In this study, Begnis et al. show that HAATI, which is a mode of telomerase-minus survival in which canonical telomeres are superseded by blocks of nontelomeric rDNA heterochromatin that have spread to all chromosome ends, is formed and maintained. Their findings demonstrate that HAATI arises when telomere loss triggers a newly recognized illegitimate recombination pathway that requires RNAi factors, uncovering novel roles for ncRNAs in assembling a telomere-free chromosome end protection device.
Keywords: RNAi, DNA repair, telomere, ALT, cancer, genome stability
Abstract
The identification of telomerase-negative HAATI (heterochromatin amplification-mediated and telomerase-independent) cells, in which telomeres are superseded by nontelomeric heterochromatin tracts, challenged the idea that canonical telomeres are essential for chromosome linearity and raised crucial questions as to how such tracts translocate to eroding chromosome ends and confer end protection. Here we show that HAATI arises when telomere loss triggers a newly recognized illegitimate translocation pathway that requires RNAi factors. While RNAi is necessary for the translocation events that mobilize ribosomal DNA (rDNA) tracts to all chromosome ends (forming “HAATIrDNA” chromosomes), it is dispensable for HAATIrDNA maintenance. Surprisingly, Dicer (Dcr1) plays a separate, RNAi-independent role in preventing formation of the rare HAATI subtype in which a different repetitive element (the subtelomeric element) replaces telomeres. Using genetics and fusions between shelterin components and rDNA-binding proteins, we mapped the mechanism by which rDNA loci engage crucial end protection factors—despite the absence of telomere repeats—and secure end protection. Sequence analysis of HAATIrDNA genomes allowed us to propose RNA and DNA polymerase template-switching models for the mechanism of RNAi-triggered rDNA translocations. Collectively, our results reveal unforeseen roles for noncoding RNAs (ncRNAs) in assembling a telomere-free chromosome end protection device.
The linearity of eukaryotic chromosomes creates two major hazards for the cell: First, chromosome ends resemble DNA double-strand breaks (DSBs) with the potential to activate an inappropriate DNA damage response (DDR). Second, the end replication problem inflicts progressive erosion of chromosome termini. Telomeres, which tame these agents of danger, comprise tandem arrays of simple DNA repeats (TTAGGG in vertebrates and related repeats in most other eukaryotes) ending in a single-stranded 3′ overhang (de Lange et al. 1990; Jain and Cooper 2010). These repeats engage six telomere-specific proteins, collectively known as shelterin and including ssDNA- and dsDNA-binding proteins (Miyoshi et al. 2008; Palm and de Lange 2008). Shelterin restrains local DDR activities and regulates the telomere repeat-specifying reverse transcriptase telomerase, which replenishes terminal sequences lost due to the end replication problem.
The life span of most human cells is limited by progressive telomere attrition due to inactivation of telomerase from embryogenesis onward. To overcome this eventual replicative senescence, cancer cells must find strategies to restore and preserve telomere length. While many cancer cells reactivate telomerase expression, telomerase stays inactive in some cancers in which alternative strategies can counteract telomere shortening. At least 10%–15% of cancers survive via a recombination-dependent mechanism known as “alternative lengthening of telomeres” (ALT), where break-induced replication, normally prevented at telomeres, is activated by a combination of replication stress and deregulation of telomeric chromatin structure (Pickett and Reddel 2015; Apte and Cooper 2017). Moreover, in specific cancer types, including liposarcomas, approximately half of samples fail to show characteristics of either telomerase activation or ALT (Costa et al. 2006; Johnson et al. 2007). Therefore, as-yet-unrecognized modes beyond ALT may be exploited by cancer cells to escape the requirement of telomerase for long-term maintenance of linear chromosomes.
In unperturbed circumstances, the fission yeast Schizosaccharomyces pombe uses the classical telomere architecture to define its chromosome ends: Approximately 300 base pairs (bp) of double-stranded TTAC(A)GG(G1–4) repeats are bound by a conserved suite of shelterin proteins whose DNA-binding lynchpins are Taz1 and Pot1. Taz1 (ortholog of human TRF1 and TRF2), which binds the double-stranded repeats, prevents lethal chromosome end fusions, promotes telomeric semiconservative replication, and enforces telomere length homeostasis, among other telomeric roles. Pot1 binds terminal overhangs, as it harbors oligonucleotide-binding (OB) folds with high affinity for single-stranded telomeric sequences and prevents rampant telomeric degradation. Shelterin is completed by Rap1, Poz1, Tpz1, and Ccq1. Rap1 binds Taz1 and mediates some of its functions (Kanoh and Ishikawa 2001; Miller et al. 2005). Poz1, Tpz1, and Ccq1, identified by purifying Pot1-associated factors (Miyoshi et al. 2008), coordinate communication between the single-stranded and double-stranded telomeric regions.
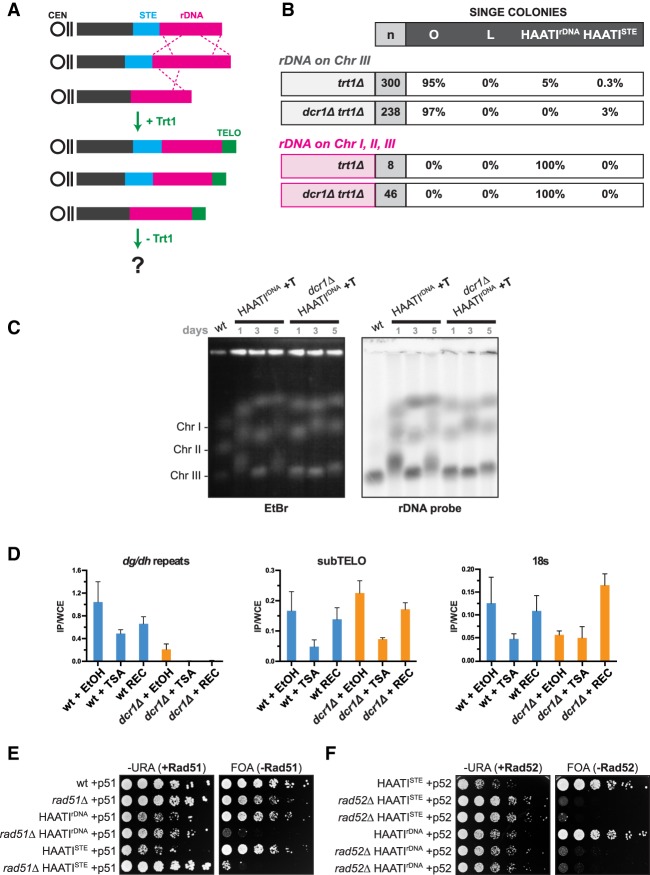
S. pombe averts the end replication problem via constitutive expression of telomerase, in whose absence three types of survivors have been described (Nakamura et al. 1998; Jain et al. 2010). First, the low fission yeast chromosome number (three per haploid genome) allows the end fusions unleashed by telomere loss to result in cells harboring three intrachromosomal end fusions with no interchromosomal fusions; such circular chromosome-containing cells (referred to here as “circulars”) lack telomeres. Second, heterogeneous telomeres can be maintained by ongoing homologous recombination—presumably break-induced replication—between eroding telomeres (“linear” survivors). Third, a survivor type that we dubbed HAATI (heterochromatin [HC] amplification-mediated and telomerase-independent) emerges under competitive culturing conditions. HAATI cells retain chromosome linearity by replacing telomeres with blocks of nontelomeric HC that acquire the ability to prevent chromosome end fusions.
