Abstract
Cryptococcus neoformans is an encapsulated pathogenic yeast that can change the size of the cells during infection. In particular, this process can occur by enlarging the size of the capsule without modifying the size of the cell body, or by increasing the diameter of the cell body, which is normally accompanied by an increase of the capsule too. This last process leads to the formation of cells of an abnormal enlarged size denominated titan cells. Previous works characterized titan cell formation during pulmonary infection but research on this topic has been hampered due to the difficulty to obtain them in vitro. In this work, we describe in vitro conditions (low nutrient, serum supplemented medium at neutral pH) that promote the transition from regular to titan-like cells. Moreover, addition of azide and static incubation of the cultures in a CO2 enriched atmosphere favored cellular enlargement. This transition occurred at low cell densities, suggesting that the process was regulated by quorum sensing molecules and it was independent of the cryptococcal serotype/species. Transition to titan-like cell was impaired by pharmacological inhibition of PKC signaling pathway. Analysis of the gene expression profile during the transition to titan-like cells showed overexpression of enzymes involved in carbohydrate metabolism, as well as proteins from the coatomer complex, and related to iron metabolism. Indeed, we observed that iron limitation also induced the formation of titan cells. Our gene expression analysis also revealed other elements involved in titan cell formation, such as calnexin, whose absence resulted in appearance of abnormal large cells even in regular rich media. In summary, our work provides a new alternative method to investigate titan cell formation devoid the bioethical problems that involve animal experimentation.
Author summary
Cryptococcus neoformans is a fungal pathogen that has a significant incidence in HIV+ patients in particular, in Sub-saharan Africa, Asia and South America. This yeast poses an excellent model to investigate fungal virulence because it develops many strategies to adapt to the host and evade the immune response. One of the adaptation mechanisms involves the formation of Titan Cells, which are yeast of an abnormal large size. However, research on these cells has been limited to in vivo studies (mainly in mice) because they were not reproducibly found in vitro. In this work, we describe several conditions that induce the appearance of cells that mimic titan cells, and that we denominated as titan-like cells. The main factor that induced titan-like cells was the addition of serum to nutrient limited media. This has allowed to easily performing new approaches to characterize several signaling pathways involved in their development. We found that the formation of these cells is regulated by quorum sensing molecules, and that pathways such as cAMP and PKC regulate the process of cellular enlargement. We have also performed transcriptomic analysis, which led to the identification of new genes that could be involved in the process. This work will open different research lines that will contribute to the elucidation of the role of these cells during infection and on the development of cryptococcal disease.
Introduction
Cryptococcus neoformans is a basidiomycetes yeast widely distributed in the environment that can behave as a pathogen in susceptible patients [1, 2]. Cryptococcus neoformans can survive in the lung, but in immunosuppressed patients it can also spread to the central nervous system and cause meningoencephalitis [2]. Cryptococcal infections are major causes of death in HIV patients. Although the incidence has significantly decreased in developed countries due to the introduction of antiretroviral therapy (ART), associated mortality remains high [3, 4]. Moreover, infections by this yeast still present a high incidence in developing areas, such as the sub-Saharan Africa and Southeast Asia [5, 6].
One of the most characteristic features of Cryptococcus is its ability to adapt to the lung environment and to evade the host immune response. Several factors contributing to cryptococcal adaptation to the lung have been described. The most important is the presence of a polysaccharide capsule [7–9], which is antiphagocytic and protects the yeasts from stress conditions [2, 10]. The size of the capsule is not constant, and it increases during the first hours of interaction with the host [11], which indicates that this process is a response that contributes to immune evasion. Furthermore, the capsular polysaccharide is also secreted to the extracellular media where it induces immunological paralysis through multiple mechanisms [9, 12–15]. Cryptococcus neoformans is also a facultative intracellular pathogen in phagocytic cells [16–19], which is another important factor that contributes to fungal survival in the host.
Cryptococcus has also developed other adaptation mechanisms that contribute to the evasion of the immune response. One of them involves the formation of titan cells, which have an abnormal large size. The average diameter of Cryptococcus cells grown in vitro ranges between 4–7 microns. In contrast, the fungal population in the lungs is very heterogeneous, and cells of even 100 microns have been described [11, 20–22]. Titan cells have been arbitrarily defined as those with a cell body diameter above 15 microns or with a total size (capsule included) over 30 microns [23]. Because of their size, titan cells cannot be phagocytosed, and they can persist in the host for long periods [24, 25]. Titan cells also contribute to virulence through other mechanisms. For example, they can divide and produce a progeny of regular size that has increased resistance to stress factors [26] as well as the ability to inhibit the phagocytosis of cells of regular size [25].
Some signals and pathways involved in titan cell formation have been characterized, and it is known that the cAMP pathway is required for this phenomenon [20, 21, 27]. Titan cell formation has been associated in vivo with anti-inflammatory Th2 type immune responses [23], but the host's factors that trigger this morphological transition remain unknown.
The investigation and characterization of titan cells has been limited by the lack of media allowing the transition in vitro, so most of the data about these cells has been obtained using animal models. Although this approach has been shown to be useful for some purposes, it does not allow obtaining a large population of titan cells. In addition, the use of mice for these purposes presents associated significant bioethical issues. In this work, we have defined in vitro conditions that induce cell enlargement in C. neoformans, which lead to the appearance of cells similar to those found in vivo, here denominated titan-like cells. We found that incubation of this fungus in low nutrient media supplemented with serum in a CO2 enriched atmosphere induced cryptococcal cell size increase. Moreover, other factors, such as oxygen limitation, or low cell density enhanced the cell growth. We have used this medium as a first step in the characterization of this transition. Our findings open future research lines that will help to define the molecular mechanisms that trigger titan cell formation and their role during infection.
Results
Cryptococcus neoformans can significantly enlarge its cell size in vitro
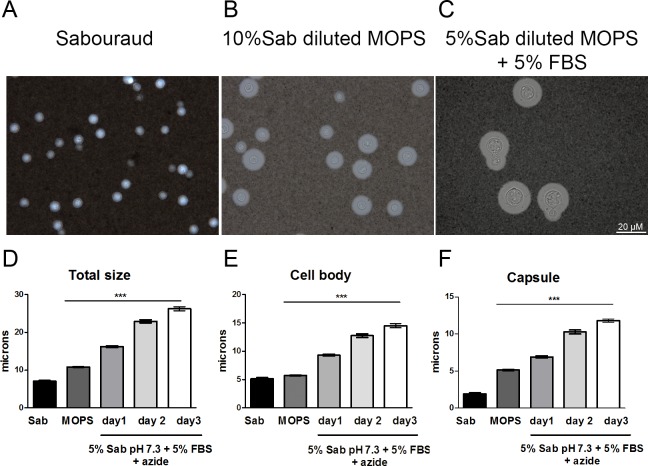
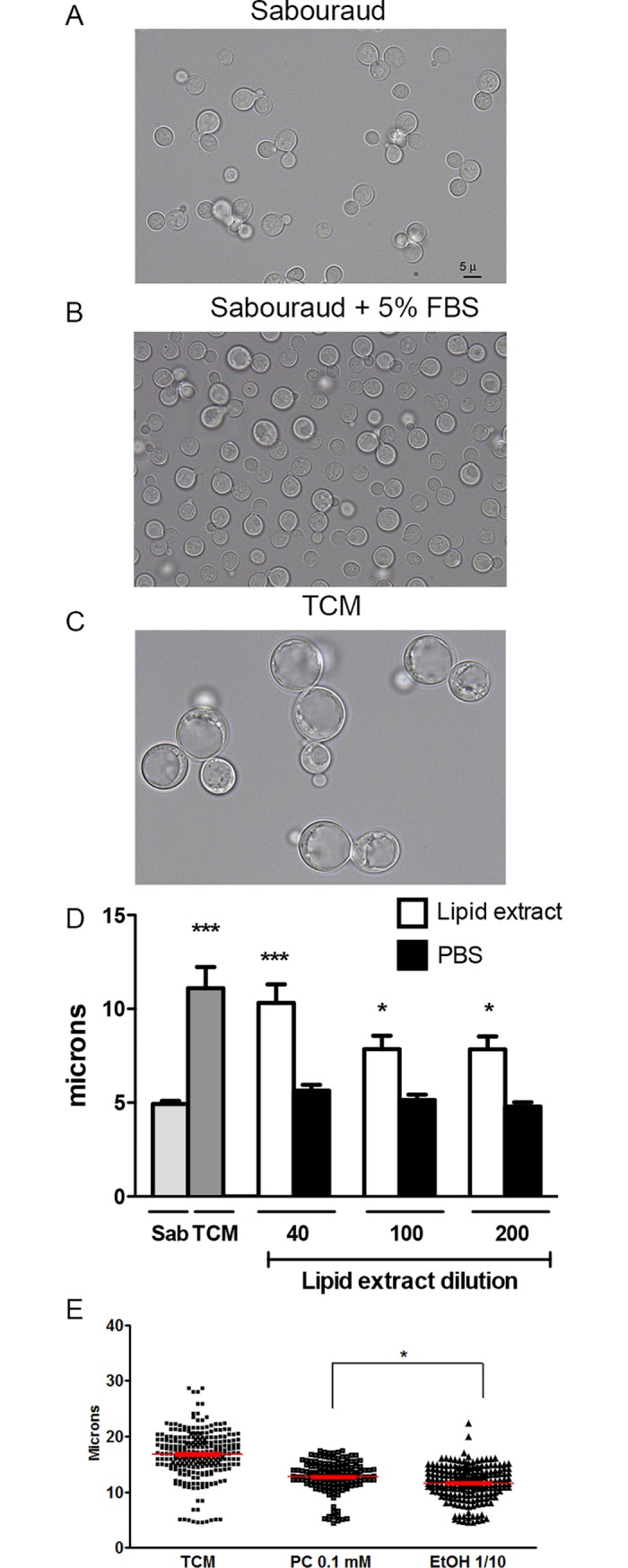
In the last years, we have characterized the phenomenon of capsule growth in vitro using a medium that contains 10% Sabouraud buffered at pH 7.3 with 50 mM MOPS. In a set of experiments, we observed that addition of serum and of the respiration inhibitor sodium azide to this medium produced not only growth of the capsule but also of the cell body (Fig 1A–1C). These cells resembled the titan cells observed in vivo, although they did not reach the same size. Despite this difference, we argued that this phenomenon might reflect the first steps of titan cell formation, so we decided to characterize the factors that induced in vitro cell growth. First, the morphology of C. neoformans was analyzed in regular growth conditions (liquid Sabouraud), in capsule inducing medium (10% Sabouraud buffered with 50 mM MOPS) and in 5% Sabouraud buffered with MOPS and 5% FBS + azide (15 μM). Titan cells have been defined as those with total diameter of 30 μm or those with a cell body diameter larger than 15 μm [22]. As shown in Fig 1D, we observed a significant increase of both the cell size and the capsule in this last medium, almost reaching the threshold for titan cells definition after three days of culture. For this reason, we decided to name the medium as TCM (Titan Cell Medium).
Fig 1. Cellular and capsular size of C. neoformans in different media.
Cells from strain H99 were inoculated into Sabouraud (A), capsule inducing medium (10% Sabouraud with MOPS at pH 7.3) (B) and 5% Sabouraud with MOPS + 5% FBS + sodium azide (C). After incubation at 37°C with shaking, total size and capsule size was visualized by suspending the cells in India ink. Total size (D), cell body size (E) and capsule size (F) distribution after incubation as described above. The asterisks indicate significant differences compared to Sabouraud control (p<0.05). Sab, Sabouraud medium.
Characterization of factors that induce cellular growth in vitro
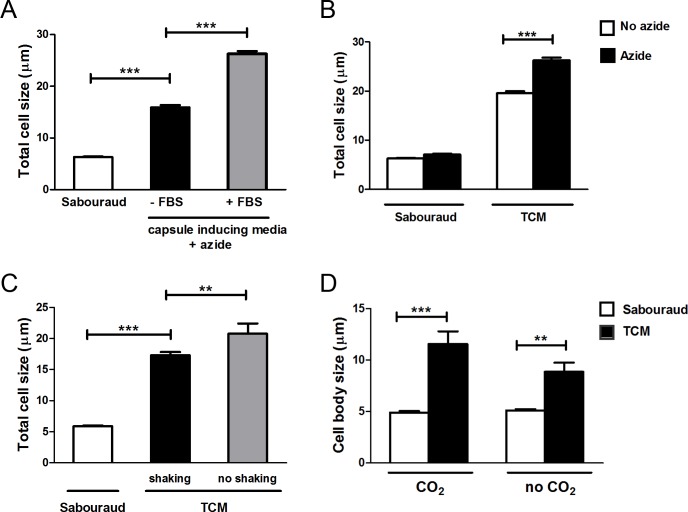
Serum was essential for cellular enlargement formation because in its absence the increase in cell size was significantly lower (p<0.05, Fig 2A). We characterized in detail the factors and conditions that favor cryptococcal cell size increase in vitro. In our initial experiments, the medium in which we first observed cells of enlarged size contained subinhibitory concentrations of the mitochondrial inhibitor sodium azide to prevent contamination. As shown in Fig 2B, the process of cell enlargement was enhanced in the presence of sodium azide. This compound is an inhibitor of complex IV of the respiratory chain, so we argued that other factors that alter respiration could induce increases in cell size. For this reason, we investigated whether cryptococcal cell growth was influenced by oxygen limitation. To assess that, we compared the yeast morphology under shaking or static conditions. As shown in Fig 2C, static incubation of the cultures resulted in a greater proportion of cells with enlarged size.
Fig 2. Effect of different factors on the cellular growth of C. neoformans.
Cells from H99 strain were incubated on Sabouraud overnight and transferred to different media to evaluate the effect of several factors on the titan-like cell formation. (A) Cells were incubated in Sabouraud or capsule inducing medium (10% Sab, pH 7.3) supplemented with 15 μM sodium azide in the presence or absence of 5% serum (FBS) and cultures were incubated at 37°C overnight. Pictures after suspension of the cells in India Ink were taken, and the total cell size was measured and plotted (B) Effect of sodium azide (black bars) on cell size in Sabouraud or TCM medium. As a control, the same media were inoculated without sodium azide (white bars). Cells were incubated at 37°C overnight. Pictures after suspension of the cells in India Ink were taken, and the total cell size was measured and plotted (C) Effect of shaking effect on titan-like cell formation. The yeasts were incubated at 37°C for 24 h in flasks with Sabouraud or TCM medium in both conditions (black bars, shaking) and (gray bars, no shaking). Pictures after suspension of the cells in India Ink were taken, and the total cell size was measured and plotted (D) Effect of CO2 on cell growth. The cells were incubated in Sabouraud (white bars) and TCM (black bars) with and without 5% of CO2. In both cases, the cells were grown at 37°C without shaking for 24 h in a 96-microdilution plate. Then, the plate was directly observed in the microscope, and the cell body size of was measured and plotted.
Cryptococcal cells sense and respond to environmental levels of CO2 and it is known that this molecule induces capsule growth. For this reason, we investigated if incubation of the cultures in a CO2-enriched environment altered cryptococcal cell size. We found that yeast size was significantly larger when the plates were placed in a 5% CO2 atmosphere in comparison to growth without CO2 (Fig 2D).
To visualize the phenomenon of cell growth, we carried out in vivo imaging by placing the cells in a 96-wells plate in TCM in 5% CO2 at 37°C under a microscope overnight and obtained videos of the cellular enlargement. As shown in S1 Video, cells actively grew and replicated in Sabouraud medium. However, in TCM, after 5–8 h of incubation the cells started to enlarge during 8–10 h (S2 Video). After this time, the cells stopped enlarging and continued budding. We also observed that cellular enlargement was associated with some intracellular phenotypic changes. For instance, a significant proportion of the cells displayed an intracellular compartment that started to divide by fission, but then fused again to render a large vesicle (S3 Video).
