Abstract
Objectives:
Firefighters have elevated cancer incidence and mortality rates. MicroRNAs play prominent roles in carcinogenesis, but have not been previously evaluated in firefighters.
Methods:
Blood from 52 incumbent and 45 new recruit nonsmoking firefighters was analyzed for microRNA expression, and the results adjusted for age, obesity, ethnicity, and multiple comparisons.
Results:
Nine microRNAs were identified with at least a 1.5-fold significant difference between groups. All six microRNAs with decreased expression in incumbent firefighters have been reported to have tumor suppressor activity or are associated with cancer survival, and two of the three microRNAs with increased expression in incumbent firefighters have activities consistent with cancer promotion, with the remaining microRNA associated with neurological disease.
Conclusion:
Incumbent firefighters showed differential microRNA expression compared with new recruits, providing potential mechanisms for increased cancer risk in firefighters.
Keywords: firefighter, cancer, microRNA
Cancer is a leading cause of death among firefighters in the United States. In a recent large study, overall cancer incidence and mortality among firefighters were 9% and 14% higher than the general public, respectively, with increased mortality rates of 30% or more for mesothelioma and cancers of the esophagus, intestine, and rectum.1 In addition, elevated incidence and/or mortality have been reported in firefighters for cancers of the bladder, kidney, lung, prostate, skin (melanoma and non-melanoma), stomach, and testes, as well as leukemia, multiple myeloma, and non-Hodgkin lymphoma.1–7
Firefighters are occupationally exposed to carcinogens and other toxicants, including benzene, polycyclic aromatic hydrocarbons, formaldehyde, arsenic, 1–3 butadiene, cadmium, chromium compounds, asbestos, flame retardants, and particulates.5,8–12 Furthermore, most firefighters work prolonged shifts associated with sleep disruption, and shiftwork with circadian disruption has been classified as a probable human carcinogen (group 2A) by the International Agency for Research on Cancer (IARC).11,13
While exposure to carcinogens and elevated cancer risk have been well established for firefighters, there is limited information on the cellular mechanisms involved. Greater understanding of these mechanisms is critical to identify potentially reversible cellular changes before the development of cancer, and to help determine causation with regard to firefighter worker's compensation cancer claims. Based in part on the lack of data regarding mechanistic changes in firefighters leading to carcinogenesis, in 2010, the IARC classified occupational exposures to firefighters as only possibly carcinogenic to humans (group 2B), despite multiple epidemiologic studies demonstrating elevated cancer incidence rates in firefighters.11,13
Epigenetic changes, including histone modifications, DNA methylation, and microRNA (miRNA) mediated pathways, play prominent roles in carcinogenesis and cancer prevention, and have been associated with activation of oncogenes or inhibition of tumor suppressor genes.14,15 MiRNAs are small (18 to 22 nucleotide) noncoding RNAs involved in regulating cell cycle progression, apoptosis, and differentiation. Some miRNAs act as oncogenes by inducing oncogene expression or tumor-suppressor genes through regulation of DNA methylation and histone modification. These epigenetic changes serve as molecular biomarkers of environmental exposures and carcinogenesis.16–19
We hypothesized that occupational exposures in firefighters would lead to changes in miRNA expression associated with activation of cancer pathways and increased cancer risk. As a first step in testing this hypothesis, we designed this study to compare miRNAs in incumbent firefighters and new recruits.
METHODS
This study was a part of larger firefighter cancer prevention study working in partnership with the Tucson Fire Department. All study protocols were approved by the University of Arizona Institutional Review Board (approval No. 1509137073). To identify epigenetic changes associated with occupational carcinogen exposures in firefighters, we recruited newly employed (new recruit) firefighters before occupational exposure to fire and smoke and incumbent firefighters. After receiving a detailed explanation of the study design and potential risks, all subjects provided written informed consent. We surveyed general characteristics using questionnaires to collect information regarding age, body weight, height, working duration as firefighters, and tobacco use. Body mass index (BMI) (kg/m2) was classified as normal (18.0 to 24.9), overweight (25.0 to 29.9), and obese (≥30) following World Health Organization (WHO) classifications.
At the time of sample selection for the current analyses, the study subjects consisted of 55 male recruits and 117 male incumbents who had completed baseline blood sampling. One recruit was excluded because of an inadequate blood draw. The 54 remaining recruits were then matched by race/ethnicity to 54 incumbents for sample processing, for a total of 108 subjects. One recruit sample was not adequate for miRNA analysis, and five subjects (four recruits and one incumbent) were later excluded for current smoking as well as another five (four recruits and one incumbent) subjects for not completing the smoking-related questions on the questionnaire, leaving 97 (45 recruits and 52 incumbents) subjects for miRNA data analysis.
Whole blood samples were collected in TempusTM Blood RNA tubes (Applied Biosystems, Foster City, California). Immediately after collection, the tube was vigorously shaken for 10 seconds and aliquoted into two 5 mL cryogenic tubes (VWR International, Radnor, Pennsylvania, Cat. # 89094–820). All aliquots were stored at −20°C until transfer under Arizona Department of Transportation guidelines to the University of Arizona for storage at −80°C for subsequent processing by the University of Arizona Genetics Core (Arizona Research Laboratories).
