Abstract
Hydrogen peroxide (H2O2) is a major reactive oxygen species (ROS) produced by various cellular sources, especially mitochondria. At high levels, H2O2 causes oxidative stress, leading to cell injury, whereas at low concentrations, this ROS acts as an important second messenger to participate in cellular redox signaling. Detection and measurement of the levels or rates of production of cellular H2O2 are instrumental in studying the biological effects of this major ROS. While a number of assays have been developed over the past decades for detecting and/or quantifying biological H2O2formation, none has been shown to be perfect. Perhaps there is no perfect assay for sensitively and accurately quantifying H2O2 as well as other ROS in cells, wherein numerous potential reactants are present to interfere with the reliable measurement of the specific ROS. In this context, each assay has its own advantages and intrinsic limitations. This article describes a highly sensitive assay for real-time detection of H2O2 formation in cultured cells and isolated mitochondria. This assay is based on the luminol/horseradish peroxidase-dependent chemiluminescence that is inhibitable by catalase. The article discusses the usefulness and shortcomings of this chemiluminometric assay in detecting biological H2O2 formation induced by beta-lapachone redox cycling with both cells and isolated mitochondria.
Keywords: Beta-Lapachone, Chemiluminescence, Hydrogen peroxide, Luminol, Oxidative stress, Reactive oxygen species, Redox cycling, Redox signaling
1. OVERVIEW
Hydrogen peroxide (H2O2) is a non-radical reactive oxygen species (ROS). In contrast to superoxide anion radical, H2O2 is relatively stable and can readily cross cell membranes via aquaporin-facilitated transmembrane diffusion [1, 2]. Although high levels of H2O2 cause oxidative stress, leading to cell and tissue injury, at low concentrations and when its formation is tightly controlled, H2O2 acts as a second messenger, participating in cell signal transduction [3, 4]. Therefore, accurate and sensitive detection and measurement of H2O2 in biological systems are of critical importance for investigating the physiological and pathophysiological roles of this ROS. Over the past decades, a number of assays have been developed for the detection and/or quantification of biological H2O2 formation. These methods can be classified into the following four categories: (1) fluorescence-based assays (e.g., the N-acetyl-3,7-dihydroxyphenoxazine oxidation assay, also known as the Amplex Red assay, which is available as a kit from various commercial sources) [5]; (2) chemiluminescence (CL)-based assays (e.g., the luminol/horseradish peroxidase-dependent CL assay described in this article) [6, 7]; (3) electrode-based assays (e.g., the H2O2 electrode assay) [8]; and (4) the most recently developed redox protein-based assays [9, 10]. Although numerous assays are available, currently there is no well-established assay for sensitive and specific detection and/or measurement of intracellular H2O2 production. The method described in this article is primarily for the detection of extracellular H2O2. Therefore, there is a strong need for developing assays that allow sensitive and selective detection of intracellular H2O2. Nevertheless, when appropriately used in well-defined biological systems, many of the currently available assays, including the one described in this article, could be used to estimate the formation of intracellular H2O2.
2. METHOD PRINCIPLES
The assay described in this article involves the use of the CL technique. Hence, it is imperative to first introduce the basic chemical principles of CL and chemiluminometry in detecting biological ROS, including H2O2.
2.1. General Principles of CL and Chemiluminometry
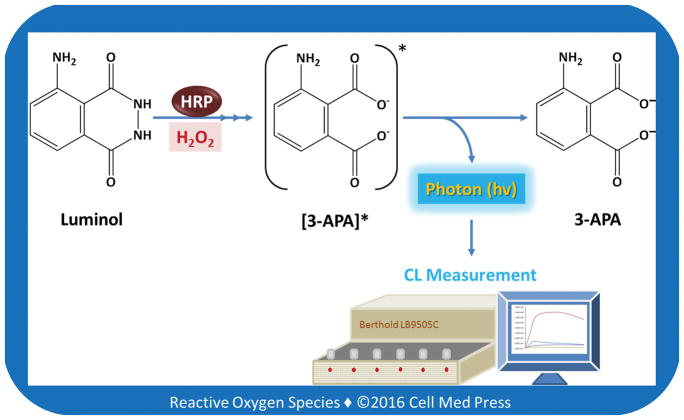
CL is the emission of light with limited release of heat as a result of a chemical reaction. Given reactants A and B, with an excited intermediate E*, the light-emitting reaction can be written as follows: [A] + [B] → [E*] → [Products] + light. For example, if [A] is luminol and [B] is H2O2 in the presence of a suitable catalyst (e.g., myeloperoxidase or horseradish peroxidase), the following reactions occur: luminol + H2O2 → [3-APA*] → 3-APA + light, where 3-APA denotes 3-aminophthalate, and 3-APA* is the excited state. The decay of 3-APA* to a lower energy level causes photon emission (Figure 1). It should be noted that CL differs from fluorescence in that the electronically excited state is derived from the product of a chemical reaction rather than the more typical way of creating electronically excited states, namely, absorption of energy that occurs in the process leading to fluorescence.
