Abstract
Glioblastoma multiforme (GBM) is an aggressive form of brain cancer that has no effective treatments and a prognosis of only 12–15 months. Microfluidic technologies deliver microscale control of fluids and cells, and have aided cancer therapy as point-of-care devices for the diagnosis of breast and prostate cancers. However, a few microfluidic devices are developed to study malignant glioma. The ability of these platforms to accurately replicate the complex microenvironmental and extracellular conditions prevailing in the brain and facilitate the measurement of biological phenomena with high resolution and in a high-throughput manner could prove useful for studying glioma progression. These attributes, coupled with their relatively simple fabrication process, make them attractive for use as point-of-care diagnostic devices for detection and treatment of GBM. Here, the current issues that plague GBM research and treatment, as well as the current state of the art in glioma detection and therapy, are reviewed. Finally, opportunities are identified for implementing microfluidic technologies into research and diagnostics to facilitate the rapid detection and better therapeutic targeting of GBM.
Keywords: glioblastoma, in vitro diagnostic devices, microfluidics
1. Overview of Glioblastoma Multiforme
Primary brain tumors arising from glial cells account for ≈3% of all new adult cancers and 25% of pediatric cancers annually.[1] Although brain tumor incidence is relatively low when compared to other systemic cancers, it carries a high mortality rate and only 34% of individuals suffering from malignant brain cancers experience a complete recovery.[2] The most commonly occurring primary intracranial tumors in adults are gliomas, which arise from the glial support cells within the brain.[2] Primary gliomas originate from astrocytes, oligodendrocytes, or a mix of the two glial cell types, with a range of stage categorizations as delineated by the World Health Organization (WHO).[3] The WHO grade system ranges from glioma stages I–IV based on malignancy and rate of tumor growth, with the most common types being astrocytoma (all grades), oligodendrocytoma (grades II and III), and oligoastrocytoma (grades II and III).[2,4] Astrocytomas arise from the dysfunction of supportive astrocytes that are essential for the maintenance of neuronal homeostasis and blood–brain-barrier (BBB) function.[5] Lower grade (I–II) gliomas are considered benign and are commonly treated with surgical resection of the affected brain area. However, higher grade (III–IV) gliomas are considered malignant and are significantly more difficult to treat. Grade III astrocytomas exhibit heterogeneous cell morphology and typically express heightened mitotic activity without evidence of necrosis or neovascularization.[6] Primary grade IV astrocytomas can begin as lower grade astrocytomas and transform into more malignant tumors.[7]
Grade IV astrocytomas are classified as glioblastoma multiforme (GBM). They comprise the majority of malignant gliomas diagnosed in the USA and are distinguished from lower grade tumors by the distinct presence of necrosis and neovasculature[2] (Figure 1). GBM is highly invasive, and glioma cells associated with GBM are known to invade through the brain parenchyma along existing blood vessels and white matter tracts, progressing outward to the meninges.[8] Typical GBM presentation is an irregular mass along or near white matter tracts, with invading cells using these white matter tracts and local vasculature to migrate into surrounding parenchyma.[8] Upon diagnosis, tumor cells are assumed to be widely dispersed in the brain parenchyma even though there is usually one central mass at that time.[9] These central masses are typically unilateral and can occupy the majority of an entire lobe before utilizing the corpus callosum to spread across the midline into the contralateral hemisphere; this movement produces the “butterfly” appearance characteristic of bilateral invasion. Secondary brain tumors most commonly occur from lung, breast, or skin metastases, with an incidence of brain metastases reportedly as high as 35% in metastatic breast cancer patients.[10] Beyond sharing some similar common mutations to GBM such as phosphatase and tensin homolog (PTEN) or epidermal growth factor receptor (EGFR), the progression of many cancers including GBM is linked to aberrant extracellular matrix (ECM) remodeling, which is implicated in promoting invasion and malignancy.[11,12] Elevated expression of chondroitin sulfate proteoglycan 4 (CSPG4) is associated with worsened prognosis for both GBM and breast cancers, with both cancers being highly invasive.[13] The tumor microenvironment (TME) is both mechanically and biologically active; thus, any alterations to the ECM composition around tumors can enhance cellular invasion or contribute to treatment resistance. Matrix metalloproteinases (MMPs) are normally involved in tissue remodeling, and are secreted by a multitude of cancer types including GBM to degrade ECM components and contribute to the release of growth factors.[12,14–16] The following increase in local ECM rigidity directly influences tumor formation and tumor cell invasion, observed in glioma, breast, and many other cancer types.[17,18]
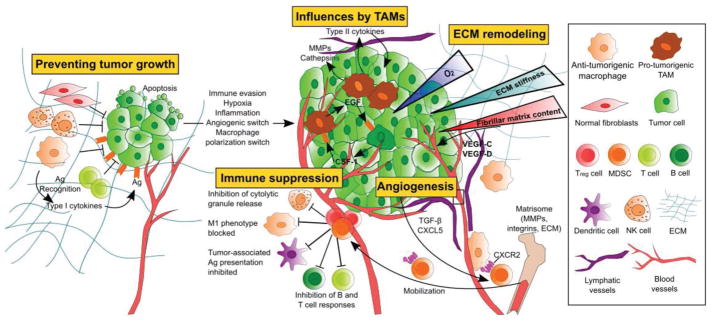
Figure 1.
Cellular influences involved in the switch from benign neoplasm to malignant growth in GBM. Dormant tumors are regulated by microenvironmental mechanisms including a lack of vasculature, limited supply of nutrients and oxygen, and recognition by nearby cell types such as fibroblasts, natural killer (NK) cells, and macrophages through tumor-associated antigen (Ag) presentation. Tumor cells can evade the body’s immune system response by recruiting immune suppressing tumor-associated macrophages (TAMs) and triggering a protumorigenic state within the microenvironment. Treg cells and myeloid-deprived suppressor cells (MDSCs) recruited by TAMs release anti-inflammatory cytokines to suppress immune cell activity. ECM remodeling by tumor cells creates a local stiffness gradient in response to aberrant ECM composition.
GBM invasion is characterized by excessive proliferation and growth of the tumor bulk, creation of hypoxic zones without access to local vasculature, and the formation of pseudopalisades. Pseudopalisades consist of hypercellular regions around necrotic tumor foci that secrete proangiogenic factors required for the formation of neovasculature that provides nutrients to the tumor bulk.[19] In order to supply oxygen and nutrients to the rapidly growing tumor, glioma cells induce the expression of angiogenesis regulator angiopoietin-2 by local endothelial cells, triggering apoptosis and hypoxia in local brain regions.[20] This change in environment, coupled with the secretion of high levels of vascular endothelial growth factor (VEGF) by tumor cells, stimulates the “budding” of new blood vessels and stimulates cell proliferation within the bulk of the tumor.[21] Constant cellular turnover leads to the formation of a necrotic core in the center of the tumor and manifests in the presence of characteristic physical symptoms of GBM such as intracranial pressure and functional impairments.[22] Depending on the area of the brain in which the tumor manifests, the patient can ultimately face death or experience serious functional deficits.
The median survival for adults with GBM who undergo standard-of-care chemo- and radiation therapy (RT) is 14 months with a 2 year survival of 30% and a 5 year survival of 5.1%.[2] Poor adjuvant treatment efficacy and high recurrence rate are often encountered due to the inability to target and stem tumor invasion, and patient outcome with recurrent GBM remains extremely high leading to many patients preferring palliative care over aggressive therapeutic intervention.[23] Several clinical trials around the world are testing novel treatment options for GBM, in the hope that combinations of novel therapies along with established treatment options will improve the current standard-of-care and prognosis for these patients.
2. The Challenge of Early Detection
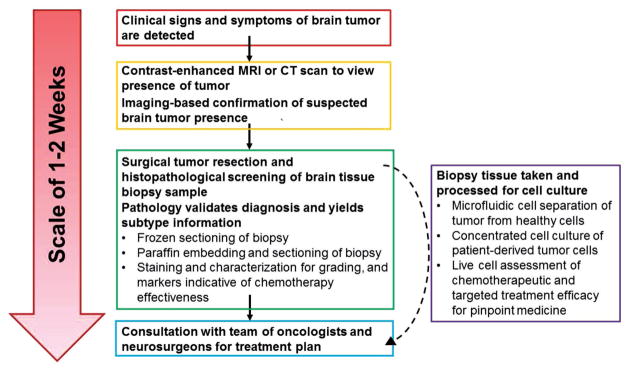
The global cancer diagnostics market is currently worth billions of dollars, and is based on technological platforms that can detect and inform the treatment of cancer.[24] Many types of cancers are diagnosed via magnetic resonance imaging (MRI). However, the equipment and technical expertise required for these scans can be cost prohibitive for most of the populace. GBM being located wholly within the brain presents another unique problem for the early detection of these tumors. GBM is detected only after the manifestation of symptoms such as extreme headaches, seizures, and dizziness. T1- and T2-weighted MRI are often performed following presentation of patient symptoms in order to confirm tumor presence. Surgeons use this information to determine the tumor margins for surgical resection, which is consistently the first course of action when dealing with primary GBM tumors. Prior to making treatment decisions, a needle biopsy is taken to correctly characterize the tumor and to determine the treatment course (Table 1). GBM is unique when compared to other lower grade brain tumors due to the presence of a highly irregular shape, centralized regions of necrosis, and extensive edema, all of which can be visualized using MRI and validated by the histopathological analysis of the tissue biopsy (Figure 2). Serial neuroimaging remains a primary method of diagnosis for glioma. However, histopathological inference of invasive tissue biopsies remains the mainstay of diagnostic testing, thereby presenting opportunities for less-invasive diagnostic measures to improve upon accuracy and time to diagnosis (Figure 2).
Table 1.
GBM diagnosis methodology.
| Preoperative imaging and assessment | Function |
|---|---|
| MR imaging/CT imaging | Provides detailed information about tumor size and location via noninvasive imaging techniques |
| Karnofsky performance status | Assessment tool for functional impairment, and can be used to assess the prognosis in individuals (0–100 scale, a score of 100 meaning normal with no evidence of disease) |
| Friedlein grading | Functional classification system considering tumor operability and prognosis based on location of the tumor being either close or within eloquent regions (Friedlein grading B, “FGB”) or at a safe distance from eloquent regions (Friedlein grading A, “FGA”) |
| Operative | |
| needle biopsy | A neurosurgeon removes a small piece of the tumor tissue, which is sent to a pathologist for review and official diagnosis. Stereotactic biopsy involves the use of computer-assisted guidance to locate and resect the tumor mass |
| Postoperative analysis | |
| Histopathology | Hematoxylin and eosin staining remains the gold standard for diagnosis and characterization of the tissue biopsy |
| World Health Organization (WHO) grading | CNS tumors are classified according to standards published by the WHO based on morphological features, growth pattern, and molecular profile of the neoplastic cells |
Figure 2.
Timescale of brain tumor detection and diagnosis methods including where microfluidic diagnostic methods could improve time to diagnosis and accuracy in prognosis. Conventionally, the process of a brain tumor diagnosis begins with presentation of symptoms and imaging-based assessment of lesion presence. Upon imaging-based confirmation of brain tumor presence, an invasive surgical procedure is scheduled to procure tissue biopsies for histopathological analyses. The pathology report containing information about the formal cancer diagnosis and staging can often take several weeks to generate, even when expedited. Microfluidic platforms provide an opportunity for the real-time analysis of tissue samples and the rapid processing of liquid biopsies (such as blood or serum samples), bypassing the potentially lengthy waiting time for diagnosis, staging information, or mutation status of GBM. MRI: Magnetic resonance imaging; CT: Computed tomography scan.
In the USA, almost 20 000 new gliomas are diagnosed every year. However, studies focused on evaluating glioma incidence rates within asymptomatic populations report that the actual number of patients with glioma in the USA is potentially 40–50 times greater than the reported incidence.[25] Due to the lack of accessible and low-cost early detection schemes, the clear majority of asymptomatic individuals will be left unawares until symptoms develop, by which time the glioma becomes lethal. Although the 1 year survival for patients receiving a 90% tumor resection is significantly higher than patients who undergo a less than 90% resection,[26] even a 90% tumor resection can prove to be insufficient, often leading to eventual recurrence by individual invasive cells that are left behind. GBM patients routinely receive adjuvant treatment in the form of RT or chemotherapy. The current standard for GBM chemotherapy is temozolomide (TMZ); however, the ability of GBM to adaptively evade treatment hampers chemotherapy efficacy. Earlier cancer detection is widely accepted as being crucial for improving patient prognosis, but a dearth of cost-efficient and accurate options to facilitate early brain cancer detection contributes to the poor prognosis of patients with malignant glioma.
3. FDA-Approved In Vitro Diagnostic Devices for Cancer Detection
In vitro diagnostic devices (IVDs) are used to detect disease or infection, and can provide additional information about a patient’s condition beyond a physical examination by their physician.[27] The United States Food and Drug Administration (FDA) classifies IVDs according to intended usage and indications for use. Intended use refers to what is specifically tested, for example, a certain mutation, while indication for use is why the patient would be tested for that mutation, such as to determine the risk of presence of disease.[27] This classification is based on the device’s risk to the patient and the subsequent information available to address the risk.[28] Class I devices are of the lowest risk and include the use of immunohistochemical reagents to diagnose the disease. Class II devices contain a moderate risk to the patient, examples being molecular tests for biomarkers or prognostic indicators.[27] IVDs that have significant importance for prevention of health impairment or for medical decision-making are considered class III devices. The majority of FDA-approved class III diagnostic devices are used to assess specific mutation status in certain cancers, such as the detection of EGFR mutations in patients.[29] IVDs intended for diagnosis or evaluation of cancer are considered class III devices after undergoing preclinical research and receiving premarket approval.
IVDs fall under two broad categories: genetic assays that indicate disease or genetic carrier status and companion assays that analyze the effectiveness of a therapeutic product on a disease.[28] Companion IVDs serve as guidance markers for the use of specific therapeutic options and anticancer reagents, eliminating a certain level of guesswork from treatment. Currently available companion IVDs for cancer assess a variety of mutation-specific cancers, and these IVDs are used after an official cancer diagnosis has been made but before deciding how to proceed with targeted treatments. These assays can evaluate a specific mutation status through either immunohistochemical or real-time polymerase chain reaction methods.[30] The resulting information can then be used to decide upon therapeutic approach for a specific cancer type. These IVDs offer a faster route to personalized treatment by facilitating a targeted treatment regime for patients with more common mutations such as lung and breast cancers among few others. Clinical evaluation of a companion IVD includes the demonstration of the device’s ability to predict treatment outcome in individuals in phase III clinical trials.[28] The companion IVD is considered for use if it adequately discriminates between patients who will likely respond to a given treatment or not. Therefore, the device is evaluated by data on the clinical sensitivity, specificity, positive predictive value, and negative predictive value. Some examples of targeted anticancer drugs that have FDA-approved companion IVDs are gefitinib, vemurafenib, crizotinib, among several others.