An aliquot for each subject was thawed for 20 to 30 minutes on ice. RNA isolation was achieved using MagMAXTM for Stabilized Blood Tubes RNA Isolation Kit (Life Technologies, Carlsbad, California, Catalog #4451893) following the manufacturer's protocol. Purified total RNA qualities and quantities were measured with the NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware) and a subset was additionally quality checked using the Advanced Analytical High Sensitivity RNA assay with the Fragment Analyzer Automated CE System (AATI, Ankeny, Iowa).
MiRNA expression was measured using the nCounter® Human v3 miRNA expression panel (NanoString Technology Inc., Seattle, Washington) with 800 miRNAs from miRBase v21 as well as 5 housekeeping genes and 20 assay controls (six positive, eight negative, and six ligation controls). The panel includes greater than 95% of human miRBase reads (https://hdmzlive.nanostring.com/application/files/7014/8943/1030/LBL-10112-01_Human_miRNA.pdf). One hundred nanogram of the purified RNA was prepared by multiplexed annealing of specific tags to each target miRNA, followed by a ligation reaction, and enzymatic purification to remove the unligated tags. Five microliters of the cleaned reaction was hybridized with the Human miRNA Code Set (Nanostring Technologies part #CSO-MIR3-12) at 65°C overnight. Purification and binding of the hybridized probes to the optical cartridge were performed on the nCounter Prep Station, and the cartridge scanned on the nCounter Digital Analyzer (NanoString Technologies, Inc., Seattle, Washington). Raw counts from each gene were normalized against background genes, and overall assay performance was assessed through evaluation of built-in positive controls.
For comparison of age and BMI between recruits and incumbents, the Chi-square test was used. The mean comparisons of age and BMI were done by the Student t test. To evaluate the correlation between age and working duration as a firefighter, Pearson correlation was used. These statistical analyses were performed using R (version 3.4.1). MiRNAs sites with mean counts that were less than 2 were filtered, leaving 821 genes for analysis. Filtered miRNAs raw counts were first transformed and quantile normalized by Voom package20 in preparation for linear modeling and then analyzed by the limma package.21 A linear model with Empirical Bayes estimator was adopted,22 with adjustment for age, ethnicity, and BMI. Probes were considered to be differentially expressed if the resulting P value was less than 0.05/m applying Bonferroni correction for multiple comparisons. The corresponding gene list was derived from the gene annotations associated with the probes.
Both K-means clustering and hierarchical clustering using the “factoextra” package in R 3.4.1 were used to discover miRNA clusters discriminating between the incumbent and new recruit groups. Both analyses were restricted to miRNAs differentially expressed between the two groups, adjusted for age, BMI, and ethnicity, with P values less than 0.05. The optimal cluster size was determined by minimizing within sum of squares in K-means clustering analysis. Hierarchal clustering was carried out based on complete linkage and person correlation.
The miRNA enrichment analysis and annotation tool miEAA (https://ccb-compute2.cs.uni-saarland.de/mieaa_tool/),23 which relies on the GeneTrail framework (https://genetrail2.bioinf.uni-sb.de/),24 was employed to investigate downstream effects of the miRNA clusters. Effects of single miRNAs on pathways and organs were determined by miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/holistic.html).25 The miRNA-disease association was evaluated using the Human microRNA Disease Database (HMDD v2.0, http://www.cuilab.cn/hmdd).26 Both miRWalk and HMDD are integrated in miEAA. Unless mentioned explicitly, all tools were used with standard parameters.
RESULTS
All subjects were white, and a similar percentage of incumbent and new recruit firefighters were of Hispanic ethnicity (15.4% and 13.3%, respectively). The subjects’ mean age in years was significantly higher in incumbents (40.8 ± 8.7) than in recruits (28.8 ± 6.2) (P < 0.001) (Table 1). The incumbent firefighters and recruits had similar BMI distributions. None of the new recruit firefighters had any previous firefighting experience. For incumbents, the mean number of years serving as a firefighter was 14.1 ± 7.3 years, and number of years of service was significantly correlated with age (Pearson r = 0.818, P < 0.001). The 45 new recruits and 52 incumbent firefighters in our study did not significantly differ in terms of age, race, ethnicity, or BMI from the larger group of 89 new recruits and 352 incumbent firefighters, respectively, currently enrolled in the larger Tucson Fire Department cancer prevention study (data not shown).
TABLE 1.