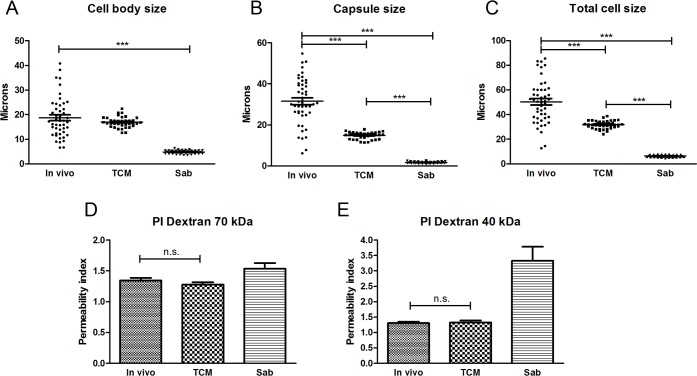
To evaluate to which extent the cells obtained in vitro resembled titan cells found in animal models, we infected mice and isolated cryptococcal cells after 14 days of infection (see Material & Methods). We first determined the total size, capsule size, and cell body size of titan cells. As shown in Fig 3, the cells obtained in vitro in TCM did not reach the size found in the cells in vivo. The main difference between both types of cells was at the capsule, being it size significantly larger in titan cells isolated from mice. For this reason, we decided to denominate the cells obtained in vitro as “titan-like” cells.
Fig 3. Comparison of titan cells obtained in vivo with enlarged cells obtained in vitro.
Measurements of cell body (A), capsule (B) and total cell sizes (C) of cells isolated from infected mice (in vivo) and cultured in TCM and Saboraud in vitro. D and E show the permeability index of 70 and 40 kDa fluorescent labelled dextrans respectively in the same type of samples (see M&M).
Titan cells also present differences in capsular features, such as the density, compared to cells obtained in vitro [21]. To investigate if the density of titan-like cells was similar to that observed in vivo, we measured the permeability index using fluorescently labeled dextrans of different molecular weights (70 and 40 kDa). As shown in Fig 3D and 3E, penetration of the dextrans in the capsule of titan-like cells was reduced compared to cells of regular size and similar to that of titan cells isolated from the lung of infected mice. This result indicated that titan-like cells had a capsule of similar density of that of titan cells generated in vivo.
Although serum was required to induce cellular enlargement in vitro, it was not sufficient for this process. Serum did not induce cellular growth in rich media (Fig 4A and 4B), in contrast to the situation in the diluted medium nutrients (Fig 4C), indicating that nutrient limitation was important for cell size enlargement.
Fig 4. Effect of serum on cryptococcal cell growth.
H99 cell suspensions were inoculated in Sabouraud (A), Sabouraud supplemented with 5% serum (B) or TCM (C). The cells were incubated overnight at 37°C in 5% CO2. Then, the cells were suspended in India Ink and pictures of the cell body size were taken. The scale in A applies to all the pictures. (D) Effect of purified serum lipids on cell growth. The cells (initial density 104 cells/mL) were grown in TCM medium in which the serum was replaced by different dilutions (40, 100 and 200) of the lipid extract. As controls, the same media was supplemented with PBS. Cultures in TCM are also included for reference. Cell body size was determined after 24 hours at 37° C with CO2 and shaking. The asterisks indicate significant differences compared to Sabouraud control. (E) Effect of phosphatydilcholine on titan-like cell formation. Cells were incubated as in (A), but serum was replaced by 0.1 mM of phosphatidylcholine (PC). A parallel control with the same concentration of ethanol was carried out in parallel. The graph shows the cell body size.
Effects of phospholipids from fetal calf serum in titan-like cell formation
Phospholipids, in particular phosphatidylcholine, can trigger the appearance of titan cells in vitro [28]. For this reason, we performed a lipid extraction of fetal calf serum, present in TCM and we incubated the cells with different amounts of these lipids (1/40, 1/100 and 1/200 dilution of the original lipid solution). As shown in Fig 4D, the lipids present in the serum induced titan-like cell formation.
Phosphatidylcholine (PC) is one of the major phospholipids contained in serum and mammalian membranes. For this reason, we investigated if this molecule had any effect on titan-like cell formation. We performed experiments in which serum was replaced by different concentrations of PC, and compared its effect with control samples containing the same concentration of solvent (ethanol). As shown in Fig 4E, PC induced an increase in cryptococcal cell size. However, this increase was lower compared to the one observed with serum, indicating that although PC seemed to induce titan-like cells, there are other serum components responsible for the cell increase.
Cell density influences titan-like cell formation
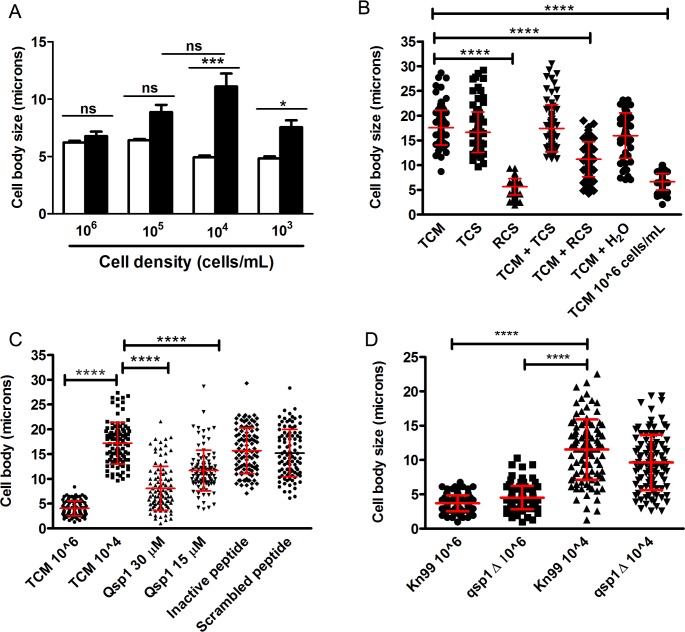
We found that titan-like cell formation depended on the cell density of the cultures. We inoculated 96-wells plates with different concentrations of cells from H99 strain (106, 105, 104 and 103 cells/mL) in TCM and Sabouraud as a control of growth. After overnight incubation at 37°C with CO2, titan-like cells were observed in the wells inoculated with 103, 104 and 105 cells/mL but were almost absent in the wells that were inoculated with the higher cell density (106 cells/mL, Fig 5A). We found that titan-like cells were observed more frequently when the cultures were inoculated with cellular concentrations around 104 cells/mL.
Fig 5. Influence of cell density on titan-like cell formation in C. neoformans.
(A). Cells from H99 strain inoculated in Sabouraud (white bars) or TCM (black bars) at different concentrations (103, 104, 105 and 106 cells/mL) in 96-well plates. Cell body size was measured after incubation overnight at 37°C with CO2. The asterisks indicate significant differences. B) Effect of conditioned media on cell body size. Cells from H99 strain were incubated in different conditioned TCM medium denominated (see material and methods) as TCS (supernatant of titan-like cells TCM cultures), RCS (supernatant of regular size TCM cultures), TCM + TCS (a 1:1 mixture of fresh TCM with TCS), TCM + RCS (a 1:1 mixture of fresh TCM with RCS). As controls we used a diluted TCM in H2O (1:1) and fresh TCM inoculated with 104 cells/mL (TCM) and with 106 cells/mL. (C) Cells from H99 strain were grown in TCM supplemented with 30 uM and 15 uM of the quorum sensing peptide Qsp1 (NFGAPGGAYPW). As controls, TCM supplemented with 30 uM of an inactive Qsp1 (NFGAPGAAYPW), with 30 uM of a scrambled Qsp1 peptide (AWAGYFPGPNG). TCM without any supplementation was used as control. Cell body size was measured after overnight incubation at 37°C with 5% CO2. D) Titan-like cell formation of qsp1 mutant. Cells from the KN99 (wild type) and qsp1 mutant were inoculated in TCM at 104 and 106 cells/mL, and cell body size was determined after 18 h of incubation at 37°C in the presence of 5% CO2.
Effect of QS molecules on titan-like cell formation
The fact that the formation of titan-like cells depends on cell density suggests that this process could be regulated by quorum sensing (QS). QS is a cell-cell communication mechanism mediated by molecules that are released directly into the medium by microorganisms. These molecules are released as a function of growth and replication rate [29, 30]. In this way, we evaluated the influence on titan-like cell formation of cell-free media obtained from titan-like and regular C. neoformans cultures. We inoculated TCM with the H99 strain at 106 cells/mL and 104 cells/mL and incubated the cultures for 18 h at 37°C in 5% CO2 to obtain cells of regular size and titan-like cells, respectively. We then collected the supernatants (named RCS and TCS, respectively). These conditioned media were added to wells that contained fresh TCM inoculated at 104 cells/mL. We found that the conditioned medium RCS significantly inhibited the formation of titan-like cells (Fig 5B) even when added to fresh TCM (TCM + RCS) (p<0.001). In contrast, the supernatant from titan-like cells cultures (TCS) did not block the formation of the titan-like cells, demonstrating a negative effect of the supernatant obtained from cells of regular sizes on titan-like cell formation (Fig 5B). The effect of the TCS conditioned medium was not explained by the dilution of the nutrients of the fresh TCM, since titan-like cells were still formed in TCM diluted with distilled water (Fig 5B).
In C. neoformans, the main QS molecule described is a short peptide (11-mer) called Qsp1 that is required for fungal virulence, replication, cell wall synthesis and protease activities [31, 32]. To investigate the influence of Qsp1 in the formation of titan-like cells, different concentrations of the peptide were added to the TCM medium and the formation of titan-like cells was evaluated. We observed that Qsp1 significantly inhibited formation of titan-like cells in a dose-dependent manner (Fig 5C). As control, we used both inactive and scrambled versions of Qsp1, and observed that none of them had any effect on titan-like cell development (Fig 5C).
It could be argued that the production of Qsp1 in TCM cultures inoculated at high cell densities was responsible for the inhibition of titan-like cell formation. To test this idea, we used a qsp1 mutant that does not produce Qsp1 [32]. Our results showed that the mutant produced titan-like cells in a similar way as the wild type strain KN99 (Fig 5D) [32], even at high cell densities (106 cells/mL). This result indicates that absence of titan-like cells in TCM cultures inoculated at high densities was not only due to Qsp1, and that most probably, other QS molecules secreted by C. neoformans might influence cellular enlargement.
Nuclear staining
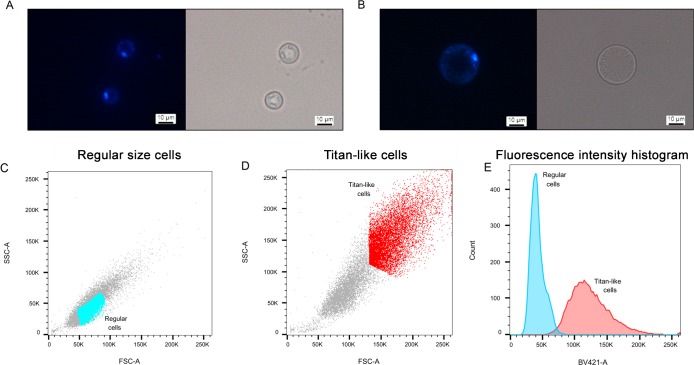
Titan cells formed in the lungs are polyploid and single-nucleated. So we investigated the morphology of the nucleus and the DNA content after staining with DAPI. As shown in Fig 6A and 6B, titan-like cells contained one nucleus. This result was confirmed using a strain that expresses a fluorescent nucleolar protein (NOP1-mCherry, [33], S1 Fig). We also quantified the fluorescence intensity of the DAPI staining by flow cytometry. As shown in Fig 6C–6E, titan-like cells emitted more fluorescence than cells of regular size. The fluorescence intensity was more heterogeneous in titan-like cells, ranging from 2 to 5-fold increase compared to cells of normal size (Fig 6E).
Fig 6. Nuclear staining of titan-like cells.
Cells of regular size and titan-like cells were obtained by inoculating H99 strain in TCM at 106 and 104 cells/mL respectively. After incubating overnight at 37°C with 5% CO2, the cells were fixed and stained with DAPI as described in material and methods. A and B show the microscopic appearance of the nucleus of cells of regular size (A), and titan-like cells (B). C-E, Analysis of nuclear staining by flow cytometry. C) FFS/SSC scatter plot of cells of regular size, D) same graph of titan-like cells. In C and E, we defined a gate to clearly separate cells of regular size (cells in blue) and titan-like cells (cells in light red). The fluorescence intensity (histograms) of the cells from these two gates is represented in panel E. Blue histogram, fluorescence from the cells shown in the gate in panel C, and light red histogram, fluorescence from the cells shown in the gate in panel D.
Inhibition of PKC signaling pathways affected titan-like cell formation
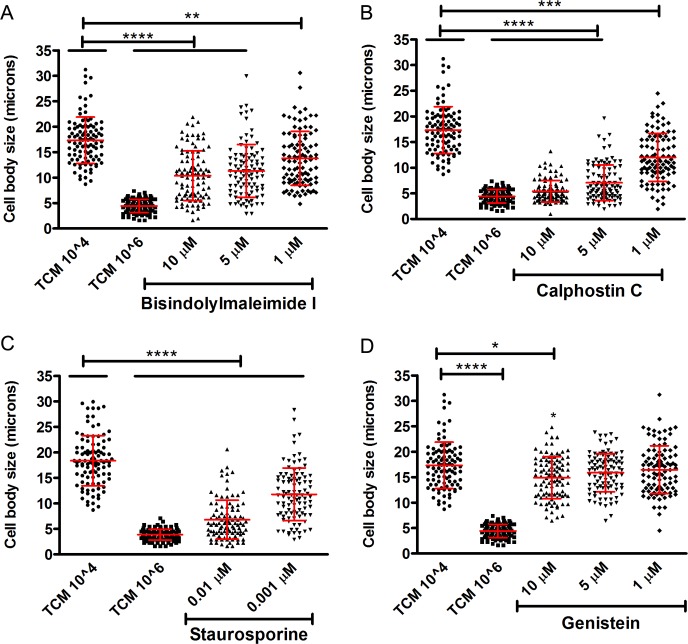
PKC is a family of protein kinases, activated by Ca2+, diacylglycerol (DAG) and phospholipids, and involved in different virulence-related aspects in C. neoformans such as melanin production [34], temperature tolerance, cell integrity [35] and fluconazole tolerance [36]. We argued that serum phospholipids could activate the PKC signaling pathway, so we tested the effect of three PKC inhibitors, calphostin C, staurosporine and bisindolylmaleimide I on titan-like cell formation. We observed that all PKC inhibitors (in particular, staurosporine and Calphostine C), impaired the formation of titan-like cells in a dose-dependent manner (Fig 7A–7C). We also included the tyrosine kinase inhibitor genistein and found that this compound had no visible effect on cellular enlargement (Fig 7D).
Fig 7. Influence of PKC pathway on titan-like cell formation.
The influence of the PKC pathway was evaluated by its inhibition with three different agents: Bisindolylmaleimide I (A), Calphostin C (B), Staurosporine (C). Genestein, a tyrosine kinase inhibitor, was used as control (D). The experiments in A, B and D were performed the same days, so they share the same control. However, for clarity, the graphs corresponding to each inhibitor have been separated. The experiments were repeated on three days, and the data from the three experiments is plotted. Cells of the H99 strain were incubated with these different agents overnight in TCM at 37°C with 5% CO2 without shaking and cell body size was measured. The asterisks indicate statistical significant differences.