FDA-approved biomarkers are currently used for monitoring disease progression or therapeutic response. However, there are several unmet clinical needs for biomarkers that aid in early detection and diagnosis of pancreatic, ovarian, and brain cancers that often do not present obvious symptoms and go undetected during routine health screenings. Most tumor markers are not screened using diagnostic device assays because of low sensitivity or low specificity, which can result in either low detection or false-positive diagnoses.[31] Tumor markers help distinguish tumor cells or from other cells in the body, and their expression positively correlates to the presence and growth of a tumor. However, in most situations, the limited specificity of tumor-associated markers means that measuring a single tumor marker is often insufficient to make a diagnosis, and studies have shown that measuring a panel of tumor markers can be of greater diagnostic value.[32] Currently available diagnostic devices that measure tumor biomarkers are also time consuming or too expensive to be used routinely during health checkups, reducing their practical application.
There is a startling lack of IVDs for the directed use with malignant glioma, with the majority of currently FDA-approved IVDs focusing on colorectal cancer, lung cancer, and melanoma. With precision medicine being the vanguard of cancer treatment, specific molecular markers of an individual’s tumor cells can be used to identify the most beneficial therapeutic approaches. More diagnostic devices beyond pathology are needed to advance personalized medicine and develop more effective therapies for malignant glioma.
4. Microfluidic Technologies for GBM
Tumor invasion and metastases involve changes in the extracellular microenvironment along with a myriad of genetic and physical intracellular changes. When compared to normal cells, cancer cells adapt quickly by changing their behavior and migration mechanisms in response to environment and extracellular stimuli. For example, conventional Boyden chamber or transwell assays have been routinely used to quantify cellular chemotaxis. However, these platforms do not facilitate the presentation of uniform chemotactic gradients to cells, and are challenging to use for live imaging of cell migration and response to chemotactic signaling.[33] There is hence a great need for cancer in vitro models that can realistically encapsulate many of the tumor microenvironmental stimuli and conditions in a precisely controlled and quantifiable platform. Microfluidic technology was developed in the early 1990s as a biological analysis tool capable of high resolution and specificity, and precise spatiotemporal control over gradients of soluble biological factors and cells.[34–36] Microfluidic devices consisting of a network of fluidic channels can be used to manipulate fluids on the order of 10−9–10−18 L, and cells down to single-cell precision.[36] They are typically fabricated using a technique called “soft photolithography” and using polydimethylsiloxane (PDMS), which is an optically transparent, air-permeable, and nontoxic polymer that enables the real-time, high-resolution optical imaging and quantification of biological entities and events.[37]
The adaptability of design and ease of production of microfluidic devices allow for their use in a variety of applications in glioma research. These include studies of migration, evaluation of biomarkers, cell sorting from tissue samples, and examination of therapeutic efficacy. Microfluidic devices are cost-effective and can be repeatedly fabricated with no loss of structural resolution.[38] Precise control over chip design, surface chemistry, flow actuation, and sample injection allow for microfluidic devices to manipulate liquids and gases within channels of dimensions between 10 and 100 μm in a high-throughput manner. Experimental goals largely influence device design, taking into account cell shape, size, deformability, and density.[39] Fluid dynamics through these devices can be regulated using micropumps and by creating hydrodynamic pressure differences by connecting inlet and outlet ports to fluid reservoirs located at differing heights.[40] Since microfluidic flow is laminar, wherein there is no mixing of adjacent fluid layers, molecular diffusion is the main method of mixing within these devices. Passive mixing can be facilitated by introducing barriers within the flow channel, splitting or combining channels, or introducing curvature into channels, whereby fluids will mix through molecular diffusion and continue traveling through the device.[39] To enhance mixing over a shorter length of time, convective mixing can be performed by the addition of bends, twists, and flattened areas to the channels, and depends on liquid properties such as surface tension, pH, non-Newtonian viscosity, and intersample variation.
Glioma tissue that is collected after surgical tumor resection is essential for the accurate histopathological and molecular determination of tumor stage and state. The discovery of novel anticancer strategies depends heavily on the ability to test drug efficacy on a representative population of patient-derived cells. Integrated microfluidic devices can be used to address this gap and to perform rapid and reproducible microscale measurements using extremely tiny amounts of cells (on the scale of a few thousand tumor cells) alongside the hallmark histopathological assessment of tumor status. Using microfluidic approaches, clinicians could potentially receive decisive information on tumor status from specific patient samples, and prescribe tailored therapy within a matter of days or weeks rather than months after detection. Four distinct applications of microfluidic technologies for glioma detection and treatment are discussed in the following sections. GBM-related microfluidic technologies discussed in this manuscript are also summarized in Table 2 for the reader’s convenience.
Table 2.
Microfluidic technologies for GBM.
| Device type | Method | Application | Ref. |
|---|---|---|---|
| CTC isolation | Tumor antigen-independent CTC enrichment | Preclinical research | [57] |
| CTC isolation | Anti-EGFR aptamer-based CTC enrichment | Preclinical research | [62] |
| CTC isolation | Anti-glial fibrillary acidic protein (GFAP) CTC enrichment | Preclinical research | [63] |
| Systems pathology | Microfluidic image cytometry for single-cell analysis | Preclinical research | [68] |
| Profiling circulating EVs | Target-specific magnetic nanoparticles combined with NMR | Preclinical research | [80,81] |
| Profiling circulating EVs | Immunoaffinity for EV isolation | Preclinical research | [78b] |
| Drug resistance monitoring | pH sensors for monitoring cell metabolism | Preclinical research | [85] |
| High-throughput drug screening | Spheroid culture system with multidrug administration and parallel testing of drug response | Preclinical research | [86] |
| Chemosensitivity testing | Parallel drug dosing in brain slice culture models | Preclinical research | [73] |
| Induction of drug resistance in culture | Microcompartments enabling exposure to ranges of drug and nutrients in culture | Preclinical research | [94] |
| Microfluidic BBB model | Endothelial and astrocyte co-culture within specialized microfluidic device | Preclinical research | [100,102,103] |
| Microfluidic BBB model | Endothelial and astrocyte co-culture within microfluidic device, with hollow fibers as artificial capillaries | Preclinical research | [101] |
| ECM regulation of invasion | Matrix stiffness and pore size are varied within microfluidic device | Preclinical research | [134] |
| Distance dependence on cell–cell interactions | Multiplex, quantitative protein assay within a microchip | Preclinical research | [136] |
4.1. Circulating Tumor Cell Isolation
The use of microfluidics for the detection of circulating tumor cells (CTCs) in patient biofluids may open the door for new, effective strategies in early cancer detection. CTCs are viable tumor cells that are shed into the blood or lymphatic vessels from the primary tumor and that circulate throughout the body spreading to new organ systems.[41] The isolation of these cells from the blood stream serves as a minimally invasive, multiple time-point liquid biopsy that can inform patient status without invasive measures. These cells are present in the blood in very low numbers (less than 100 cells mL−1 of whole blood), are heterogeneous, and are very difficult to isolate as pure populations, inspiring the design and fabrication of novel microfluidic platforms to improve the efficacy of CTC capture through both affinity-based and affinity-free technologies[42–45] (Figure 3). Affinity-based cell separation relies on the presence of unique biomarkers on the cancer cell surface that can be recognized by a magnetic or a polymer bead coupled to a high-affinity ligand to selectively separate bound cells of interest. Affinity-free cell separation relies on intrinsic physical differences between the cell type of interest and other cell types in a heterogeneous suspension.
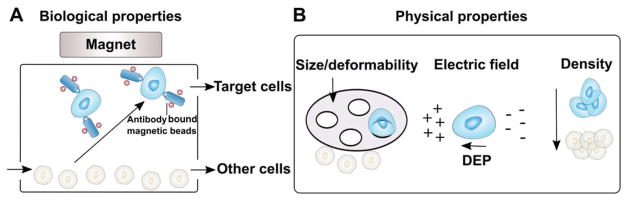
Figure 3.
A,B) Microfluidic cell separation schemes using either biological properties or physical properties of the desired cell types. When separating a population of cells out of a tissue sample, target cells can be enriched based on differences in the expression of cell surface markers, or via physical properties such as size, deformability, electric charge (dielectrophoresis, or DEP), and density.
Microfluidic devices have been used successfully thus far for cell-affinity chromatographic separation, cellular biophysics-based separation, and magnetically activated cell sorting across a variety of cancer cell types or for separating the different components of blood.[46,47] Cell characteristics such as differences in cell size can be used for microfluidic cancer cell sorting without other accommodations for biochemical characteristics.[47,48] Biophysical properties such as size work well when isolating cancer cells from blood due to the slightly larger size of CTCs when compared to other cells found in whole blood. Similar criteria can be incorporated in microfluidic platforms designed to screen out glioma cells to assess the invasive potential or to inform prognosis of the patient. With the ability to isolate pure populations of patient-derived cells, malignant glioma patients can benefit from informed treatment according to therapeutic sensitivities or gene analysis that can also be performed within a microfluidic chip. Cell-affinity chromatography selectively quarantines cancer cells from a heterogeneous cell population in suspension through high-affinity ligand binding to the cancer cells.[49] The first antibody-based microfluidic chromatography system captured cervical cancer cells through α6-integrin binding onto the surface of PDMS microchannels, and has been successfully adapted for other cancer cell lines for high cell capture and identification yield.[50] This technique was further adjusted for extracting CTCs from blood samples of patients, capable of accurately processing milliliters of whole blood in short periods of time using essential parameters such as flow velocity and shear force to influence efficiency of separation.[43,47] Cells thus sorted can subsequently be analyzed for mutations, secreted proteins, and drug resistance pathways to better inform treatment schemes or contribute to the design of novel therapeutic agents.
Affinity-based CTC separation technologies using the cell surface biomarkers such as the epithelial cell adhesion molecule (EpCAM) can be used to selectively enrich a subset of CTCs from blood circulation[43,44] (Figure 4). CTC separation devices using tumor-specific labeling technologies have been used successfully to separate CTCs from blood samples from pancreatic, prostate, breast, colon, melanoma, and lung cancer patients to provide prognostic information.[42,51] However, the innate heterogeneity of biomarker expression and the uncertainty introduced by epithelial-to-mesenchymal transitions of CTCs could limit the efficacy of affinity-based methods. Alternatively, affinity-free methods including those based on filtration,[52] acoustophoresis,[53] dielectrophoresis,[54] dean flow,[55] and vortex technology[56] exploit the intrinsic physical differences among cell types to deplete non-CTCs from blood and enrich cancer cells. Negative selection has been used to successfully enrich for GBM CTCs from patient blood samples, without the use of costly tumor-specific capture antibodies.[57] Difference in cell size is the most frequently used physical marker for enrichment. Most CTCs of epithelial origin have a size range between 15 and 25 μm, and are larger than red blood cells (RBCs, 6–9 μm), and the majority of white blood cells (WBCs, 8–14 μm).[58] However, CTCs of smaller sizes found in blood[59] and large WBCs such as monocytes that may have overlapping sizes with CTCs could limit the accuracy of label-free separation methods.[45,60]
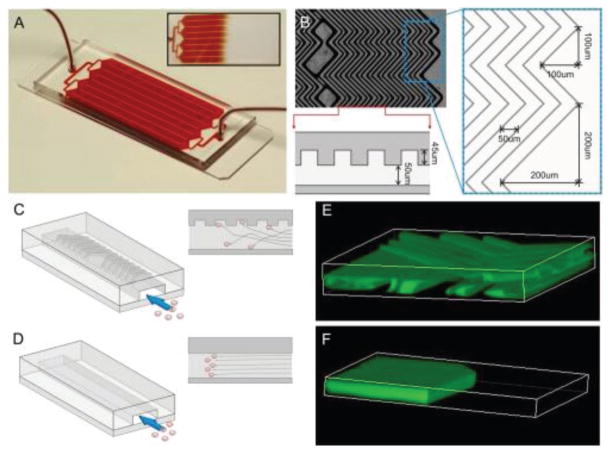
Figure 4.
A herringbone microfluidic device (HB-Chip) for the isolation of CTCs from blood samples, using EpCAM antibody-coated microposts. A) The HB-Chip consists of a microfluidic array of channels with a single inlet and exit. Inset illustrates the uniform blood flow through the device. B) A microscopic image of the grooved surface illustrates the asymmetry and periodicity of the herringbone grooves. Cartoon illustrating the cell–surface interactions in C) the HB-Chip and D) a traditional flat-walled microfluidic device. Flow visualization studies using two paired streams of the same viscosity (one stream is green and the other is clear) demonstrate E) the chaotic microvortices generated by the herringbone grooves, and the lack of mixing in F) traditional flat-walled devices. Reproduced with permission.[44] Copyright 2010, Proceedings of the National Academy of Sciences (PNAS).
When compared to other cancers, the levels of CTCs in GBM patients have not been extensively studied due to the lack of observed extracranial metastases and the unique brain microenvironment that limits the migration of glioma cells into circulation.[61] However, GBMs have been shown to shed CTCs into circulation in over one-third of patient samples, allowing an opportunity for the better development of CTC enriching strategies, and a less-invasive method of characterizing patient tumors and monitoring treatment efficacy.[57] The ability to selectively enrich primary human GBM cells from blood using aptamers for EGFR mutations has been demonstrated within affinity interaction-based cell sorting microfluidic devices.[62] However, CTC assays using patient-derived samples have not demonstrated the same level of success, likely due to the lack of expression of common cell surface biomarkers required to facilitate the separation through these devices.[62] Despite these drawbacks, the quantification of CTCs in GBM patients represents an opportunity for the development of diagnostic devices that can facilitate early detection and intervention.[63] Novel microfluidic devices for the isolation of GBM CTCs systematically remove RBCs and platelets via size-based exclusion, then align nucleated cells into single file arrangement using inertial flow dynamics to allow for the sorting of magnetically tagged leukocytes into a waste channel.[57] This scheme leaves the untagged CTCs free in solution for downstream processing or cell culture while excluding other contaminating cell types.[57] With improvements in the sensitivity of these platforms, GBM CTC analyses could aid in noninvasive disease monitoring during chemotherapy or RT treatment as well as help collect information related to the biology of GBM invasion via expression profiling of these circulating cells compared to bulk tumor tissue.