General Characteristics of Subjects
| Variables | New Recruits (n = 45) | Incumbents (n = 52) | P |
| Race/Ethnicity | |||
| White, non-Hispanic | 39 (86.7%) | 44 (84.6%) | 1.0 |
| White, Hispanic | 6 (13.3%) | 8 (15.4%) | |
| Age, years | |||
| ≤29 | 26 (57.8%) | 4 (7.7%) | <0.001 |
| 30–39 | 14 (31.1%) | 16 (30.8%) | |
| ≥40 | 5 (11.1%) | 32 (61.5%) | |
| Mean (standard deviation) | 28.8 (6.21) | 40.8 (8.70) | <0.001 |
| BMI | |||
| Normal | 14 (31.1%) | 10 (19.2%) | 0.39 |
| Overweight | 24 (53.3%) | 32 (61.5%) | |
| Obese | 7 (15.6%) | 10 (19.2%) | |
Comparing incumbents to recruits and adjusting for multiple comparisons, nine miRNAs demonstrated statistically significant differences in expression at a level of at least 1.5-fold (Table 2). Among these, only two miRNAs differed significantly by age group and none by BMI group. Expression levels of all nine miRNAs remained significantly different between incumbents and recruits after adjusting for age, BMI, and ethnicity. Comparing within incumbent and recruit groups, there were no significant differences by age for these nine miRNAs (data not shown).
TABLE 2.
Fold Change (FC) of miRNAs Between Groups by Job Status, Age, and Body Mass Index (BMI)
| Incumbents vs Recruits | Age: ≥40 vs 21–39 | BMI: Overweight and Obese vs Normal | Incumbents vs Recruits Adjusted for Age, BMI, and Ethnicity | |||||||||
| 95% CI | 95% CI | 95% CI | 95% CI | |||||||||
| Gene Name | FC | Lower | Upper | FC | Lower | Upper | FC | Lower | Upper | FC | Lower | Upper |
| hsa-miR-1260a | 0.54 | 0.44 | 0.66 | 0.69 | 0.55 | 0.87 | 0.94 | 0.71 | 1.24 | 0.55 | 0.43 | 0.71 |
| hsa-miR-548h-5p | 0.55 | 0.43 | 0.72 | 0.82 | 0.60 | 1.12 | 0.84 | 0.60 | 1.18 | 0.59 | 0.51 | 0.69 |
| hsa-miR-145-5p | 0.57 | 0.51 | 0.65 | 0.75 | 0.64 | 0.87 | 0.93 | 0.77 | 1.12 | 0.44 | 0.32 | 0.61 |
| hsa-miR-4516 | 0.59 | 0.52 | 0.66 | 0.75 | 0.64 | 0.87 | 0.96 | 0.80 | 1.16 | 0.56 | 0.48 | 0.65 |
| hsa-miR-331-3p | 0.59 | 0.52 | 0.67 | 0.72 | 0.62 | 0.84 | 0.87 | 0.72 | 1.06 | 0.60 | 0.52 | 0.70 |
| hsa-miR-181a-5p | 0.60 | 0.53 | 0.68 | 0.76 | 0.65 | 0.89 | 0.96 | 0.79 | 1.15 | 0.62 | 0.53 | 0.72 |
| hsa-miR-5010-3p | 1.56 | 1.41 | 1.72 | 1.25 | 1.10 | 1.41 | 1.12 | 0.96 | 1.30 | 1.59 | 1.41 | 1.81 |
| hsa-miR-374a-5p | 1.57 | 1.33 | 1.85 | 1.09 | 0.90 | 1.32 | 1.18 | 0.95 | 1.47 | 1.72 | 1.40 | 2.13 |
| hsa-miR-486-3p | 3.51 | 2.88 | 4.28 | 2.22 | 1.64 | 3.01 | 1.23 | 0.85 | 1.78 | 3.35 | 2.59 | 4.33 |
Significantly differentially expressed genes (after Bonferroni correction) are highlighted in bold font.
BMI, body mass index; CI, confidence interval; FC, fold change.
Three incumbent firefighters reported a previous diagnosis of non-melanoma skin cancer, while none of the new recruits reported this diagnosis. To address this difference, we performed a sensitivity analysis by excluding the three firefighters with skin cancer, and all nine miRNAs remained significant (data not shown). Although all firefighters in the study were current nonsmokers, significantly more new recruits (11) than incumbents (four) previously smoked over 100 cigarettes (Pearson Chi-square P = 0.034).
Cluster analysis was based on 234 miRNAs differentially expressed between incumbent and new recruit firefighters. The optimal number of clusters for K-mean analysis was determined to be three, with centroids hsa-miR-525-3p (cluster 1), hsa-miR-52 (cluster 2), and hsa-miR-376b-3p (cluster 3) (Fig. 1). There were 103, 80, and 51 miRNAs in these three clusters, respectively. An enrichment analysis was performed to investigate whether the miRNA sets within the three clusters belonged to a pathway, gene ontology, organ, or other functional category, with FDR adjusted P value less than 0.05 (Table 3). MiRNAs in the first cluster were associated with stem cells and three pathways: inflammation mediated by chemokine and cytokine signaling; cytokine-cytokine receptor interaction; and cell adhesion molecules. MiRNAs in the second cluster were also associated with stem cells. The third cluster yielded miRNAs associated with carcinoma, Burkitt lymphoma, melanoma, and 10 targeted genes.