Titan-like cell formation did not correlate with the serotype or mating type of the strain
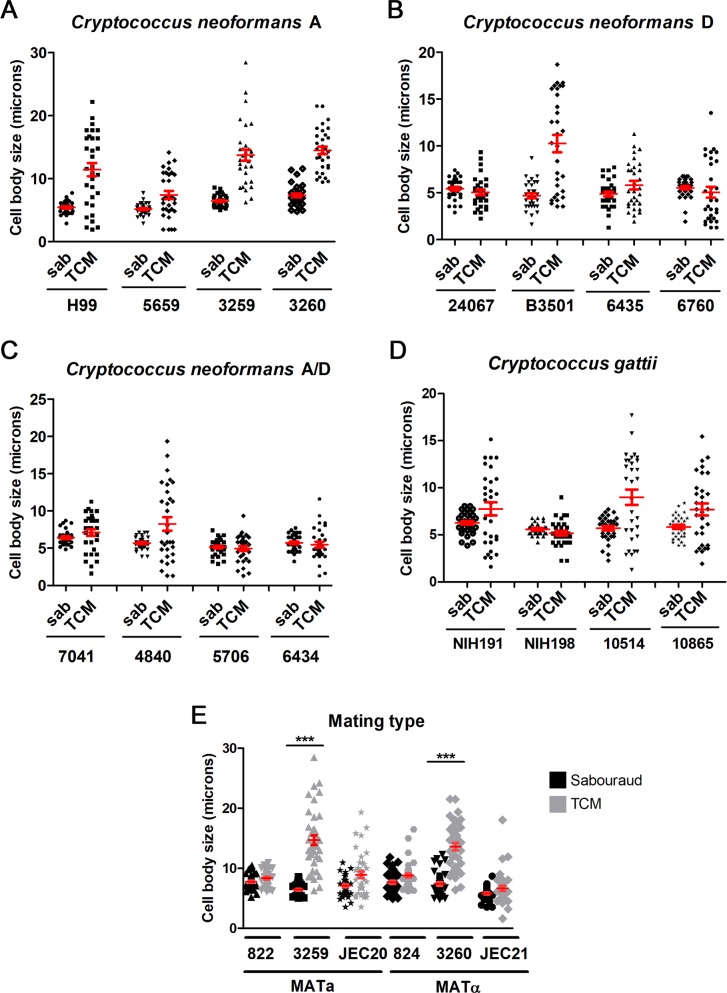
Cryptococcus neoformans is divided in different serotypes and varieties: variety grubii (serotype A), variety neoformans (serotype D), and A/D hybrids. It has been proposed that these groups should be divided into different species [37], although recent reports argue against this differentiation and suggest the term of species complexes [38]. We investigated if there was a correlation between the main species complex of the strain and the formation of titan-like cells. As shown in Fig 8A–8C, for each serotype/species complex, there were strains with high and low capacity to induce titan-like cells, which indicates that there was not a direct association between the serotype and cell growth.
Fig 8. Titan-like cell induction in different strains.
Four strains of each species (serotypes) were inoculated into Sabouraud (black symbols) or TCM (grey symbols) at a density of 104 cells/mL and grown overnight at 37°C with CO2. (A) C. neoformans var. grubii (serotype A); (B) C. neoformans var neoformans (serotype D); (C) C. neoformans A/D; (D) C. gattii. (E) MATa and MATα strains: NE822/NE824 (serotype D), 3259/3260 (serotype A) and JEC20/JEC21 (serotype D). Cell body size was measured in all cases. The red lines represent the mean and standard error.
We also investigated the behavior of strains from the related species C. gattii, which can infect immunocompetent patients. As shown in Fig 8D, there were strains that had high and low ability to induce titan-like cells. We next examined if the hyper (CBS10514, R265) and hypovirulent (CBS10865, R272) strains isolated at the Vancouver outbreak [39, 40] formed titan-like cells in a different way. Interestingly, the hypervirulent strain tended to produce more titan-like cells compared to the strain with reduced virulence (Fig 8D). We also tested other C. gattii strains (NIH 191 and NIH198), which also presented differences in their capacity in producing titan-like cells. In summary, the many inter-strain differences in the capability to form titan-like cells did not allow associating this ability to the serotype/genotype of the isolates.
Coinfection with a and α strains results in a higher proportion of titan cells in the lungs [20], so we investigated whether strains from different mating type had different ability to form titan-like cells. We studied three pair of strains with different mating type JEC20/JEC21, NE822/NE824 and 3259/3260 (KN99), and we found that the pair 3259/3260 increased the size of the cell body in the TCM medium compared to the Sabouraud significantly (p<0.05, Fig 8E). In NE822/NE824 and JEC20/JEC21 pairs, there was a small increase in cell size in TCM and we observed a small amount of titan-like cells in this medium although this difference was not statistically significant. These results indicate that titan-like cell formation can occur in vitro independently of the mating type of the strains. However, co-incubation of both mating types did not result in a higher proportion of titan cells in TCM (S2 Fig).
Titan-like cell formation in different mutants
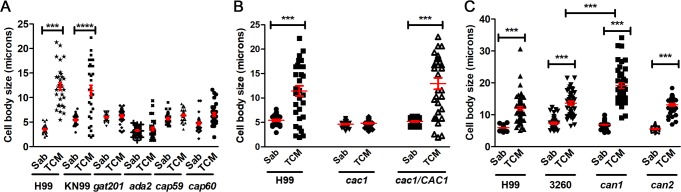
To identify genes that play an important role in the formation of titan-like cells, we studied the phenotype of mutants with problems to induce capsule growth, such as gat201 or ada2 [41, 42] or acapsular strains (cap59 and cap60). As shown in Fig 9A, none of these mutants induced cellular enlargement in TCM.
Fig 9. Titan-like cell formation in different mutants.
(A) Cells from the capsule deficient mutants gat201, cap59, cap60 and ada2 (H99 background) were inoculated in Sabouraud and TCM medium at a density of 104 cells/mL in 96-well plates. Cell body size was determined after overnight incubation at 37°C with CO2. (B) Role of adenylate cyclase on cell growth of C. neoformans. Cells from the wild-type strain H99, the adenylate cyclase mutant (cac1) and the reconstituted strain (cac1/CAC1) were incubated as described above. (C) Role of carbonic anhydrases on cell growth. Cells from the wild type strain H99 and KN99 (3260) and the can1 and can2 mutants were used for this purpose. Cell body size was determined after overnight incubation at 37°C with CO2 in all cases. The asterisks indicate significant differences (see M&M). The red lines represent the mean and standard error.
Titan cell formation is regulated by the cAMP pathway [21, 27]. Moreover, the CO2 activates adenylate cyclase [43, 44]. For this reason, we investigated the formation of these cells in cac1 mutant (which lack the enzyme adenylate cyclase) and in the reconstituted strain cac1/CAC1. As shown in Fig 9B, the cac1 mutant was defective to produce cellular enlargement, whereas the reconstituted strain produced titan-like cells as the wild type.
CO2 is transformed into HCO3- by the action of carbonic anhydrases (Can). In C. neoformans, there are two genes encoding these enzymes (CAN1 and CAN2) [44, 45], being Can2 the most abundant and physiologically active. Since can2 mutant can only grow in a CO2 enriched environment, these strains were maintained always in 5% CO2. Deletion of CAN2 did not have any effect of titan-like cell formation. Strikingly, in the absence of CAN1, titan-like cell size was even larger than that observed for the WT strain in TCM (Fig 9C).
Interaction of titan-like cells with macrophages
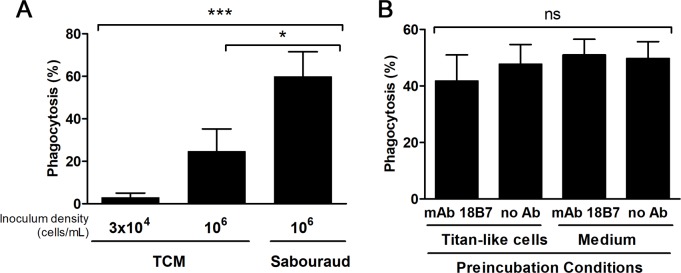
Titan cells are not phagocytosed in vivo [21, 25]. For this reason, we examined the interaction between macrophage-like cell lines and titan-like cells obtained in vitro. We compared the phagocytosis of titan-like cells (incubated in TCM inoculated at 3x104 cells/mL), and of cells of regular size obtained in TCM inoculated at 106 cells/mL or in Sabouraud. Phagocytosis was quantified both by microscopic observation and by flow cytometry using a cryptococcal strain that expressed GFP (see Material and Methods). In all cases, we found that titan-like cells obtained in vitro were not phagocytosed (Fig 10A and S4 Video). Interestingly, cells of regular size were not equally internalized, since cells cultivated in Sabouraud medium inoculated at high cellular density were more efficiently phagocytosed than the same cells inoculum grown in TCM (S5 and S6 Videos, Fig 10A). This difference was not due to difference in binding to the antibody used as opsonin in this experiment (18B7, S3 Fig), but correlated with the increase in cell size (in particular, due to capsule enlargement) found in cells inoculated in TCM at high density compared to the cells cultivated in Sabouraud (S3 Fig).
Fig 10. Interaction of titan-like cells with murine-like macrophages.
A) Cells from H99-GFP strain were incubated in TCM inoculated at 3x104 cells/mL (titan-like cells), 106 cells/mL (regular cells in TCM), and Sabouraud. Then, phagocytosis experiments with RAW264.7 cells were performed and quantified by flow cytometry as described in material and methods. Statistical differences are highlighted. B) Effect of preincubation of macrophages with titan-like cells on the phagocytosis of regular cells. Titan-like cells were obtained by incubation of C. neoformans in TCM as described in material and methods, and were exposed to RAW264.7 macrophages at 1:1 ratio. The incubation was performed in media containing the opsonizing mAb 18B7 or in its absence. As control, macrophages were preincubated with growth medium with or without the mAb. The plates were incubated for 1h at 37°C in the presence of 5% CO2, and then the cells were washed to remove titan-like cells. Next, C. neoformans cells (H99-GFP) of regular size grown in Sabouraud were added to the macrophages at 1:1 ratio in the presence of mAb 18B7, and phagocytosis was performed for 1 h at 37°C in CO2. Phagocytosis percentage was quantified by flow cytometry as described in M&M. ns: no statistical difference. The experiment was performed in triplicates in three different days.
It has been shown that titan cells can also prevent phagocytosis in vivo of cryptococcal cells of regular size [25], so we tested if titan-like cells induced a similar phenomenon in vitro. For this purpose, RAW264.7 macrophages were preincubated with titan-like cells for 1 h with mAb 18B7. As control, parallel samples were incubated only with titan-like cells without mAb or with medium without yeast cells (with or without mAb). After this time, the plate was washed to remove titan-like cells, and H99-GFP cells of normal size cultivated in liquid Sabouraud were added to the macrophages. As shown in Fig 10B, preincubation of the macrophages with titan-like cells did not affect the phagocytosis of regular size yeasts (Fig 10B).
Analysis of gene expression profile in titan-like cells
To gain insights about the molecular mechanisms involved in titan-like cell formation, we compared their gene expression profile with that of cells of regular size.
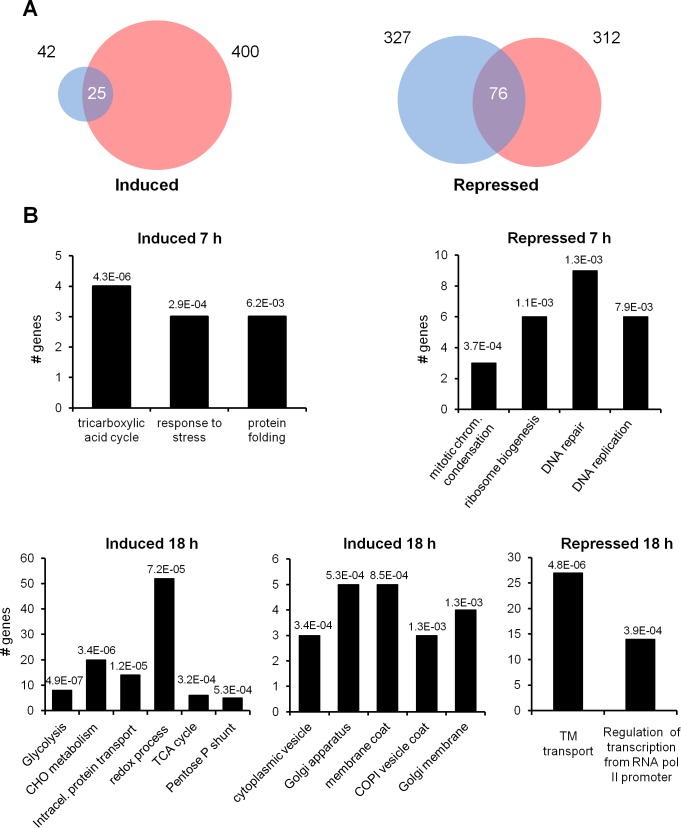
To this end, cryptococcal cells were inoculated in TCM at low (104 cells/mL, titan-like cells) and high densities (106 cells/mL, regular cells) at 37°C with CO2, and total RNA was isolated after 7 and 18 h of incubation. Differences in gene expression were investigated by RNAseq (see material and methods). After mapping the reads and subsequent analysis of differentially expressed genes using the DESeq2 algorithm, we found 42 genes induced at least 1.6-fold in titan-like cells after 7 h and 400 genes induced at least 2-fold after 18 h (Fig 11). A lower expression threshold was set for the 7h data to carry out a comparative GO analysis. Interestingly, the number of repressed genes (after 7h) was relatively high (327) and did not increase after 18 h (312).
Fig 11. Gene expression changes by RNAseq in titan-like cells obtained in vitro.
Transcriptomic profiling of the transition to titan-like cells. A) Number of genes induced or repressed after 7 h (blue) or 18 h (red). The number of genes whose expression is modified at both times is denoted at the intersection of the circles. Scale is only approximated. B) Gene Ontology analysis of genes induced or repressed at 7 h (upper panel) or 18 h (lower panel). All classifications correspond to “biological function” terms, except the central graph in the lower panel, which correspond to “cellular components”. Numbers in the histograms denote the p-value for each assignment. See Materials and Methods for details.
Gene Ontology analysis of genes induced at 7 h revealed a moderate increase in genes involved in the tricarboxylic acid (TCA)/glyoxylate cycle and in response to stress and protein folding. The effect of the transition was more evident after 18 h (Fig 11B and Table 1), with the induction of numerous genes involved in carbohydrate metabolism. This included diverse genes related to glycolysis, such as CNAG_03769 and CNAG_05480, encoding two isoforms of hexokinase, CNAG_03916 (glucose-6-phosphate isomerase), CNAG_04676 (6-phosphofructokinase), CNAG_06770 (fructose-bisphosphate aldolase 1), CNAG_02035 (triose-phosphate isomerase) and two isoforms of enolase (CNAG_03072 and CNAG_06868). The TCA cycle was also affected, with induction of mitochondrial citrate synthase (CNAG_00061), E1 and E2 components of ketoglutarate deshydrogenase (CNAG_03674 and CNAG_03596), the two subunits of isocitrate dehydrogenase (CNAG_07363 and CNAG_07851), and CNAG_05653, encoding malate synthase. In addition, genes belonging to the oxidative segment of the pentose-phosphate shunt were also induced, such as glucose-6-phosphate dehydrogenase (CNAG_03245), 6-phosphogluconolactonase (CNAG_02133), and two isoforms of 6-phosphogluconate dehydrogenase (CNAG_04099 and CNAG_07561). Finally, genes involved in trehalose metabolism, such as CNAG_03113, CNAG_03765, and CNAG_05292, encoding a trehalose phosphatase and two trehalose synthases, respectively, were upregulated.