HAATI encompasses two subtypes that harbor different repeats at their noncanonical chromosome ends (Jain et al. 2010). By far, the most common is HAATIrDNA, in which the ribosomal DNA (rDNA) repeat region spreads from its wild-type loci just inside either telomere of chromosome III to all of the termini of all three chromosomes. In the vastly rarer second subtype, HAATISTE, the rDNA remains at its original loci on chromosome III, while the so-called subtelomeric elements (STEs) are amplified from the subterminal regions of chromosomes I and II to all chromosome termini as well as multiple internal genomic loci. In both cases, the newly acquired termini engage Pot1, which is required for preservation of chromosome linearity despite the absence of terminal telomeric repeats. The terminal rDNA (or, rarely, STE) tracts undergo continual expansion and contraction. However, upon reintroduction of telomerase to HAATI cells, canonical telomere repeats are added to HAATI chromosome ends, stabilizing the rearranged genomes; expansion/contraction of terminal rDNA or STE tracts ceases, essentially “freezing” the sequence arrangement at the moment of telomere addition.
As HAATIrDNA can be reliably obtained under competitive conditions, much has been learned about the underlying genetic requirements. HC generally comprises repetitive sequences packed in a repressive structure in which nucleosome turnover rates are low and histones are decorated with specific modifications (Choi et al. 2005; Aygun et al. 2013). These modifications tend to exclude histone acetylation and include histone H3 Lys9 dimethylation or trimethylation (H3K9me2/3), which provides a binding site for the chromodomain HP1 (HC protein 1) proteins. Accordingly, histone deacetylases (HDACs) such as those in SHREC (Snf2/HDAC-containing repressor complex) are recruited to fission yeast HC, as are the histone H3K9 methyltransferase Clr4 and the HP1 ortholog Swi6 (Cowieson et al. 2000; Nakayama et al. 2001; Sugiyama et al. 2007). Notably, while telomeric DNA is embedded in HC across all species studied, the heterochromatic nature of telomeres is dispensable for the crucial telomere functions of preventing rampant DNA degradation and chromosome end fusions (Tuzon et al. 2004; Khair et al. 2010). On the contrary, HAATIrDNA cells rely on Clr4, Swi6, and SHREC to maintain protected chromosome ends; they also rely on the SHREC-interacting shelterin component Ccq1 (Jain et al. 2010). These data led us to propose that the SHREC-bound HC at HAATIrDNA chromosome termini recruits Ccq1, which in turn promotes local accumulation of Pot1.
The discovery of HAATI overturned the notion that fission yeast telomere sequences per se are essential for chromosome linearity. Moreover, HAATI resembles the approach taken by Drosophila melanogaster to maintain linear chromosomes. Flies lack both telomere repeats and telomerase; instead, their terminal sequences consist of the retrotransposons HET-A, TART, and TAHRE (referred to collectively as HTT arrays) (Mason et al. 2008). HP1 binds these terminal HTT arrays and is required for recruitment of specific end protection factors (including HOAP, HipHop, and Ver) (Cenci et al. 2003; Gao et al. 2010). Remarkably, the protection of fly chromosome ends is sequence-independent, as stable terminally deleted chromosomes lacking HTT repeats can be isolated. As at HTT-containing chromosome ends, such cells harboring terminal deletions require HP1 and its interactors for chromosome linearity (Fanti et al. 1998). Hence, the stability of linear chromosomes in flies is determined epigenetically. The analogy between fission yeast HAATI and Drosophila telomeres suggests that epigenetic chromosome end maintenance strategies are widely conserved and likely to represent alternative means for telomerase-minus cancer cells to attain unlimited proliferative capacity.
Although HC was traditionally considered transcriptionally inactive, a low level of RNA polymerase II (Pol II)-mediated transcription is needed for RNAi-induced silencing mechanisms (Djupedal et al. 2005). These mechanisms generate small interfering RNAs (siRNAs) from transcripts via the RNA-dependent RNA polymerase Rdp1 and the RNase III ribonuclease Dicer (Dcr1) (Colmenares et al. 2007). These siRNAs are loaded onto the nuclear Argonaute (Ago1) complex RITS, which is targeted by the siRNA to cognate nascent RNA sequences (Irvine et al. 2006); such targeting is sufficient to initiate Swi6/Clr4-dependent silencing (Buhler et al. 2006). Similar mechanisms of RNAi-based HC assembly have been identified in both plants and animals (Moazed 2009; Castel and Martienssen 2013). Dicer also promotes HC maintenance by displacing RNA Pol II from highly transcribed and repetitive DNA regions, including the rDNA, where Dicer-mediated release of RNA Pol II serves to limit collisions between the transcription and replication machineries, safeguarding both genome stability and HC reassembly following replication (Kloc et al. 2008; Zaratiegui et al. 2011; Castel et al. 2014).
The HC nature of HAATI chromosome ends led us to investigate the role of RNAi. Here we report that RNAi and the HC assembly machinery perform remarkably separate functions in assisting HAATI survival. While HC components are required for the continual recruitment of Pot1 to HAATI termini, the RNAi pathway is dispensable for HAATI maintenance. We found that Dcr1, but not the other canonical RNAi components, actively inhibits the amplification of STE sequences upon loss of telomerase and thereby prevents HAATISTE formation, revealing an RNAi-independent role of Dcr1 in preventing STE mobilization. Most surprisingly, the canonical RNAi pathway is essential for the genomic rearrangements that lead to HAATIrDNA formation. Hence, we define a previously unrecognized RNAi-based translocation pathway; this pathway is prohibited by canonical telomeres.