FIGURE 1.
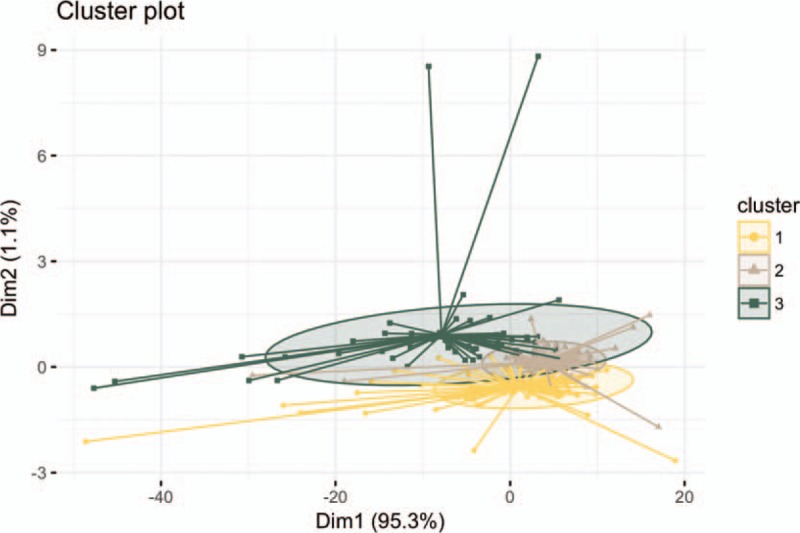
K-mean clustering using 234 MiRNAs.
TABLE 3.
MiRNA Enrichment Analysis Results for Three K-Mean Clusters
| Cluster | Category | Subcategory | N | Observed miRNAs | P (FDR) |
| 1 | Organs | Stem Cells | 8 | miR-126-3p; miR-133b; miR-15a-5p; miR-195-5p; miR-29b-3p; miR-302a-3p; miR-302b-3p; miR-326 | 0.048 |
| Pathways | Inflammation mediated by chemokine and cytokine signaling (P00031) | 19 | miR-126-3p; miR-135a-5p; miR-15a-5p; miR-193b-3p; miR-195-5p; miR-196b-5p; miR-218-5p; miR-22-3p; miR-29b-3p; miR-302a-3p; miR-302b-3p; miR-30e-3p; miR-320c; miR-326; miR-337-3p; miR-451a; miR-7-5p; miR-935; miR-96-5p | 0.048 | |
| Cytokine–cytokine receptor interaction (hsa04060) | 9 | miR-126-3p; miR-133b; miR-15a-5p; miR-193b-3p; miR-195-5p; miR-22-3p; miR-29b-3p; miR-302b-3p; miR-7-5p | 0.048 | ||
| Cell adhesion molecules (hsa04514) | 6 | miR-126-3p; miR-15a-5p; miR-193b-3p; miR-196b-5p; miR-218-5p; miR-29b-3p | 0.048 | ||
| 2 | Organs | Stem cells | 6 | miR-137; miR-138-5p; miR-214-3p; miR-224-5p; miR-27a-3p; miR-302d-3p | 0.021 |
| 3 | Diseases | Burkitt lymphoma | 2 | let-7a-5p; let-7b-5p | 0.033 |
| Carcinoma | 19 | let-7a-5p; let-7b-5p; let-7c-5p; miR-106b-5p; miR-1226-3p; miR-125a-5p; miR-141-3p; miR-145-5p; miR-151a-5p; miR-181a-5p; miR-181c-5p; miR-185-5p; miR-197-3p; miR-19a-3p; miR-19b-3p; miR-21-5p; miR-223-3p; miR-376b-3p; miR-93-5p | 0.033 | ||
| Melanoma | 2 | let-7a-5p; let-7b-5p | 0.033 | ||
| Target genes | AIDA | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | |
| ANKRD17 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| CCNB2 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| CSNK2A1 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| DHX9 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| IPO7 | 5 | let-7a-5p; let-7b-5p; let-7c-5p; miR-106b-5p; miR-1226-3p | 0.010 | ||
| LTA4H | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| NME4 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| PTGES2 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 | ||
| RPS24 | 3 | let-7a-5p; let-7b-5p; let-7c-5p | 0.019 |
DISCUSSION
The study results supported our hypothesis that incumbent firefighters, compared with new recruits, would show differences in expression of miRNAs associated with cancers or cancer pathways. This adds to the scarce published literature on epigenetic effects in firefighters, to our knowledge limited to hypomethylation of dual specificity phosphatase 22 promoter,27 and suggests potential mechanisms for the association between firefighting and cancer.