FIGURE 1. Schematic illustration of the major chemical reactions leading to luminol-dependent CL.
As shown, in the presence of HRP, luminol reacts with hydrogen peroxide, leading to the eventual formation of the excited state of 3-aminophthalate (3-APA*), whose decay to a lower energy level results in photon emission, which is measured luminometrically as the chemiluminescence (CL) response.
By measuring the photon emission using a luminometer, the rate of H2O2 formation in a biological system can be determined. Because the above CL response is dependent on an exogenously added peroxidase, such as horseradish peroxidase (HRP), which does not cross cell membranes, the luminol/HRP-dependent CL response is indicative of the level of H2O2 released into the extracellular milieu.
For the detection of a particular ROS, a CL probe (e.g., luminol) along with other required reagents (e.g., HRP) is incubated with the ROS-generating system (e.g., enzymes, cells, tissues), and the light emission resulting from the chemical reactions between the probe and the ROS is measured by a luminometer (e.g., the Berthold LB9505 Multi-Channel Biolumat or LB953 AutoLumat) or a liquid scintillation counter. CL imaging can also be applied to visualize ROS formation in situ in cultured cells or tissues [11, 12].
Similar to fluorescence-based methods, CL assays represent a highly sensitive way of detecting ROS in biological systems. However, many CL probes have limited specificity for different ROS, and this limitation also applies to many other ROS-detecting probes, such as fluorescence probes.
2.2. Assay Principle of the Luminol/HRP-Dependent CL
As noted earlier in Section 2.1, the CL probe luminol is oxidized by H2O2 in the presence of HRP to yield light emission. Hence, luminol-derived CL in the presence of exogenously added HRP can be used to sensitively and selectively detect extracellular H2O2 [6]. The kinetic mode of modern luminometers allows the real-time detection of the CL responses over time at various temperatures, which may yield important information on the rate of H2O2 flux in the extracellular milieu. Because H2O2 like water can readily diffuse across cell membranes via an aquaporin-dependent mechanism [1, 2], intracellularly generated H2O2 can be instantly released into the extracellular space. Hence, under well-defined conditions, by detecting extracellular H2O2 release, intracellular H2O2 formation can be estimated. This is particularly useful for studying chemical redox cycling inside cells to form H2O2 (Figure 2). It is noteworthy that in addition to H2O2, other oxidants might also be involved in the reactions leading to luminol/HRP-dependent CL. To determine the specificity of the luminol/HRP-dependent CL for detecting H2O2 formation, catalase can be added to the reaction mix to determine catalase-inhibitable luminol/HRP-dependent CL.
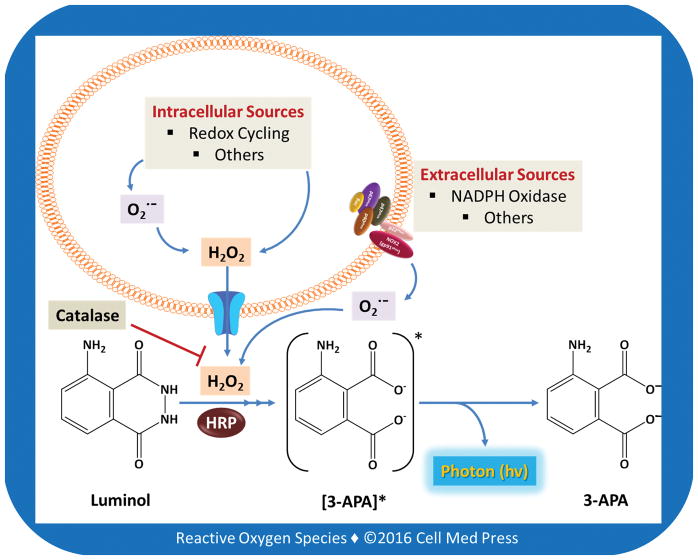
FIGURE 2. Detection of extracellular hydrogen peroxide by the luminol/HRP-dependent CL in cells.