Results
HC engages Pot1 at HAATIrDNA terminal repeats
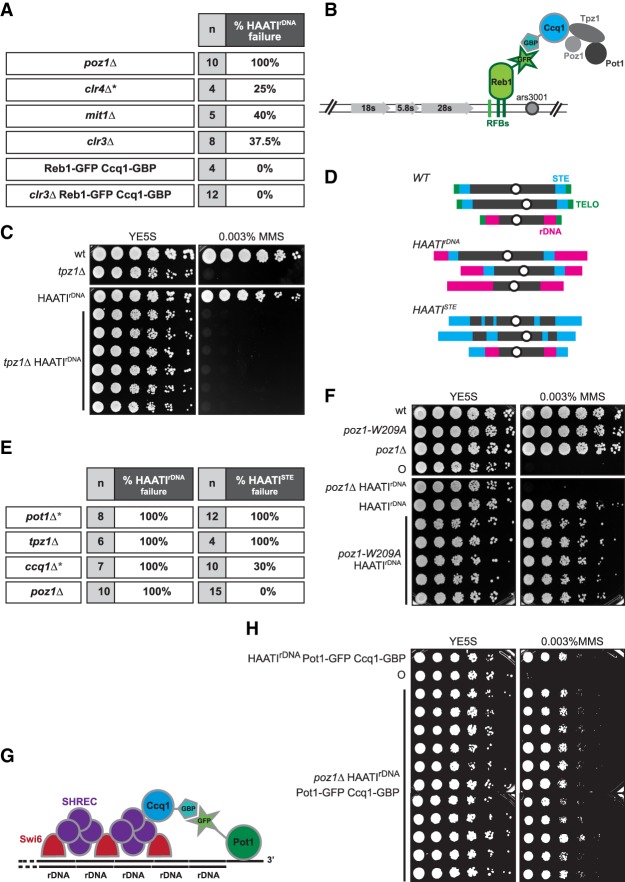
A defining difference between the end protection strategies of HAATI and wild-type telomeres is the dependence of the former, but not the latter, on the heterochromatic nature of the terminal sequences. HAATIrDNA cells rely on Clr4 to maintain protected chromosome ends; they also require the SHREC-interacting shelterin component Ccq1 (Jain et al. 2010). The SHREC HC machinery consists of four core proteins—Clr1, Clr2, Clr3, and Mit1—all of which are dispensable for the maintenance of linear or circular trt1Δ survivors (Sugiyama et al. 2007; data not shown). To assess their role in HAATIrDNA maintenance, we scored the loss of HAATI maintenance via chromosome circularization, which occurs when HAATIrDNA cells lose end protection capability; circularization is assessed via the exquisite sensitivity of circular strains to low concentrations of the alkylating agent methyl methanesulfonate (MMS) and by pulsed-field gel electrophoretic (PFGE) analysis of chromosome arrangement (Jain et al. 2010; see below). Deletion of clr3+ (encoding the SHREC HDAC) or mit1+ (encoding the SHREC chromatin remodeler) compromises HAATIrDNA maintenance, as continued survival depends on chromosome circularization in ∼40% of HAATIrDNA cultures deleted for either of these genes (Fig. 1A); this decline in HAATIrDNA maintenance resembles the effects seen upon clr4+ deletion (Jain et al. 2010).
Figure 1.
Pot1 engagement at HAATI chromosome ends. (A) Table showing the requirement of shelterin and SHREC proteins for maintenance of HAATIrDNA chromosomes. The gene deletions indicated in the table (poz1Δ, mit1Δ, and clr3Δ) were constructed by one-step gene replacement in already formed haploid HAATIrDNA survivors. Previously reported data for clr4+ deletion (Jain et al. 2010) are marked by an asterisk and shown for comparison. “n” indicates the total number of HAATIrDNA transformants analyzed for each gene deletion; percentages indicate the frequency at which HAATIrDNA capability is lost, leading to chromosome circularization. This was scored first by exposure to MMS, as circulars display extreme hypersensitivity to this agent (Jain et al. 2010); for a subset of isolates, the chromosome circularity was further confirmed by PFGE (explained in detail in Fig. 3). (B) Schematic illustrating the strategy for tethering Ccq1 to rDNA arrays in the absence of SHREC by forcing interaction between Reb1 and Ccq1 via the GFP-binding protein (GBP)–GFP interaction. (C) Fivefold serial dilutions of the indicated (107 cells per milliliter) cultures on medium lacking or containing MMS. (D) Schematic depicting wild-type and HAATI chromosome organization. (Top) In wild-type cells, the telomeres on chromosomes I and II are flanked by the STE regions, comprising ∼20 kb of imperfect ∼86-bp repeats. The telomeres of chromosome III are flanked by the rDNA repeats, which comprise ∼1 Mb of the 3.6-Mb chromosome. (Middle) HAATIrDNA contains rDNA at the ends of all chromosomes. (Bottom) In HAATISTE cells, STE sequences localize to the ends of all chromosomes as well as multiple sites in the chromosomal interiors. (E) Table summarizing the requirement of telomere-associated proteins for maintenance of HAATIrDNA versus HAATISTE. The genes indicated in the table were deleted by one-step gene replacement in already formed haploid HAATIrDNA or HAATISTE survivors. Percentages indicate the frequency of chromosome circularization in each HAATI subtype. Chromosome circularization was measured by assessing MMS sensitivity. The asterisks mark published results (Jain et al. 2010) shown for comparison. (F) Dilution assay performed as in C. (G) Schematic illustrating the strategy for tethering Pot1 to Ccq1 (via GBP–GFP interaction) in the absence of Poz1. (H) Dilution assay performed as in C. (○) Circular strain.
Why is HC important for HAATIrDNA maintenance? As both Pot1 and Ccq1 are essential for HAATI (Jain et al. 2010), we hypothesized that Ccq1 links Pot1 to rDNA termini through SHREC (Jain et al. 2010). To test this hypothesis and assess whether artificial Pot1 recruitment would be sufficient to guarantee HAATIrDNA maintenance in the absence of SHREC activity, we developed an ectopic tethering strategy. Ccq1 was fused with the GFP-binding protein (GBP) in cells in which Reb1, a protein that binds the intergenic spacer of each rDNA unit, was endogenously tagged with GFP. With this system, Ccq1 is recruited to the rDNA in a SHREC-independent manner (Fig. 1B). Telomere length is not significantly affected by simultaneous tagging of Ccq1-GBP and Reb1-GFP (Supplemental Fig. S1A); moreover, coexpression of these fusion proteins never compromises HAATIrDNA maintenance (Fig. 1A). In contrast to the chromosome circularization seen in 40% of HAATIrDNA cells upon clr3+ deletion, 100% of clr3Δ reb1-GFP ccq1-GBP HAATIrDNA transformants retain chromosome linearity (Fig. 1A). Hence, SHREC becomes dispensable for HAATIrDNA maintenance when Pot1 is artificially recruited to the rDNA repeats. This observation demonstrates that the principle function of terminal HC in HAATIrDNA maintenance is engaging Ccq1 to then capture Pot1.
