast, cross-resistance between two antibiotics was independent of whether they showed synergistic effects in combination. Last, transcriptome profiling of laboratory-evolved resistant bacteria showed that the expression signature of a handful of genes is also predictive of antibiotic cross-resistance [37]. Such considerations could pave the way towards in silico methods for estimating the cross-resistance propensity of novel antimicrobial compounds before entry into clinical usage.
More generally, compound screening could focus on the identification of small-molecule inhibitors that differentially target susceptible and resistant pathogens. This strategy has been used by Lukens and coworkers by focusing on the malaria parasite, P. falciparum [34]. The authors screened a chemical library against chloroquine-sensitive and chloroquine-resistant lines, and found a compound (IDI-3783) that killed the resistant lines only. Strikingly, laboratory evolution of chloroquine-resistant clinical lines in the presence of this compound restored chloroquine susceptibility. This result implies that usage of IDI-3783 (or a derivative thereof) could render chloroquine useful again in treating malaria. More generally, the concept of collateral sensitivity could be useful for the identification of novel small-molecule inhibitors which render drug-resistant microbes susceptible to existing antimicrobial agents.
Concluding remarks and future directions
Although there has been much progress in our understanding of collateral sensitivity, there are several key questions that remain unanswered and should be studied in the future (Box 1). As most systematic studies have focused on a single species, the evolutionary conservation of collateral sensitivity remains largely unknown. This issue will certainly depend on the extent of overlap in the mechanisms of resistance and trade-offs across related species. Furthermore, most prior laboratory evolution studies focused on genomic mutations and largely ignored the acquisition of resistance genes by horizontal transfer. Therefore, studying and exploiting the fitness costs of plasmid-mediated antibiotic resistance mechanisms are of paramount importance. Lastly, it will be crucial to decipher the long-term impact of collateral sensitivity on resistance evolution. It may well be the case that the associated costs that render microbes vulnerable to certain antibiotic stresses are only temporary, and that compensatory evolution can rapidly restore fitness. Future works should elucidate to what extent, and how, mutations ameliorating the fitness cost of resistance under drug-free conditions re-wire the collateral-sensitivity interactions between antibiotics. Alternatively, collateral sensitivity may have a long-lasting effect with a substantial impact on reaching clinically significant resistance levels. This issue can readily be tested by comparing the mutant-prevention concentration for antibiotic-resistant and antibiotic-sensitive strains. We anticipate that collateral sensitivity will contribute to the sustainable use of drugs in the clinic by mitigating the rate of resistance evolution.
Box 1. Outstanding questions.
What is the best strategy to exploit collateral sensitivity in vivo? Concurrent application or cycling of drug pairs?
How rapidly do collateral-sensitivity interactions diverge during long-term bacterial evolution? Do broad and nearly universal mechanisms of collateral sensitivity exist?
To what extent do compensatory mutations that ameliorate the fitness cost of drug resistance re-wire collateral-sensitivity networks?
Does the frequent horizontal transfer of certain resistance genes shape their propensity to induce collateral sensitivity?
Acknowledgments
This work was supported by the Lendu¨ let Programme of the Hungarian Academy of Sciences, the Wellcome Trust (B.P. and C.P.), European Research Council (C.P.) and the Hungarian Academy of Sciences Postdoctoral Fellowship Programme (V.L.). We are also grateful to Reka Spohn for her help with the figures.