As depicted, H2O2 in the extracellular milieu may be either generated by membrane-associated enzymes such as NADPH oxidases or derived from intracellular sources (e.g., chemical redox cycling). In this context, intracellularly generated H2O2 can diffuse out of cells via an aquaporin-facilitated mechanism. Inclusion of exogenous catalase in the assay, i.e., detection of catalase-inhibitable luminol/HRP-dependent CL, makes it possible to specifically determine the flux of H2O2 in the extracellular milieu.
3. MATERIALS AND INSTRUMENTS
This article describes the catalase-inhibitable luminol/HRP-dependent CL assay for detecting H2O2 formation by two biological systems: cultured Lewis lung carcinoma (LLC) cells and intact mitochondria isolated from the LLC cells.
3.1. Major Materials
The major materials for the catalase-inhibitable luminol/HRP-dependent CL assay are listed alphabetically in Table 1.
Table 1.
Major materials for the catalase-inhibitable luminol/HRP-dependent CL assay
| Major Material | Vendor and Catalog Number | Note |
|---|---|---|
| Bio-Rad protein assay dye reagent concentrate | Bio-Rad; 5000006 | For measurement of mitochondrial protein |
| Catalase | Sigma-Aldrich; C9322 | Source: bovine liver; 2,300 units/mg |
| Complete phosphate-buffered saline (CPBS) | Investigator-prepared | Components: 8.1 mM Na2HPO4, 1.47 mM KH2PO4, 138 mM NaCl, 2.67 mM KCl, 0.5 mM MgCl2, 0.7 mM CaCl2, and 0.1% glucose, pH 7.4 |
| Horseradish peroxidase (HRP) | Sigma-Aldrich; P8250 | Type II; source: horseradish roots (Amoracia rusticana); molecular mass: ~44 kDa |
| Beta-Lapachone | Sigma-Aldrich; L2037 | 5 mg size |
| Lewis lung carcinoma (LLC) cells | ATCC; CRL-1642 | LLC cells are derived from Lewis lung carcinoma in C57BL/6 mice. |
| Luminol (sodium salt) | Sigma-Aldrich; A4685 | Luminol sodium salt is soluble in water (50 mg/ml), yielding a clear solution. |
| Mitochondrial isolation buffer | Investigator-prepared | Components: 250 mM sucrose, 10 mM Hepes, 1 mM EGTA, and 0.5% bovine serum albumin, pH 7.4 |
| Mitochondrial respiration buffer | Investigator-prepared | Components: 70 mM sucrose, 220 mM mannitol, 2 mM Hepes, 2.5 mM KH2PO4, 2.5 mM MgCl2, 0.5 mM EDTA, and 0.1% bovine serum albumin, pH 7.4 |
| Phosphate-buffered saline (PBS) | Investigator-prepared | Components: 8.1 mM Na2HPO4, 1.47 mM KH2PO4, 138 mM NaCl, and 2.67 mM KCl, pH 7.4 |
| Succinate | Sigma-Aldrich; S2378 | Sodium succinate dibasic hexahydrate |
3.2. Major Instruments
Luminometers: Berthold LB9505 Multi-Channel Biolumat or Berthold LB950 AutoLumat
Beckman Coulter DU-800 ultraviolet/visible (UV/Vis) spectrophotometer
Dounce tissue grinder
Water bath or dry bath
Cell culture equipment (CO2 incubator, tissue culture hood, centrifuge, microscope, etc.)
4. PROTOCOLS AND STEPS
This section describes the detailed protocols and steps involved in the use of the catalase-inhibitable luminol/HRP-dependent CL assay to detect H2O2 generated from cultured LLC cells as well as isolated mitochondria in the presence of beta-lapachone, a potential anticancer compound that undergoes redox cycling to generate superoxide and H2O2 [13].
4.1. Cultured Cells
4.1.1. Assay Layout
Figure 3 illustrates the assay layout for determination of the catalase-inhibitable luminol/HRP-dependent CL resulting from H2O2 generated by redox cycling of beta-lapachone in LLC cells in CPBS at 37°C for 30 min. It should be noted that the incubation time can vary from a few seconds to a few hours depending on the experimental design.