Of the nine differentially expressed miRNAs identified in our study, all six (miR-548h-5p, miR-145-5p, miR-4516, miR-331-3p, miR-181a-5p, and miR-1260a) with decreased expression in incumbent firefighters have been reported to have tumor suppressor activity or are associated with cancer survival, and two (miR-374a-5p and miR-486-3p) of the three miRNAs with increased expression in incumbent firefighters have activities consistent with cancer promotion. The miR-548 family suppresses tumor cell growth and development by increasing apoptosis and regulating reactive oxygen species.28 MiR-4516 and miR-145-5p play a role in tumor suppression by controlling p53.29–31 MiR-331-3p acts as a tumor suppressor in colorectal and gastric cancer.32–34 MiR-181a-5p has a tumor suppressor effect in nonsmall cell lung cancer through reduction of K-ras expression.35 Increased levels of miR-1260a are associated with survival in glioblastoma patients.36 MiR-374a-5p promotes cell proliferation, migration, and invasion in esophageal and gastric cancer,37,38 and is overexpressed in colorectal cancer.39 Upregulation of miR-486-3p is associated with K-ras mutation in colorectal cancer.40 The one miRNA for which we did not find an association with cancer, miR-5010-3p, is increased in patients with Alzheimer disease.41 In addition to their previously described roles in cancer, miR-374a-5p is overexpressed with high altitude, hypoxia, and oxidative stress,42 miR-486-3p is increased in ST-elevated acute myocardial infarction,43 and miR-1260a is increased in children with asthma.44
Aging and obesity are major risk factors for cancer,45 and they are also associated with epigenetic changes.46–53 Specific to miRNAs, age, BMI, and sex are associated with miRNA expression.54 In our study, only two miRNAs showed significant differences in expression by age group, and there were no significant differences in miRNA expression by BMI group. Furthermore, all nine miRNAs continued to show significant differences between incumbents and new recruits following adjustment for age and BMI.
In addition to documented associations with cancer pathways, some of the miRNAs identified in our study have also been evaluated for relationships with exposures common to firefighters. In a study examining gaseous formaldehyde exposure and miRNA expression in human bronchial epithelial cells, miR-181a was one of the most significantly downregulated.55 MiR-181a-5p expression, decreased in incumbent firefighters in our study, was downregulated in Lin-c-Kit+ cells obtained from mice exposed to benzene.56 MiR-313-3p expression, decreased in incumbent firefighters in our study, was reduced following short-term PM10 exposure in a population of overweight/obese subjects,57 but was increased in lung adenocarcinoma patients exposed to asbestos compared with nonexposed patients with adenocarcinoma.58 MiR-4516 expression was decreased in our incumbent firefighters, but was upregulated in a study of A549 cells exposed to PM2.5 as well as the serum of persons living in a Chinese city with moderate air pollution.59 MiR-145-5p expression was decreased in our incumbent firefighters, but increased in service members with polychlorinated dibenzodioxin (PCDD) and polychlorinated dibenzofuran (PCDF) exposures from open air burn pits comparing pre- and post-deployment.60 However, to evaluate for dose–response relationships specific to firefighters, longitudinal studies including exposure assessment and measurement of miRNA changes are needed.
Cluster analysis comparing the incumbent firefighters to new recruits identified miRNA groupings associated with stem cells, inflammation, cytokine–cytokine receptor interactions, cell adhesion molecules, cancers, and a number of target genes. MiRNAs control stem cell self-renewal and differentiation,61 and through this role have been implicated in the etiology of a variety of cancers.62,63 For example, hsa-miR-302a-3p and hsa-miR-302b-3p, enriched in cluster 1, and hsa-miR-302d-3p, enriched in cluster 2, belong to the miR-302 family, which has important roles relative to stem cells.64 MiR-302 inhibits human pluripotent stem cell tumorigenicity by enhancing multiple G1 phase arrest pathways.65 Moreover, the miR-302 family functions to reprogram skin cancer cells into a stem cell-like pluripotent state.66 MiR-137, enriched in cluster 2, is downregulated in colon cancer stem cells compared with normal colon stem cells.67 MiR-124 and 137 also regulate the differentiation and proliferation of neural stem cells and glioblastoma-multiforme tumor cells.68
Beyond stem cells, all three pathways identified in cluster 1 were related to inflammation. Chronic inflammation has long been linked with cancers.69 Inflammation mediated by the chemokine and cytokine signaling pathway (P00031) includes chemokine-induced adhesion and migration of leukocytes70,71 and miRNAs differentially expressed in lung cancer,72,73 bladder cancer,74 senescence, and aging.75 The cytokine–cytokine receptor interaction pathway (CCRI, hsa04060) is a large pathway that includes 270 related genes.76 Cytokines that act through receptors are released in response to infection, inflammation, and immunity, and cytokines and cytokine receptors can function to inhibit tumor development and progression. Cancer cells also respond to host-derived cytokines that promote growth, attenuate apoptosis, and facilitate invasion and metastasis.77,78 Cell adhesion molecules (CAMs, hsa04514), constituting the third pathway, play a critical role in a wide array of biologic processes, including immune response and inflammation, contributing to cancer development.79–81