To further investigate the interplay between HC and Pot1-mediated end protection, we tested the requirement for two Pot1 partners: Tpz1 and Poz1. In wild-type cells with canonical telomeres, the N-terminal domain of Tpz1 forms a heterodimer with Pot1, and the C-terminal domain of Tpz1 interacts with Poz1 and Ccq1 (Harland et al. 2014). Loss of Tpz1 in a wild-type setting leads to immediate chromosome end fusions, with chromosome circularization being the sole survival mode, as seen upon pot1+ deletion. Similarly, deletion of tpz1+ in HAATIrDNA results in chromosome circularization (Fig. 1C). Thus, as at wild-type telomeres, Pot1 acts in concert with Tpz1 to protect HAATI chromosome ends.
Poz1 regulates telomere length and silencing but is dispensable for preventing chromosome circularization in a trt1+ setting. In contrast, 100% of HAATIrDNA cells circularize their chromosomes upon Poz1 loss (Fig. 1D). This crucial end protection role of Poz1 is specific to HAATIrDNA, as 100% of poz1Δ HAATISTE cells maintain linear chromosomes (Fig. 1D; data not shown). To determine whether Poz1 acts in HAATIrDNA by promoting local HC, we used the poz1-W209A mutation, which affects telomere silencing without significantly modifying telomere length (Tadeo et al. 2013). As poz1-W209A HAATIrDNA cells robustly maintain chromosome linearity (Fig. 1E) and since all poz1-W209A trt1Δ cells establish HAATIrDNA survival (Supplemental Fig. S1B; data not shown), we infer that Poz1 is required for HAATIrDNA in some capacity other than HC promotion. To address the alternative idea that Poz1 stabilizes interactions between Ccq1 and Pot1, we constructed strains expressing Ccq1-GBP and Pot1-GFP. This coexpression does not perturb telomere maintenance in a wild-type background (although telomeres are slightly elongated in all strains carrying Pot1-GFP) (Fig. 1F; Supplemental Fig. S1A); likewise, HAATIrDNA is efficiently maintained in the presence of Ccq1-GBP and Pot1-GFP (Fig. 1G). All poz1Δ ccq1-GBP pot1-GFP HAATIrDNA transformants maintain the HAATIrDNA survival mode (Fig. 1G). Therefore, the essential role of Poz1 in HAATIrDNA is to ensure interaction between Ccq1 and Pot1. As wild-type cells do not require Poz1 to reinforce the Ccq1–Pot1 association, we infer that Ccq1–Pot1 interaction is augmented by the presence of canonical telomere repeats, which bind Pot1 with high affinity, in wild-type settings (Supplemental Fig. S1C).
RNAi is not involved in the HC dynamics that protect HAATIrDNA chromosomes
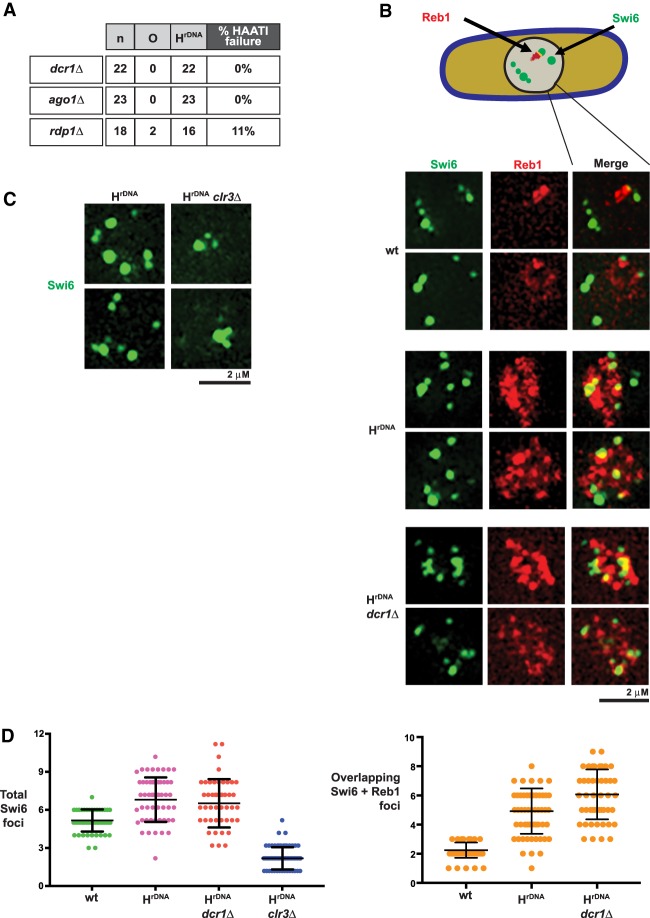
As HC is required for the maintenance of Pot1 association with HAATIrDNA chromosome termini, we investigated the determinants of HC over the rDNA ends. At wild-type telomeres, HC is nucleated by Taz1 and its interacting partners (Cooper et al. 1997; Kanoh et al. 2005); however, HAATI chromosomes lack telomere repeats, and Taz1 is dispensable for HAATI maintenance (Jain et al. 2010). As the RNAi pathway is also implicated in driving HC formation at subtelomeres (Kanoh et al. 2005; Buhler et al. 2006), we investigated its role in HAATI maintenance. Loss of Dcr1 in already formed HAATIrDNA cells has no effect on HAATI maintenance (Fig. 2A); likewise, ago1+ is dispensable for HAATI maintenance (Fig. 2A). Hence, the RNAi pathway is dispensable for HAATI maintenance.
Figure 2.
RNAi is dispensable for the HC that maintains HAATIrDNA. (A) Table summarizing the requirement of dcr1+ and ago1+ for maintenance of HAATIrDNA chromosomes. “n” indicates the total number of HAATIrDNA transformants analyzed for each gene deletion; percentage of HAATIrDNA failure was scored via chromosome circularization in already formed HAATIrDNA, as described in Figure 1A. (B) Dicer is dispensable for Swi6 localization in HAATIrDNA. Two representative examples of cells harboring endogenously GFP-tagged Swi6 and endogenously tdTomato-tagged Reb1 are shown for each genotype. Cells were photographed as described in the Materials and Methods. The rDNA is tightly localized in wild-type cells and barely overlaps with Swi6; in contrast, the rDNA expands in HAATIrDNA cells and contains domains harboring intense Swi6 fluorescence. This is consistent with localization of Swi6 to the amplified rDNA sequences, which is retained even after deletion of dcr1+ (shown in the bottom two rows). (C) Clr3 is required for Swi6 localization in HAATIrDNA. Two representative examples of HAATIrDNA and clr3Δ HAATIrDNA cells harboring endogenously GFP-tagged Swi6 are shown. (D) Quantitation of Swi6 foci (left) and Swi6–Reb1 colocalization (right). The total number of Swi6 foci from 50–80 cells (from two to three independent experiments) for each genetic background is shown. A standard t-test was used to compute mean ± SE. HAATIrDNA shows an increased number of Swi6 foci relative to wild type. This increase is Dcr1-independent but Clr3-dependent. While Swi6 foci rarely colocalize with Reb1 in wild type, they do in HAATIrDNA. HAATIrDNA cells also show enhanced Reb1 signal with or without Dcr1.