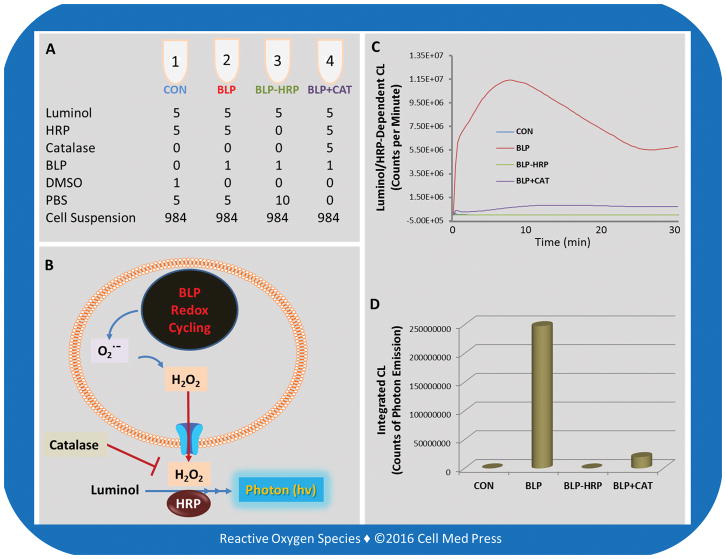
FIGURE 3. Assay layout for the catalase-inhibitable luminol/HRP-dependent CL responses in LLC cells treated with beta-lapachone.
Redox cycling of beta-lapachone (BLP) in LLC cells generates superoxide which undergoes spontaneous or superoxide dismutase-catalyzed dismutation to form H2O2. The H2O2 formed can readily defuse out of cells and react with luminol in the presence of HRP, resulting in CL responses. Addition of catalase (CAT) inhibits the CL response by >92%, indicating that the assay under the present experimental conditions selectively detects extracellular H2O2. Notably, the CL response is completely abolished in the absence of added HRP, indicating that the CL response is dependent on the presence of HRP, thus confirming that the CL response occurs in the extracellular milieu. The unit in panel A is μl.
4.1.2. Assay Description
The catalase-inhibitable luminol/HRP-dependent CL assay is used to detect extracellular H2O2 released from cultured LLCs. Briefly, LLC cells are cultured in Dulbecco’s modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 μg/ml of streptomycin in 150 cm2 tissue culture flasks at 37°C in a humidified atmosphere of 5% CO2. The cells are fed every 2–3 days, and subcultured once they reach 80–90% confluence. For the CL measurement, LLC cells are harvested from the cultures and washed once in CPBS. As many as 1 × 106 cells are suspended in 1 ml CPBS in a CL tube containing 1 μM beta-lapachone, 10 μg/ml of HRP, and 10 μM luminol in the presence or absence of 500 units/ml of catalase, and the CL tube is immediately transferred to a Berthold LB9505 luminometer for recording the CL response at 37°C for 30 min. The results are expressed as the real-time photon emission curve (CL response) and the integrated CL response (the area under the curve, representing the total counts of photon emission over the 30 min of incubation time).
4.1.3. Preparation of Reagents
Luminol solution (2 mM in PBS): 7.96 mg luminol sodium (molecular mass = 199.14) dissolved in 2 ml PBS (aliquot into microfuge tubes and store at −20°C).
HRP solution (2 mg/ml in PBS): 4 mg HRP dissolved in 2 ml PBS (aliquot into microfuge tubes and store at −20°C).
Beta-Lapachone solution (1 mM in dimethyl sulfoxide): first prepare 10 mM beta-lapachone (molecular mass = 242.27) (add 2.06 ml dimethyl sulfoxide to a vial containing 5 mg beta-lapachone), and then dilute this 10 mM beta-lapachone solution in dimethyl sulfoxide to yield a 1 mM solution.
Catalase solution (100,000 units/ml in PBS): 50 mg catalase (2,300 units/mg) dissolved in 1.15 ml PBS (aliquot into microfuge tubes and store at −20°C).
4.1.4. Steps
Aliquot 1 × 106 cells into each of the 4 microfuge tubes followed by centrifugation to pellet the cells and discard of the medium. Keep the cell pellets on ice.
Add 5 μl of 2 mM luminol to each of the 4 CL tubes.
Add 5 μl HRP (2 mg/ml) to the CL tubes 1, 2, and 4, and 5 μl PBS to the CL tube 3.