Table 1. Genes that were significantly overexpressed (fold>2 with P<0.05) at 7 or 18 h in titan-like cells were subjected to Gene Ontology analysis.
For unassigned genes, additional Blastp comparisons against fungal data bases were carried out. T: Titan-like cells (cells incubated in TCM at 104 cells/mL); S (cells of regular size, incubated in TCM at 106 cells/mL).
| 7h | 18 h | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Process | Gene ID | Description | T/S ratio | p-val. | T/S ratio | p-val. | |||
| Carbohydrate Metabolism | |||||||||
| Glycolysis (p-value: 4.9E-07) | |||||||||
| CNAG_03769 | hexokinase | 0.7 | 2.9 | 2.1E-05 | |||||
| CNAG_05480 | hexokinase, variant | 1.0 | 1.9E-03 | 2.6 | 2.5E-03 | ||||
| CNAG_03916 | glucose-6-phosphate isomerase | 0.9 | 3.6E-02 | 2.7 | 1.5E-04 | ||||
| CNAG_04676 | 6-phosphofructokinase | 1.2 | 6.0E-03 | 2.1 | 2.6E-02 | ||||
| CNAG_06770 | fructose-bisphosphate aldolase 1 | 0.8 | 2.0 | 2.3E-02 | |||||
| CNAG_02035 | triose-phosphate isomerase | 0.8 | 2.3E-02 | 2.1 | 1.4E-02 | ||||
| CNAG_03072 | enolase (ENO1) | 1.3 | 2.6E-04 | 2.2 | 6.5E-03 | ||||
| CNAG_06868 | phosphopyruvate hydratase (ENO2) | 0.4 | 3.2 | 3.3E-02 | |||||
| TCA cycle (p-value: 3.2E-04) | |||||||||
| CNAG_00061 | citrate synthase, mitochondrial | 1.3 | 2.5E-03 | 2.4 | 1.4E-02 | ||||
| CNAG_03674 | oxoglutarate deHase, E1 component | 0.9 | 3.3E-02 | 2.3 | 4.4E-03 | ||||
| CNAG_03596 | 2-oxoglutarate deHase E2 component | 0.6 | 2.0 | 2.4E-02 | |||||
| CNAG_07363 | isocitrate deHase, NAD-dependent | 1.0 | 3.4E-03 | 2.8 | 2.0E-03 | ||||
| CNAG_07851 | isocitrate deHase, NAD-dependent | 2.1 | 1.8E-09 | 2.7 | 5.7E-04 | ||||
| CNAG_05653 | malate synthase A (MLS1) | 1.8 | 2.4E-05 | 7.8 | 1.7E-11 | ||||
| Pentose-phosphate shunt (p-value: 5.3E-04) | |||||||||
| CNAG_03245 | glucose-6-phosphate deHase | 0.6 | 2.8 | 3.2E-04 | |||||
| CNAG_02133 | 6-phosphogluconolactonase | 0.6 | 2.6 | 8.7E-04 | |||||
| CNAG_01492 | hypothetical 6-phosphogluconate deHase | 0.5 | 2.7 | 1.0E-03 | |||||
| CNAG_04099 | 6-phosphogluconate deHase, decarboxylating | 1.2 | 1.2E-02 | 3.3 | 3.8E-05 | ||||
| CNAG_07561 | 6-phosphogluconate deHase, decarboxylating 1 | 0.5 | 2.3 | 8.9E-03 | |||||
| Others | |||||||||
| CNAG_06313 | phosphoglucomutase | 1.2 | 4.0E-04 | 2.7 | 1.9E-04 | ||||
| CNAG_06923 | phosphoketolase | 0.8 | 3.0 | 1.6E-04 | |||||
| CNAG_01155 | glycerol kinase | 1.3 | 6.6E-05 | 4.5 | 2.4E-06 | ||||
| CNAG_03113 | trehalose synthase | 1.1 | 3.7E-04 | 3.7 | 3.2E-03 | ||||
| CNAG_03765 | trehalose-phosphatase (TPS2) | 0.8 | 3.1 | 4.9E-03 | |||||
| CNAG_05292 | α,α-trehalose-phosphate synthase (TPS1) | 0.5 | 2.7 | 2.4E-03 | |||||
| CNAG_00373 | glucan 1,3-β-glucosidase | 0.3 | 2.0 | 4.7E-02 | |||||
| CNAG_05138 | glucan 1,3-β-glucosidase | 0.7 | 2.9 | 7.8E-03 | |||||
| CNAG_05652 | glucan 1,3-β-glucosidase, putative | 0.7 | 2.3 | 2.3E-02 | |||||
| CNAG_04033 | α1,3-glucosidase | 0.6 | 2.5 | 4.8E-03 | |||||
| CNAG_00393 | 1,4-α-glucan-branching enzyme | 0.4 | 2.1 | 2.6E-02 | |||||
| CNAG_04879 | glycogen debranching enzyme | 0.9 | 2.2E-02 | 3.4 | 2.8E-05 | ||||
| CNAG_01164 | glutamine-fructose-6-phosphate transaminase | 0.9 | 2.2 | 1.6E-02 | |||||
| CNAG_01239 | chitin deacetylase | 1.5 | 3.3 | 1.9E-02 | |||||
| CNAG_02445 | phosphoacetylglucosamine mutase | 0.8 | 2.5 | 2.5E-02 | |||||
| CNAG_06098 | glucosamine-6-phosphate deaminase | 0.9 | 2.1E-02 | 2.4 | 6.4E-03 | ||||
| CNAG_06659 | hexosaminidase | 0.7 | 6.2 | 2.4E-06 | |||||
| Intracellular protein transport (p-value: 1.2E-05) | |||||||||
| COPI and COPII vesicles | |||||||||
| CNAG_03554 | coatomer, subunit α (COP1, SEC33, RET1) | 0.9 | 2.2E-02 | 2.1 | 6.0E-03 | ||||
| CNAG_03299 | coatomer beta subunit (SEC26) | 1.2 | 1.1E-03 | 2.4 | 7.8E-03 | ||||
| CNAG_04074 | coatomer beta' subunit (SEC27) | 1.2 | 9.5E-03 | 2.2 | 1.2E-02 | ||||
| CNAG_01274 | coatomer subunit gamma (SEC21) | 1.1 | 1.0E-03 | 2.1 | 2.3E-02 | ||||
| CNAG_01211 | putative coatomer subunit epsilon (SEC28) | 0.7 | 2.0 | 1.5E-02 | |||||
| CNAG_06773 | protein transporter SEC24 (SEC24) | 1.2 | 2.9E-02 | 2.8 | 5.5E-03 | ||||
| CNAG_04803 | protein transporter SEC31 (SEC31) | 1.3 | 5.6E-03 | 2.7 | 7.9E-03 | ||||
| CNAG_00148 | COPII coat assembly protein Sec16 | 1.1 | 1.8E-02 | 2.8 | 6.1E-03 | ||||
| clathrin-dependent transport from the TNG to endosomes | |||||||||
| CNAG_07318 | AP-1 complex subunit gamma-1 (APL4) | 0.7 | 2.1 | 2.4E-02 | |||||
| CNAG_00977 | ADP-ribosylation factor-binding protein (GGA2) | 0.8 | 2.0 | 2.4E-02 | |||||
| CNAG_04499 | clathrin light chain | 0.8 | 2.2 | 8.0E-03 | |||||
| CNAG_04904 | clathrin heavy chain | 1.0 | 2.5E-03 | 2.3 | 5.6E-03 | ||||
| Others | |||||||||
| CNAG_01653 | cytokine inducing-glycoprotein Cig1 | 1.0 | 1.4E-02 | 3.5 | 2.0E-03 | ||||
| CNAG_00815 | Siderophore-iron transporter | 1.2 | 1.1E-03 | 4.4 | 6.3E-11 | ||||
| CNAG_02500 | calnexin | 3.1 | 1.0E-12 | 3.6 | 2.0E-05 | ||||
Diverse genes encoding closely related enzymes involved in amino sugar metabolism were also induced, including glutamine-fructose-6-phosphate transaminase (CNAG_01164), phosphoacetylglucosamine mutase (CNAG_02445), glucosamine-6-phosphate deaminase (CNAG_06098), and N-acetylglucosamine-6-phosphate deacetylase (CNAG_06098), plus CNAG_02355, coding for a UDP-xylose/UDP-N-acetylglucosamine transporter. The expression of two additional genes related to chitin degradation (CNAG_01239, encoding chitin deacetylase, and CNAG_06659, hexosaminidase), was also increased.
Gene Ontology analysis did not report any significantly enriched category for the set of gene repressed at 7h when the Benjamini-Hochberg adjustment method was used. However, when a non-adjusted search was conducted, we found several genes related to ribosome biogenesis, DNA repair and DNA replication. In particular, three genes (CNAG_00681, CNAG_01959, and CNAG_03148) involved in the nuclear condensin complex, required for establishment and maintenance of chromosome condensation, and 3 out of 4 components of the GINS complex (CNAG_03374, CNAG_02884, and CNAG_04682) which participates in both initiation and elongation of DNA replication. Similarly, the expression of two cyclin-encoding genes (CNAG_03385 and CNAG_00442) belonging to the G1-phase Pcl1,2 family, was clearly repressed (Table 2). These assignations were confirmed by exploring the FunCat classification ontology. Analysis of the repressed genes after 18 h incubation yielded two main GO categories: genes related to transmembrane transport and to the regulation of RNA polymerase II promoters. In the former category there were included diverse sugar transporters (CNAG_01862, CNAG_02733, CNAG_05662, CNAG_06292, and CNAG_06932), plus a likely glycerol proton symporter similar to budding yeast Stl1 (CNAG_04784). The latter category contained 14 putative transcription factors, most of them belonging to the zinc finger transcriptional activator family. However, in most cases their sequence similarity with previously characterized transcription factors was rather low, thus precluding a tentative functional assignment.
Table 2. Genes significantly repressed (>2.0-fold and p<0.05) at 7 or 18h during the formation of titan-like cells.
Functional assignments were based on Gene Ontology. For unassigned genes, additional Blastp comparisons against fungal data bases were carried out. T: Titan-like cells (cells incubated in TCM at 104 cells/mL); S (cells of regular size, incubated in TCM at 106 cells/mL).
| 7 h | 18 h | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Process | Gene ID | Description | T/S ratio | p-val. | T/S ratio | p-val. | |||
| DNA Replication adn DNA Repair (p-value: 1.3E-03) | |||||||||
| CNAG_00680 | kinetochore protein Nuf2, variant | 0.1 | 3.6E-04 | 0.4 | 6.9E-03 | ||||
| CNAG_00681 | condensin complex subunit 3, variant | 0.2 | 6.7E-03 | 0.6 | |||||
| CNAG_00991 | flap endonuclease 1 | 0.1 | 2.4E-03 | 0.7 | |||||
| CNAG_01144 | replication factor A1 | 0.3 | 4.1E-02 | 0.8 | |||||
| CNAG_01430 | hypothetical protein | 0.1 | 2.8E-03 | 0.6 | |||||
| CNAG_01916 | DNA mismatch repair protein Msh6 | 0.3 | 4.8E-02 | 0.6 | |||||
| CNAG_01959 | condensin complex subunit 1 | 0.2 | 1.3E-02 | 0.4 | 6.6E-03 | ||||
| CNAG_02491 | nuclear protein | 0.1 | 3.4E-02 | 0.2 | 2.4E-03 | ||||
| CNAG_02884 | DNA replication complex GINS protein Psf2 | 0.2 | 2.8E-02 | 0.7 | |||||
| CNAG_02963 | anaphase-promoting complex subunit 8 | 0.1 | 2.6E-02 | 0.4 | |||||
| CNAG_03148 | nuclear condensin complex protein | 0.2 | 2.5E-02 | 0.5 | 4.6E-02 | ||||
| CNAG_04231 | hypothetical protein | 0.2 | 3.9E-02 | 0.5 | |||||
| CNAG_04682 | DNA replication complex GINS protein Psf3 | 0.1 | 6.5E-04 | 0.2 | 4.5E-03 | ||||
| CNAG_05177 | DNA polymerase kappa subunit | 0.1 | 6.0E-04 | 0.3 | 3.4E-03 | ||||
| CNAG_05468 | AP endonuclease 1 | 0.2 | 3.2E-02 | 0.4 | |||||
| CNAG_05891 | TDG/mug DNA glycosylase | 0.2 | 2.9E-02 | 0.8 | |||||
| CNAG_06273 | hypothetical protein | 0.2 | 2.8E-02 | 0.5 | |||||
| CNAG_06588 | hypothetical protein, variant | 0.2 | 3.9E-02 | 0.5 | |||||
| CNAG_06634 | DNA polymerase epsilon subunit B | 0.2 | 1.5E-02 | 0.5 | |||||
| CNAG_06793 | exonuclease 1 | 0.2 | 2.6E-02 | 0.6 | |||||
| CNAG_07657 | U3 small nucleolar RNA-associated protein 19 | 0.2 | 4.6E-02 | 0.5 | |||||
| Ribosome biogenesis (p-value: 1.1E-03) | |||||||||
| CNAG_01104 | ribosomal RNA-processing protein 8 (RRP8) | 0.2 | 7.3E-04 | 0.5 | |||||
| CNAG_01550 | pre-rRNA-processing protein (TSR3) | 0.1 | 6.1E-04 | 0.5 | |||||
| CNAG_01715 | ribosome biogenesis protein (BMS1) | 0.2 | 1.2E-02 | 0.7 | |||||
| CNAG_01812 | large subunit ribosomal protein L24e | 0.2 | 4.8E-02 | 0.4 | |||||
| CNAG_02672 | nonsense-mediated mRNA decay 3 (NMD3) | 0.2 | 2.5E-02 | 0.5 | |||||
| CNAG_03724 | ribosomal RNA methyltransferase Nop2 | 0.3 | 3.3E-02 | 0.5 | |||||
| CNAG_03954 | ribosomal RNA-processing protein 17 | 0.2 | 1.9E-02 | 0.4 | |||||
| CNAG_04259 | H/ACA ribonucleoprotein complex subunit 1 | 0.2 | 4.5E-02 | 1.0 | |||||
| Transmembrane transport (p-value: 4.8E-6) | |||||||||
| CNAG_00726 | Allantoate permease (DAL5) | 0.0 | 0.0 | 3.4E-02 | |||||
| CNAG_00898 | multidrug efflux pump, YOR378W-related | 0.1 | 0.0 | 4.9E-03 | |||||
| CNAG_00904 | aflatoxin efflux pump Aflt | 0.1 | 0.0 | 7.5E-03 | |||||
| CNAG_01118 | putative dicarboxylic amino acid permease | 0.1 | 1.0E-03 | 0.2 | 4.2E-02 | ||||
| CNAG_01545 | hypothetical protein, variant | 0.2 | 0.3 | 3.3E-02 | |||||
| CNAG_01862 | hexose transporter (STL1, HXT8, HXT13,. family) | 0.3 | 0.2 | 4.7E-02 | |||||
| CNAG_02595 | hypothetical protein, variant | 0.1 | 2.7E-02 | 0.2 | 1.5E-02 | ||||
| CNAG_02733 | hexose transporter (STL1, HXT8, HXT13,. family) | 0.3 | 0.0 | 6.9E-03 | |||||
| CNAG_02777 | Pi transporter PHO84 | 0.4 | 0.5 | 1.4E-02 | |||||
| CNAG_04567 | putative TPO-related spermine transporter | 0.2 | 0.3 | 2.8E-02 | |||||
| CNAG_04597 | hypothetical protein | 0.2 | 0.3 | 1.8E-02 | |||||
| CNAG_04784 | Sugar transporter (STL1, SNF3 family) | 0.1 | 0.2 | 3.3E-02 | |||||
| CNAG_04794 | spermine transporter (TPO2, TPO3 family) | 0.2 | 5.3E-04 | 0.5 | 4.4E-02 | ||||
| CNAG_04818 | MFS monocarboxylate transporter, putative | 0.1 | 2.2E-02 | 0.2 | 2.4E-02 | ||||
| CNAG_05662 | hexose transporter (HXT8, HXT13, HXT17 family) | 0.1 | 1.2E-02 | 0.3 | 2.2E-02 | ||||
| CNAG_05867 | L-fucose permease, putative | 0.2 | 0.2 | 1.1E-05 | |||||
| CNAG_05992 | Allantoate permease | 0.1 | 0.0 | 1.9E-02 | |||||
| CNAG_06102 | ADP,ATP carrier protein, variant 1 | 0.1 | 6.6E-05 | 0.2 | 7.5E-03 | ||||
| CNAG_06292 | sugar transporter | 0.1 | 0.2 | 2.5E-02 | |||||
| CNAG_06557 | N-acetylglucosamine-specific transporter | 0.6 | 0.0 | 3.2E-03 | |||||
| CNAG_06561 | allantoate transporter | 0.0 | 0.0 | 2.6E-02 | |||||
| CNAG_06758 | multidrug efflux pump, YOR378W-related | 0.3 | 0.2 | 2.9E-04 | |||||
| CNAG_06932 | sugar transporter (STL1, HXT17, HXT13 family) | 0.2 | 0.2 | 6.7E-03 | |||||
| CNAG_07449 | General amino acid permease (AGP2) | 0.1 | 0.2 | 2.8E-02 | |||||
Interestingly, several genes encoding proteins involved in Golgi-related protein traffic were induced, specifically five out of the seven components of the COPI-coated vesicles, also known as coatomer (α, β, β', γ, and ε, encoded by CNAG_03554, CNAG_03299, CNAG_04074, CNAG_01274, and CNAG_01211, respectively). COPI-coated vesicles are found associated with Golgi membranes, are involved in Golgi to endoplasmic reticulum (retrograde) vesicle transport, and also likely in intra-Golgi transport. The expression of components of the COPII coat, such as CNAG_06773 (SEC24) and CNAG_04803 (SEC31), as well as that of the interacting protein CNAG_00148 (SEC16) also increased. In addition, genes encoding proteins required for cargo-selective, clathrin-dependent transport from the TNG to endosomes in yeast were induced. These are CNAG_00977, encoding a likely homolog of the budding yeast monomeric clathrin adaptor proteins GGA1 or GGA2, CNAG_07318, encoding the AP-1 complex subunit gamma-1, CNAG_06564, coding for an AP-1 interacting protein, or the genes coding for clathrin light and heavy chains (CNAG_04499 and CNAG_04904).