The dispensability of RNAi for HAATI maintenance stands in stark contrast to the important role played by Clr4. To clarify the distinction between general HC factors and RNAi in HAATI dynamics, we inspected the pattern of Swi6-GFP distribution as an initial proxy for HC organization; rDNA was viewed via mCherry tagging of Reb1. HAATIrDNA cells show enhanced Reb1 fluorescence compared with wild-type cells, in line with the average sixfold increase in rDNA levels in these cells (Jain et al. 2010). Swi6-containing HC foci are also brighter and more numerous in HAATIrDNA than in wild-type cells (Fig. 2B; Jain et al. 2010). Moreover, while only a small proportion of Swi6 intensity overlaps with rDNA in wild-type cells, a substantial proportion of the intense Swi6 foci colocalizes with Reb1 in HAATIrDNA cells (Fig. 2B,D). This increase in Swi6 foci is lost in that subset of clr3Δ mutants that maintains HAATIrDNA chromosomes (Fig. 2C), indicating that SHREC and/or hypoacetylated nucleosomes are important for localizing Swi6 to HAATI rDNA tracts. In contrast, dcr1Δ HAATIrDNA mutants preserve the amplified Swi6 pattern (Fig. 2B). Furthermore, while it has been reported that pericentromeric HC levels are further reduced in dcr1Δ clr3Δ cells compared with single mutants (Buscaino et al. 2013), in no case did double deletion of dcr1+ and clr3+ lead to additive levels of HAATIrDNA chromosome circularization relative to clr3+ loss alone (data not shown). Thus, RNAi is dispensable for HAATIrDNA maintenance even under conditions of reduced HDAC availability. We conclude that while HC assembly factors such as Clr4 and SHREC safeguard the rDNA HC and therefore ensure Pot1 association with HAATIrDNA termini, the RNAi machinery does not share this role in HAATIrDNA maintenance.
RNAi is required for HAATIrDNA formation
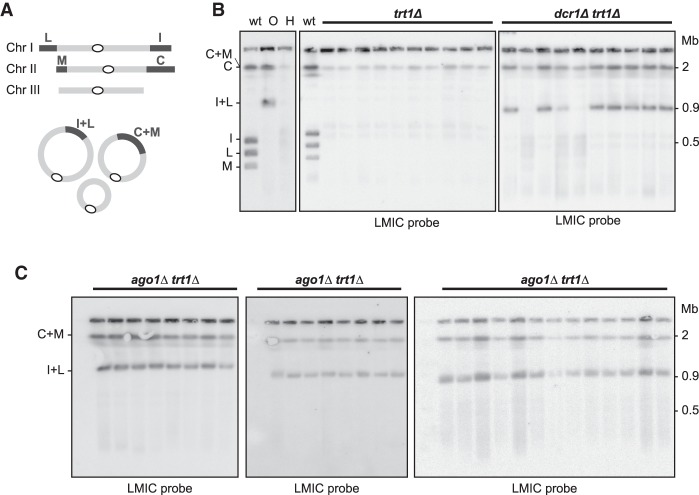
Having ruled out a contribution to HAATI maintenance, we investigated the involvement of RNAi in HAATI formation. We generated trt1Δ mutants carrying or lacking dcr1+ and propagated them under conditions in which virtually all trt1Δ survivors use HAATIrDNA. As described above, survivor type was initially assessed by exposure to MMS (Jain et al. 2010) and then verified by analyzing chromosome organization via PFGE. Digestion of linear chromosomes with the rare cutting restriction enzyme NotI releases four terminal fragments—referred to as L, M, I, and C—from linear chromosomes I and II (Fig. 3A), while circular chromosomes yield fused terminal fragments (L+I and C+M) (Fig. 3A,B). In contrast to DNA from both linear and circular survivors, the terminal fragments of HAATI chromosomes fail to enter gels, resulting in retention of LMIC hybridization signal in the well (Fig. 3B); this is presumably due to the continual expansion and contraction (and hence the continual presence of branched recombination intermediates) of the rDNA or STE tracts at HAATI chromosome termini (Jain et al. 2010). In an otherwise wild-type setting, 10 out of 10 trt1Δ populations survive using the HAATI mode (Fig. 3B). Remarkably, however, the majority of dcr1Δtrt1Δ populations yields cells with circular chromosomes (Fig. 3B). Thus, the absence of Dcr1 severely compromises HAATI formation. Moreover, deletion of ago1+ completely abolishes HAATI formation (Fig. 3C). Collectively, we conclude that while RNAi is dispensable for HAATI maintenance, it is crucial for the processes involved in HAATI establishment.
Figure 3.
RNAi is essential for HAATIrDNA formation. (A) NotI digestion of wild-type chromosomes releases four terminal fragments—referred to as L, M, I, and C—from the ends of chromosomes I and II; in circulars, these are replaced by fusion fragments L+I and C+M. (B) Dcr1 is important for HAATI formation. Heterozygous dcr1Δ/dcr1+ trt1Δ/trt1+ diploids were sporulated, and the indicated progeny (10 trt1Δ and 10 dcr1Δtrt1Δ progeny) were grown for 30 d under competitive conditions that favor HAATIrDNA formation. PFGE of NotI-digested chromosomes from each of the resulting survivor populations was performed. All 10 trt1Δ survivors use HAATI, as indicated by retention of hybridization signal in the well; in contrast, seven out of 10 dcr1Δtrt1Δ progeny yield a circular hybridization pattern. (Left) As a reference, NotI digests of wild-type (“WT”), circular (“O”), and HAATIrDNA (“H”) cells are shown. (C) HAATI survival requires Ago1. ago1Δtrt1Δ progeny were raised from heterozygous ago1Δ/ago1+ trt1Δ/trt1+ diploids as in B. All ago1Δtrt1Δ progeny are circulars.