The third cluster identified in our analyses included specific cancer types and genes. Of the 19 miRNAs enriched in carcinomas, three of them were from the miRNA let-7 family and are reported to be downregulated in human lung carcinomas, where reduced let-7 expression is associated with a poor cancer prognosis.82 Also among these 19 miRNAs, miR-21 and miR-181 are potential diagnostic or prognostic biomarkers for nonsmall cell lung cancer,83 and miR-106 and miR-93 are expressed in hepatocellular carcinoma.84 The enriched let-7 family in cluster 3 targets 10 genes, and let-7 is highly conserved in human organs and associated with colon adenocarcinoma, kidney renal clear cell carcinoma, esophageal carcinoma, lung squamous cell carcinoma, and liver hepatocellular carcinoma, among others.23 Specifically, CSNK2A1, IP07, and DHX9 are coexpressed in kidney renal clear cell carcinoma and lung squamous cell carcinoma.24
Our study has a number of limitations. The number of firefighter participants was limited and cross-sectional in nature, and larger prospective longitudinal studies are needed to validate the epigenetic changes observed. Although we adjusted for age, there were only a limited number of new recruits older than 40 years of age, and similarly only a limited number of incumbent firefighters less than 30 years old. We did not validate the miRNA findings using a second technique, given the strong correlation reported among multiple miRNA platforms including NanoString.85 Although we did not have complete information on toxic exposures outside of firefighting, previous use of cigarettes was lower in the incumbent firefighters, so we do not believe that this past exposure could explain the miRNA findings. We were not able to adjust for other potential confounders due to incomplete responses to survey questions such as but not limited to diet and exercise. Additional studies in other geographic regions are also needed to determine whether the results of our study are generalizable to firefighters elsewhere.
In conclusion, this study identified multiple miRNAs with significantly different expression levels comparing incumbent firefighters with new recruits. These findings suggest potential mechanisms for development of cancer in firefighters.
Footnotes
Kyoung Sook Jeong and Jin Zhou contributed equally to this work.
US Federal Emergency Management Agency Assistance to Firefighters Grant program, EMW-2014-FP-00200.
The authors have no conflicts of interest.
REFERENCES
- 1.Daniels RD, Kubale TL, Yiin JH, et al. Mortality and cancer incidence in a pooled cohort of US firefighters from San Francisco, Chicago and Philadelphia (1950–2009). 2014; 71:388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn YS, Jeong KS, Kim KS. Cancer morbidity of professional emergency responders in Korea. 2012; 55:768–778. [DOI] [PubMed] [Google Scholar]
- 3.Daniels RD, Bertke S, Dahm MM, et al. Exposure-response relationships for select cancer and non-cancer health outcomes in a cohort of U.S. firefighters from San Francisco, Chicago and Philadelphia (1950–2009). 2015; 72:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass DC, Pircher S, Del Monaco A, et al. Mortality and cancer incidence in a cohort of male paid Australian firefighters. 2016; 73:761–771. [DOI] [PubMed] [Google Scholar]
- 5.Ide CW. Cancer incidence and mortality in serving whole-time Scottish firefighters 1984–2005. 2014; 64:421–427. [DOI] [PubMed] [Google Scholar]
- 6.LeMasters GK, Genaidy AM, Succop P, et al. Cancer risk among firefighters: a review and meta-analysis of 32 studies. 2006; 48:1189–1202. [DOI] [PubMed] [Google Scholar]
- 7.Pukkala E, Martinsen JI, Weiderpass E, et al. Cancer incidence among firefighters: 45 years of follow-up in five Nordic countries. 2014; 71:398–404. [DOI] [PubMed] [Google Scholar]
- 8.Bastes MN. Registry-based case-control study of cancer in California firefighters. 2007; 50:339–344. [DOI] [PubMed] [Google Scholar]
- 9.Bolstad-Johnson DM, Burgess JL, Crutchfield CD, et al. Characterization of firefighter exposures during fire overhaul. 2000; 61:636–641. [DOI] [PubMed] [Google Scholar]
- 10.Driscoll TR, Carey RN, Peters S, et al. The Australian work exposures study: prevalence of occupational exposure to formaldehyde. 2016; 60:132–138. [DOI] [PubMed] [Google Scholar]
- 11.IARC (International Agency for Research on Cancer). Painting, firefighting, and shiftwork. IARC Monogr Eval Carcinog Risk Hum 2010; 98:395–764. [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw SD, Berger ML, Harris JH, et al. Persistent organic pollutants including polychlorinated and polybrominated dibenzo-p-dioxins and dibenzofurans in firefighters from Northern California. 2013; 91:1386–1394. [DOI] [PubMed] [Google Scholar]
- 13.Straif K, Baan R, Grosse Y, et al. Carcinogenicity of shift-work, painting, and fire-fighting. 2007; 8:1065e6. [DOI] [PubMed] [Google Scholar]
- 14.Biswas S, Rao CM. Epigenetics in cancer: fundamentals and beyond. 2017; 173:118–134. [DOI] [PubMed] [Google Scholar]