4.2. Molecular Diagnostics
Novel targets for early diagnosis and prevention are highly sought after to replace the rising costs associated with treatment of end-stage disease, and novel tumor biomarkers represent a critical area of research to improve both early detection and prognosis of malignant glioma. Monotherapies are often inadequate to address the multiple pathways driving tumor growth and spread of GBM; therefore, a personalized approach to treating GBM should target both patient-to-patient differences and clonal diversity within individual tumors. Currently, mutation analysis is performed as a prognostic measure, but histopathological grading of diffuse gliomas is still regarded as the best predictor of survival time in current clinical practice. Although histopathology is reproducible, inaccurate tumor classification using this technique can negatively influence treatment decisions.[64]
Patient therapeutic decisions rely on accurate histopathological grading; however, these methods yield little insight into the molecular pathways of glioma drug resistance or spread and cannot be solely used to guide novel targeted therapies. Genomic profiling has been used to further subcategorize infiltrative glioma by molecular subtype to better understand origin of the tumor cells and better predict response to targeted therapies.[65,66] Verhaak et al.[66] classified diffuse gliomas into four molecular subtypes: classic, mesenchymal, proneural, and neural, all based on similarities within genomic expression signatures. Mutations and gene expression of EGFR, neurofibromin 1, and isocitrate dehydrogenase 1 (IDH1) were used to define the classic, mesenchymal, and proneural subtypes respectively, with the neural subtype relying on expression of specific neuron markers.[66] The identified sets of genes that define the subclassification of gliomas provide insights into the design of targeted molecular therapies and can predict prognosis, but ultimately do not address patient-to-patient variability common to GBM.[67] Promising microfluidic technologies for diagnostics and subclassification are capable of characterizing heterogeneity at the single-cell level in primary brain tumor biopsies.[68] Microfluidic cytometry can detect chromosomal translocations, measure protein expression and phosphorylation, as well as quantify biomarker measurements using either serum or saliva, and more recently, tumor tissue.[34,68,69]
There are currently no identified biomarkers that can be used to detect or diagnose GBM. MRI and histopathology of biopsied tissue are the only accepted measures of confirmation of tumor presence, and are routinely used to inform diagnosis. Though information on invasive triggers and susceptibility to anticancer treatment is widely reported, there are few studies identifying molecular signatures of cancer that can be used for early detection and diagnosis of tumors. Different cancers have unique biomarkers that can help early detection, determine risk, and predict response to treatments. Brevegen is an example of a DNA-based assay that recognizes single nucleotide polymorphisms associated with breast cancer, and along with analysis of the tumor suppressor BRCA1 and 2 gene statuses can predict breast cancer risk in a patient.[70] However, these IVDs are not sufficient to make a diagnosis due to patient-to-patient variability. The discovery and inclusion of novel biomarkers in prospective clinical trials could therefore aid in the development of effective personalized therapies for cancer. There are common mutations across GBM, including EGFRvIII amplification, O6-methylguanine–DNA methyltransferase (MGMT) promoter methylation, and IDH1 mutations that are tested in clinical practice and that are used to inform therapeutic strategy.[71] However, these results are often not the primary line of confirmation of tumor status due to significant endemic intratumoral heterogeneity.[72]
Since glioma is a heterogeneous disease, the evaluation of cell-to-cell variations is important in understanding tumor progression and therapeutic response. The combination of cell separation technology with single-cell imaging and analysis techniques can tremendously improve screening and diagnosis of cancers.[73,74] Microfluidics present several advantages for these types of assays including minimal reagent usage, high-throughput screening, and single-cell analysis using lesser amounts of sample on the scale of 1–3000 cells alone.[68] Processing strategies such as microfluidic flow cytometry allows for single-cell measurements that confirm intratumoral heterogeneity of common tumor signaling pathways and can be validated by immunohistochemical processing of the same tissue samples. The evolution of single-cell analysis platforms, such as microfluidic devices for single-cell proteomics, can simultaneously measure the expression of multiple proteins or signaling molecules using small numbers of cells from patient tissue, as well as enable profiling of GBM patient serum or blood samples for potential biomarkers or indicators of drug efficacy.[68,75] These platforms can also accommodate high-resolution imaging for the real-time assessment of the effects of therapeutic strategies on tumor cells. Microfluidic platforms have been used to measure expression levels of EGFR, PTEN, phosphorylated protein kinase B (pAkt), and pS6 from single cells, and can be used to characterize the heterogeneity in brain tumor biopsies.[68] Live single-cell behavior studies suggest that the dynamic physical properties of tumor cells characterize normal or disease state, and thus should be included in the study of tumor biomarkers.[76] Cellular phenotypic biomarkers of interest include cell morphology, motility, contractibility, and cytoskeletal dynamics. This depth of data combined with the ability to perform bioinformatics analyses allows clinicians to stratify patients according to tumor progression and survival prospects, enabling the personalized medical treatment of these patients.[68]
Micro- and nanoscale vesicles (extracellular vesicles, EVs) shed from cancer cells into the peripheral blood can provide clinicians with valuable information about the genetic status and progression of the tumor. EVs actively secreted by healthy mammalian or tumor cells contain unique proteins and nucleic acids that are indicative of their cell of origin.[77] Recent efforts focused on the isolation and molecular analysis of tumor-secreted EVs have led to the successful development of microfluidic platforms for immunoaffinity-based isolation and molecular analysis of EVs with high yield and efficiency.[78] The size-based isolation of EVs from whole blood using a micro fluidic device containing a tunable filtration system has also been reported.[79] The combination of size- and immuno affinity-based separation has been implemented into microfluidic platforms for the rapid and sensitive detection of glioma-secreted EVs that can be used to inform and monitor therapeutic response by examining EV messenger RNA (mRNA) levels for biomarkers including MGMT.[75,80] Although real-time monitoring of GBM cell-shed EVs has diagnostic value, novel microfluidic devices that integrate exosome isolation with real-time RNA analysis would help identify markers predictive of TMZ resistance and epigenetic status of the tumor at large, thereby enhancing the prognostic value of these devices in the clinic.[75,80,81] The development and use of microfluidic platforms designed to facilitate the analysis of glioma-related mutations in circulating EVs also present an opportunity to obtain noninvasive diagnostic information. Two-step microfluidic devices that rapidly isolate EVs from circulation and analyze them using imaging-compatible or biosensor-equipped chambers enable the quantitative detection and protein expression analysis of liquid biopsy samples faster than histopathological staining of tissue biopsies[75] (Figure 5). The low cost and reproducibility of microfluidics for the processing of circulating EVs allow for easy integration into the clinical setting, and rapid detection and evaluation of individual-specific mutations and response to treatment.
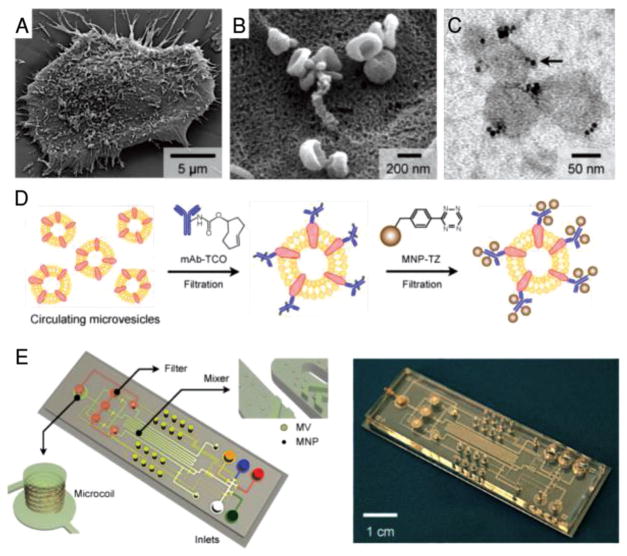
Figure 5.
Human glioblastoma cells produce abundant microvesicles (MVs), which can be analyzed by micronuclear magnetic resonance (μNMR). A) Scanning electron microscopic image of a primary human glioblastoma cell (GBM20/3) grown in culture, releasing abundant MVs. B) High-magnification image shows that many of the MVs on the cell surface assumed typical saucer-shaped characteristics of exosomes. C) Transmission electron microscopic image of MVs (≈80 nm) targeted with magnetic nanoparticles (MNPs) via CD63 antibody. The samples were purified by membrane filtration to collect small MVs. The MNPs appear as black dots (indicated by an arrow). D) Labeling procedure for extravesicular markers. The two-step BOND-2 assay configuration uses bioorthogonal amplification chemistry to maximize MNP binding onto target proteins on MVs (not drawn to scale). E) Microfluidic system for on-chip detection of circulating MVs. The system was designed to (i) allow MNP targeting of MVs, (ii) concentrate MNP-tagged MVs while removing unbound MNPs, and (iii) provide in-line μNMR detection. Reproduced with permission.[75] Copyright 2012, Nature Publishing Group.
4.3. Drug Efficacy Screening
Currently approved anticancer agents fail to facilitate the progression-free survival of GBM patients, and glioma recurrence is often encountered within 6–9 months of initial diagnosis.[82] There are currently no robust preclinical models for testing the susceptibility of GBM subtypes to anticancer agents. At the heart of the problem of treating GBM is the uncontrollable cellular invasion and drug resistance, which are thought to be linked to the presence of glioma stem cells (GSCs). The mechanisms underlying GSC invasive capacity are not well understood and are difficult to study in traditional cell culture models due to the diffuse single-cell migration that is characteristic of GBM.[83] It is well documented that cancer cells respond differently to drug treatments within 3D culture systems when compared to 2D cell culture substrates. Therefore, drug metabolism, penetration, and elimination can be more realistically appraised within 3D microfluidic systems.[84] In order to more successfully screen GBM cells for drug resistance in vitro, cell array platforms for the monitoring of cultured GBM cells in response to infused anticancer drugs have been developed for rapid drug screening. Patient cells or cell lines can be stably cultured within microfluidic chips, allowing for the tunable exposure of anticancer drugs, and the dynamic monitoring and dose-dependent response of cultured cells[85] (Figure 6). 3D models using poly(ethylene) glycol diacrylate hydrogels provide a relevant microenvironment and facilitate the evaluation of cell–cell or cell–matrix interactions as well as control over spatiotemporal and biological conditions in a high-throughput manner.[86,87] These “brain cancer chips” enable diffusion of drugs through cell or spheroid culture via microfluidic inlets, and can be adapted for parallel testing of multiple drugs to mimic combination therapies for GBM patients.[86]
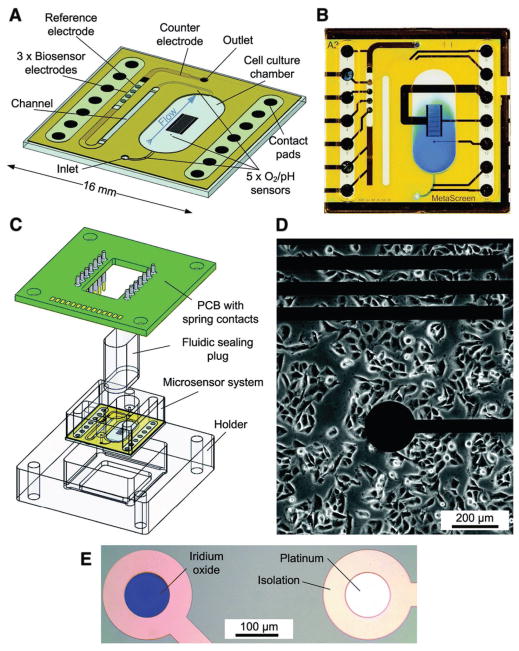
Figure 6.
Schematic of a microfluidic chip system for drug efficacy screening. A transparent glass chip allows for adherent cell culture on D) the chip surface, with A–C) a sealable chamber that constrains the height and confines the liquid volume over cells. Microfluidics allow for medium exchange within cell culture areas and transport of medium to downstream sensors. E) Five oxygen and pH sensors, within iridium oxide (pH) or thin-film platinum (O2) electrodes, are located within the inlet channel, inside cell culture areas, and in outlet areas. Three bio-sensor electrodes are located within the downstream outlet channels to measure glucose and lactate. Metabolic rates are acquired over several hours in stop/flow cycles with cell culture medium. As drug compounds are added to the medium, changes in cellular metabolism can be quantified. Reproduced with permission.[85] Copyright 2013, Royal Society of Chemistry.
The ability to perform preclinical studies in a small, cost-effective package is indispensable for the high-throughput and personalized assessment of drug efficacy. Novel microfluidic devices can maintain the TME and use interstitial perfusion for the real-time assessment of chemotherapeutic efficacy on needle biopsy-derived tumor samples.[88] Other devices incorporating slice cultures into multiwell platforms can be used to develop personalized therapies and evaluate the effects of different drugs and treatment conditions on tumor progression.[73] Compartmentalized or multichannel microfluidic devices incorporating gradient generators, regulators of fluid flow, and other tunable parameters enable the detailed characterization of the migratory behavior of tumor cells, help study tumor recurrence and metastasis, and facilitate the efficacy testing of glioma-specific anticancer agents.[89] In another example, microenvironment-specific chemokine gradients were constructed within a microfluidic device to study the anticancer drug efficacy and penetration into the tumor bulk.[90] The diffusion coefficient of common chemotherapeutics was examined by creating a chemokine gradient with surrounding blood vessels in tumors. 3D cell–cell and cell–matrix interactions were facilitated through a perfusion-culture system to study the 3D cytoarchitecture and cell differentiation.[91] Moving forward, the implementation of cluster and spheroid culture within soluble gradient microfluidic platforms could provide realistic information on tumor bulk penetration and response to fluid-driven chemokine and growth factor presentation, as well as influence on migratory cells in the periphery of the tumor[92] (Figure 7).
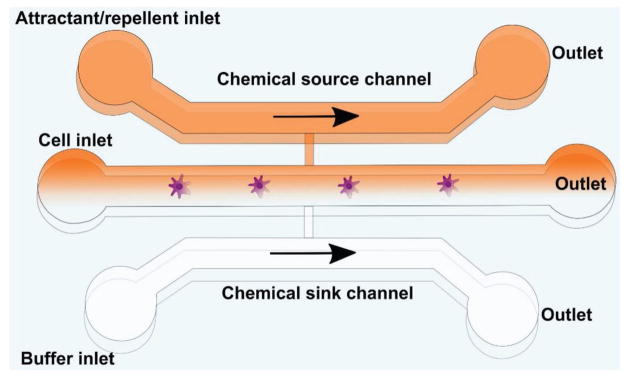
Figure 7.
General template of a microfluidic platform for chemotaxis studies. In the body, cells are exposed to extracellular chemoattractant gradients in 3D space and time, which influence cell behavior and migration. Simple microfluidic channels as depicted here can be used to present defined gradients of chemokines to cell types of interest, using either diffusion or laminar flow mixing to introduce chemokines into the outlets through a source channel in order to study glioma cell response to chemoattractants.