Dicer acts independently of the RNAi pathway to inhibit HAATISTE formation
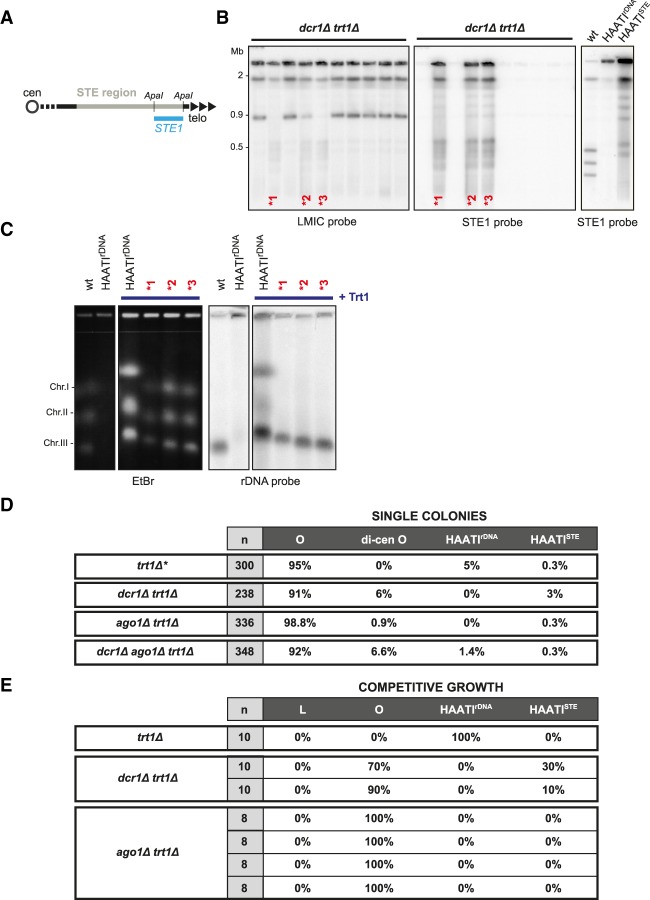
As three out of 10 dcr1Δtrt1Δ populations yield PFGE patterns common to HAATIrDNA and HAATISTE (Fig. 3B), we investigated their sequence arrangement in more detail. Remarkably, the STE1 probe hybridizes to all of the internally located NotI fragments from these three dcr1Δtrt1Δ isolates, demonstrating that STE1 sequences have spread to internal genomic loci; this pattern is diagnostic of HAATISTE (Fig. 4A,B). To determine whether these isolates also undergo rDNA rearrangement, we analyzed chromosome arrangement following reintroduction of telomerase, which results in telomere addition and cessation of terminal chromosome rearrangements; the resulting “HAATI+Trt1” cells have stable telomere-containing chromosome ends that enter gels despite retaining rDNA repeats just inside from the added telomeres at all chromosome ends (Fig. 4C). In all three dcr1Δtrt1Δ isolates, the rDNA remains localized exclusively on chromosome III. We conclude that in the absence of Dcr1, only the HAATISTE subtype can arise.
Figure 4.
Dicer inhibits HAATISTE formation. (A) Schematic of ApaI restriction sites and STE1 probe in the subtelomeric (STE) region. (B) The LMIC-probed membrane from Figure 3B was stripped and reprobed for STE1 sequences. While the circular chromosomes in seven out of 10 isolates lack STE1 sequences, the STE1 probe hybridizes strongly with every NotI restriction fragment derived from the three dcr1Δtrt1Δ genomes indicated by asterisks, demonstrating that STE1 sequences have spread to internal sites on chromosomes I and II (Jain et al. 2010). As a reference, the STE1 hybridization pattern of NotI digests from wild-type, HAATIrDNA, and HAATISTE cells is shown at the right. (C) Trt1 reintroduction to HAATI cells results in telomere addition and chromosomal gel entry, as seen by PFGE of whole undigested chromosomes. Southern blot analysis of the genomes from the dcr1Δtrt1Δ isolates indicated by asterisks reveals that the rDNA is contained only on chromosome III. The restriction of rDNA to chromosome III along with the appearance of STEs at internal sites on chromosomes I and II are diagnostic of HAATISTE (Jain et al. 2010). (D) Single-colony analysis of survivor formation reveals survivor type preferences under noncompetitive growth conditions. “n” indicates the total number of colonies screened. Data are compared with those obtained for trt1Δ single colony isolates (asterisk marks published results from Jain et al. 2010). (E) Analysis of trt1Δ survivor formation under competitive conditions. “n” indicates the number of independent populations raised. The table summarizes the results from experiments in Figure 3, B and C, and additional repeats.
The elevated level of HAATISTE formation in the absence of Dcr1 suggests that the rarity of HAATISTE stems from an active role for Dcr1 in barring establishment of this survival mode. To explore this idea, we assessed the dcr1Δ survivor profile under single-colony conditions in which competition between survivors does not drive the choice of survivor pathway. Under these conditions, 3% of dcr1Δtrt1Δ colonies harbor HAATISTE chromosomes (Fig. 4D). This is in marked contrast to the survivor distribution seen in a dcr1+ setting, in which only 0.3% of trt1Δ colonies survive as HAATISTE. This 10-fold increase in the frequency of HAATISTE formation demonstrates that upon telomerase loss, Dcr1 inhibits the occurrence of HAATISTE.
Surprisingly, we were never able to isolate HAATISTE survivors from ago1Δtrt1Δ populations under competitive growth conditions (Fig. 4E). Moreover, when analyzing HAATI formation in single-colony conditions, HAATISTE arose at the same low frequency in an ago1Δtrt1Δ background as in a trt1Δ background, confirming that Ago1 does not share Dcr1's role in blocking HAATISTE formation (Fig. 4D). We also observed that in some instances, RNAi mutants give rise to survivors displaying dicentric dichromosome circles (Fig. 4D). This phenomenon has been described previously in trt1Δclr4Δ survivors (Jain et al. 2010) and is most likely made possible by a deficit in pericentric function that confers a higher probability of centromere inactivation. These dicentric chromosomes are stably maintained, as the corresponding restriction pattern is conserved over many generations (data not shown).
Hence, Dcr1 actively inhibits the establishment of HAATISTE survival through mechanisms that are independent of the canonical RNAi pathway. At the same time, Dcr1 is required via canonical RNAi to promote HAATIrDNA establishment.