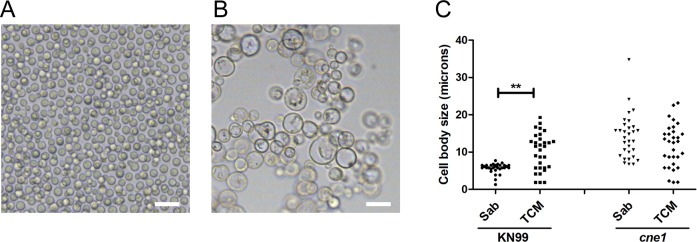
One of the genes clearly overexpressed at both 7 and 24 h encoded Calnexin (CNAG_02500), which is a chaperone located at the ER that contributes to the proper folding of glycoproteins. To test if this protein had any effect on titan cell formation, we obtained the mutant and investigated its phenotype. As shown in Fig 12, in the absence of calnexin, the cells had significantly larger size in regular rich medium compared to the wild type strain (Fig 12), and no change was detected when the cells were transferred to TCM. This result confirmed that calnexin was involved in processes that regulate cell size in C. neoformans.
Fig 12. Phenotype of the mutant lacking calnexin (cne1).
Pictures of the WT (KN99 strain,A) and a cne1 mutant (lacking calnexin, CNAG_02500, B) in liquid Sabouraud medium. The scale bars denote 20 μm. (C) Measurement of cell body size of the WT strain (KN99) and the mutant lacking calnexin. The cells were incubated in liquid Sabouraud at 30°C or TCM (overnight) at 37°C with 5% CO2.
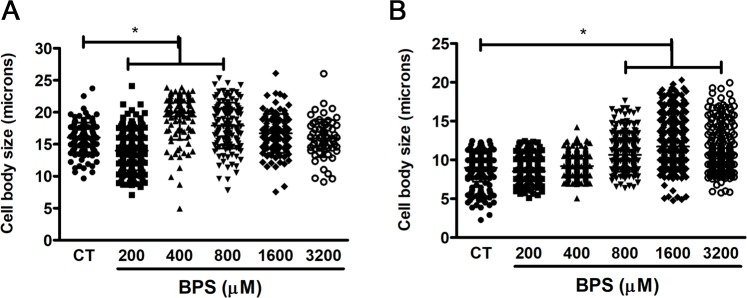
We also noticed that some genes related to iron metabolism were also induced in titan-like cells compared to cells of regular size. One of them (CNAG_01653) encoded Cig1, (initially annotated as cytokine inducing glycoprotein), which is involved in iron uptake from heme groups and is overexpressed during iron limitation conditions [46, 47]. Iron is an important element required by yeasts, and in C. neoformans, its limitation also induces capsular enlargement. For this reason, we investigated whether iron concentration had any effect on titan-like cell formation. As shown in Fig 13, depletion of iron using the chelator BPS induced the appearance of cells of large size when the cultures were inoculated at high cell densities (106 cells/mL), suggesting that iron limitation also contributes to cellular enlargement.
Fig 13. Effect of iron chelation on cryptococcal morphology.
The effect of iron chelation on cell body size was observed by the supplementation of TCM with the iron chelator Bathophenanthroline disulfonic acid (BPS) in TCM cultures inoculated at 104 cells/mL cultures (A) and 106 cells/mL cultures (B). The cells were incubated at 37°C overnight with 5% CO2 without shaking. CT, control cells without BPS.
Discussion
Cryptococcus neoformans is an exceptional model to understand mechanisms induced by pathogenic fungi to adapt to the host and cause disease. Some of the most important changes are related to changes in cell size, which can occur by growth of the capsule, or growth of both the cell body and capsule. In this last case, C. neoformans induces a specific cell type that has been denominated titan cells [20, 21]. At the moment, only the cAMP-dependent and the mating pathways have been described as relevant in titan cell formation [20, 21, 27]. It has been shown that increased expression of PKA induces cell enlargement in C. neoformans and the ploidy of the cells [48, 49]. One of the limitations to study titan cells is the difficulty of reproducing this phenomenon in vitro. Originally, we described that a small proportion of titan cells could be observed in minimal media [21]. Then, it was described that these cells appear in the presence of phospholipids [28]. In this work, we have defined a medium in which we have consistently replicated this phenomenon in vitro. This medium has several characteristics: limitation of nutrients at neutral pH, inclusion of mammalian serum and a CO2 enriched atmosphere. In these conditions, we could obtain a high proportion of cells of a size around 30 μm (capsule included). This diameter is smaller than the average size measured in cells isolated from mouse lungs [11, 20, 22], where average size is around 40–50 μm, so we argue that in our TCM we could be examining the initial steps of this morphological transition. It is possible that in vitro the cells do not reach the same size than cells obtained from in vivo infections because the nutrients of the medium or the factors that favor this morphological change are consumed. Instead, in vivo infections are maintained for a longer time, with a stable nutrient concentration allowing cells to reach significantly larger sizes. Despite this limitation, the availability of in vitro conditions that, at least in part mimic titan cell formation is a key contribution to understand the biology of these cells.
In general, our results indicate that titan-like cell formation is induced by multiple factors, being some of them necessary, but not sufficient, to trigger the transition. This is the case of serum which only induced cell growth under nutrient limiting conditions. These results indicate that titan-like cells are formed in response to some elements of the host present in the serum in the context of the stress produced by the limitation of nutrients in the medium. These findings allow the dissection of the intracellular pathways that are triggered during cellular growth.
It has been described that serum and the fraction of polar lipids induce cell enlargement in C. neoformans. Our results agree with previous findings [28], because polar lipids were also able to induce the formation of titan cells in TCM. The most plausible mechanism for this effect is that phospholipids are degraded by phospholipase C, which produces diacylglycerol (DG), or by phospholipase B, which produces arachidonic acid (AA) [50]. DG activates human as well as C. neoformans PKC [34]. To investigate the role of this signaling pathway, we blocked the activity of this kinase with several pharmacological inhibitors, and found that both staurosporine and calphostine C had a dramatic effect on titan-like cell formation. This protein participates in multiples processes, such as maintenance of the integrity of the cell wall [51–53] and polarized growth [54]. Mutants lacking the PKC1 gene present many cellular alterations, such as osmotic instability and susceptibility to temperature [35]. We tried to evaluate the effect of these inhibitors at a lower temperature and in the presence of sorbitol in an attempt to overcome the cellular defects associated with the absence of Pkc1. For this reason, further studies using mutants affected in signaling pathway components are required to understand in detail how this pathway participates in titan-like cell formation.
We also observed that other important factor for titan-like cell formation in vitro is CO2. The capsule is induced in response to CO2 concentrations present in the host [55], which is consistent with this factor also favoring the growth of the cell body. Carbonic anhydrase converts CO2 into HCO3-, which activates adenylate cyclase [44, 56], so our results are consistent with the hypothesis that CO2 induces cell growth through the cAMP pathway. In C. neoformans, there are two carbonic anhydrases encoding genes, CAN1 and CAN2, with CAN2 being the most important [44]. Interestingly, deletion of CAN1 resulted in an enhancement of the production of titan-like cells. Although we do not know the molecular mechanism for this phenomenon, we postulate that in the absence of Can1, there might be a compensatory overexpression of Can2 that could induce the hyperactivation of the cAMP pathway.
We also observed that subinhibitory concentrations of azide have a modest, but reproducible positive effect on titan-like cell development. We argue that a partial inhibition of the respiratory chain can trigger a stress signal that results in a stop of the cell cycle and allows cellular size increase. In this regard, it could be assumed that a limitation in the respiratory capacity might lead the organism to generate energy, at least in part, by fermentative metabolism, a situation that has been associated to increased PKA activity in many fungi [57]. Although C. neoformans is mainly a respiratory yeast, it has been shown that it can produce both ethanol and acetate in vitro and in vivo [58, 59], so a similar activation of the PKA activity could also occur in these conditions. However, the effect of the mitochondrial inhibitor was not significant in the presence other factors (in particular, serum and CO2). Since CO2 is an activator of the adenylate cyclase, it is reasonable to argue that in these conditions the effect of azide would be not significant.
One of the most striking results of this work is the effect of cell density on the formation of titan-like cells, suggesting that this transition is regulated by quorum sensing (QS) phenomena. In this way, a higher cell density results in a higher concentration of released molecules and greater effect on the cells [29, 60, 61]. A major QS signal for C. neoformans is a 11-mer peptide (Qsp1) [31, 32]. We tested the effect of this peptide on titan-like cell formation, and found that it inhibited the transition. Qsp1 promotes cell division and replication. Although further studies are required to understand the molecular mechanism by which Qsp1 regulates this transition it is plausible to propose that, since titan-like cells are formed in the absence of budding, Qsp1 blocks this development due to its positive effect of cell division and replication. The biological role of QS phenomena on titan-like cells is not known. However, it is worth noting that a cell-dose relationship similar to that found here has been described in vivo, since a low number of cryptococcal CFUs in the lung correlates with a higher proportion of titan cells [21]. Therefore, it will be necessary to elucidate the role of Qsp1 in the context of lung colonization in the future. However, this peptide does not seem to be the only factor that represses cellular growth at high densities, since qsp1 mutants form titan-like cells similarly to the wild type strain. In agreement with this notion, C. neoformans produces QS molecules that are not susceptible to high temperature, proteinase, trypsin, pronase, DNAse, RNAse and glucosidase [30]. These authors also described that farnesol, tyrosol and Qsp1 did not replicate the effects observed with their conditioned media, and demonstrated that pantothenic acid could in part reproduce QS phenomena. In summary, we hypothesize that several QS molecules negatively regulate titan-like cell formation.
The formation of titan cells has been observed in clinical samples [62–64], and despite being a mechanism that confers advantages to C. neoformans against the host during infection, we observed that this process is not a universal phenotype. Our results showed that there is great variability among different isolates, and not all the strains did form titan-like cells in vitro. Our work provides a new way to investigate the genetic differences between strains with high and low capacity to form titan-like cells using genomic approaches. However, the strains of C. neoformans var. grubii (serotype A) exhibited the highest proportion of titan-like cells, suggesting that this serotype has a greater capacity of adaptation to the lung. This result is in agreement with the literature, since this serotype is the one most frequently isolated in infected patients [10], suggesting that there is a correlation between the ability to form titan cells and development of the disease. We believe that this correlation should be confirmed in future clinical studies.
Previous reports demonstrate that during capsule growth, the size of this structure correlates with cell body size [65]. In addition, capsule growth mainly occurs in G1 [66], which is also the cell cycle phase in which the growth of the cell body occurs. However, although titan-like cells increased capsule thickness, the size of this structure was significantly smaller than the one found in titan cells isolated from animals, indicating that the process that occurs in vitro is, at least quantitatively, not fully equivalent to the one observed in vivo. We argue that the phenomenon that we observed in vitro in this work describes the first steps of titan cell formation. In addition, it is reasonable to argue that when the cell body enlarges significantly, there could be a metabolic limitation affecting the growth of the capsule, since its volume increases with the cube value of the radius.
Titan cells contribute to virulence through different mechanisms, such as polarization of Th2-type immune responses [23, 24], replication [26, 67], resistance to oxidative damage [20, 21] and phagocytosis avoidance [21, 25]. In our case, titan-like cells obtained in vitro were not phagocytosed, most probably due to their large size. In our case, these cells did not prevent the phagocytosis of cryptococcal cells of regular size, as it is the case in experiments performed in vivo [25]. We believe that activation of the macrophages in vivo is different from in vitro, so in the lungs there could be multiple factors that affect the phagocytic activity of macrophages, explaining why the effect of titan cells in vivo on macrophages are not identical from the effects induced in vitro. Furthermore, it is also possible that titan cells in vivo express virulence factors or epitopes that are not produced in vitro. Further studies are required to fully understand the response of immune cells to cryptococcal titan-like cells.
The isolation of titan-like cells in vitro opens new perspectives and research lines. For example, we investigated gene expression changes associated with titan cells development. In our approach, we compared cells of different size incubated in the same medium (TCM), so the cells were exposed to the same nutritional environment and inducing factors (CO2 and serum). The difference in cell size was obtained due to the different initial cellular density of the cultures (104 vs 106 cells/mL). In this way, our approach would also detect those changes due to quorum sensing phenomena. Despite the cells were incubated in the same medium, we detected changes in many genes involved in metabolism at both times (7 and 18 h). This suggests that the increase in cell size requires a metabolic adaptation that allows the efficient use of extracellular and intracellular sources of nutrients. The induction of many genes involved glycolysis and TCA cycle suggest a higher demand of energy, whereas the increase in the expression most of the components of the oxidative branch of the pentose phosphate shunt (including glucose-6P dehydrogenase, which catalyzes the rate-limiting step) suggest an increased need for NADPH, a requirement for anabolic biosynthetic pathways. Such requirement could be explained by a higher demand for fatty acid synthesis to provide structural components for an increasing membrane surface. Alternatively, it could be considered that induction of a large number of metabolic enzymes would be simply required to maintain its normal concentration in response to a growing cellular volume.