Drug resistance is among the most critical problems in GBM treatment, with GSCs developing resistance to chemotherapeutics within 48 h of treatment in vitro.[93] A major obstacle in drug discovery is the lack of understanding of drug resistance at the molecular level, and resistant cell lines can require several months of treatment and culture to ensure a robust resistant phenotype. Microfluidic devices can be used to rapidly induce drug resistance in GBM cell lines without the need for patient-derived tissues for the study of resistance development.[94,95] Previous studies found that induction of bacterial resistance to antibiotics was accelerated by a platform containing thousands of connected microscale chambers with the antibiotic administered in a concentration gradient, and the addition of fluid perfusion to a similar platform proved able to administer concentrations of doxorubicin to create drug-resistant breast cancer and multiple myeloma cancer cells.[96] These chips allow for the mass production of resistant cancer cells and the high-throughput analysis of causal mutations that will inform subsequent drug design efforts to better combat resistant cell populations, as well as provide guidance for clinicians on the efficacious use of specific drug combinations for specific cancer subtypes.[94] Resistant cell-producing microfluidic platforms combined with single-cell analysis techniques would provide additional insight into the molecular pathways of resistance to better inform drug development. In addition to creating therapy-resistant cell lines, the use of microfluidic platforms also confers an advantage of integrating 3D culture systems into the experiment for more realistic studies. By combining tumor spheroids in a 3D hydrogel scaffold and co-culturing of an endothelial monolayer in a microfluidic system, the antimetastatic drug responses were found to better inhibit epithelial-to-mesenchymal transition and a significant difference in drug response was observed between 2D and 3D models.[97] In a microfluidic study of non-small-cell lung cancer, multiple growth factors and inhibitors were assayed within 3D basement membrane extract matrix to reveal that invadopodia formation is largely related to EGFR signaling, which could be inhibited by the presence of matrix metalloproteinase inhibitors, prompting clinically relevant future directions.[98]
The unique aspects of microfluidic devices make them promising platforms to investigate potential disease treatments where site specificity is crucial to the delivery and pharmacokinetics of the tested drug. By culturing primary mammalian cells around an array of micropillars or in a complex thin 3D matrix layer, the maximal cell–cell interactions can be accessed for potential drug metabolite study. Multiple organs were interconnected through the individual compartments to simulate the body’s response to multidrug-resistant cancer treatment as a preclinical “brain-on-a-chip.”[99] In the same vein as drug interactions, microfluidic models of the BBB have been consistently improved upon in the recent years to accurately replicate preclinical modeling of drug delivery.[100–102] Previous techniques to replicate the BBB in vitro relied upon transwell assays to study barrier permeability. However, these platforms inaccurately represent microenvironmental characteristics. The incorporation of endothelial cells into microfluidic chambers result in the formation of tight junctions that more accurately replicate BBB permeation as detected by biochemical analysis.[102] Subsequent iterations of these microfluidic devices incorporate fluid shear stress and astrocyte/endothelial cell cultures to better inform preclinical testing methods for BBB drug permeability screening.[100,103] Microfluidic platforms presenting 3D environmental conditions, fluid flow, and oxygen gradients, and incorporating organ-level complexity can be used to rapidly screen treatment efficacy in targeting glioma.[104] With the complex microenvironment that accompanies GBM tumors, organ-on-a-chip devices present an expeditious way to discern the physiological effects of novel therapeutics on the BBB, on the tumor cells, and on the TME.
In addition to drug efficacy, successful drug delivery to malignant gliomas remains low due to a lack of efficient targeting agents and the presence of the BBB, which serves as a significant obstacle to drug transport into the brain. Nanoparticles help overcome the limitations of systemic chemotherapy owing to their ability to carry targeting agents, facilitate site-specific controlled release, and ability to pass through the BBB with relatively high efficiency.[105] Nanocarrier-encapsulated drugs can efficiently target brain tumors due to their ability to accumulate in the tumor vasculature, which is facilitated by the enhanced permeation and retention effect.[106] A range of magnetic, metal, and polymer nanoparticles can be synthesized for medical imaging and targeting of glioma. The translation of nanoparticle drug carriers into clinical trials is slow when compared to other small molecule drugs, due to the difficulties with reproducibly synthesizing nanoparticles that possess identical properties, with known pharmacokinetics and off-target effects, and in sufficient quantity for clinical use.[107] Microfluidic platforms can help address this gap by serving as model systems to quickly and efficiently synthesize drug-loaded nanoparticles, and to perform distribution and targeting assays when compared to conventional approaches. Microfluidic systems can mix reagents rapidly at a controlled temperature, and allow addition of reagents at exact times for the most efficient nanoparticle synthesis.[108] Precise conditions and timing allow for narrow size distributions, batch reproducibility, and efficient drug loading into nanoparticle constructs.[109] The advantages of microfluidic preparation of drug-loaded nanoparticles for cancer treatment also extend to rapid screening of the nanoparticles to identify the ideal formulation required to target-specific cancer cells.[110]
4.4. Microfluidic-Based Assays to Study GBM Progression
The measurement of mechanical phenotype characteristics such as modifications in cell structure, processing of micro-mechanical cues, or cell-influenced remodeling of the ECM is essential for understanding tumor pathology.[111] Cellular phenotypic changes are associated with genetic drivers of cell proliferation, tumor heterogeneity, and chemoresistance.[112] However, the inability to characterize tumor progression in in vitro models makes is difficult to quantify the relationship between physiological phenotype and tumor behavior.
Numerous in silico models have been formulated to capture information about individual cell behavior and predict gross tumor behavior, but these models lack the depth of information and sample set required to make meaningful predictions that can better inform patient treatment decisions.[113] Conventional migration assays are helpful to understand broad trends in cell populations, but are limited in their ability to capture visual detail at the single-cell level. Microfluidic devices can be designed to present microchannels that can simulate the individual cellular spread of GBM cells through interstitial spaces of brain tissue, thereby offering a precise window into understanding specific mechanisms of tumor progression.[89] Microfluidic cancer-on-a-chip models recreate cancer cell micro-environments in a simplified manner, overcoming limitations for reconstructing in vitro cancer systems with precision control over all variables.[114] Cancer-on-a-chip models are capable of high-resolution real-time imaging, quantitative measurement of cellular response, and precisely mimicking complex 3D organ-level microarchitectures to help quantify tumor cell invasion and migration,[115] intravasation and extravasation,[116] and angiogenesis[117] (Figure 8).
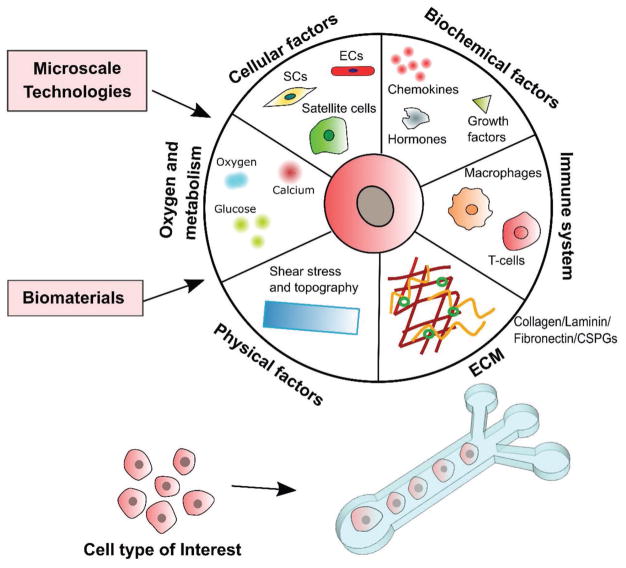
Figure 8.
Tunable microfluidics for the multifactorial in vitro study of brain tumors. Microfluidic platforms better mimic the in vivo brain tumor microenvironment by simultaneously integrating multiple factors that influence cancer progression. The adaptation of microchannels and microwells allows for the mixing or separation of multiple cell types to study effects of cell–cell interactions on tumor behavior at the individual or bulk levels. Biochemical factors can be perfused through the TME in realistic gradients, allowing for the evaluation of tumor response to external cues for growth, proliferation, or migration. Oxygen and glucose metabolism can be closely monitored and altered to allow for studies within hypoxic or normoxic conditions, representing different areas of the tumor or different states in tumor pathology. Physical factors and ECM deposition can be controlled to display tumor behavior under different conditions of shear and topography or in response to changes in extracellular environment.
4.4.1. ECM Signaling
Unlike other malignant tumors, GBM tumors do not kill by metastatic spreading to secondary organ sites. They are lethal due to their aggressive spread through brain tissue until the patient eventually succumbs to intracranial pressure and edema. Invasive ability is predominantly regulated by distinctive biological interactions with the ECM, and dynamic ECM remodeling by tumor cells in turn facilitates invasion.[118] Many in vitro platforms including transwell assays and wound healing assays can be used to measure cell motility.[119] However, these assays do not accurately evaluate cell invasion, which includes the migration of cells through tissue barriers. Invasion requires cell movement through 3D matrix, accompanied by the modification of the cell’s shape and the surrounding environment to accommodate the moving cell body.[120] A common method of tumor cell invasion, mesenchymal migration, involves the formation of strong cellular focal adhesions with the ECM in order for the cell to migrate through a tissue.[121] GBM cells migrate individually in a traction-dependent manner, concentrating integrins to generate strong adhesion forces at the focal contacts on ECM.[16] Beyond solely responding to ECM cues, glioma cells also regularly remodel local ECM for optimal growth and invasion through the secretion of proteases, correlating with poor patient prognosis.[12,15] Gliomas dynamically disrupt normal tissue composition in a variety of ways, including haphazard vascular proliferation, generation of hypoxic and necrotic areas, and the remodeling of ECM components.[122]
Healthy brain ECM is a highly regulated mixture of glia, signaling molecules, and scaffolding molecules, comprising between 10% and 20% of total brain volume and allowing for neuronal support and homeostasis.[123] The ECM components are divided among three components: basement membrane, perineuronal nets, and neuronal interstitial matrix.[124] Basement membrane contains type IV collagen, fibronectin and laminin secreted by local endothelial cells, existing between cerebral blood vessels and the rest of brain connective tissue, and aids in the formation of the BBB.[124] Perineuronal nets are mostly composed of CSPGs, tenascin R, and hyaluronan that aggregate around neuronal cell bodies to protect synapses and enhance neuroplasticity.[125] The remaining interstitial matrix consists of CSPGs, tenascin R, and other fibrous proteins.[124] Around GBM, the ECM contains an increased amount of fibrillary collagens compared to healthy brain ECM, particularly in the basement membrane around blood vessels.[126] Increased rigidity is also observed around white matter tracts, where GBM cells are known to invade preferentially as they concurrently degrade other unwanted ECM components.[17,127]
Invading cells secrete MMPs to remodel local ECM during mesenchymal migration.[15,16] Targeting these proteolytic enzymes involved in migration of GBM cells has been proposed as an anti-invasion therapy, but any attempts to treat GBM with MMP inhibitors have resulted in severe side effects including musculoskeletal pain and inflammation.[128] Therapeutic strategies that target MMP-dependent migration have largely been ineffective at treating cancer metastasis. The use of microfluidic devices to study these and other physical interactions with the ECM can provide useful information on how tumor cells take advantage of physical confinement to change shape and spread.[129,130] Further enhancement of these platforms involves the use of hydrogels with MMP-degradable sites to study realistic ECM remodeling by tumor cells in vitro.[131]
Evidence suggesting that tumors are often more rigid than surrounding normal tissue, and that stiffening of tissue stimulates tumorigenesis and invasion, further highlights the important role of the ECM in tumor progression.[17,132] 3D hydrogel matrices of different stiffness gradients can be generated using microfluidics to study glioma cell behavior and to evaluate the relationship between cell migration and channel width or pore size.[129,133] Hydrogel composites composed of tumor-relevant ECM components offer an attractive means of representing the extracellular TME in microfluidic devices. The mechanical strength of hydrogel scaffolds can be tuned to mimic the stiffness of the tumor ECM by controlling the crosslinking density of the backbone polymer. Other physical properties of the hydrogel matrix such as pore size, and retention and binding of bioactive molecules can be similarly controlled to influence fluid flow and cellular mobility within the scaffold. Integrating collagen or other tumor-associated ECM components within the hydrogel along with precisely controlled stiffness, pore size, concentration of ECM components, pH, and temperature helps create finely controlled, physiologically relevant environments to study tumor cell behavior.[134]
4.4.2. Cell–Cell Interactions
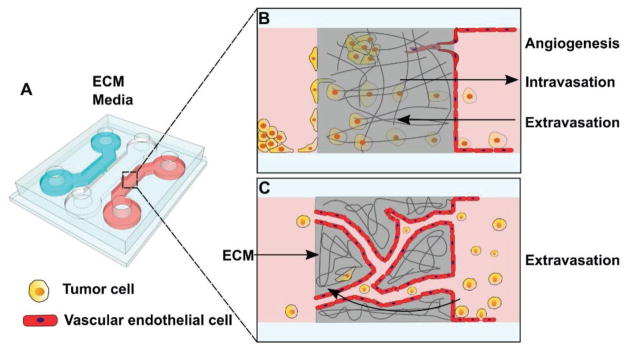
Microfluidic devices designed for observation of cell–cell interactions can yield insight into how GBM cells self-organize, and provide information on how pairwise cell interactions can influence the cellular architecture at the tumor level.[135] Micropatterned surfaces can also be incorporated for studies of cell–cell interactions and soluble factor signaling, using antibody arrays to immobilize secreted, cytoplasmic, or membrane proteins that might play a role in GBM cell migration.[136] Microfluidics can be adapted to examine any tumor–endothelial cell interactions, which are critical to cancer metastasis and contribute to the population of CTCs in the bloodstream.[137] GBM cell movement through endothelial barrier walls represents an integral part of the invasion cascade and is essentially impossible to study in 2D cell culture systems. Devices accommodating the manipulation of stromal cells have been expanded upon in recent years to better understand external contributors to glioma invasion through brain parenchyma, many taking after the classic transwell invasion chamber assays with modifications to evaluate components of stromal cell involvement such as gap junctions between cells.[138] Specifically designed devices for the visualization of fibrosarcoma cell actions on endothelial monolayers display the intravasation process on the cellular scale and allow for modulation of permeability of the endothelial barrier in response to soluble biochemical factors[139] (Figure 9). Another widely used platform involves two different but adjacent hydrogel channels, with breast cancer cells within one channel and the stromal cells within the other, allowing for the observation of cancer-associated fibroblasts and their influence on cancer invasion and progression.[140] A similar arrangement can be adapted to evaluate intravasation or extravasation by GBM cells into or out from artificial lymph tissue blood vessels (Figure 10). Tumor cell movement through endothelial barrier walls represents an integral part of the invasion cascade and is essentially impossible to study in 2D cell culture systems. The exact mechanisms by which tumor cells enter circulation are unknown, prompting the development of novel in vitro platforms that can aid in bridging this gap in knowledge.
Figure 9.
A) Representation of a microfluidic platform for the study of cancer cell intravasation or extravasation through blood-vessel walls. 3D schematic of a microfluidic device, cross sections demonstrating the usage for the study of tumor cell migration through endothelial cell culture and 3D ECM components. B) Tumor cells moving through a seeded endothelial cell culture wall provide information on how tumor cells move through blood vessel walls to migrate, and how tumor cells can influence angiogenic budding. C) Endothelial cells grown with ECM can self-assemble into networks of microvessels, yielding an environment in which to study how tumor cells can circulate through the network and invade. Adapted with permission.[139] Copyright 2012, National Academy of Sciences.
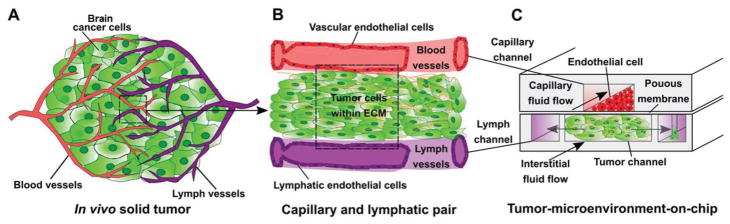
Figure 10.
A–C) Schematic of considerations for the design of “organ-on-a-chip” microfluidic platforms. The in vivo TME is complex, but aspects such as vasculature, ECM components, and lymph fluid flow can be incorporated into an in vitro microfluidic scheme for the study of cancer cell behavior. In vitro models of vascular flow are imperative to understanding how cancer cells move through endothelial junctions into blood vessels or into lymph nodes. Adapted with permission.[145] Copyright 2011, National Academy of Sciences. Adapted with permission.[146] Copyright 2015, Elsevier.