The catalytic activity of Dicer is required for its roles in HAATI regulation
To explore the mechanism by which Dcr1 acts independently of RNAi to inhibit HAATISTE formation, we used two mutant alleles. The dcr1-cd allele harbors two mutations (D937A and D1127A) that abolish Dicer's RNase III catalytic activity; this allele abrogates the genesis of siRNA (Colmenares et al. 2007). The dcr1-ΔCTD allele encodes a C-terminally truncated protein that prevents nuclear import and abolishes RNAi (Emmerth et al. 2010). We raised telomerase-minus survivors from cells in which dcr1+ was replaced with either the dcr1-ΔCTD or dcr1-cd allele. In both cases, HAATIrDNA formation was abolished (Supplemental Fig. S2A). Hence, rDNA jumping requires the endoribonuclease activity of Dicer. Moreover, both dcr1-ΔCTD and dcr1-cd confer HAATISTE survival to an extent resembling that of dcr1Δ (Supplemental Fig. S2A). Hence, catalytic activity is required for Dicer's role in prohibiting HAATISTE formation, notwithstanding the fact that Ago1 is dispensable for this prohibition. These results suggest that HAATISTE formation is elicited by a transcript whose generation follows telomere loss and whose role in STE mobilization is inactivated by Dcr1-mediated cleavage.
One of the phenotypes associated with loss of RNAi components is a defect in telomere clustering during mitotic interphase (Hall et al. 2003); although telomeres remain localized to the nuclear periphery in RNAi-defective settings, the number of Taz1 foci increases, indicating dispersion (Supplemental Fig. S2B; Hall et al. 2003). We considered the possibility that telomere clustering promotes the spreading of rDNA tracts from chromosome III to other chromosome ends, thus explaining the need for RNAi factors. Quantitation of telomere foci, of which there are two to three in most wild-type cells, shows a modest dispersion of telomeres upon dcr1+ deletion; this telomere clustering defect is retained and slightly exacerbated by the dcr1-ΔCTD and dcr1-cd alleles (Supplemental Fig. S2B,C). At first glance, these effects of Dicer inactivation on telomere clustering appear disproportionately small relative to the complete loss of HAATIrDNA formation in RNAi-defective settings; however, as we discuss below, the role of RNAi in bringing disparate chromosome ends together may be intensified under conditions in which telomeres are eroding and chromosome mobility is increasing.
RNAi is required only for the rDNA translocations necessary to initiate HAATI
Formation of HAATIrDNA minimally comprises a two-step process: First, the rDNA repeats must spread to all chromosomal termini. Second, the new nontelomeric chromosome ends must acquire the ability to engage Pot1. To delineate which of these steps requires RNAi, we used HAATIrDNA+Trt1 cells, in which translocation of rDNA to all chromosomal termini has already occurred (prior to the addition of telomere repeats by reintroduced Trt1). We forced the loss of Trt1 in a HAATIrDNA+Trt1 setting to induce progressive telomere attrition against this backdrop of rDNA repeat tracts occupying every subtelomere (Fig. 5A). Remarkably, in this scenario, HAATI becomes the exclusive mode of survival regardless of whether cells are grown under single-colony or competitive conditions (Fig. 5B; Supplemental Fig. S3). Hence, the rarity of HAATIrDNA survival stems solely from the rDNA translocations required for its establishment; once the rDNA has spread to every subtelomere, HAATIrDNA arises readily.
Figure 5.
RNAi-dependent pathways confer illegitimate rDNA translocation. (A) Schematic of HAATIrDNA+Trt1 cells forced into subsequent Trt1 loss. (B) Table summarizing the frequency of survivor formation under noncompetitive (single-colony) growth conditions. Single-colony data for “rDNA on chromosome III” (i.e., wild-type cells), taken from Figure 4D, are shown for comparison. (C) PFGE analysis of whole chromosomes hybridized with rDNA probe. Cultures of the indicated genotypes were propagated in liquid for 5 d. Aliquots were sampled at days 1, 3, and 5 for transformation with a plasmid encoding trt1+, which confers telomere addition and gel entry and thus a snapshot of chromosome length over time in the HAATIrDNA population. (D) Levels of H3K9me enrichment in wild-type and dcr1Δ (trt1+) cells before (+EtOH), during (+TSA [trichostatin A]), and after (+REC, for recovery) TSA treatment. Levels of the indicated genome regions in H3K9me2 immunoprecipitates were quantified by RT–PCR using primers for pericentromeric dg/dh repeats, subtelomeric (subTELO) regions, and the 18S rDNA. Means are shown; error bars represent standard deviations of three biological repeats. (E) Rad51 is essential for HAATI maintenance. Fivefold serial dilutions of the indicated strains on medium selecting for (FOA) or against (−URA) loss of plasmid expressing Rad51 (“+p51”). (F) Rad52 is essential for HAATI maintenance. Fivefold serial dilutions of the indicated strains on medium selecting for (FOA) or against (−URA) loss of plasmid expressing Rad52 (“+p52”).
Is RNAi required for HAATIrDNA formation upon Trt1 loss in the HAATIrDNA+Trt1 setting? Strikingly, trt1+ deletion in cells bearing rDNA at all subtelomeres yields HAATIrDNA regardless of the presence or absence of Dcr1 (Fig. 5B; Supplemental Fig. S3). Hence, RNAi is dispensable for HAATIrDNA establishment when the rDNA has already been transferred to chromosome ends. These observations point to an unanticipated role for the RNAi pathway in promoting the transfer of rDNA repeats from their original sites on chromosome III to all chromosome ends lacking telomeres.
A conspicuous feature of HAATI chromosomes is their continual size alteration, which reflects the constant expansion and contraction of rDNA tracts; this size variation ceases as soon as canonical telomeres are added back via telomerase reintroduction (Jain et al. 2010). Notably, the size variation persists after dcr1+ deletion (Fig. 5C), indicating that continual rDNA rearrangement occurs independently of RNAi. Hence, an RNAi-dependent pathway is required only for the initial translocation or “jumping” that places rDNA at all chromosome ends.