Our results also highlight other processes that might be involved in titan cell formation. In particular, we found that several genes encoding components of COPI and COPII vesicles, which belong to the coatomer complex. These proteins cover the surface of vesicles involved in intracellular protein sorting and early secretory pathway. Interestingly, we found that clathrin- encoding genes were also induced. Although we do not know how these proteins contribute to cryptococcal cellular enlargement, we argue that titan cells might recycle, import and export proteins at a higher rate than regular cells. Interestingly, we also found that the expression of genes encoding some chaperones, such as calnexin, involved in folding of glycosylated proteins increased in titan-like cells, and mutants lacking this protein have an abnormal large size, even in regular rich medium. This result was somehow unexpected, because the expression of this gene increased in titan-like cells. In consequence, we argue that calnexin plays a role, not only in proper folding of glycosylated proteins, but also regulates cell size in conditions of active protein synthesis and replication. In addition, it is possible that it absence results in an stress signal that results in cell cycle changes and G1 arrest, that would produce an increase in cell size too. In this sense, mutants lacking Atg7 (which is part of the ubiquitin-activating enzyme (E1) family and is required for proper authophagy [68]) have also increased cell size and higher DNA content, supporting the idea that titan formation is affected by the trafficking and recycling of intracellular proteins.
Our results indicated that iron depletion from the medium can also regulate the induction of titan-like cells. Interestingly, iron limitation also induces the growth of the cryptococcal capsule [69]. Iron is an important cofactor required for multiple enzymatic activities, so its absence is an important stress signal that results in the expression different acquisition mechanisms, involving siderophores, Cig1 and transporters [70]. Our findings are in agreement with the idea that titan-like cell formation is a response to nutritional stress.
Titan cells isolated from mice are polyploid, so presumably the factors and signaling pathways must alter the normal progression of cell cycle. Interestingly, we found that in titan-like cells, genes related to DNA replication were repressed. In agreement, G1/S cyclins, like Pcl1, were repressed, which suggests that the process requires an elongation of the G1 phase. This result is in agreement with previous findings that demonstrated that pcl1 mutants have increased capacity to form titan-like cells in vivo [27]. In our titan-like cells, we found that they have a higher content of DNA compared to cells of regular size. However, the DNA content of titan-like cells is lower than the content in titan cells isolated from mice, indicating that cell cycle changes that occur in vitro are different from in vivo.
Finally, we would like to acknowledge that other groups (Dr. Alanio, Pasteur Institute, France, and Dr. Ballou, University of Birmingham, UK) have identified in parallel to our work other conditions in which titan-like cells are formed. We believe that these works together with our findings provide significant advances to the scientific community because they will allow the design of multiple research lines that will facilitate the characterization of the factors and signaling pathways that are involved in titan cell development. The data presented by different groups also indicate that C. neoformans may induce titan-like cells in vitro in response to multiple factors. In addition, the ability to obtain titan cells in vitro will also have a positive bioethical impact because we will be able to significantly reduce the number of experimental animals compared to what was required previously to characterize these cells.
Material and methods
Strains and media
In most of the experiments, we used C. neoformans var. grubii (serotype A) H99 strain [71], but we also included C. neoformans var. neoformans (C. deneoformans, serotype D), A/D hybrids and C. gattii, and different mutants obtained from the library described by Liu and coworkers [41] and from the Fungal Genetic Stock Centre. All strains are described in Table 3. Strains were preserved in Sabouraud medium containing 30% glycerol at -80% and were recovered at 30°C in Sabouraud solid medium (Oxoid LTD, UK).
Table 3. List of strains used.
| Species | Strain | Reference or Source |
|---|---|---|
|
C. neoformans var. grubii (C. neoformans, serotype A) |
H99 | [71] |
| CL-5659 | Mycology Reference Laboratory (CNM) | |
| 3259 KN99 (MATa) | Institut Pasteur | |
| 3260 KN99 (MATα) | Provided by Guilhem Janbon (Institut Pasteur) | |
| H99-GFP | [77] | |
| gat201 | [41] | |
| can1 (NE411) | [44] | |
| can2 (NE 417) | [44] | |
| RPC3 (cac1) | [82] | |
| RPC7 (cac1::CAC1) | [82] | |
| cne1 | This work | |
| cap60 | Obtained from collection described in [41] | |
| qsp1 | [32] obtained from FGSC | |
| ada2 | [42] obtained from FGSC | |
|
C. neoformans var. neoformans (C. deneoformans, serotype D) |
ATCC 24067 | American Type Culture Collection |
| B3501 | [83] | |
| CL-6435 | Mycology Reference Laboratory (CNM) | |
| CL-6760 | Mycology Reference Laboratory (CNM) | |
| JEC20 (MATa) | [84] | |
| JEC21 (MATa) | [84] | |
| NE 822 (MATa) | [85] | |
| NE 824 (MATα) | [85] | |
| C536 (cap59) | [86] | |
|
C.neoformans hybrids (A/D) |
CL-4041 | Mycology Reference Laboratory (CNM) |
| CL-4840 | Mycology Reference Laboratory (CNM) | |
| CL-5706 | Mycology Reference Laboratory (CNM) | |
| CL6434 | Mycology Reference Laboratory (CNM) | |
| C. gattii | NIH 191 | NIH, Bethesda, Maryland |
| NIH 198 | NIH, Bethesda, Maryland | |
| CBS-10514 (R265) | [40] | |
| CBS-10865 (R272) | [40] |
For clarity, we have specified the different suggested nomenclatures for the Cryptococcus species complex, specifying the varieties, serotypes and species name. In the case of C. gattii, we preferred to use the classical nomenclature that groups all the genotypes, serotypes and species because we did not include different strains from all these different groups.
In order to delete the CNE1 (CNAG_2500), a gene a disruption cassette was constructed by overlapping PCR [72] using PCR fragments amplified from either the plasmid pHYG (kindly given by Prof. Jennifer Lodge, Washington University in St Louis, USA) or genomic DNA extracted from the strain KN99α [73].
The C. neoformans strain KN99α was then transformed by biolistic delivery [74] and transformants were screened on YPD medium containing 200 U ml-1 of hygromycin (Calbiochem). The correct deletion of the gene was tested using primers within and outside the cassette. Finally, the absence of additional ectopic integration of the cassette was checked by Southern blot experiment. Two independent cne1Δ::HYG strains (NE375 and NE376) were selected for further studies.
The primers used in these PCR experiments are listed below
CNE1seroAex
CCATCTCTTCTTCGGAATCCG
CNE1seroA-5’5
TAGCACTGTGAATCGATCCCG
CNE1seroA-5’3
GTCATAGCTGTTTCCTGGGATGGGATGAATGGAAGACG
MKRrCNE1seroA
CGTCTTCCATTCATCCCATCCCAGGAAACAGCTATGAC
CNE1seroA-3’5
TACAACGTCGTGACTGGGGAGATTCCTGCTGAAGGCTCG
MKRfCNE1seroA
CGAGCCTTCAGCAGGAATCTCCCCAGTCACGACGTTGTA
CNE1seroA-3’3
TGTTACGTTCGACTTGACGCTG
CNE1seroAex2
ACAACGCTTCGACATCTGCAG
The yeasts were routinely grown in liquid Sabouraud medium at 30°C or 37°C with shaking (150 r.p.m.). To induce titan-like cells, strains were grown in a medium that we have defined as Titan Cells Medium (TCM), which is based in the medium described to induce capsule growth (10% Sabouraud buffered at pH 7.3 with 50 mM MOPS, [75]). TCM contains 5% Sabouraud and 5% inactivated fetal calf serum (FCS, Biological Industries) diluted in MOPS 50 mM at pH 7.3 plus 15 μM sodium azide (Sigma Aldrich). Cultures were grown in tissue culture flasks or 96-wells plates at 37°C in an atmosphere enriched with CO2 for 18 hours. For iron depletion different concentrations of bathphenanthroline disulfonate (BPS, Sigma Aldrich) were prepared directly in TCM. Cells were then inoculated at 104 or 106 per mL and incubated at 37°C with 5% CO2 for 18 hours.
India Ink staining and measurement of cell size and capsule permeability
To observe and measure the size of the cells, 10 μL of a cell suspension were mixed with a drop of India Ink drop (Remel Bactidrop, Lenexa, Kansas) and observed under a Leica DMI 3000B microscope. Pictures were taken with a Leica DFC 300FX camera using the Leica Application Suite (Leica Microsystems) and processed with Adobe Photoshop 7.0 (San Jose, CA) or ImageJ (https://imagej.nih.gov/ij/) [75]. In some experiments, the permeability index was measured using fluorescently-labeled dextrans as described in [67].
Infections of mice with C. neoformans and yeast isolation from lungs
Six to eight weeks-old male mice from C57BL/6J (in house bred at the National Centre for Microbiology) were used in all experiments. The animals were kept in ventilated racks at 22–24°C with proper environmental enrichment (cupboard houses and hollow cylinders).
Yeast cells were incubated overnight in liquid Sabouraud medium at 30°C, centrifuged at 2830 g, washed and suspended in sterile PBS. The cell density was determined using a TC20 cell counter (BioRad) and a suspension of 3.3x107 cells/mL was prepared in sterile PBS. The animals were anesthetized with a mixture of ketamine (Imalgene 1000, 50 mg/Kg) and xylazine (Xilagesic 2%, 20 mg/Kg) and infected intranasally with 30 μL of the yeast suspension (106 cells per mouse) as previously described [23]. Animals were sacrificed after 14 days of infection by exposure to a high CO2 enriched environment.
We excised and homogenized the lungs in 10 mL of sterile water using cell strainers (100 μm size pore, BD Falcon) and a 5 mL syringe plunge in Petri plates. This process disrupts the mammalian cells without significantly affecting the integrity of the cryptococcal cells. Then the cell suspension was centrifuged at 2,830 g and washed with sterile water three times to fully break and remove the mouse cells. Finally, the yeasts were suspended in PBS.
Lipids extraction of fetal bovine serum (FBS)
Polar lipids from serum were obtained as described in [28]. Briefly, aliquots of 1 mL of FBS were shaken with a mixture of chloroform and methanol (2:1) (v:v) for 3 hours at room temperature. The samples were centrifuged for 10 minutes at 2,830 gs for phase partitioning. The upper phase was collected in a 1.5 mL tube and dried during 1 hour in a SpeedVac concentrator. The pellet was suspended in 200 μL of PBS and conserved at 4°C. To evaluate the effect of serum polar lipids on titan-like cell formation, we performed experiments with different amounts of the extraction solution described above (1/40; 1/100 and 1/200 dilution) in 5% Sabouraud buffered at pH 7.3 with 50 mM MOPS and 15 μM sodium azide. As a control, the same medium with PBS was used. In parallel, cells were grown in Sabouraud and TCM as growth control and titan-like cell formation respectively.
To evaluate the role of phosphatidylcholine (Sigma Aldrich), a 20 mM stock of this phospholipid was prepared in EtOH 100%. Then, 1/10 dilutions were done in distilled water to yield 0.1, 0.01 and 0.001 mM (final concentration) in TCM without serum. Same dilutions of EtOH 100% were added to the medium as control.
Visualization of titan-like cell formation by real-time microscopy
Yeast cells were inoculated in TCM at 104 cells/mL as detailed above. One hundred seventy μL from the yeast suspension were placed in a 96 well plate and incubated at 37°C with 5% CO2 under a Leica DMI 4000B microscope. Photographs were taken every 3 min using the 20x objective. The videos generated by the Leica software were exported as AVI documents and processed with ImageJ software. In all cases, the videos were assembled with 12 frames per second, so one second of video corresponds to 36 minutes of real time. Time was included in each frame using the Time Stamper plugin from ImageJ.
Effect of conditioned media and Qsp1 on titan-like cell formation
Cell suspensions were prepared at a concentration 106 cells/mL in parallel in Sabouraud with 15 μM sodium azide and TCM. Serial 1/10 dilutions were made up to 103 cells/mL. A volume of 170 μL of these suspensions was incubated in a 96 well plate at 37°C overnight with 5% CO2 without shaking. The plates were observed with a Leica DMI 3000B microscope. Pictures were taken with a Leica DFC 300FX camera using Leica Mycrosystems software. The cell body diameter of thirty to fifty cells was measured with Adobe Photoshop 7.0.
Cryptococcus neoformans was inoculated at 104 and 106 cells/mL in TCM as described above and incubated at 37°C in a 5% CO2 enriched atmosphere. After 18 h of incubation, the cultures were centrifuged and the supernatants collected to yield Titan-like Cell Supernatant (TCS) and regular cells supernatant (RCS). To evaluate the influence of these supernatants on the titan-like cell formation, C. neoformans cultures inoculated at 104 cells/mL in 96 wells plates were prepared in different growth conditions: 1) Fresh TCM medium (TCM), 2) Supernatant from cultures of titan-like cells (TCS), 3) Supernatant from cultures of cells of regular size (RCS). These conditioned media were mixed with fresh TCM (1:1 proportion v/v). As control, we carried out a culture in which fresh TCM was diluted with the same volume of distilled sterile H2O. After the inoculation of the different media and mixtures with C. neoformans at 104 and 106 cells/mL and incubation at 37°C in a CO2 incubator for 18 h, the cell size was measured by microscopy as described above.
Chemical synthesis of the peptides was done by the proteomic facility of the National Centre for Biotechnology (CSIC, Spain) using an Multipep automatic synthesizer (Intavis, Köln, Germany) and Fmoc-Amino Acid Wang resins (Merck, Darmstadt, Germany). After release from the resin, the peptides were purified by reverse-phase chromatography in a semipreparative HPLC system (Jasco, Tokio, Japan) with a C18 Kromaphase column (Scharlab, Barcelona, Spain). The fractions were analyzed by mass spectrometry and lyophilized until their use. We synthesized peptides described in [32]: Qsp1 (NFGAPGGAYPW), an inactive version of this peptide (NFGAPGAAYPW) and a scrambled Qsp1 peptide (AWAGYFPGPNG). The peptides were dissolved in sterile PBS at 1 mM, and their effect on titan-like cell formation was tested at 30 μM and 15 μM in TCM. The samples were incubated for 18 hours at 37°C with 5% of CO2 in 96 wells plates. After the incubation period, the cells were observed by optical microscopy and the body cell sizes were measured.
Nuclei analysis in titan-like cells
Cryptococcus neoformans cells from H99 strain were cultured overnight in TCM at 104 and 106 cells/mL at 37°C with CO2 without shaking. After the confirmation of titan-like cell formations by optical microscopy, cells were washed with dH2O and fixed with 70% ethanol at 24°C for one hour followed by an overnight incubation at 4°C. After fixing, the cells were washed twice with RNAse A buffer (0.2 M Tris, pH 7.5, 20 mM EDTA) and treated with 10 μg/mL of RNAse A for 4 hours at 37°C. After the incubation, cells were washed twice with PBS, suspended in PBS and incubated overnight at 4°C. Next day, the cells were centrifuged and suspended in a 200 ng/mL solution of 4',6-diamidino-2-phenylindole (DAPI) and incubated in the dark for 10 minutes at room temperature to stain the nucleus. Then, the cells were washed, suspended in PBS and the fluorescence intensity of the nucleus were analyzed by flow cytometry. Cells were examined for cell size by forward scatter parameter (FCS) and granularity by side scatter parameter (SSC) using the BD LSRFortessa X-20 cytometer (BD, Bioscience). Two populations of titan-like and regular cells were delimited and, in each population, the fluorescence intensity of the DAPI staining in 10,000 cells was measured. Data obtained were analyzed with the BD FACSDiva (BD, Bioscience) and FlowJo 10.4.2 (Tree Star Inc, Ashland, Oregon) softwares. The nuclei of the C. neoformans H99 titan-like and regular cells stained with DAPI were also observed by conventional fluorescence in a Leica DMI3000B microscope and confocal microscopy using a Leica SP5 confocal microscope.