4.4.3. Vascular Flow and Angiogenesis
The incorporation of fluid flow is a crucial parameter in a realistic in vitro investigation of glioma. Within and around the periphery of the tumor, cells are influenced by both the interstitial fluid flow and the vascular fluid flow. Tumors typically demonstrate elevated interstitial fluid flow compared to healthy brain tissue, due to the permeability of newly synthesized tumor blood vessels combined with faulty drainage of excess fluid.[141] Interstitial fluid flow through brain parenchyma has been shown to be involved in cell migration around brain tumors due to the local increase in interstitial pressure and subsequent high interstitial pressure gradient at the tumor margins, potentially guiding tumor cell migration outward.[142] This increase in fluid flux combined with the breakdown of healthy vasculature around brain tumors contributes to an elevation in fluid shear stress on cells throughout the tumor.[143]
GBM is characterized by microvascular hyperplasia, a hallmark believed to be linked to the regions of pseudopalisading cells at the tumor periphery that in turn stimulates cell migration.[144] Microfluidic devices can evaluate vascular perfusion by exercising control over flow rates and the accompanying shear stresses (Figure 10).[145,146] Interstitial flow has also been shown to affect cancer cells differently depending on their location within or around the tumor boundaries.[145,147] As GBM tumors grow, local interstitial pressure builds up around the tumor bulk and creates a gradient of increased interstitial pressure between the tumor and healthy tissue.[148] Cancer cells have demonstrated a preference for directionally migrating with interstitial flow as opposed to against it.[149] Perfusable vascular networks created via microchannels using either collagen or stromal cells can also replicate important aspects of the BBB and the TME and, as medium is perfused through the system, can result in in vivo like vasculature.[146,150] In addition to creating realistic in vivo conditions, perfusable microvascular networks within microfluidic devices allow for the real-time imaging of tumor cell interactions with fluid flow in order to understand how tumor cells use vascular flow to migrate through tissues.[151] Cerebrospinal fluid (CSF) fluid flow patterns maintain the neuronal microenvironment and homeostasis in healthy brain tissue, but perturbation around brain tumors leads to disrupted flow patterns and heightened interstitial pressures. Altered interstitial flow around GBM is thought to modulate glioma cell invasion pathways, and is a crucial parameter of study in a brain-mimetic in vitro model of glioma.
The finely tuned control innate to microfluidic platforms allows for the study of cell response to environmental conditions, specifically with respect to conditions that drive invasive behavior such as hypoxia. Normoxic conditions within the brain can range from 0.5% to 8% O2 depending on region, but intra- and peritumoral tissue oxygenation is consistently less oxygenated (1.25% O2 for intratumoral and 2.5% O2 for peritumoral tissues).[152] Low-oxygen conditions within cancer arise as a result of the uncontrolled proliferation of tumor cells and the subsequent exhaustion of nutrient and oxygen supplies from healthy vasculature, which in turn triggers the release of angiogenic factors from hypoxic tumor cells.[153] Angiogenic factors secreted from tumor cells create haphazardly developed new blood vessels, contributing to vascular leakage and non-laminar blood flow through the area.[154] These tumor-created blood vessels also possess loose endothelial cell junctions that do not carry blood efficiently, leading to the creation of additional hypoxic areas.[155] A loss of oxygen tension contributes to the enhanced activity of hypoxia-inducible factors (HIFs), which are degraded by cells during normal oxygen conditions.[156] The HIF transcription factors regulate expression of many invasion-relevant gene targets involved in angiogenesis, survival, and migration.[153,157] Increased HIF activity is directly correlated with the malignant tumor cell phenotype, making the hypoxic environment an important driver of tumor cell invasion to study.[158] These conditions can be replicated in vitro using mixed cell populations representing the tumor bulk, the external stroma, and the endothelial cells lining blood vessels. Microfluidic platforms offer finely tuned control over the cell–cell interactions that contribute to angiogenic budding, and influence over conditions that mediate oxygen tension.[159]
Conventional gas variable incubators are commonly used for the study of oxygen levels on cell behavior using premixed ratios of oxygen, nitrogen, and carbon dioxide to control the environment. Although valuable information can be gleaned from those specific environmental contexts within a cell culture system, these incubators do not allow for the generation of oxygen gradients that exist in the in vivo TME.[160] One improvement over these cell culture platforms is the implementation of gas microchannels into the cell culture plates that could generate a hypoxic environment, but these devices were limited to use within 2D cell cultures.[161] In order to culture cells in a 3D microenvironment under hypoxic conditions, microfluidic platforms, containing gas channels positioned above and below the cell chamber, can be used to introduce the desired gas conditions adjacent to cell chambers to control oxygen concentration.[162] Hybrid PDMS and polycarbonate films have also been used in these devices, wherein the gas impermeable polycarbonate film is patterned above cell channels to reduce diffusion of oxygen from the atmosphere while PDMS enables oxygen diffusion from gas channels.[163] Biomaterials can also be used to study hypoxia more easily within microfluidic platforms, for example, hypoxia-inducing gelatin and ferulic acid scaffolds derived from oxygen consumption control oxygen levels and form oxygen gradients within the hydrogels.[164] Though these hydrogels are not physiologically relevant and do not mimic the brain ECM, they can serve as reliable microenvironments to study hypoxia-related cell behavior. More work with microfluidic platforms is necessary to improve the utility of these devices in translational research, and to provide opportunities for real-time studies of cell behavior under controlled oxygen conditions when compared to conventional assay methods.[160]
4.4.4. Immune Cell Interactions
Immunotherapy is a recent approach that has shown promise in early clinical trials for GBM treatment, but immune-based treatments face several obstacles in targeting brain tumors. In order for the therapeutic agent to reach the tumor, the agent has to be able to target the tumor regardless of irregular disruption of normal BBB and despite the complex presence of both immune-activating and immune-dampening cells.[165] It has been demonstrated that inflammation in the brain can cause the BBB to allow immune cells’ access to brain tissue, but the timing and mechanisms of immune response to tumor formation are not well understood, and more information is necessary for the successful design of immunotherapies for glioblastoma. In vitro cell culture experiments show that patient-derived immune cells can be adapted to recognize and destroy autologous glioma cells, but the ability to target these activated immune cells to specific tumor cells among healthy tissue is challenging to investigate in 2D culture platforms.[166]
The cellular interplay in the body between tumor cells, immune cells, stromal cells, and ECM components composes a diverse ecosystem, each contributing to and affecting disease progression. While cancer cells remodel local ECM to create the ideal TME, these changes create inflammation and hypoxic conditions which in turn influence inter- and intracellular signaling.[167] GBM tumors are poorly immunogenic relative to other solid tumors, which is potentially due to its localization within brain tissue, which harbors an immunosuppressive environment.[168] GBM functionally suppresses the native immune system by producing suppressive cytokines, inhibiting T-cell proliferation, and inducing tissue hypoxia, all of which dampen the body’s antitumor immune response.[169] Malignant glioma simultaneously recruits microglia to the TME, leading to chronic inflammation that promotes ECM deposition, angiogenesis, and tissue remodeling.[170] Understanding the context of inflammation and the immune system’s contributions to tumor growth and spread is essential to the development of novel targeted therapies.
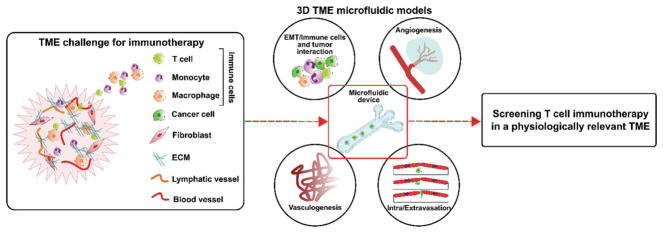
Models of immune system–cancer interactions have been designed to advance cancer research, with the ability to study cancer–immune cell communication in general or in specific environmental contexts.[171] Preclinical testing currently relies on animal models before human studies, which are expensive and time consuming, and lack direct application to human physiology.[172] Recent studies incorporating microfluidics into the study of immune–tumor cell interactions have revealed the need for studying several cell types at the same time to evaluate the physiologically relevant interplay during the metastatic cascade, as the immune system dynamic changes over the course of tumor progression[172,173] (Figure 11). Conventional in vitro culture methods do not accommodate for the spatiotemporal dynamics of immune cells targeting cancer cells within solid tissue as 3D microfluidic models can. A 3D microfluidic device designed to test whether T cells can overcome both physical and metabolic barriers to target cancer cells within a 3D culture incorporates both oxygen and chemokine gradients to observe their effects on engineered T-cell function under immunosuppressive conditions.[174] Several organ-on-a-chip models designed to study the inflammatory responses could serve as templates to further study the role of immune and inflammatory cells and molecules on glioma.[175] Microfluidic assays can prove effective in these contexts and facilitate the high-throughput assessment of combination therapies based on molecular profiling of GBM patient biopsy. These platforms can also be used to evaluate the efficacy of potential immunotherapies and their ability to bolster the adoptive immune response to cancer. These types of assays will be necessary in the future to address the current gaps in standard-of-care treatment and to give the patient the best possible chance at long-term progression-free and overall survival.
Figure 11.
The adaptability of microfluidics to address challenges for in vitro studies of brain cancer immunotherapies.[176] The complexity of the brain tumor microenvironment (TME) represents a large challenge for developing and studying models of immunotherapies with the presence of the BBB, unique immune cell interactions and recruitment, and the rampant angiogenesis associated with glioma growth. Major limitations for immunotherapy against brain tumors include the lymph and blood vessels restraining T-cell interactions with the targeted cancer cells, which severely lessens the ability of T cells to both destroy the glioma cells and limits T-cell proliferation in the area of the tumor. 3D microfluidic models that address these key parts of the TME can aid in tailoring immunotherapies for both successful delivery and targeting of cancer cells without adverse effects for healthy tissue.
5. Future Perspectives
GBM remains one of the most malignant primary central nervous system (CNS) tumors in humans, with no effective treatments or cure. Fifty years of research and observations have not yet yielded any significant advancements to patient survival after diagnosis with GBM. The aggressive cellular invasion that is characteristic to GBM tumors has so far thwarted all surgical and therapeutic interventions, with patients often left with tumors cells within brain tissue which repopulate into novel neoplasms within months of surgery. These tumors are continuously diagnosed in the late stages due to a lack of early-stage symptoms and early-stage diagnostic tools or indicators of risk. Investigational drug studies are often found lacking due to the cellular and genetic heterogeneity that plagues drug efficacy and the presence of the BBB that limits delivery to the already precious affected structures. Though GBM continues to be predominantly diagnosed by histopathological analyses, evidence from patient-derived tumor tissues continues to demonstrate the immense cellular, genomic, proteomic, and extracellular heterogeneity, which require complex multiscale parallel analysis methods. As monotherapies are derailed by resistance, combination therapies for GBM are consistently being pushed forward into new clinical trials. Patients receiving either antiangiogenic drugs or small molecule inhibitors in combination with chemotherapy have reported longer progression-free survival compared to chemotherapy alone,[177] but the current area of interest for combination therapy revolves around immunotherapy approaches such as checkpoint inhibitors, gene therapies, and vaccine therapies (Table 3).
Table 3.
Emerging GBM treatments under clinical trial investigation.
| Intervention | Mechanism | Clinical trials/ongoing studies | Advantages | Disadvantages |
|---|---|---|---|---|
| Ipilimumab (Yervoy) | Monoclonal antibody targeting CTLA-4, enhances immune response |
NCT02017717 NCT02311920 |
Potential to be non-cancer-type specific | Potential severe immunogenic adverse events Success is patient specific depending on the tumor’s mutations and antigen profile |
| Nivolumab (Opdivo) | Monoclonal antibody targeting PD-1, enhances immune response |
NCT02017717 NCT02311920 NCT02667587 NCT02617589 NCT02658981 NCT02335918 NCT02327078 |
Potential to be non-cancer-type specific Amenable for use in combination approaches |
Side effects include severe inflammation of organs |
| Heat shock protein peptide complex-96 vaccine | Vaccine with peptides that bind proteins involved in antigen- presenting pathway, creating an antitumor immune response |
NCT02722512 NCT01814813 NCT00905060 NCT00293423 |
Efficient production, storage, and distribution Specific targeting of tumor cells |
Only a small population of GBM patients would be eligible, due to the requirement of patients having undergone complete surgical resection before treatment The vaccine is created from the patient’s tumor tissue, leading to a potential delay in administration of treatment |
| Anti-EGFRvIII CAR-T- cell therapy | Adoptive cellular therapy, CAR-T cells recognize EGFRvIII tumor antigens and activate an anti- tumor response |
NCT02664363 NCT02209376 NCT03283631 NCT01454596 |
Ability to target-specific cells, without antigen presentation via MHC 1 | Potential severe immunogenic adverse effects Only useful against known antigens Time of production can be limiting |
| Natural killer (NK) cell therapy | Autologous NK cell infusions use patient-derived, activated lymphocytes to create an antitumor immune response |
NCT00005813 NCT00003067 |
Targets the tumor cells without needing to identify specific antigens on the tumor cell Short expansion time |
Expansion of NK cells on a large scale is difficult Risk of graft-versus-host disease, even with well- matched donors Nonspecific killing of cells |
| Rindopepimut (Rintega) | Peptide vaccine targeting the EGFRvIII deletion mutation |
NCT01480479 NCT01498328 NCT00458601 |
Target-specific tumor cells | May only affect a portion of tumor cells with the targeted mutation |
| Dendritic cell (DC) vaccines | Autologous DCs are treated with tumor lysates and reintroduced to stimulate the immune system |
NCT01808820 NCT00323115 NCT02010606 NCT01957956 NCT01204684 NCT03014804 NCT00639639 |
Large-scale isolation and expansion of DCs are feasible, taken from peripheral blood of the patient Tumor–antigen pulsed DCs can stimulate immune effector cells potently and rapidly |
Cost of treatment can be prohibitive due to complex vaccine design No consensus yet on preferred target molecules |
Along with new treatment schemes, innovative research combining biological assays with tissue engineered constructs is consistently producing new information about GBM cell behavior, migration, and genetics. Molecular information is integral to developing clinical treatments to gain more targeted and tailored therapies that can be used for precision medicine. There is no clear single genetic profile for GBM or GSCs, prompting the need for developing personalized therapeutic interventional strategies. Integrative new tools into existing detection and treatment paradigms can help enhance diagnosis and treatment efficacy every step along the way, beginning with biomarker discovery and ending with rapid multiscale analysis of patient-derived cells. This review has illustrated the use of microfluidics in all aspects of GBM research, notably for the development of cell-sorting techniques for single-cell analysis, the recapitulation of the TME in vitro, and drug development and testing. The design of implantable drug delivery devices using additive manufacturing techniques and 3D printing could present novel opportunities for the localized delivery of chemotherapeutic agents.[178] Using these techniques, hydrogel-based devices with micrometer-scale features can be fabricated and finely tuned to facilitate the “on demand” release of a regulated dose of chemotherapeutic agent into the local tumor area after implantation.[178] Implantable drug delivering devices allow for adjustment of treatment and dosing depending on the tumor growth and without additional surgeries, while avoiding many of the adverse events associated with systemic chemotherapy infusion such as peripheral organ toxicity.[178]
There are a few clinically used IVDs for glioma diagnosis and molecular testing, representing areas of translational research with immense room for growth and development. Since research into microfluidic technology began, microfluidic devices have become immensely popular within the research space but less integrated into clinical application. Acquiring and testing biopsied tissue samples remains the gold standard, due to operational challenges with the design of successful IVDs including high individual cost, additional equipment needed to interpret the results of the IVD, or inability to test for multiple genetic or molecular markers at once. Currently approved IVDs successfully aid treatment decision-making by detecting relevant biomarkers in a variety of cancers. However, the same level of accuracy has not yet been achieved for GBM due to the high degree of heterogeneity and the lack of identified biomarkers for GBM. A thorough characterization of prognostic and predictive GBM biomarkers is needed before a clinical IVD can be implemented for early GBM diagnosis. The highly invasive surgical resection of GBM tissue samples for analyses is an additional limitation, which can be overcome by the development of high CTC yielding microfluidic enrichment devices that can take advantage of patient blood samples.