Add 5 μl catalase (100,000 units/ml) to the CL tube 4, and 5 μl PBS to the CL tubes 1, 2, and 3.
Add 1 μl of 1 mM beta-lapachone to the CL tubes 2, 3, and 4, and 1 μl dimethyl sulfoxide (DMSO) to the CL tube 1.
Add 984 μl of 37°C-prewarmed air-saturated CPBS to the microfuge tube 1, as indicated in Step (1) above, that contains 1 × 106 cells to resuspend the cells. Transfer the entire cell suspension to the CL tube 1 immediately followed by mixing of the sample and recording of the CL response at 37°C for 30 min. Do the same for the remaining 3 samples. The background CL levels of the instrument are measured with the CL tubes containing all reactants/components but cells. The background CL levels are subtracted from the CL responses of the tubes containing cells so that the reported CL responses in the figures are due to cellular activity.
4.1.5. Calculations
The Berthold LB9505 multi-channel luminometer enables real-time measurement of the CL responses from each of the 4 samples (CL tubes) which are reported as the CL response curves (see Figure 3C) with the unit of the Y and X axis being counts of photon emission per minute (CPM) and minute, respectively. The luminometer automatically calculates the area under the curve for each of the CL response curves from the 4 samples. This is designated as the integrated CL (see Figure 3D) with the unit being total counts of photon emission over the 30 min of incubation time per 1 × 106 cells.
4.1.6. Other Considerations
The Berthold LB9505 multi-channel luminometer can measure up to 6 samples simultaneously, whereas the newer model of Berthold LB953 luminometer with auto-injectors can measure much more samples simultaneously under its kinetic mode. However, our laboratories have consistently noticed that the Berthold LB9505 luminometer is probably one of the most sensitive luminometers for detecting CL responses from biological systems, including tissues, intact cells, and isolated mitochondria, as well as other subcellular components. Both the Berthold LB9505 and LB953 luminometers are equipped with a temperature control unit, allowing the measurement of the CL responses at various temperatures including 37°C. Although this article describes the use of the Berthold LB9505 luminometer in detecting biological H2O2formation, the protocols can be easily adopted to detect biological H2O2 formation with other commercially available luminometers if the detection sensitivity is not a major concern. As many of the commercially available luminometers are not equipped with the kinetic and temperature control units, to detect the time- and temperature-dependent CL responses, the sample CL tubes can be incubated at various temperatures in a water bath or dry bath and then transferred to the luminometer for the measurement of the CL responses at different time intervals to obtain a time-dependent response.
4.2. Isolated Mitochondria
4.2.1. Assay Layout
Figure 4 illustrates the assay layout for determination of the catalase-inhibitable luminol/HRP-dependent CL responses resulting from H2O2 generated by redox cycling of beta-lapachone in isolated mitochondria at 37°C for 30 min. It should be noted that the incubation time can vary from a few seconds to a few hours depending on the experimental requirement and the luminometer used.
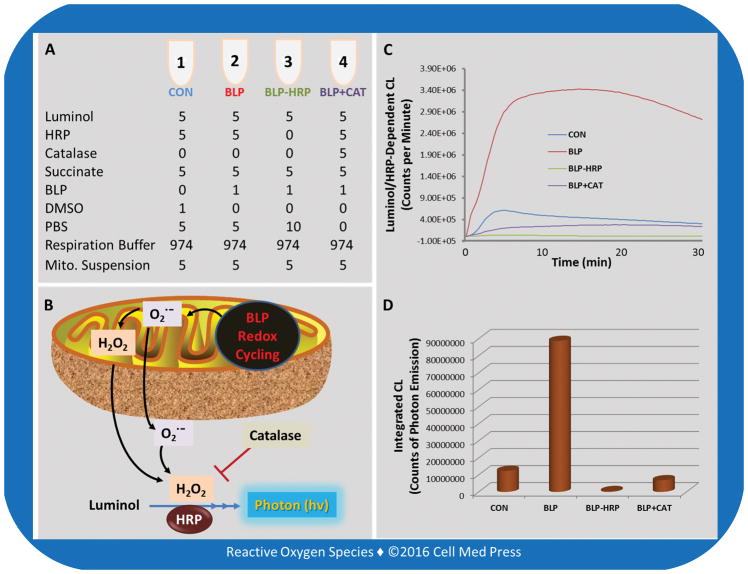
FIGURE 4. Assay layout for the catalase-inhibitable luminol/HRP-dependent CL responses in isolated mitochondria treated with beta-lapachone.