- 15.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. 2010; 80:1771–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang JC, Jones PA. Epigenetics and microRNAs. 2007; 61:24R–29R. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. 2002; 3:415–428. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The epigenomics of cancer. 2007; 128:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozek LS, Dolinoy DC, Sartor MA, Omenn GS. Epigenetics: relevance and implications for public health. 2014; 35:105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. 2014; 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. 2004; 3:Article3. [DOI] [PubMed] [Google Scholar]
- 23.Zhan C, Yan L, Wang L, et al. Identification of reference miRNAs in human tumors by TCGA miRNA-seq data. 2014; 453:375–378. [DOI] [PubMed] [Google Scholar]
- 24.Kim P, Cheng F, Zhao J, Zhao Z. ccmGDB: a database for cancer cell metabolism genes. 2016; 44:D959–D968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweep H, Sticht C, Pandey P, Gretz N. MiRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. 2011; 44:839–847. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Qiu C, Tu J, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. 2014; 42:D1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang B, Baxter CS, Lam HM, et al. Hypomethylation of dual specificity phosphatase 22 promoter correlates with duration of service in firefighters and is inducible by low-dose benzo[a]pyrene. 2012; 54:774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu B, Ying X, Wang J, et al. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. 2014; 74:2283–2294. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhari S, Saini N. hsa-miR-4516 mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA induced apoptosis in keratinocytes. 2014; 229:1630–1638. [DOI] [PubMed] [Google Scholar]
- 30.Ozen M, Karatas OF, Gulluoglu S, et al. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. 2015; 33:251–258. [DOI] [PubMed] [Google Scholar]
- 31.Samulin Erdem J, Skaug V, Bakke P, et al. Mutations in TP53 increase the risk of SOX2 copy number alterations and silencing of TP53 reduces SOX2 expression in non-small cell lung cancer. 2016; 16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epis MR, Giles KM, Barker A, et al. MiR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. 2009; 284:24696–24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epis MR, Giles KM, Kalinowski FC, et al. Regulation of expression of deoxyhypusine hydroxylase (DOHH), the enzyme that catalyzes the activation of eIF5A, by miR-331-3p and miR-642-5p in prostate cancer cells. 2012; 287:35251–35259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao D, Sui Y, Zheng X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. 2016; 35:1075–1082. [DOI] [PubMed] [Google Scholar]
- 35.Ma Z, Qui X, Wang D, et al. MiR-181a-5p inhibits cell proliferation and migration by targeting Kras in non-small cell lung cancer A549 cells. 2015; 47:630–638. [DOI] [PubMed] [Google Scholar]
- 36.Herman A, Gruden K, Bleject A, et al. Analysis of glioblastoma patients’ plasma revealed the presence of microRNAs with a prognostic impact on survival and those of viral origin. 2015; 10:e0125791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Xin H, Han Z, et al. MicroRNA-374a promotes esophageal cancer cell proliferation via Axin2 suppression. 2015; 34:1988–1994. [DOI] [PubMed] [Google Scholar]
- 38.Xu X, Wang W, Su N, et al. MiR-374a promotes cell proliferation, migration and invasion by targeting SRCIN1 in gastric cancer. 2015; 589:407–413. [DOI] [PubMed] [Google Scholar]
- 39.Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. 2015; 137:428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosakhani N, Sarhadi VK, Borze I, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. 2012; 51:1–9. [DOI] [PubMed] [Google Scholar]
- 41.Leidinger P, Backer C, Deutscher S, et al. A blood based 12-miRNA signature of Alzheimer disease patients. 2013; 14:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buroker NE, Ning XH, Zhou QN, et al. Circulating miRNAs from dried blood spots are associated with high altitude sickness. 2013; 2:1000125. [Google Scholar]
- 43.Wei T, Folkersen L, Ehrenborg E, Gabrielsen A. MicroRNA 486-3p as a stability marker in acute coronary syndrome. 2016; 36:e00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Yang L, Li P, et al. Circulating microRNA signatures associated with childhood asthma. 2015; 61:467–474. [DOI] [PubMed] [Google Scholar]
- 45.Steele CB, Thomas CC, Henley SJ, et al. Vital signs: trends in incidence of cancers associated with overweight and obesity- United States, 2005–2014. 2017; 66:1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunet A, Berger SL. Epigenetics of aging and aging-related disease. 2014; 69 (Suppl 1):S17–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. 2007; 23:413–418. [DOI] [PubMed] [Google Scholar]
- 48.IARC. GLOBOCAN 2012; Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. Lyon: WHO; 2012. [Google Scholar]
- 49.Karavidas A, Lazaros G, Tsiachris D, Pyrgakis V. Aging and the cardiovascular system. 2010; 51:421–427. [PubMed] [Google Scholar]
- 50.Bacos K, Gillberg L, Volkov P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. 2016; 7:11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almen MS, Nilsson EK, Jacobsson JA, et al. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. 2014; 548:61–67. [DOI] [PubMed] [Google Scholar]