Development of microfluidic platforms for rapid detection of tumor-related biomarkers and processing of tumor tissue can improve time to diagnosis and treatment planning, and pave the way for more glioma-targeting FDA-approved IVDs that can help researchers and clinicians parse through the cellular and genetic heterogeneity, which makes these tumors difficult to treat. These diagnostics tools and methods are particularly important with respect to GBM due to the unique molecular profile that varies from patient to patient, and this information is necessary for the application of precision medicine for the patient’s best chance at survival. Microfluidic platforms can be tailored to specific environmental conditions or incorporate features for a system’s level analysis, paving the way for a massive expansion of our current cancer biology knowledge base and, ideally, increase the speed at which we develop and evaluate potential therapeutic options for malignant disease like GBM. The continued research and development of targeted therapies, combined with advancements in microfluidic-based advanced screening and detection tools, hold immense potential for increasing survival and prognosis of GBM patients.
Acknowledgments
L.K. acknowledges funding from the National Institutes of Health (NIH 1R01NS099596-01A1), and the National Science Foundation (NSF) Engineering Research Center for Cell Manufacturing Technologies (CMaT; EEC-1648035). M.L. acknowledges funding from the Achievement Rewards for College Scientists Foundation, Inc. (ARCS Foundation). L.M. and W.Z. acknowledge funding from the National Institutes of Health (NIH R21GM104528, UL1TR002378), and the National Science Foundation (Award No. 1150042).
Biographies

Meghan Logun is currently pursuing her Ph.D. in neuroscience at the University of Georgia under the supervision of Dr. Lohitash Karumbaiah. She received her B.S. in biology from Emory University in 2014, and her M.S. in animal science from the University of Georgia in 2016. Her current research focuses on understanding mechanisms behind extracellular matrix-mediated glioma cell invasion.

Leidong Mao is an Associate Professor in the College of Engineering at the University of Georgia, USA. He received his B.S. degree in materials science from Fudan University, Shanghai, China, in 2001. He received his M.S. and Ph.D. degrees in electrical engineering from Yale University, New Haven, CT, in 2002 and 2007, respectively. His current research interests are microfluidic technologies for circulating-tumor-cell and single-cell research.

Lohitash Karumbaiah is an Assistant Professor of regenerative medicine in the Regenerative Bioscience Center, University of Georgia. Research in his laboratory is focused on the development of novel brain tissue mimicking glycomaterials to study stem cell behavior in invasive glioma. He is using these tools to develop personalized medicine approaches and technologies that can help rapidly isolate invasive subpopulations of glioma cells and improve therapeutic screening efficacy.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Meghan Logun, Regenerative Bioscience Center, ADS Complex, University of Georgia, 425 River Road, Athens, GA 30602-2771, USA.
Wujun Zhao, Department of Chemistry, University of Georgia, Athens, GA 30602-2771, USA.
Dr. Leidong Mao, School of Electrical and Computer Engineering, College of Engineering, University of Georgia, Athens, GA 30602-2771, USA
Dr. Lohitash Karumbaiah, Regenerative Bioscience Center, ADS Complex, University of Georgia, 425 River Road, Athens, GA 30602-2771, USA
References
- 1.Butowski NA. Continuum. 2015;21:301. doi: 10.1212/01.CON.0000464171.50638.fa. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. Neuro-Oncology. 2014;16:iv1. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. Acta Neuropathol. 2007;114:97. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barchana M, Margaliot M, Liphshitz I. Asian Pac J Cancer Prev. 2012;13:5857. doi: 10.7314/apjcp.2012.13.11.5857. [DOI] [PubMed] [Google Scholar]
- 5.a) Alvarez JI, Katayama T, Prat A. Glia. 2013;61:1939. doi: 10.1002/glia.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Daneman R. Ann Neurol. 2012;72:648. doi: 10.1002/ana.23648. [DOI] [PubMed] [Google Scholar]
- 6.Castillo M, Smith JK, Kwock L. AJNR Am J Neuroradiol. 2000;21:1645. [PMC free article] [PubMed] [Google Scholar]
- 7.Biernat W, Tohma Y, Yonekawa Y, Kleihues P, Ohgaki H. Acta Neuropathol. 1997;94:303. doi: 10.1007/s004010050711. [DOI] [PubMed] [Google Scholar]
- 8.Cuddapah VA, Robel S, Watkins S, Sontheimer H. Nat Rev Neurosci. 2014;15:455. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foty RA. Future Oncol. 2013;9:1121. doi: 10.2217/fon.13.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Eichler AF, Chung E, Kodack DP, Loeffler JS, Fukumura D, Jain RK. Nat Rev Clin Oncol. 2011;8:344. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn KA, Adkins CE, Roberts A, Thorsheim HR, Gaasch JA, Huang S, Palmieri D, Steeg PS, Smith QR. Clin Cancer Res. 2010;16:5664. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Cox TR, Erler JT. Dis Models Mech. 2011;4:165. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Timar J, Lapis K, Dudas J, Sebestyen A, Kopper L, Kovalszky I. Semin Cancer Biol. 2002;12:173. doi: 10.1016/S1044-579X(02)00021-4. [DOI] [PubMed] [Google Scholar]; c) Insua-Rodriguez J, Oskarsson T. Adv Drug Delivery Rev. 2016;97:41. doi: 10.1016/j.addr.2015.12.017. [DOI] [PubMed] [Google Scholar]; d) Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. Science. 1997;275:1943. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]; e) Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]; f) Frederick L, Wang XY, Eley G, James CD. Cancer Res. 2000;60:1383. [PubMed] [Google Scholar]; g) Sharma SV, Bell DW, Settleman J, Haber DA. Nat Rev Cancer. 2007;7:169. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 12.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Hsu NC, Nien PY, Yokoyama KK, Chu PY, Hou MF. Biochem Biophys Res Commun. 2013;441:514. doi: 10.1016/j.bbrc.2013.10.093. [DOI] [PubMed] [Google Scholar]; b) Svendsen A, Verhoeff JJ, Immervoll H, Brogger JC, Kmiecik J, Poli A, Netland IA, Prestegarden L, Planaguma J, Torsvik A, Kjersem AB, Sakariassen PO, Heggdal JI, Van Furth WR, Bjerkvig R, Lund-Johansen M, Enger PO, Felsberg J, Brons NH, Tronstad KJ, Waha A, Chekenya M. Acta Neuropathol. 2011;122:495. doi: 10.1007/s00401-011-0867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Chabottaux V, Noel A. Clin Exp Metastasis. 2007;24:647. doi: 10.1007/s10585-007-9113-7. [DOI] [PubMed] [Google Scholar]; b) Place AE, Jin Huh S, Polyak K. Breast Cancer Res. 2011;13:227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao JS. Nat Rev Cancer. 2003;3:489. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 16.Zhong J, Paul A, Kellie SJ, O’Neill GM. J Oncol. 2010;2010:430142. doi: 10.1155/2010/430142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulrich TA, de Juan Pardo EM, Kumar S. Cancer Res. 2009;69:4167. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Kauppila S, Stenback F, Risteli J, Jukkola A, Risteli L. J Pathol. 1998;186:262. doi: 10.1002/(SICI)1096-9896(1998110)186:3<262::AID-PATH191>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; b) Zaman MH, Kamm RD, Matsudaira P, Lauffenburger DA. Biophys J. 2005;89:1389. doi: 10.1529/biophysj.105.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lopez JI, Kang I, You WK, McDonald DM, Weaver VM. Integr Biol. 2011;3:910. doi: 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Brat DJ, Castellano-Sanchez A, Kaur B, Van Meir EG. Adv Anat Pathol. 2002;9:24. doi: 10.1097/00125480-200201000-00004. [DOI] [PubMed] [Google Scholar]; b) Brat DJ, Castellano-Sanchez AA, Hunter SB, Pecot M, Cohen C, Hammond EH, Devi SN, Kaur B, Van Meir EG. Cancer Res. 2004;64:920. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- 20.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Cancer Res. 2006;66:7843. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 21.Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Brain Pathol. 2005;15:297. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker FG, 2nd, Davis RL, Chang SM, Prados MD. Cancer. 1996;77:1161. doi: 10.1002/(sici)1097-0142(19960315)77:6<1161::aid-cncr24>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 23.a) Appelbaum PS. N Engl J Med. 2007;357:1834. doi: 10.1056/NEJMcp074045. [DOI] [PubMed] [Google Scholar]; b) De Bonis P, Fiorentino A, Anile C, Balducci M, Pompucci A, Chiesa S, Sica G, Lama G, Maira G, Mangiola A. Clin Neurol Neurosurg. 2013;115:883. doi: 10.1016/j.clineuro.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Eckhouse S, Lewison G, Sullivan R. Mol Oncol. 2008;2:20. doi: 10.1016/j.molonc.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris Z, Whiteley WN, Longstreth WT, Jr, Weber F, Lee YC, Tsushima Y, Alphs H, Ladd SC, Warlow C, Wardlaw JM, Al-Shahi Salman R. BMJ. 2009;339:b3016. doi: 10.1136/bmj.b3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orringer D, Lau D, Khatri S, Zamora-Berridi GJ, Zhang K, Wu C, Chaudhary N, Sagher O. J Neurosurg. 2012;117:851. doi: 10.3171/2012.8.JNS12234. [DOI] [PubMed] [Google Scholar]
- 27.Mansfield E, O’Leary TJ, Gutman SI. J Mol Diagn. 2005;7:2. doi: 10.1016/S1525-1578(10)60002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuzery AK, Levin J, Chan MM, Chan DW. Clin Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown P. Future Oncol. 2016;12:451. doi: 10.2217/fon.15.311. [DOI] [PubMed] [Google Scholar]
- 30.Koch WH. Nat Rev Drug Discovery. 2004;3:749. doi: 10.1038/nrd1496. [DOI] [PubMed] [Google Scholar]
- 31.Sun Z, Fu X, Zhang L, Yang X, Liu F, Hu G. Anticancer Res. 2004;24:1159. [PubMed] [Google Scholar]
- 32.a) Carpelan-Holmstrom M, Haglund C, Lundin J, Jarvinen H, Roberts P. Eur J Cancer. 1996;32A:1156. doi: 10.1016/0959-8049(96)00030-5. [DOI] [PubMed] [Google Scholar]; b) Louhimo J, Carpelan-Holmstrom M, Alfthan H, Stenman UH, Jarvinen HJ, Haglund C. Int J Cancer. 2002;101:545. doi: 10.1002/ijc.90009. [DOI] [PubMed] [Google Scholar]
- 33.Zigmond SH. J Cell Biol. 1977;75:606. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 35.Hong JW, Quake SR. Nat Biotechnol. 2003;21:1179. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- 36.Whitesides GM. Nature. 2006;442:368. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 37.a) Xia YN, Whitesides GM. Angew Chem, Int Ed. 1998;37:550. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]; b) Xia YN, Whitesides GM. Annu Rev Mater Sci. 1998;28:153. [Google Scholar]
- 38.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 39.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 40.Shevkoplyas SS, Yoshida T, Munn LL, Bitensky MW. Anal Chem. 2005;77:933. doi: 10.1021/ac049037i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riquet M, Rivera C, Gibault L, Pricopi C, Mordant P, Badia A, Arame A, Le Pimpec Barthes F. Rev Pneumol Clin. 2014;70:16. doi: 10.1016/j.pneumo.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR. ACS Nano. 2015;9:62. doi: 10.1021/nn5056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, FloydJr FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Natl Acad Sci USA. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alix-Panabieres C, Pantel K. Nat Rev Cancer. 2014;14:623. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 46.a) Chang WC, Lee LP, Liepmann D. Lab Chip. 2005;5:64. doi: 10.1039/b400455h. [DOI] [PubMed] [Google Scholar]; b) Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Proc Natl Acad Sci USA. 2006;103:14779. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. Nat Biotechnol. 1999;17:1109. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]; d) Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 47.Zhao W, Cheng R, Miller JR, Mao L. Adv Funct Mater. 2016;26:3916. doi: 10.1002/adfm.201504178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HT, Lee W, Amini H, Di Carlo D. Anal Bioanal Chem. 2010;397:3249. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Didar TF, Tabrizian M. Lab Chip. 2010;10:3043. doi: 10.1039/c0lc00130a. [DOI] [PubMed] [Google Scholar]
- 50.a) Du Z, Colls N, Cheng KH, Vaughn MW, Gollahon L. Biosens Bioelectron. 2006;21:1991. doi: 10.1016/j.bios.2005.09.005. [DOI] [PubMed] [Google Scholar]; b) Wankhede SP, Du Z, Berg JM, Vaughn MW, Dallas T, Cheng KH, Gollahon L. Biotechnol Prog. 2006;22:1426. doi: 10.1021/bp060127d. [DOI] [PubMed] [Google Scholar]
- 51.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA. Science. 2015;349:1351. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Clin Cancer Res. 2010;16:5011. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang PH, Truica CI, Drabick JJ, El-Deiry WS, Dao M, Suresh S, Huang TJ. Proc Natl Acad Sci USA. 2015;112:4970. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.a) Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Electrophoresis. 2009;30:1388. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang SB, Wu MH, Lin YH, Hsieh CH, Yang CL, Lin HC, Tseng CP, Lee GB. Lab Chip. 2013;13:1371. doi: 10.1039/c3lc41256c. [DOI] [PubMed] [Google Scholar]; c) Moon HS, Kwon K, Kim SI, Han H, Sohn J, Lee S, Jung HI. Lab Chip. 2011;11:1118. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 55.a) Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA, Lim CT. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Warkiani ME, Guan G, Luan KB, Lee WC, Bhagat AA, Chaudhuri PK, Tan DS, Lim WT, Lee SC, Chen PC, Lim CT, Han J. Lab Chip. 2014;14:128. doi: 10.1039/c3lc50617g. [DOI] [PubMed] [Google Scholar]; c) Warkiani ME, Khoo BL, Wu L, Tay AK, Bhagat AA, Han J, Lim CT. Nat Protoc. 2016;11:134. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 56.a) Che J, Yu V, Dhar M, Renier C, Matsumoto M, Heirich K, Garon EB, Goldman J, Rao J, Sledge GW, Pegram MD, Sheth S, Jeffrey SS, Kulkarni RP, Sollier E, Di Carlo D. Oncotarget. 2016;7:12748. doi: 10.18632/oncotarget.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dhar M, Pao E, Renier C, Go DE, Che J, Montoya R, Conrad R, Matsumoto M, Heirich K, Triboulet M, Rao J, Jeffrey SS, Garon EB, Goldman J, Rao NP, Kulkarni R, Sollier-Christen E, Di Carlo D. Sci Rep. 2016;6:35474. doi: 10.1038/srep35474. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sollier E, Go DE, Che J, Gossett DR, O’Byrne S, Weaver WM, Kummer N, Rettig M, Goldman J, Nickols N, McCloskey S, Kulkarni RP, Di Carlo D. Lab Chip. 2014;14:63. doi: 10.1039/c3lc50689d. [DOI] [PubMed] [Google Scholar]