Redox cycling of beta-lapachone (BLP) in succinate-driven mitochondria generates superoxide which undergoes spontaneous or superoxide dismutase-catalyzed dismutation to form H2O2. The H2O2formed can readily diffuse out of mitochondria and react with luminol in the presence of HRP, resulting in CL responses. Superoxide may also exit mitochondria and then undergo spontaneous dismutation, contributing to the extramitochondrial H2O2 formation. Addition of catalase (CAT) inhibits the CL response by >92%, indicating that the assay under the present experimental conditions selectively detects the formation of extramitochondrial H2O2. The CL response is completely abolished in the absence of added HRP, indicating that the CL response is dependent on the presence of HRP, confirming that the CL response occurs in the extramitochondrial milieu. The unit in panel A is μl. Mito. denotes mitochondrial.
4.2.2. Assay Description
The catalase-inhibitable luminol/HRP-dependent CL assay is used to detect H2O2 released from intact mitochondria isolated from LLC cells. Briefly, LLC cells are cultured in DMEM supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin in 150 cm2 tissue culture flasks at 37°C in a humidified atmosphere of 5% CO2. The cells are fed every 2–3 days, and subcultured once they reached 80–90% confluence. Mitochondria are isolated from the freshly harvested LLC cells according to the method described before [14]. Briefly, LLC cells (~3 × 107 cells) are harvested and washed once with PBS. The cell pellet is resuspended in 6 ml of sucrose buffer (250 mM sucrose, 10 mM Hepes, 1 mM EGTA, and 0.5% bovine serum albumin, pH 7.4) and homogenized in a Dounce tissue grinder (40 strokes with the tight pestle) on ice. The homogenate is centrifuged at 1,500 × g for 10 min at 4°C. The supernatant is collected and centrifuged at 10,000 × g for 10 min at 4°C. The resulting mitochondrial pellet is washed twice with the sucrose buffer and then resuspended in bovine serum albumin (BSA)-free sucrose buffer (250 mM sucrose, 10 mM Hepes, and 1 mM EGTA, pH 7.4). The mitochondrial protein is measured with Bio-Rad protein assay dye based on the method of Bradford [15] with BSA as the standard. The mitochondrial suspension is diluted to 1 mg/ml in the above BSA-free sucrose buffer and kept on ice for experiments within 3 hours. For the CL measurement, 5 μl of mitochondrial suspension (giving a final concentration of 0.05 mg mitochondrial protein/ml) is added to a CL tube containing the various reactants (250 μM succinate, 1 μM beta-lapachone, 10 μg/ml of HRP, and 10 μM luminol in the presence or absence of 500 units/ml of catalase) in the respiration buffer (in a final volume of 1 ml; 70 mM sucrose, 220 mM mannitol, 2 mM Hepes, 2.5 mM KH2PO4, 2.5 mM MgCl2, 0.5 mM EDTA, and 0.1% BSA, pH 7.4), and the CL tube is immediately transferred to the Berthold LB9505 luminometer for recording the CL responses at 37°C for 30 min. The results are expressed as the real-time photon emission (CL response curve) and the integrated CL response (the area under the curve, representing the total counts of photon emission over the 30 min of incubation time).
4.2.3. Preparation of Reagents
Luminol solution (2 mM in PBS): see Section 4.1.3 for preparation.
HRP solution (2 mg/ml in PBS): see Section 4.1.3 for preparation.
Beta-Lapachone solution (1 mM in dimethyl sulfoxide): see Section 4.1.3 for preparation.
Catalase solution (100,000 units/ml in PBS): see Section 4.1.3 for preparation.
Succinate solution (0.5 M in deionized water): 1.35 g sodium succinate dibasic hexahydrate (molecular mass = 270.14) dissolved in 10 ml deionized water (aliquot into microfuge tubes and store at −20°C).
4.2.4. Steps
Add 5 μl of 0.5 M succinate to each of the 4 CL tubes.
Add 5 μl of 2 mM luminol to each of the 4 CL tubes.
Add 5 μl of HRP (2 mg/ml) to the CL tubes 1, 2, and 4, and 5 μl PBS to the CL tube 3.
Add 5 μl catalase (100,000 units/ml) to the CL tube 4, and 5 μl PBS to the CL tubes 1, 2, and 3.