- 52.Almen MS, Jacobsson JA, Moschonis G, et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. 2012; 99:132–137. [DOI] [PubMed] [Google Scholar]
- 53.WHO. Obesity: Preventing and Managing the Global Epidemic. Report of a WHO Consultation. Geneva: WHO; 2000. [PubMed] [Google Scholar]
- 54.Ameling S, Kacprowski T, Chilukoti RK, et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. 2015; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rager JE, Smeester L, Jaspers I, et al. Epigenetic changes induced by air toxics: formaldehyde exposure alters miRNA expression profiles in human lung cells. 2011; 119:494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei H, Zhang J, Tan K, Sun R, Yin L, Pu Y. Benzene-induced aberrant miRNA expression profile in hematopoietic progenitor cells in C57BL/6 mice. 2015; 16:27058–27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pergoli L, Cantone L, Favero C, et al. Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. 2017; 14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nymark P, Guled M, Borze I, et al. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. 2011; 50:585–597. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Lv Y, Hao J, et al. Role of microRNA-4516 involved autophagy associated with exposure to fine particulate matter. 2016; 7:45385–45397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woeller CF, Thatcher TH, Van Twisk D, et al. MicroRNAs as novel biomarkers of deployment status and exposure to polychlorinated dibenzo-p-dioxins/dibenzofurans. 2016; 58:S89–S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuchs E, Chen T. A matter of life and death: self-renewal in stem cells. 2013; 14:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tay Y, Zhang J, Thomson AM, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. 2008; 455:1124–1128. [DOI] [PubMed] [Google Scholar]
- 63.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. 2008; 331:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuppusamy KT, Sperber H, Ruohola-Baker H. MicroRNA regulation and role in stem cell maintenance, cardiac differentiation and hypertrophy. 2013; 13:757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. 2010; 70:9473–9482. [DOI] [PubMed] [Google Scholar]
- 66.Lin SL, Chang DC, Chang-Lin S, et al. MiR-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. 2008; 14:2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakaguchi M, Hisamori S, Oshima N, et al. MiR-137 regulates the tumorigenicity of colon cancer stem cells through the inhibition of DCLK1. 2016; 14:354–362. [DOI] [PubMed] [Google Scholar]
- 68.Silber J, Lim DA, Petritsch C, et al. MiR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. 2008; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. 2008; 454:436–444. [DOI] [PubMed] [Google Scholar]
- 70.Vicente-Manzanares M, Sancho D, Yanez-Mo M, Sanchez-Madrid F. The leukocyte cytoskeleton in cell migration and immune interactions. 2002; 216:233–289. [DOI] [PubMed] [Google Scholar]
- 71.PANTHER. Inflammation Mediated by Chemockine and Cytokine Signaling Pathway. 2017. Available at: http://www.pantherdb.org/pathway/pathDetail.do?clsAccession=P00031 Accessed November 8, 2017. [Google Scholar]
- 72.Vosa U, Vooder T, Kolde R, et al. Meta-analysis of microRNA expression in lung cancer. 2013; 132:2884–2893. [DOI] [PubMed] [Google Scholar]
- 73.Donnem T, Fenton CG, Lonvik K, et al. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. 2012; 7:e29671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren R, Tyryshkin K, Graham CH, et al. Comprehensive immune transcriptomic analysis in bladder cancer reveals subtype specific immune gene expression patterns of prognostic relevance. 2017; 8:70982–71001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowe R, Overhoff MG, Ramagopalan SV, et al. The senescent methylome and its relationship with cancer, ageing and germline genetic variation in humans. 2015; 16:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Genome. KEGG Pathway: hsa04060. 2017. Available at: http://www.genome.jp/dbget-bin/www_bget?hsa04060 Accessed November 8, 2017. [Google Scholar]
- 77.Heaney ML, Golde DW. Soluble receptors in human disease. 1998; 64:135–146. [DOI] [PubMed] [Google Scholar]
- 78.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. 2004; 4:11–22. [DOI] [PubMed] [Google Scholar]
- 79.Genome. KEGG Cell Adhesion Molecules: hsa04514. 2017. Available at: http://www.genome.jp/kegg-bin/show_pathway?hsa04514 Accessed November 8, 2017. [Google Scholar]
- 80.Montoya MC, Sancho D, Vicente-Manzanares M, Sanchez-Madrid F. Cell adhesion and polarity during immune interactions. 2002; 186:68–82. [DOI] [PubMed] [Google Scholar]
- 81.Elangbam CS, Qualls CW, Jr, Dahlgren RR. Cell adhesion molecules-update. 1997; 34:61–73. [DOI] [PubMed] [Google Scholar]
- 82.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. 2004; 64:3753–3756. [DOI] [PubMed] [Google Scholar]
- 83.Gao W, Yu Y, Cao H, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. 2010; 64:399–408. [DOI] [PubMed] [Google Scholar]
- 84.Li Y, Tan W, Neo TW, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. 2009; 100:1234–1242. [DOI] [PubMed] [Google Scholar]
- 85.Kolbert CP, Feddersen RM, Rakhshan F, et al. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. 2013; 8:e52517. [DOI] [PMC free article] [PubMed] [Google Scholar]