Influence of the protein kinase C (PKC) inhibitors and rapamycin on the formation of titan-like cells
The influence of the PKC pathway in the formation of the titan-like cells was evaluated by the addition of four different inhibitors: calphostin C, staurosporine and bisindolylmaleimide I, and genistein (all from Calbiochem) that inhibits tyrosine kinase as a control. For this purpose, 10 μM, 5 μM and 1 μM of calphostin C, bisindolylmaleimide I and Genistein and 0.01 μM and 0.001 μM of staurosporine. In all cases, the final concentration of DMSO was 0.1%, which did not inhibit titan-like cell formation.
Phagocytosis of titan-like cells in vitro with RAW 264.7 macrophages
RAW 264.7 macrophages were maintained in DMEM medium supplemented with heat-inactivated 10% fetal bovine serum (FBS, Hyclone-Perbi), 10% NCTC medium and 1% non-essential amino acids (Sigma-Aldrich, Steinheim, Germany). The day before the experiment, the macrophage monolayer was separated from the plate by pipetting and the cells were centrifuged at 1,265 g. Macrophage suspensions were prepared at 2.5x105 cells/mL. Two hundred μL per well were inoculated into 96 well plates and incubated overnight at 37° C and 5% CO2. The next day, different types of cells (titan-like cells obtained in vitro, and cells of regular size) at a final concentration of 5x105 cells/mL with 5 μg/mL of monoclonal antibody 18B7 [76] were added to the macrophages for 2 hours. Phagocytosis was quantified by two different methods. First, the plates were observed with a Leica DMI 3000B microscope, and the percentage of infected macrophages was determined visually. In some cases, the plate was visualized with a Leica 4000B with a chamber that allowed incubation at 37°C and 5% CO2, and videos were taken as explained above. Alternatively, we quantified the phagocytosis percentage by flow cytometry. In this approach, we performed phagocytosis assays as above, but using larger volumes in 24-well plates containing 1 mL of medium. To differentiate yeast cells, we used a H99 strain that expresses the green fluorescence protein (H99-GFP) [77]. In some experiments, macrophages were exposed for 1 h to titan-like cells (H99 strain, 1:1 ratio) with mAb 18B7. As control, macrophages were also preincubated with the same cells, but without mAb, and also with medium alone (with and without mAb). Then, the wells were washed with fresh medium, and cryptococcal cells of regular size (H99-GFP, grown overnight in Sabouraud at 30°C) were added at 1:1 ratio for 2 h. After the incubation, we washed the plates, and separated the macrophages by continuous pipetting. The cell suspensions were washed and suspended in PBS containing 1% FCS. Macrophages were blocked with anti-Fc mAb (2.4G2, BD Biosciences, 5 μg/mL) for 10 min at 4°C. After that, the macrophages were washed with PBS/FCS, and then incubated with an anti-Mac1 mAb (anti-CD11b/Mac1-PE/Cy7, BioLegend, 1 μg/mL) for 20 min at 4°C in the dark. Finally, the cells were washed and suspended in 4% p-formaldehyde prepared in PBS. The cells were analyzed in a FACS Canto cytometer (Biosciences, California, EEUU) using FASCDiva software (versión 6.1). The phagocytosis percentage was calculated as followed: (number of PE-Cy7+/GFP+)/(total number of the PE/ PE-Cy7+ cells)*100. Experiments were performed in triplicate in three times on different days.
Confocal microscopy
In some experiments, cryptococcal cells were labeled with mAb 18B7 labeled with Alexa-488 [78] at 1 μg/mL to label the capsule, and calcofluor (10 μg/mL, Sigma Aldrich) to label the cell wall. The cells were incubated for 1 h at 37, and then, they were washed and observed in a confocal SP5 Leica microscope.
RNA preparation for RNASeq and transcriptomic data analysis
RNA extraction was performed using Trizol (TRI Reagent, Sigma Aldrich) with some modifications. After incubation of the cells, Trizol was added to the samples immediately and maintained in ice. Cells were broken during 5 minutes with FastPrep -24 (MPTM), alternating 20 seconds beating with 1 minute on ice. The RNAs concentration and quality was determined with a Nanodrop 8000 Spectrophotometer (Thermo scientific). RNA samples (0.1 μg/μL) were treated with DNase using the DNA-free kit (Thermo Fisher Scientific). Then, the RNA samples were purified using RNeasy Mini Kit (Qiagen).
Total RNA samples (0.5–1 μg) were treated to remove rRNA using Ribo-Zero Magnetic Kit (Epicentre, Illumina, San Diego, CA) according to the manufacturer's instructions. Then, mRNA was processed for library preparation using ScriptSeq v2 RNA-Seq Library Preparation kit (Epicentre, Illumina, San Diego, CA). Libraries were quantified using the QuantiFluor RNA System (Promega) and the quality and average size was determined using an Agilent 2100 Bioanalyzer. An Illumina NextSeq 500 High Output (400 M reads, 1x75 cycles) was using for sequencing. The FastQ files generated by the equipment were analyzed for low quality reads and those with Q<30 were removed. Original FastQ documents were uploaded to GEO repository (series record GSE111400). Mapping was carried out with the Bowtie2 software [79] using the C. neoformans var. grubii H99 genome as reference. The SAM files generated by Bowtie2 were analyzed using the software SeqMonk v0.39.0 (Babraham Bioinformatics). Three biological replicates for cells treated for 7 h (comprising 8.3 to 9.5E+06 reads each) and two for 18 h (6.9 and 7.9E+06 reads each) were combined, subjected to the RNA-Seq pipeline and evaluated for differential expression using the DESeq2 algorithm [80] with a FDR of p<0.05. The products of this test were recovered as Annotated Probe Report, which included the coding sequences (overlapping option), and processed in Excel format. To identify functional families significantly overrepresented among the differentially expressed genes, the lists of genes was submitted to Gene Ontology (GO) and FunCat analyses at the FungiFun server (https://elbe.hki-jena.de/fungifun/fungifun.php) [81]. To this end, unless otherwise stated, a Fisher's exact test with significance level set to 0.05 and the Benjamini-Hochberg adjustment method were used. Due to the large number of cryptococcal gene products lacking functional annotation in the databases, a Blastp search was conducted against the proteomes of several different, relatively well-annotated fungi, in order to expand the available functional information on the relevant genes.
Ethics statement
All the animal procedures were approved by the Bioethical Committee and Animal Welfare of the Instituto de Salud Carlos III (CBA2014_PA51) and of the Comunidad de Madrid (PROEX 330/14) and followed the current Spanish legislation (Real Decreto 53/2013).
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5. Before comparison among groups, the normality of each simple was assessed using the Kolmogorov-Smirnov test (non-normal distribution when p<0.1). When normal distribution was assumed, differences were estimated using ANOVA and T-Student. For non-parametric distributions, the Kruskal-Wallis and Mann-Whitney tests were used. Statistical significant is highlighted with asterisks in the figures as follows: p>0.05, not significant (ns); p<0.05 and >0.01 (*); p<0.01 and p>0.001 (**); p<0.001 and p>0.0001 (***); p<0.0001 (****).
Supporting information
In this case, we prepared suspensions of C. neoformans cells at 104 and 106 cells/mL in 30 mL of TCM cell. After the incubation period, the cells were collected by centrifugation, fixed with 4% p-formaldehyde and analyzed by flow cytometry using the BD LSRFortessa X-20cytometer (BD, Bioscience). Two populations of titan-like and regular cells were delimited and, in each population, the fluorescence intensity of the NOP1-mCherry protein in 10,000 cells was measured. Data obtained were analyzed with the software BD FACSDiva (BD, Bioscience) and FlowJo 7.6.1 software (Tree Star Inc, Ashland, Oregon). Nuclei of the C. neoformans SL305 titan-like and regular cells were also observed by confocal microscopy using a Leica SP5 confocal microscope. In this case, cells were stained with mAb 18B7 conjugated to Alexa-488. A) Titan-like cells; B) cells of regular size. C. Histogram analysis of the NOP1-mCherry fluorescence intensity from cells of regular size (blue) or titan-like cells.
(TIF)
Cells from different a and α pairs (A, 3259/3260; B, JEC20/JEC21; and C, NE822/NE824) were placed in TCM at 104 cells/mL. In addition, parallel cultures in which the medium was inoculated with a mixture of same amount of cells (yielding a 104 cells/mL concentration too) of both mating types were examined. The plates were incubated at 37°C with 5% CO2 without shaking for 18 h. Then, pictures were taken, and the size of around 50–100 cells was measured and plotted.
(TIF)
Then, the cells were observed in a confocal microscope, and brightfield and fluorescence images were taken. The bars denote 10 microns in all pictures. At the right panel, cells grown and labeled in the same way were analysed by flow cytometry to quantify the fluorescence intensity of the mAb. The corresponding dot plots and fluorescence histograms are shown. In the histograms, the fluorescence of control cells (without mAb) or with mAb Alexa-488 (18B7) are shown with arrows. The fluorescence image and cytometry analysis for each sample are aligned in the same rows.
(TIF)
Videos were obtained as described in Material and Methods. Pictures were taken every 3 minutes, and video was assembled at 12 frames per second, so one second of video corresponds to 36 minutes of real time. Time in each frame reflects the incubation time in TCM.
(AVI)
The yeasts were inoculated in TCM at and incubated overnight at 37°C with 5% CO2. Previous to the start of the acquisition, the cells had been in TCM for 8 h, so the time shown in each frame reflects the total incubation time since the cells were placed in TCM. Videos were assembled as described in S1 Video.
(AVI)
Previous to the start of the video, the cells had been in TCM for 7h 30 min, so the time shown in each frame reflects the total incubation time since the cells were placed in TCM. Images were collected and processed as described above.
(AVI)
Videos of the interaction between titan-like cells and macrophages were performed as described in Material and Methods. Arrow highlights a titan-like cell. Time in the upper right part reflects the time since the beginning of the phagocytosis in minutes.
(AVI)
Cryptococcal cells were incubated in TCM at a cell density of 106 cells/mL, and after one night of incubation at 37°C with 5% CO2, phagocytosis was performed as described in material and methods. Arrow highlights phagocytosis events. Time in the upper right part reflects the time (min) since the beginning of the phagocytosis.
(AVI)
Cryptococcal cells were incubated in liquid Sabouraud and after one night of incubation at 37°C with 5% CO2, phagocytosis was performed as described in Material and Methods. Arrow highlights phagocytosis events. Time in the upper right part reflects the time since the beginning of the phagocytosis in minutes.
(AVI)
Gene expression changes in titan-like cells isolated in vitro. Titan-like cells were obtained in vitro as described in Material and Methods, and transcriptomic analysis was performed by RNAseq. The table contains the genes that were significantly upregulated (using DESeq2 statistics) at 7 or 18 h of incubation in TCM. P value and fold change are also shown.
(XLSX)
Same as S1 Table, but repressed genes (p<0.05) are listed.
(XLSX)
Acknowledgments
We want to thank Dr. Joseph Heitman (Duke University Medical Center, Durham, North Carolina, United States of America), Dr. Robin May (Birmingham University, UK) and Dr. June Kwon-Chung (NIH, Bethesda, USA), for the gift of strains. I.L. participated in this work in the frame of the master in “Microbiology Applied to Public Health and Research on Infectious Diseases” (Alcalá University and Instituto de Salud Carlos III). We would like to thank Pilar Jiménez Sancho (Genomics Unit, Core Scientific Services, Instituto de Salud Carlos III) for her technical assistance in the RNAseq experiments. We also would like to thank Dr. Manuel Lombardía (Proteomics Facility, National Centre for Biotechnology, Madrid, Spain), for his technical assistance in peptide synthesis.