- 57.Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, Chi AS, Wakimoto H, Rothenberg SM, Sequist LV, Kapur R, Shah K, Iafrate AJ, Curry WT, Loeffler JS, Batchelor TT, Louis DN, Toner M, Maheswaran S, Haber DA. Cancer Discovery. 2014;4:1299. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Li P, Huang PH, Xie Y, Mai JD, Wang L, Nguyen NT, Huang TJ. Lab Chip. 2014;14:626. doi: 10.1039/c3lc90136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marrinucci D, Bethel K, Lazar D, Fisher J, Huynh E, Clark P, Bruce R, Nieva J, Kuhn P. J Oncol. 2010;2010:861341. doi: 10.1155/2010/861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Y, Skelley AM, Merdek KD, Sprott KM, Jiang C, Pierceall WE, Lin J, Stocum M, Carney WP, Smirnov DA. J Mol Diagn. 2013;15:149. doi: 10.1016/j.jmoldx.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 61.a) Fonkem E, Lun M, Wong ET. J Clin Oncol. 2011;29:4594. doi: 10.1200/JCO.2011.39.0187. [DOI] [PubMed] [Google Scholar]; b) Kalokhe G, Grimm SA, Chandler JP, Helenowski I, Rademaker A, Raizer JJ. J Neuro-Oncol. 2012;107:21. doi: 10.1007/s11060-011-0731-1. [DOI] [PubMed] [Google Scholar]; c) Piccirilli M, Brunetto GM, Rocchi G, Giangaspero F, Salvati M. Tumori. 2008;94:40. doi: 10.1177/030089160809400109. [DOI] [PubMed] [Google Scholar]
- 62.Wan Y, Tan J, Asghar W, Kim YT, Liu Y, Iqbal SM. J Phys Chem B. 2011;115:13891. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 63.Muller C, Holtschmidt J, Auer M, Heitzer E, Lamszus K, Schulte A, Matschke J, Langer-Freitag S, Gasch C, Stoupiec M, Mauermann O, Peine S, Glatzel M, Speicher MR, Geigl JB, Westphal M, Pantel K, Riethdorf S. Sci Transl Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 64.Prayson RA, Agamanolis DP, Cohen ML, Estes ML, Kleinschmidt-DeMasters BK, Abdul-Karim F, McClure SP, Sebek BA, Vinay R. J Neurol Sci. 2000;175:33. doi: 10.1016/s0022-510x(00)00274-4. [DOI] [PubMed] [Google Scholar]
- 65.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Proc Natl Acad Sci USA. 2005;102:5814. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Cancer Cell. 2010;17:98. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Proc Natl Acad Sci USA. 2004;101:9309. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR. Cancer Res. 2010;70:6128. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.a) Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O’Neill R, Padua RA, Xu L, Taketa D, Shirer AE, Beer S, Yee AX, Voehringer DW, Felsher DW. Nat Med. 2009;15:566. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jokerst JV, Raamanathan A, Christodoulides N, Floriano PN, Pollard AA, Simmons GW, Wong J, Gage C, Furmaga WB, Redding SW, McDevitt JT. Biosens Bioelectron. 2009;24:3622. doi: 10.1016/j.bios.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) VanDijken J, Kaigala GV, Lauzon J, Atrazhev A, Adamia S, Taylor BJ, Reiman T, Belch AR, Backhouse CJ, Pilarski LM. J Mol Diagn. 2007;9:358. doi: 10.2353/jmoldx.2007.060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.a) Campeau PM, Foulkes WD, Tischkowitz MD. Hum Genet. 2008;124:31. doi: 10.1007/s00439-008-0529-1. [DOI] [PubMed] [Google Scholar]; b) Easton DF. Breast Cancer Res. 1999;1:14. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cancer Genome Atlas Research Network. Nature. 2008;455:1061. [Google Scholar]
- 72.a) Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Nat Rev Cancer. 2015;15:302. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nakamura M, Watanabe T, Yonekawa Y, Kleihues P, Ohgaki H. Carcinogenesis. 2001;22:1715. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]; c) Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. N Engl J Med. 2009;360:765. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang TC, Mikheev AM, Huynh W, Monnat RJ, Rostomily RC, Folch A. Lab Chip. 2014;14:4540. doi: 10.1039/c4lc00642a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.a) Jonas O, Landry HM, Fuller JE, Santini JT, Jr, Baselga J, Tepper RI, Cima MJ, Langer R. Sci Transl Med. 2015;7:284ra57. doi: 10.1126/scitranslmed.3010564. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Otto O, Rosendahl P, Golfier S, Mietke A, Herbig M, Jacobi A, Topfner N, Herold C, Klaue D, Girardo S, Winzi M, Fischer-Friedrich E, Guck J. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:1861. doi: 10.1109/EMBC.2015.7318744. [DOI] [PubMed] [Google Scholar]
- 75.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Nat Med. 2012;18:1835. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darling EM, Di Carlo D. Annu Rev Biomed Eng. 2015;17:35. doi: 10.1146/annurev-bioeng-071114-040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.a) Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10:1470. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Thery C, Zitvogel L, Amigorena S. Nat Rev Immunol. 2002;2:569. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 78.a) Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Lab Chip. 2015;15:2388. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]; b) Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Lab Chip. 2010;10:505. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Lab Chip. 2012;12:5202. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 80.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Nat Biotechnol. 2014;32:490. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorlia T, Stupp R, Brandes AA, Rampling RR, Fumoleau P, Dittrich C, Campone MM, Twelves CC, Raymond E, Hegi ME, Lacombe D, van den Bent MJ. Eur J Cancer. 2012;48:1176. doi: 10.1016/j.ejca.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 83.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Cell. 2009;138:822. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 84.a) Bauer M, Su G, Beebe DJ, Friedl A. Integr Biol. 2010;2:371. doi: 10.1039/c0ib00001a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Nat Methods. 2007;4:855. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]; c) Pickl M, Ries CH. Oncogene. 2009;28:461. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- 85.Weltin A, Slotwinski K, Kieninger J, Moser I, Jobst G, Wego M, Ehret R, Urban GA. Lab Chip. 2014;14:138. doi: 10.1039/c3lc50759a. [DOI] [PubMed] [Google Scholar]
- 86.Fan Y, Nguyen DT, Akay Y, Xu F, Akay M. Sci Rep. 2016;6:25062. doi: 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.a) Gao B, Wang L, Han S, Pingguan-Murphy B, Zhang X, Xu F. Crit Rev Biotechnol. 2016;36:619. doi: 10.3109/07388551.2014.1002381. [DOI] [PubMed] [Google Scholar]; b) Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Anal Chem. 2007;79:8557. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]; c) Ohno K, Tachikawa K, Manz A. Electrophoresis. 2008;29:4443. doi: 10.1002/elps.200800121. [DOI] [PubMed] [Google Scholar]
- 88.a) Altiok S, Mezzadra H, Jagannath S, Tsottles N, Rudek MA, Abdallah N, Berman D, Forastiere A, Gibson MK. Int J Oncol. 2010;36:19. [PMC free article] [PubMed] [Google Scholar]; b) Holton AB, Sinatra FL, Kreahling J, Conway AJ, Landis DA, Altiok S. PLoS One. 2017;12:e0169797. doi: 10.1371/journal.pone.0169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Y, Agrawal B, Clark PA, Williams JC, Kuo JS. J Visualized Exp. 2011;58:e3297. doi: 10.3791/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. Lab Chip. 2009;9:545. doi: 10.1039/b810571e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toh YC, Zhang C, Zhang J, Khong YM, Chang S, Samper VD, van Noort D, Hutmacher DW, Yu HR. Lab Chip. 2007;7:302. doi: 10.1039/b614872g. [DOI] [PubMed] [Google Scholar]
- 92.McCutcheon S, Unachukwu U, Thakur A, Majeska R, Redenti S, Vazquez M. Cell Adhes Migr. 2017;11:1. doi: 10.1080/19336918.2015.1131388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, De Maria R. Cell Death Differ. 2006;13:1238. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 94.Han J, Jun Y, Kim SH, Hoang HH, Jung Y, Kim S, Kim J, Austin RH, Lee S, Park S. Proc Natl Acad Sci USA. 2016;113:14283. doi: 10.1073/pnas.1614898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK, Pourmand N, Austin RH. Science. 2011;333:1764. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 96.a) Wu A, Loutherback K, Lambert G, Estevez-Salmeron L, Tlsty TD, Austin RH, Sturm JC. Proc Natl Acad Sci USA. 2013;110:16103. doi: 10.1073/pnas.1314385110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wu A, Zhang Q, Lambert G, Khin Z, Gatenby RA, Kim HJ, Pourmand N, Bussey K, Davies PC, Sturm JC, Austin RH. Proc Natl Acad Sci USA. 2015;112:10467. doi: 10.1073/pnas.1512396112. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang XL, Tian LL, Huang J, Ma JH, Zhang H, Feng QJ, Chen WF. Nanfang Yike Daxue Xuebao. 2011;31:1974. [Google Scholar]
- 97.Aref AR, Huang RYJ, Yu WM, Chua KN, Sun W, Tu TY, Bai J, Sim WJ, Zervantonakis IK, Thiery JP, Kamm RD. Integr Biol. 2013;5:381. doi: 10.1039/c2ib20209c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang S, Li E, Gao Y, Wang Y, Guo Z, He J, Zhang J, Gao Z, Wang Q. PLoS One. 2013;8:e56448. doi: 10.1371/journal.pone.0056448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tatosian DA, Shuler ML. Biotechnol Bioeng. 2009;103:187. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- 100.Booth R, Kim H. Lab Chip. 2012;12:1784. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 101.Cucullo L, Marchi N, Hossain M, Janigro D. J Cereb Blood Flow Metab. 2011;31:767. doi: 10.1038/jcbfm.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prabhakarpandian B, Shen MC, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K. Lab Chip. 2013;13:1093. doi: 10.1039/c2lc41208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang YI, Abaci HE, Shuler ML. Biotechnol Bioeng. 2017;114:184. doi: 10.1002/bit.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.a) Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Lab Chip. 2012;12:2156. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]; b) Mahto SK, Yoon TH, Rhee SW. Biomicrofluidics. 2010;4 doi: 10.1063/1.3486610. pii: 034111. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kwapiszewski R, Skolimowski M, Ziolkowska K, Jedrych E, Chudy M, Dybko A, Brzozka Z. Biomed Microdevices. 2011;13:431. doi: 10.1007/s10544-011-9511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hobbs SK, Monsky WL, Yuan F, Roberts WG, Griffith L, Torchilin VP, Jain RK. Proc Natl Acad Sci USA. 1998;95:4607. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maeda H, Nakamura H, Fang J. Adv Drug Delivery Rev. 2013;65:71. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 107.a) Chou LY, Ming K, Chan WC. Chem Soc Rev. 2011;40:233. doi: 10.1039/c0cs00003e. [DOI] [PubMed] [Google Scholar]; b) Murday JS, Siegel RW, Stein J, Wright JF. Nanomedicine. 2009;5:251. doi: 10.1016/j.nano.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 108.deMello J, deMello A. Lab Chip. 2004;4:11N. doi: 10.1039/b403638g. [DOI] [PubMed] [Google Scholar]
- 109.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG. J Am Chem Soc. 2012;134:6948. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 110.Valencia PM, Pridgen EM, Rhee M, Langer R, Farokhzad OC, Karnik R. ACS Nano. 2013;7:10671. doi: 10.1021/nn403370e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar S, Weaver VM. Cancer Metastasis Rev. 2009;28:113. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.a) Brown DV, Filiz G, Daniel PM, Hollande F, Dworkin S, Amiridis S, Kountouri N, Ng W, Morokoff AP, Mantamadiotis T. PLoS One. 2017;12:e0172791. doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. Cell Cycle. 2009;8:3274. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zeng AL, Yan W, Liu YW, Wang Z, Hu Q, Nie E, Zhou X, Li R, Wang XF, Jiang T, You YP. Oncogene. 2017;36:5369. doi: 10.1038/onc.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.a) Anderson AR. Math Med Biol. 2005;22:163. doi: 10.1093/imammb/dqi005. [DOI] [PubMed] [Google Scholar]; b) DiMilla PA, Barbee K, Lauffenburger DA. Biophys J. 1991;60:15. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ferreira SC, Jr, Martins ML, Vilela MJ. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2002;65:021907. doi: 10.1103/PhysRevE.65.021907. [DOI] [PubMed] [Google Scholar]
- 114.a) Portillo-Lara R, Annabi N. Lab Chip. 2016;16:4063. doi: 10.1039/c6lc00718j. [DOI] [PubMed] [Google Scholar]; b) Young EWK. Integr Biol. 2013;5:1096. doi: 10.1039/c3ib40076j. [DOI] [PubMed] [Google Scholar]
- 115.a) Aijian AP, Garrell RL. JALA. 2015;20:283. doi: 10.1177/2211068214562002. [DOI] [PubMed] [Google Scholar]; b) Bender BF, Aijian AP, Garrell RL. Lab Chip. 2016;16:1505. doi: 10.1039/c5lc01569c. [DOI] [PubMed] [Google Scholar]
- 116.Chaw KC, Manimaran M, Tay EH, Swaminathan S. Lab Chip. 2007;7:1041. doi: 10.1039/b707399m. [DOI] [PubMed] [Google Scholar]