Add 1 μl of 1 mM beta-lapachone to the CL tubes 2, 3, and 4, and 1 μl dimethyl sulfoxide (DMSO) to the CL tube 1.
Add 974 μl of 37°C-prewarmed air-saturated mitochondrial respiration buffer to the CL tube 1 followed by the addition of 5 μl of the mitochondrial suspension (1 mg/ml). Upon mixing, the CL tube is transferred to the LB9505 luminometer for recording the CL response at 37°C for 30 min. Do the same for the remaining 3 samples. It is worth mentioning that the background CL levels of the instrument are measured with the CL tubes containing all reactants/components but mitochondria. The background CL levels are subtracted from the CL responses of the tubes containing mitochondria so that the reported data of CL responses in the figures are due to mitochondrial activity.
4.2.5. Calculations
The Berthold LB9505 multi-channel luminometer enables real-time measurement of the CL responses from each of the 4 samples (CL tubes) which are reported as the CL response curves (see Figure 4C) with the unit of the Y and X axis being CPM and minute, respectively. The luminometer automatically calculates the area under the curve for each of the CL response curves from the 4 samples. This is designated as the integrated CL (see Figure 4D) with the unit being total counts of photon emission over the 30 min of incubation time per 0.05 mg mitochondria.
4.2.6. Other Considerations
In addition to those mentioned in Section 4.1.6, the protocols described in this article can be easily adopted to study the effects of other mitochondrial substrates, such as pyruvate/malate, which provide NADH, thus feeding the electrons through the mitochondrial complex I. It is noteworthy that in addition to using this assay to study chemical redox cycling in mitochondria, the assay can also be employed to detect H2O2 formation by isolated mitochondria under various conditions. In this context, mitochondria can be isolated from cells or tissues under certain pathophysiological conditions to determine the altered formation of mitochondrial H2O2. Notably, mitochondria are the major source of cellular H2O2under diverse physiological and pathophysiological conditions and mitochondria-derived H2O2 plays an important role in biology and medicine [16–18].
5. DISCUSSION OF ADVANTAGES AND LIMITATIONS
5.1. Advantages
As with other CL-based techniques, the biggest advantage of the luminol/HRP-dependent CL assay is its high sensitivity for detecting biological H2O2 formation. With the kinetic mode of the Berthold LB9505 (as well as the Berthold LB953) luminometer, the relative rate of formation of H2O2 in a biological system can be sensitively detected in a real-time manner. Examining the shapes of the CL response curves may also yield important insight into the dynamics of H2O2formation in a biological system under a particular physiological or pathophysiological condition.
5.2. Limitations
The luminol/HRP-dependent CL assay for detecting biological H2O2 formation is not without limitations. As noted earlier, this assay only detects the rate of H2O2 flux in the extracellular or extramitochondrial milieu. Because intracellularly formed H2O2 can readily penetrate the biomembranes to enter the extracellular/extramitochondrial milieu, detection of the rate of extracellular/extramitochondrial H2O2 flux can, however, be used to reflect the dynamics of H2O2 formation from the intracellular sources. Another shortcoming of the assay is related to its limited ability to directly quantify the absolute amounts of H2O2 formed in a biological system. To circumvent this limitation, a concurrently run experiment using known amounts of H2O2 may be considered to obtain a standard curve (i.e., a linear relationship between the integrated luminol/HRP-dependent CL and the amounts of exogenously supplemented H2O2). The exogenously supplemented H2O2 may be in the form of either a H2O2 solution of known concentration or preferentially a known rate of H2O2 formation by a standard enzymatic system, such as the glucose/glucose oxidase. Nevertheless, it should be borne in mind that due to the complexity of reactions involved in detecting H2O2 as well as other ROS in biological systems, the reliability of such a standard curve for quantifying biological H2O2 should be rigorously tested and validated.
6. CONCLUSION
As with many other assays and techniques for detecting biological H2O2 as well as other members of the ROS family, the luminol/HRP-dependent CL assay has its own advantages and limitations. The biggest advantage of this assay is its high sensitivity and real-time detection of H2O2 formation. On the other hand, a major limitation of the assay is its inability to directly detect intracellular H2O2 formation. In addition, the assay does not allow quantification of H2O2 in biological systems unless a concurrently run standard curve is employed. Nevertheless, as mentioned earlier, if appropriately used under controlled conditions along with the results being correctly interpreted, detection of H2O2 by the luminol/HRP-dependent CL assay can yield important information on the real-time formation of this ROS in various biological systems under different physiological and pathophysiological conditions. In fact, detecting the changes in the relative rate of H2O2formation, rather than the absolute amounts of H2O2 formed, under different conditions, is frequently the goal of the experiments towards understanding the role of this important ROS in biology and medicine.