Dedicatory: Óscar Zaragoza would like to dedicate this article to the memory of Dr. Teresa Gárate Ormaechea and Dr. José Antonio Melero Fondevila, colleagues from the Spanish National Centre for Microbiology who tragically passed away recently. They both will always be remembered, not only for their great contribution to the scientific development of our Institution, but also for their support, kindness and friendship.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
OZ is funded by grant SAF2014-54336-R and SAF2017-86192-R1 from the Spanish Ministry for Economics, Industry and Competitivity. JA is funded by grants BFU2014-54591-C2-1-P and BFU2017-82574-P (Spanish Ministry for Economics, Industry and Competitivity) and an “Ajut 2014SGR-4” (Generalitat de Catalunya). NT-C was supported by a FPI fellowship (reference BES-2012-051837). SAR was supported by a fellowship from Coordenação de aperfeiçoamento de pessoal de nivel superior, CAPES, program Ciências Sem Fronteiras (202436/2015-2). HCdO is funded by postdoctoral fellowship from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-BEPE 2016/20631-3). RG-R is funded by a "Juan de la Cierva" Contract from the Spanish Ministry for Economics, Industry and Competitivity (reference: IJCI-2015-25683). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lazera MS, Salmito Cavalcanti MA, Londero AT, Trilles L, Nishikawa MM, Wanke B. Possible primary ecological niche of Cryptococcus neoformans. Medical mycology. 2000;38(5):379–83. [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A, Perfect J. . CAP JR, editor. Washington DC: ASM; 1998. 541 p. [Google Scholar]
- 3.Chottanapund S, Singhasivanon P, Kaewkungwal J, Chamroonswasdi K, Manosuthi W. Survival time of HIV-infected patients with cryptococcal meningitis. J Med Assoc Thai. 2007;90(10):2104–11. [PubMed] [Google Scholar]
- 4.Dromer F, Mathoulin-Pelissier S, Launay O, Lortholary O. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med. 2007;4(2):e21 doi: 10.1371/journal.pmed.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–30. doi: 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- 6.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17(8):873–81. doi: 10.1016/S1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annual review of microbiology. 2009;63:223–47. doi: 10.1146/annurev.micro.62.081307.162753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clinical microbiology reviews. 2012;25(3):387–408. doi: 10.1128/CMR.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol. 2009;68:133–216. doi: 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heitman J, Kozel TR, Kwon-Chung KJ, Perferct JR, Casadevall A. Cryptococcus. From Human pathogen to model yeast. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Washington DC: ASM Press; 2011. 620 p. [Google Scholar]
- 11.Feldmesser M, Kress Y, Casadevall A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology. 2001;147(Pt 8):2355–65. doi: 10.1099/00221287-147-8-2355 [DOI] [PubMed] [Google Scholar]
- 12.Ellerbroek PM, Hoepelman AI, Wolbers F, Zwaginga JJ, Coenjaerts FE. Cryptococcal glucuronoxylomannan inhibits adhesion of neutrophils to stimulated endothelium in vitro by affecting both neutrophils and endothelial cells. Infection and immunity. 2002;70(9):4762–71. doi: 10.1128/IAI.70.9.4762-4771.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipovsky MM, Tsenova L, Coenjaerts FE, Kaplan G, Cherniak R, Hoepelman AI. Cryptococcal glucuronoxylomannan delays translocation of leukocytes across the blood-brain barrier in an animal model of acute bacterial meningitis. Journal of neuroimmunology. 2000;111(1–2):10–4. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell TG, Friedman L. In vitro phagocytosis and intracellular fate of variously encapsulated strains of Cryptococcus neoformans. Infection and immunity. 1972;5(4):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy JW, Cozad GC. Immunological unresponsiveness induced by cryptococcal capsular polysaccharide assayed by the hemolytic plaque technique. Infection and immunity. 1972;5(6):896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infection and immunity. 2000;68(7):4225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker SC, Casadevall A. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc Natl Acad Sci U S A. 2002;99(5):3165–70. doi: 10.1073/pnas.052702799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelz K, Johnston SA, May RC. Intracellular replication and exit strategies In: Joseph Heitman TRK, Kyung J Kwon-Chung, John R Perferct, and Arturo Casadevall, editor. Cryptococcus From human pathogen to model yeast. Washington: ASM; 2011. p. 441–51. [Google Scholar]
- 19.Garcia-Rodas R, Zaragoza O. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS immunology and medical microbiology. 2012;64(2):147–61. doi: 10.1111/j.1574-695X.2011.00871.x [DOI] [PubMed] [Google Scholar]
- 20.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, et al. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS pathogens. 2010;6(6):e1000953 doi: 10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. Fungal cell gigantism during mammalian infection. PLoS pathogens. 2010;6(6):e1000945 doi: 10.1371/journal.ppat.1000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaragoza O, Nielsen K. Titan cells in Cryptococcus neoformans: cells with a giant impact. Current opinion in microbiology. 2013;16(4):409–13. doi: 10.1016/j.mib.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Barbazan I, Trevijano-Contador N, Rueda C, de Andres B, Perez-Tavarez R, Herrero-Fernandez I, et al. The formation of titan cells in Cryptococcus neoformans depends on the mouse strain and correlates with induction of Th2-type responses. Cellular microbiology. 2016;18(1):111–24. doi: 10.1111/cmi.12488 [DOI] [PubMed] [Google Scholar]
- 24.Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K. Titan cell production enhances the virulence of Cryptococcus neoformans. Infection and immunity. 2012;80(11):3776–85. doi: 10.1128/IAI.00507-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okagaki LH, Nielsen K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryotic cell. 2012;11(6):820–6. doi: 10.1128/EC.00121-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, Fraser JA, et al. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio. 2015;6(5):e01340–15. doi: 10.1128/mBio.01340-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okagaki LH, Wang Y, Ballou ER, O'Meara TR, Bahn YS, Alspaugh JA, et al. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryotic cell. 2011;10(10):1306–16. doi: 10.1128/EC.05179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chrisman CJ, Albuquerque P, Guimaraes AJ, Nieves E, Casadevall A. Phospholipids trigger Cryptococcus neoformans capsular enlargement during interactions with amoebae and macrophages. PLoS pathogens. 2011;7(5):e1002047 doi: 10.1371/journal.ppat.1002047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. Journal of the Royal Society, Interface / the Royal Society. 2009;6(40):959–78. doi: 10.1098/rsif.2009.0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albuquerque P, Nicola AM, Nieves E, Paes HC, Williamson PR, Silva-Pereira I, et al. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. mBio. 2014;5(1):e00986–13. doi: 10.1128/mBio.00986-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H, Chang YC, Nardone G, Kwon-Chung KJ. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Molecular microbiology. 2007;64(3):591–601. doi: 10.1111/j.1365-2958.2007.05666.x [DOI] [PubMed] [Google Scholar]
- 32.Homer CM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, et al. Intracellular Action of a Secreted Peptide Required for Fungal Virulence. Cell host & microbe. 2016;19(6):849–64. doi: 10.1016/j.chom.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SC, Heitman J. Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryotic cell. 2012;11(6):783–94. doi: 10.1128/EC.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. The Journal of biological chemistry. 2004;279(20):21144–53. doi: 10.1074/jbc.M312995200 [DOI] [PubMed] [Google Scholar]
- 35.Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryotic cell. 2008;7(10):1685–98. doi: 10.1128/EC.00146-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Khanal Lamichhane A, Garraffo HM, Kwon-Chung KJ, Chang YC. Involvement of PDK1, PKC and TOR signalling pathways in basal fluconazole tolerance in Cryptococcus neoformans. Molecular microbiology. 2012;84(1):130–46. doi: 10.1111/j.1365-2958.2012.08016.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 38.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, et al. The Case for Adopting the "Species Complex" Nomenclature for the Etiologic Agents of Cryptococcosis. mSphere. 2017;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrnes EJ, 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS pathogens. 2010;6(4):e1000850 doi: 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101(49):17258–63. doi: 10.1073/pnas.0402981101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135(1):174–88. doi: 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, et al. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS pathogens. 2011;7(12):e1002411 doi: 10.1371/journal.ppat.1002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Current biology: CB. 2005;15(22):2021–6. doi: 10.1016/j.cub.2005.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mogensen EG, Janbon G, Chaloupka J, Steegborn C, Fu MS, Moyrand F, et al. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryotic cell. 2006;5(1):103–11. doi: 10.1128/EC.5.1.103-111.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Current biology: CB. 2005;15(22):2013–20. doi: 10.1016/j.cub.2005.09.047 [DOI] [PubMed] [Google Scholar]
- 46.Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, et al. The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. The Journal of infectious diseases. 2013;207(8):1339–47. doi: 10.1093/infdis/jit029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lian T, Simmer MI, D'Souza CA, Steen BR, Zuyderduyn SD, Jones SJ, et al. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Molecular microbiology. 2005;55(5):1452–72. doi: 10.1111/j.1365-2958.2004.04474.x [DOI] [PubMed] [Google Scholar]
- 48.Choi J, Vogl AW, Kronstad JW. Regulated expression of cyclic AMP-dependent protein kinase A reveals an influence on cell size and the secretion of virulence factors in Cryptococcus neoformans. Molecular microbiology. 2012;85(4):700–15. doi: 10.1111/j.1365-2958.2012.08134.x [DOI] [PubMed] [Google Scholar]
- 49.Geddes JM, Caza M, Croll D, Stoynov N, Foster LJ, Kronstad JW. Analysis of the Protein Kinase A-Regulated Proteome of Cryptococcus neoformans Identifies a Role for the Ubiquitin-Proteasome Pathway in Capsule Formation. mBio. 2016;7(1):e01862–15. doi: 10.1128/mBio.01862-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noverr MC, Cox GM, Perfect JR, Huffnagle GB. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infection and immunity. 2003;71(3):1538–47. doi: 10.1128/IAI.71.3.1538-1547.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Molecular microbiology. 1999;32(4):671–80. [DOI] [PubMed] [Google Scholar]
- 52.Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9(13):1559–71. [DOI] [PubMed] [Google Scholar]
- 53.Davenport KR, Sohaskey M, Kamada Y, Levin DE, Gustin MC. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. The Journal of biological chemistry. 1995;270(50):30157–61. [DOI] [PubMed] [Google Scholar]
- 54.Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. The EMBO journal. 1996;15(1):83–91. [PMC free article] [PubMed] [Google Scholar]
- 55.Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. The Journal of clinical investigation. 1985;76(2):508–16. doi: 10.1172/JCI112000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Molecular biology of the cell. 2005;16(5):2285–300. doi: 10.1091/mbc.E04-11-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuller KK, Rhodes JC. Protein kinase A and fungal virulence: a sinister side to a conserved nutrient sensing pathway. Virulence. 2012;3(2):109–21. doi: 10.4161/viru.19396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Himmelreich U, Allen C, Dowd S, Malik R, Shehan BP, Mountford C, et al. Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes and infection / Institut Pasteur. 2003;5(4):285–90. [DOI] [PubMed] [Google Scholar]
- 59.Wright L, Bubb W, Davidson J, Santangelo R, Krockenberger M, Himmelreich U, et al. Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes and infection / Institut Pasteur. 2002;4(14):1427–38. [DOI] [PubMed] [Google Scholar]
- 60.Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. Journal of bacteriology. 1970;104(1):313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annual review of cell and developmental biology. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- 62.Love GL, Boyd GD, Greer DL. Large Cryptococcus neoformans isolated from brain abscess. J Clin Microbiol. 1985;22(6):1068–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cruickshank JG, Cavill R, Jelbert M. Cryptococcus neoformans of unusual morphology. Appl Microbiol. 1973;25(2):309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang JM, Zhou Q, Cai HR, Zhuang Y, Zhang YF, Xin XY, et al. Clinicopathological features of pulmonary cryptococcosis with cryptococcal titan cells: a comparative analysis of 27 cases. Int J Clin Exp Pathol. 2014;7(8):4837–46. [PMC free article] [PubMed] [Google Scholar]
- 65.Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Molecular microbiology. 2006;59(1):67–83. doi: 10.1111/j.1365-2958.2005.04928.x [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Rodas R, Cordero RJ, Trevijano-Contador N, Janbon G, Moyrand F, Casadevall A, et al. Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. mBio. 2014;5(3):e00945–14. doi: 10.1128/mBio.00945-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Rodas R, Casadevall A, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PloS one. 2011;6(9):e24485 doi: 10.1371/journal.pone.0024485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliveira DL, Fonseca FL, Zamith-Miranda D, Nimrichter L, Rodrigues J, Pereira MD, et al. The putative autophagy regulator Atg7 affects the physiology and pathogenic mechanisms of Cryptococcus neoformans. Future Microbiol. 2016;11:1405–19. doi: 10.2217/fmb-2016-0090 [DOI] [PubMed] [Google Scholar]
- 69.Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, Jacobson ES. Regulation of cryptococcal capsular polysaccharide by iron. The Journal of infectious diseases. 1993;167(1):186–90. [DOI] [PubMed] [Google Scholar]
- 70.Kronstad JW, Hu G, Jung WH. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends in microbiology. 2013;21(9):457–65. doi: 10.1016/j.tim.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101(1):177–94. [PMC free article] [PubMed] [Google Scholar]
- 72.Moyrand F, Janbon G. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37 degrees C and for capsule biosynthesis. Eukaryotic cell. 2004;3(6):1601–8. doi: 10.1128/EC.3.6.1601-1608.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infection and immunity. 2003;71(9):4831–41. doi: 10.1128/IAI.71.9.4831-4841.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. Journal of bacteriology. 1993;175(5):1405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaragoza O, Casadevall A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online. 2004;6:10–5. doi: 10.1251/bpo68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Casadevall A, Mukherjee J, Devi SJ, Schneerson R, Robbins JB, Scharff MD. Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. The Journal of infectious diseases. 1992;165(6):1086–93. [DOI] [PubMed] [Google Scholar]
- 77.Voelz K, Johnston SA, Rutherford JC, May RC. Automated analysis of cryptococcal macrophage parasitism using GFP-tagged cryptococci. PloS one. 2010;5(12):e15968 doi: 10.1371/journal.pone.0015968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trevijano-Contador N, Herrero-Fernandez I, Garcia-Barbazan I, Scorzoni L, Rueda C, Rossi SA, et al. Cryptococcus neoformans induces antimicrobial responses and behaves as a facultative intracellular pathogen in the non mammalian model Galleria mellonella. Virulence. 2015;6(1):66–74. doi: 10.4161/21505594.2014.986412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics. 2015;31(3):445–6. doi: 10.1093/bioinformatics/btu627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryotic cell. 2002;1(1):75–84. doi: 10.1128/EC.1.1.75-84.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68(4):821–33. [PubMed] [Google Scholar]
- 84.Sun S, Xu J. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics. 2007;177(3):1475–86. doi: 10.1534/genetics.107.078923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wollschlaeger C, Trevijano-Contador N, Wang X, Legrand M, Zaragoza O, Heitman J, et al. Distinct and redundant roles of exonucleases in Cryptococcus neoformans: Implications for virulence and mating. Fungal Genet Biol. 2014;73:20–8. doi: 10.1016/j.fgb.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang YC, Wickes BL, Kwon-Chung KJ. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene. 1995;167(1–2):179–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In this case, we prepared suspensions of C. neoformans cells at 104 and 106 cells/mL in 30 mL of TCM cell. After the incubation period, the cells were collected by centrifugation, fixed with 4% p-formaldehyde and analyzed by flow cytometry using the BD LSRFortessa X-20cytometer (BD, Bioscience). Two populations of titan-like and regular cells were delimited and, in each population, the fluorescence intensity of the NOP1-mCherry protein in 10,000 cells was measured. Data obtained were analyzed with the software BD FACSDiva (BD, Bioscience) and FlowJo 7.6.1 software (Tree Star Inc, Ashland, Oregon). Nuclei of the C. neoformans SL305 titan-like and regular cells were also observed by confocal microscopy using a Leica SP5 confocal microscope. In this case, cells were stained with mAb 18B7 conjugated to Alexa-488. A) Titan-like cells; B) cells of regular size. C. Histogram analysis of the NOP1-mCherry fluorescence intensity from cells of regular size (blue) or titan-like cells.
(TIF)
Cells from different a and α pairs (A, 3259/3260; B, JEC20/JEC21; and C, NE822/NE824) were placed in TCM at 104 cells/mL. In addition, parallel cultures in which the medium was inoculated with a mixture of same amount of cells (yielding a 104 cells/mL concentration too) of both mating types were examined. The plates were incubated at 37°C with 5% CO2 without shaking for 18 h. Then, pictures were taken, and the size of around 50–100 cells was measured and plotted.
(TIF)
Then, the cells were observed in a confocal microscope, and brightfield and fluorescence images were taken. The bars denote 10 microns in all pictures. At the right panel, cells grown and labeled in the same way were analysed by flow cytometry to quantify the fluorescence intensity of the mAb. The corresponding dot plots and fluorescence histograms are shown. In the histograms, the fluorescence of control cells (without mAb) or with mAb Alexa-488 (18B7) are shown with arrows. The fluorescence image and cytometry analysis for each sample are aligned in the same rows.
(TIF)
Videos were obtained as described in Material and Methods. Pictures were taken every 3 minutes, and video was assembled at 12 frames per second, so one second of video corresponds to 36 minutes of real time. Time in each frame reflects the incubation time in TCM.
(AVI)
The yeasts were inoculated in TCM at and incubated overnight at 37°C with 5% CO2. Previous to the start of the acquisition, the cells had been in TCM for 8 h, so the time shown in each frame reflects the total incubation time since the cells were placed in TCM. Videos were assembled as described in S1 Video.
(AVI)
Previous to the start of the video, the cells had been in TCM for 7h 30 min, so the time shown in each frame reflects the total incubation time since the cells were placed in TCM. Images were collected and processed as described above.
(AVI)
Videos of the interaction between titan-like cells and macrophages were performed as described in Material and Methods. Arrow highlights a titan-like cell. Time in the upper right part reflects the time since the beginning of the phagocytosis in minutes.
(AVI)
Cryptococcal cells were incubated in TCM at a cell density of 106 cells/mL, and after one night of incubation at 37°C with 5% CO2, phagocytosis was performed as described in material and methods. Arrow highlights phagocytosis events. Time in the upper right part reflects the time (min) since the beginning of the phagocytosis.
(AVI)
Cryptococcal cells were incubated in liquid Sabouraud and after one night of incubation at 37°C with 5% CO2, phagocytosis was performed as described in Material and Methods. Arrow highlights phagocytosis events. Time in the upper right part reflects the time since the beginning of the phagocytosis in minutes.
(AVI)
Gene expression changes in titan-like cells isolated in vitro. Titan-like cells were obtained in vitro as described in Material and Methods, and transcriptomic analysis was performed by RNAseq. The table contains the genes that were significantly upregulated (using DESeq2 statistics) at 7 or 18 h of incubation in TCM. P value and fold change are also shown.
(XLSX)
Same as S1 Table, but repressed genes (p<0.05) are listed.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.