- 117.a) Nguyen DHT, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA, Chen CS. Proc Natl Acad Sci USA. 2013;110:6712. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim C, Kasuya J, Jeon J, Chung S, Kamm RD. Lab Chip. 2015;15:301. doi: 10.1039/c4lc00866a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.a) Friedl P, Alexander S. Cell. 2011;147:992. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]; b) Friedl P, Wolf K. Nat Rev Cancer. 2003;3:362. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 119.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschlager M, Dolznig H. Mutat Res. 2013;752:10. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 120.Friedl P, Wolf K. J Cell Biol. 2010;188:11. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Friedl P, Zanker KS, Brocker EB. Microsc Res Tech. 1998;43:369. doi: 10.1002/(SICI)1097-0029(19981201)43:5<369::AID-JEMT3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 122.Oliver L, Olivier C, Marhuenda FB, Campone M, Vallette FM. Curr Mol Pharmacol. 2009;2:263. doi: 10.2174/1874467210902030263. [DOI] [PubMed] [Google Scholar]
- 123.a) Nicholson C, Sykova E. Trends Neurosci. 1998;21:207. doi: 10.1016/s0166-2236(98)01261-2. [DOI] [PubMed] [Google Scholar]; b) Ou KL, Hosseinkhani H. Int J Mol Sci. 2014;15:17938. doi: 10.3390/ijms151017938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW. Nat Rev Neurosci. 2013;14:722. doi: 10.1038/nrn3550. [DOI] [PubMed] [Google Scholar]
- 125.a) Deepa SS, Carulli D, Galtrey C, Rhodes K, Fukuda J, Mikami T, Sugahara K, Fawcett JW. J Biol Chem. 2006;281:17789. doi: 10.1074/jbc.M600544200. [DOI] [PubMed] [Google Scholar]; b) Hensch TK. Nat Rev Neurosci. 2005;6:877. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]; c) Koppe G, Bruckner G, Hartig W, Delpech B, Bigl V. Histochem J. 1997;29:11. doi: 10.1023/a:1026408716522. [DOI] [PubMed] [Google Scholar]
- 126.Huijbers IJ, Iravani M, Popov S, Robertson D, Al-Sarraj S, Jones C, Isacke CM. PLoS One. 2010;5:e9808. doi: 10.1371/journal.pone.0009808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller K, Chinzei K, Orssengo G, Bednarz P. J Biomech. 2000;33:1369. doi: 10.1016/s0021-9290(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 128.Coussens LM, Fingleton B, Matrisian LM. Science. 2002;295:2387. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 129.a) Irimia D, Toner M. Integr Biol. 2009;1:506. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. PLoS One. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mak M, Reinhart-King CA, Erickson D. Lab Chip. 2013;13:340. doi: 10.1039/c2lc41117b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.a) Kievit FM, Florczyk SJ, Leung MC, Wang K, Wu JD, Silber JR, Ellenbogen RG, Lee JS, Zhang M. Biomaterials. 2014;35:9137. doi: 10.1016/j.biomaterials.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sarkar S, Nuttall RK, Liu S, Edwards DR, Yong VW. Cancer Res. 2006;66:11771. doi: 10.1158/0008-5472.CAN-05-0470. [DOI] [PubMed] [Google Scholar]
- 132.Lam WA, Cao L, Umesh V, Keung AJ, Sen S, Kumar S. Mol Cancer. 2010;9:35. doi: 10.1186/1476-4598-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heuze ML, Collin O, Terriac E, Lennon-Dumenil AM, Piel M. Methods Mol Biol. 2011;769:415. doi: 10.1007/978-1-61779-207-6_28. [DOI] [PubMed] [Google Scholar]
- 134.a) Pathak A, Kumar S. Proc Natl Acad Sci USA. 2012;109:10334. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Peyton SR, Kalcioglu ZI, Cohen JC, Runkle AP, Van Vliet KJ, Lauffenburger DA, Griffith LG. Biotechnol Bioeng. 2011;108:1181. doi: 10.1002/bit.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang YL, Motte S, Kaufman LJ. Biomaterials. 2010;31:5678. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 135.Kravchenko-Balasha N, Wang J, Remacle F, Levine RD, Heath JR. Proc Natl Acad Sci USA. 2014;111:6521. doi: 10.1073/pnas.1404462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J, Tham D, Wei W, Shin YS, Ma C, Ahmad H, Shi Q, Yu J, Levine RD, Heath JR. Nano Lett. 2012;12:6101. doi: 10.1021/nl302748q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.a) Nguyen DX, Bos PD, Massague J. Nat Rev Cancer. 2009;9:274. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]; b) Steeg PS. Nat Med. 2006;12:895. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 138.Hong X, Sin WC, Harris AL, Naus CC. Oncotarget. 2015;6:15566. doi: 10.18632/oncotarget.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Proc Natl Acad Sci USA. 2012;109:13515. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.a) Liu T, Lin B, Qin J. Lab Chip. 2010;10:1671. doi: 10.1039/c000022a. [DOI] [PubMed] [Google Scholar]; b) Ma H, Liu T, Qin J, Lin B. Electrophoresis. 2010;31:1599. doi: 10.1002/elps.200900776. [DOI] [PubMed] [Google Scholar]; c) Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Integr Biol. 2011;3:439. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.a) Liu LJ, Brown SL, Ewing JR, Ala BD, Schneider KM, Schlesinger M. PLoS One. 2016;11:e0140892. doi: 10.1371/journal.pone.0140892. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Qazi H, Shi ZD, Tarbell JM. PLoS One. 2011;6:e20348. doi: 10.1371/journal.pone.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Heldin CH, Rubin K, Pietras K, Ostman A. Nat Rev Cancer. 2004;4:806. doi: 10.1038/nrc1456. [DOI] [PubMed] [Google Scholar]
- 143.a) Huynh GH, Deen DF, Szoka FC., Jr J Controlled Release. 2006;110:236. doi: 10.1016/j.jconrel.2005.09.053. [DOI] [PubMed] [Google Scholar]; b) Jain RK, Tong RT, Munn LL. Cancer Res. 2007;67:2729. doi: 10.1158/0008-5472.CAN-06-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rong Y, Durden DL, Van Meir EG, Brat DJ. J Neuropathol Exp Neurol. 2006;65:529. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 145.Polacheck WJ, Charest JL, Kamm RD. Proc Natl Acad Sci USA. 2011;108:11115. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Duinen V, Trietsch SJ, Joore J, Vulto P, Hankemeier T. Curr Opin Biotechnol. 2015;35:118. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 147.a) Buchanan C, Rylander MN. Biotechnol Bioeng. 2013;110:2063. doi: 10.1002/bit.24944. [DOI] [PubMed] [Google Scholar]; b) Munson JM, Bellamkonda RV, Swartz MA. Cancer Res. 2013;73:1536. doi: 10.1158/0008-5472.CAN-12-2838. [DOI] [PubMed] [Google Scholar]
- 148.a) Gritsenko PG, Ilina O, Friedl P. J Pathol. 2012;226:185. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]; b) Munson JM, Shieh AC. Cancer Manage Res. 2014;6:317. doi: 10.2147/CMAR.S65444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Haessler U, Teo JC, Foretay D, Renaud P, Swartz MA. Integr Biol. 2012;4:401. doi: 10.1039/c1ib00128k. [DOI] [PubMed] [Google Scholar]
- 150.a) Hsu YH, Moya ML, Hughes CC, George SC, Lee AP. Lab Chip. 2013;13:2990. doi: 10.1039/c3lc50424g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD. Proc Natl Acad Sci USA. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen MB, Whisler JA, Frose J, Yu C, Shin Y, Kamm RD. Nat Protoc. 2017;12:865. doi: 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.a) Beppu T, Kamada K, Yoshida Y, Arai H, Ogasawara K, Ogawa A. J Neuro-Oncol. 2002;58:47. doi: 10.1023/a:1015832726054. [DOI] [PubMed] [Google Scholar]; b) Erecinska M, Silver IA. Respir Physiol. 2001;128:263. doi: 10.1016/s0034-5687(01)00306-1. [DOI] [PubMed] [Google Scholar]
- 153.Semenza GL. Trends Pharmacol Sci. 2012;33:207. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Carmeliet P, Jain RK. Nature. 2000;407:249. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 155.a) Dvorak HF, Nagy JA, Feng D, Brown LF, Dvorak AM. Curr Top Microbiol Immunol. 1999;237:97. doi: 10.1007/978-3-642-59953-8_6. [DOI] [PubMed] [Google Scholar]; b) Hashizume H, Baluk P, Morikawa S, McLean JW, Thurston G, Roberge S, Jain RK, McDonald DM. Am J Pathol. 2000;156:1363. doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang BH, Semenza GL, Bauer C, Marti HH. Am J Physiol. 1996;271:C1172. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 157.a) Liao D, Johnson RS. Cancer Metastasis Rev. 2007;26:281. doi: 10.1007/s10555-007-9066-y. [DOI] [PubMed] [Google Scholar]; b) Shah T, Krishnamachary B, Wildes F, Mironchik Y, Kakkad SM, Jacob D, Artemov D, Bhujwalla ZM. Oncotarget. 2015;6:28104. doi: 10.18632/oncotarget.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.a) Dewhirst MW, Cao Y, Moeller B. Nat Rev Cancer. 2008;8:425. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Eales KL, Hollinshead KE, Tennant DA. Oncogenesis. 2016;5:e190. doi: 10.1038/oncsis.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stroock AD, Fischbach C. Tissue Eng, Part A. 2010;16:2143. doi: 10.1089/ten.tea.2009.0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Allen CB, Schneider BK, White CW. Am J Physiol: Lung Cell Mol Physiol. 2001;281:L1021. doi: 10.1152/ajplung.2001.281.4.L1021. [DOI] [PubMed] [Google Scholar]
- 161.a) Lo JF, Sinkala E, Eddington DT. Lab Chip. 2010;10:2394. doi: 10.1039/c004660d. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Oppegard SC, Nam KH, Carr JR, Skaalure SC, Eddington DT. PLoS One. 2009;4:e6891. doi: 10.1371/journal.pone.0006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Oppegard SC, Eddington DT. Biomed Microdevices. 2013;15:407. doi: 10.1007/s10544-013-9737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Funamoto K, Zervantonakis IK, Liu Y, Ochs CJ, Kim C, Kamm RD. Lab Chip. 2012;12:4855. doi: 10.1039/c2lc40306d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Park KM, Gerecht S. Nat Commun. 2014;5:4075. doi: 10.1038/ncomms5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.a) Gimsa U, Mitchison NA, Brunner-Weinzierl MC. Mediators Inflammation. 2013;2013:320519. doi: 10.1155/2013/320519. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Domingues P, Gonzalez-Tablas M, Otero A, Pascual D, Miranda D, Ruiz L, Sousa P, Ciudad J, Goncalves JM, Lopes MC, Orfao A, Tabernero MD. Brain, Behav, Immun. 2016;53:1. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]; c) Dubois LG, Campanati L, Righy C, D’Andrea-Meira I, Spohr TC, Porto-Carreiro I, Pereira CM, Balca-Silva J, Kahn SA, DosSantos MF, de Oliveira MA, Ximenes-da-Silva A, Lopes MC, Faveret E, Gasparetto EL, Moura-Neto V. Front Cell Neurosci. 2014;8:418. doi: 10.3389/fncel.2014.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yang I, Han SJ, Sughrue ME, Tihan T, Parsa AT. J Neurosurg. 2011;115:505. doi: 10.3171/2011.4.JNS101172. [DOI] [PubMed] [Google Scholar]
- 166.Jacobs SK, Wilson DJ, Kornblith PL, Grimm EA. J Neurosurg. 1986;64:114. doi: 10.3171/jns.1986.64.1.0114. [DOI] [PubMed] [Google Scholar]
- 167.a) Bloom AB, Zaman MH. Physiol Genomics. 2014;46:309. doi: 10.1152/physiolgenomics.00170.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Malik R, Lelkes PI, Cukierman E. Trends Biotechnol. 2015;33:230. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Roth P, Junker M, Tritschler I, Mittelbronn M, Dombrowski Y, Breit SN, Tabatabai G, Wick W, Weller M, Wischhusen J. Clin Cancer Res. 2010;16:3851. doi: 10.1158/1078-0432.CCR-10-0705. [DOI] [PubMed] [Google Scholar]
- 169.a) Hao C, Parney IF, Roa WH, Turner J, Petruk KC, Ramsay DA. Acta Neuropathol. 2002;103:171. doi: 10.1007/s004010100448. [DOI] [PubMed] [Google Scholar]; b) Jackson C, Ruzevick J, Phallen J, Belcaid Z, Lim M. Clin Dev Immunol. 2011;2011:732413. doi: 10.1155/2011/732413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.a) Badie B, Schartner JM. Neurosurgery. 2000;46:957. doi: 10.1097/00006123-200004000-00035. [DOI] [PubMed] [Google Scholar]; b) da Fonseca AC, Badie B. Clin Dev Immunol. 2013;2013:264124. doi: 10.1155/2013/264124. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Finn OJ. Ann Oncol. 2012;23:viii6. doi: 10.1093/annonc/mds256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Businaro L, De Ninno A, Schiavoni G, Lucarini V, Ciasca G, Gerardino A, Belardelli F, Gabriele L, Mattei F. Lab Chip. 2013;13:229. doi: 10.1039/c2lc40887b. [DOI] [PubMed] [Google Scholar]
- 172.Feder-Mengus C, Ghosh S, Reschner A, Martin I, Spagnoli GC. Trends Mol Med. 2008;14:333. doi: 10.1016/j.molmed.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 173.Labelle M, Begum S, Hynes RO. Cancer Cell. 2011;20:576. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, Kamm RD, Bertoletti A. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89762. pii: 89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.a) Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]; b) Nesmith AP, Agarwal A, McCain ML, Parker KK. Lab Chip. 2014;14:3925. doi: 10.1039/c4lc00688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Adriani G, Pavesi A, Tan AT, Bertoletti A, Thiery JP, Kamm RD. Drug Discovery Today. 2016;21:1472. doi: 10.1016/j.drudis.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.a) Chamberlain MC, Johnston SK. J Neuro-Oncol. 2010;96:259. doi: 10.1093/neuonc/noq011. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fenton TR, Nathanson D, Ponte de Albuquerque C, Kuga D, Iwanami A, Dang J, Yang H, Tanaka K, Oba-Shinjo SM, Uno M, Inda MM, Wykosky J, Bachoo RM, James CD, DePinho RA, Vandenberg SR, Zhou H, Marie SK, Mischel PS, Cavenee WK, Furnari FB. Proc Natl Acad Sci USA. 2012;109:14164. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WK, Paleologos N, Nicholas MK, Jensen R, Vredenburgh J, Huang J, Zheng M, Cloughesy T. J Clin Oncol. 2009;27:4733. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]; d) Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA. J Clin Oncol. 2009;27:740. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Mancuso MR, Davis R, Norberg SM, O’Brien S, Sennino B, Nakahara T, Yao VJ, Inai T, Brooks P, Freimark B, Shalinsky DR, Hu-Lowe DD, McDonald DM. J Clin Invest. 2006;116:2610. doi: 10.1172/JCI24612. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Narita Y. Ther Clin Risk Manage. 2015;11:1759. doi: 10.2147/TCRM.S58289. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Raizer JJ, Grimm S, Chamberlain MC, Nicholas MK, Chandler JP, Muro K, Dubner S, Rademaker AW, Renfrow J, Bredel M. Cancer. 2010;116:5297. doi: 10.1002/cncr.25462. [DOI] [PubMed] [Google Scholar]
- 178.Chin SY, Poh YC, Kohler AC, Compton JT, Hsu LL, Lau KM, Kim S, Lee BW, Lee FY, Sia SK. Sci Rob. 2017;2:eaah6451. doi: 10.1126/scirobotics.aah6451. [DOI] [PMC free article] [PubMed] [Google Scholar]