Acknowledgments
The work was supported in part by a grant from the U.S. National Institutes of Health/National Cancer Institute (CA192936).
ABBREVIATIONS
- 3-APA
3-aminophthalate
- BSA
bovine serum albumin
- CL
chemiluminescence
- CPBS
complete phosphate-buffered saline
- CPM
counts of photon emission per minute
- DMEM
Dulbecco’s modified Eagle medium
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- H2O2
hydrogen peroxide
- HRP
horseradish peroxidase
- PBS
phosphate buffered saline
- ROS
reactive oxygen species
References
- 1.Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta. 2014;1840(5):1596–604. doi: 10.1016/j.bbagen.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Hara-Chikuma M, Satooka H, Watanabe S, Honda T, Miyachi Y, Watanabe T, et al. Aquaporin-3-mediated hydrogen peroxide transport is required for NF-kappaB signalling in keratinocytes and development of psoriasis. Nat Commun. 2015;6:7454. doi: 10.1038/ncomms8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312(5782):1882–3. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 4.Li YR, Jia Z, Trush MA. Defining ROS in biology and medicine. Reactive Oxygen Species. 2016;1(1):9–21. doi: 10.20455/ros.2016.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253(2):162–8. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Trush MA. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem Biophys Res Commun. 1998;253(2):295–9. doi: 10.1006/bbrc.1998.9729. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Stansbury KH, Zhu H, Trush MA. Biochemical characterization of lucigenin (bis-N-methylacridinium) as a chemiluminescent probe for detecting intramitochondrial superoxide anion radical production. Biochem Biophys Res Commun. 1999;262(1):80–7. doi: 10.1006/bbrc.1999.1174. [DOI] [PubMed] [Google Scholar]
- 8.Cox JA, Jaworski RK. Voltammetric reduction and determination of hydrogen peroxide at an electrode modified with a film containing palladium and iridium. Anal Chem. 1989;61(19):2176–8. doi: 10.1021/ac00194a012. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht SC, Barata AG, Grosshans J, Teleman AA, Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14(6):819–29. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Ermakova YG, Bilan DS, Matlashov ME, Mishina NM, Markvicheva KN, Subach OM, et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat Commun. 2014;5:5222. doi: 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WT, Tung CH, Weissleder R. Imaging reactive oxygen species in arthritis. Mol Imaging. 2004;3(3):159–62. doi: 10.1162/1535350042380290. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Shen H, Zhu H, Trush MA, Jiang M, Wang G. In situ real-time chemiluminescence imaging of reactive oxygen species formation from cardiomyocytes. Int J Biomed Imaging. 2008;2008:941729. doi: 10.1155/2008/941729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li JZ, Ke Y, Misra HP, Trush MA, Li YR, Zhu H, et al. Mechanistic studies of cancer cell mitochondria- and NQO1-mediated redox activation of beta-lapachone, a potentially novel anticancer agent. Toxicol Appl Pharmacol. 2014;281(3):285–93. doi: 10.1016/j.taap.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zhu H, Kuppusamy P, Roubaud V, Zweier JL, Trush MA. Validation of lucigenin (bis-N-methylacridinium) as a chemilumigenic probe for detecting superoxide anion radical production by enzymatic and cellular systems. J Biol Chem. 1998;273(4):2015–23. doi: 10.1074/jbc.273.4.2015. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zhu H, Kuppusamy P, Zweier JL, Trush MA. Mitochondrial electron transport chain-derived superoxide exits macrophages: implications for mononuclear cell-mediated pathophysiological processes. Reactive Oxygen Species. 2016;1(1):81–98. doi: 10.20455/ros.2016.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chouchani ET, Pell VR, James AM, Work LM, Saeb-Parsy K, Frezza C, et al. A unifying mechanism for mitochondrial superoxide production during ischemia-reperfusion injury. Cell Metab. 2016;23(2):254–63. doi: 10.1016/j.cmet.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Shadel GS, Horvath TL. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163(3):560–9. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]