References
- 1.Bennett AF, Lenski RE. An experimental test of evolutionary trade-offs during temperature adaptation. Proc Natl Acad Sci USA. 2007;104(Suppl. 1):8649–8654. doi: 10.1073/pnas.0702117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Velicer GJ, et al. Application of traditional and phylogenetically based comparative methods to test for a trade-off in bacterial growth rate at low versus high substrate concentration. Microb Ecol. 1999;38:191–200. doi: 10.1007/s002489900169. [DOI] [PubMed] [Google Scholar]
- 3.Szybalski W, Bryson V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J Bacteriol. 1952;64:489–499. doi: 10.1128/jb.64.4.489-499.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagneux S, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 5.Pluchino KM, et al. Collateral sensitivity as a strategy against cancer multidrug resistance. Drug Resist Updat. 2012;15:98–105. doi: 10.1016/j.drup.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazar V, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5:204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 8.Oz T, et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol Biol Evol. 2014;31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock RE. Collateral damage. Nat Biotechnol. 2014;32:66–68. doi: 10.1038/nbt.2779. [DOI] [PubMed] [Google Scholar]
- 10.Lazar V, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun. 2014;5:4352. doi: 10.1038/ncomms5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayers DL, et al. Antimicrobial Drug Resistance. Humana Press; New York: 2009. [Google Scholar]
- 12.Palmer AC, Kishony R. Opposing effects of target overexpression reveal drug mechanisms. Nat Commun. 2014;5:4296. doi: 10.1038/ncomms5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 15.Dragosits M, et al. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol. 2013;9:643. doi: 10.1038/msb.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshima T, et al. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesh B, et al. The Erwinia chrysanthemi 3937 PhoQ sensor kinase regulates several virulence determinants. J Bacteriol. 2006;188:3088–3098. doi: 10.1128/JB.188.8.3088-3098.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webber MA, et al. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio. 2013;4:e00273–e313. doi: 10.1128/mBio.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damper PD, Epstein W. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob Agents Chemother. 1981;20:803–808. doi: 10.1128/aac.20.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumert N, et al. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb Drug Resist. 2002;8:253–260. doi: 10.1089/10766290260469507. [DOI] [PubMed] [Google Scholar]
- 21.Merlin TL, et al. Aminoglycoside uptake increased by tet gene expression. Antimicrob Agents Chemother. 1989;33:1549–1552. doi: 10.1128/aac.33.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad S, et al. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1996;38:443–455. doi: 10.1093/jac/38.3.443. [DOI] [PubMed] [Google Scholar]
- 24.Besier S, et al. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol Microbiol. 2003;47:463–469. doi: 10.1046/j.1365-2958.2003.03307.x. [DOI] [PubMed] [Google Scholar]
- 25.Macvanin M, et al. Fusidic acid-resistant EF-G perturbs the accumulation of ppGpp. Mol Microbiol. 2000;37:98–107. doi: 10.1046/j.1365-2958.2000.01967.x. [DOI] [PubMed] [Google Scholar]
- 26.Nagaev I, et al. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol Microbiol. 2001;40:433–439. doi: 10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- 27.Macvanin M, Hughes D. Hyper-susceptibility of a fusidic acid-resistant mutant of Salmonella to different classes of antibiotics. FEMS Microbiol Lett. 2005;247:215–220. doi: 10.1016/j.femsle.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Hastings MD, Sibley CH. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc Natl Acad Sci USA. 2002;99:13137–13141. doi: 10.1073/pnas.182295999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent BM, et al. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogtay N, et al. Artemisinin-based combination therapy for treating uncomplicated Plasmodium vivax malaria. Cochrane Database Syst Rev. 2013;10 doi: 10.1002/14651858.CD008492.pub3. CD008492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchison DA. Prevention of drug resistance by combined drug treatment of tuberculosis. Handb Exp Pharmacol. 2012;211:87–98. doi: 10.1007/978-3-642-28951-4_6. [DOI] [PubMed] [Google Scholar]
- 32.Thompson MA, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society–USA panel. JAMA. 2012;308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 33.Lehar J, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukens AK, et al. Harnessing evolutionary fitness in Plasmodium falciparum for drug discovery and suppressing resistance. Proc Natl Acad Sci USA. 2014;111:799–804. doi: 10.1073/pnas.1320886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munck C, et al. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci Transl Med. 2014;6:262ra156. doi: 10.1126/scitranslmed.3009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, et al. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc Natl Acad Sci USA. 2014;111:14494–14499. doi: 10.1073/pnas.1409800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki S, et al. Prediction of antibiotic resistance by gene expression profiles. Nat Commun. 2014;5:5792. doi: 10.1038/ncomms6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui L, et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]