In principle, our observation that RNAi is essential for rDNA jumping could reflect a role in stimulating the translocation itself or in stabilizing the translocation by driving HC establishment at the new rDNA ends. The RNAi pathway has been variably reported to promote silencing of a LEU2 reporter inserted within rDNA arrays (Cam et al. 2005), have no influence on H3K9 methylation in this region (Castel et al. 2014), and suppress H3K9 methylation in the region specifically in quiescent cells (Roche et al. 2016). To further address the role of RNAi in rDNA silencing, we performed HC establishment assays on telomerase-positive cells using trichostatin A (TSA), an inhibitor of class I/II HDACs that promotes hyperacetylation of nucleosomes and loss of H3K9 methylation (Vanhaecke et al. 2004). Treatment of wild-type cells with TSA causes a clear drop in H3K9me2 levels genome-wide, shown by chromatin immunoprecipitation (ChIP) using an antibody against H3K9me2 (Fig. 5D). As shown previously (Hall et al. 2003; Volpe et al. 2003; Kanoh et al. 2005), Dcr1 is required for robust H3K9me2 maintenance at pericentric dg/dh repeats and is essential for H3K9me2 re-establishment following TSA treatment. Dcr1 is dispensable for H3K9me2 maintenance and re-establishment at subtelomeres (Fig. 5D), where shelterin confers HC formation; indeed, subtelomeric H3K9me2 levels are slightly higher in a dcr1Δ background due to the loss of pericentric HC, which liberates limiting HC factors to associate with subtelomeres (Tadeo et al. 2013). In contrast, the release of pericentric HC conferred by dcr1+ deletion does not increase H3K9me2 at the rDNA region; rather, Dicer is required for wild-type levels of H3K9me2 at the rDNA (Fig. 5D). Intriguingly, however, re-establishment of H3K9me2 at rDNA following TSA washout is independent of Dicer. Thus, we propose that while RNAi is dispensable for HC establishment at rDNA, the failure to evict RNA Pol II from the rDNA intergenic spacer regions in the absence of Dicer (Castel et al. 2014) leads to a moderate loss of HC in this region over time, leading to the reduced HC seen in unperturbed dcr1Δ cells.
These observations demonstrate two principles regarding the role of RNAi in rDNA jumping. First, the reduction in rDNA HC upon dcr1+ deletion precludes a model in which RNAi promotes rDNA jumping by channeling HC away from rDNA (thereby facilitating rDNA rearrangements). Second, the dispensability of Dcr1 for HC nucleation in rDNA regions precludes a model in which rDNA jumping occurs independently of RNAi but RNAi is required for HC formation over the translocated rDNA. Reinforcing this argument is the distinction between general HC assembly mutants and RNAi mutants; while the latter lead to complete loss of HAATIrDNA, the former only partially reduce HAATIrDNA formation. Moreover, rDNA translocation is not detected in dcr1Δ HAATISTE or circular survivors (Fig. 4C; data not shown), arguing that jumping itself fails to occur in a dcr1Δ background. Hence, our observations strongly favor a role for RNAi in the translocation reaction itself.
Rad51 and Rad52 are vital for HAATI perpetuation
The observation that RNAi is dispensable for HAATI chromosome size variation (Fig. 5C) indicates that the mechanism of this size variation is distinct from that of the initial rDNA jumping. We found previously that deletion of rad51+ completely abolishes HAATI formation (Jain et al. 2010) and inferred that this Rad51 requirement stems from its role in recombination. However, Rad51 may instead (or in addition) play a role in transferring nontelomeric 3′ overhangs at HAATI termini to Pot1 (Schlacher et al. 2011); in the latter scenario, Rad51 would be necessary for prompt HAATI maintenance as well as establishment. To determine whether Rad51 is required for HAATI maintenance, we attempted to delete rad51+ in already formed HAATIrDNA and HAATISTE cells; however, we failed to isolate any such transformants. Therefore, we used a plasmid-shuffling approach. The rad51+ gene was introduced into a pREP4 vector carrying the selectable ura4+ marker (referred to here as p51). The efficacy of plasmid expression was verified via its ability to complement rad51Δ phenotypes (Supplemental Fig. S4A). p51 was then introduced into HAATI cells, and the genomic copy of rad51+ was deleted. Immediately upon forced loss of p51, the viability of these rad51Δ HAATI cells is lost (Fig. 5E). Therefore, Rad51 carries out an essential role in HAATI maintenance. Rad52 assists in Rad51 loading and is similarly vital for cells harboring HAATI chromosomes (Fig. 5F; Supplemental Fig. S4B). While the persistent recombination and amplification of the repeats may be important for insulating coding sequences from the end replication problem, this function alone would not be expected to confer such drastic death upon loss of Rad51 from HAATI cells. Rather, we propose that persistent Rad51-mediated processes facilitate the disposition of HAATI termini toward Pot1 loading following DNA replication.
Sequencing of HAATIrDNA genomes confirms the illegitimate nature of rDNA jumping
The requirement of RNAi for rDNA jumping suggests a previously unforeseen role of RNAi in genomic rearrangements. To determine whether sequence homology could underlie the jumping reaction as well as whether additional genomic rearrangements invisible to our Southern blot analyses (Jain et al. 2010; data not shown) characterize HAATIrDNA, we performed whole-genome paired-end Illumina sequencing on three wild-type isolates and three independent HAATIrDNA isolates derived therefrom; preliminary analysis of three additional HAATIrDNA isolates suggests that the conclusions drawn from the first three are applicable to all HAATIrDNA isolates. In addition to mapping the rearranged chromosome ends, we performed a comparative single-nucleotide polymorphism (SNP) analysis (see the Materials and Methods).
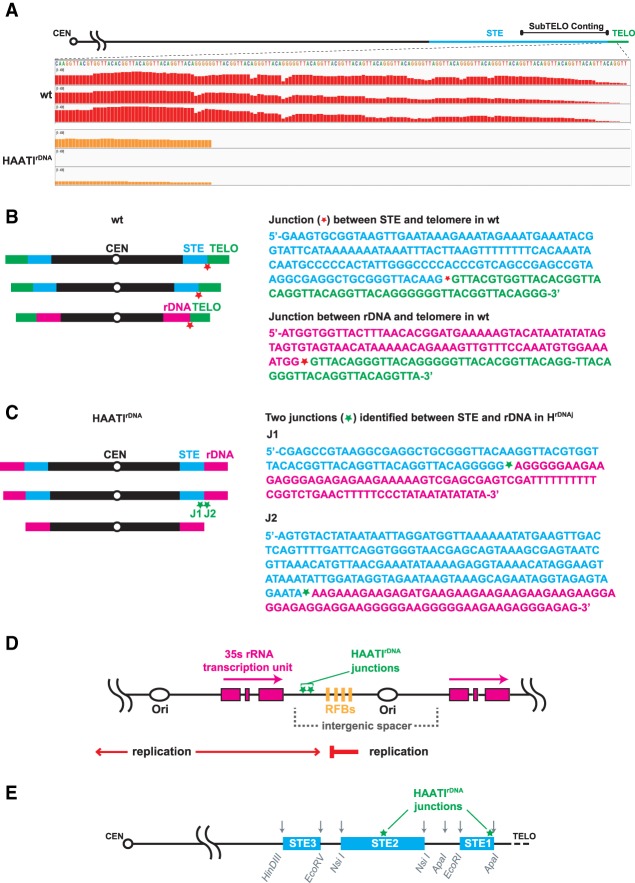
The sequencing data reveal several salient points. First, telomere sequences are indeed missing from HAATIrDNA chromosome termini (Fig. 6A). Second, the rDNA regions flanking the junctions with centromere-proximal regions of each HAATI chromosome end lie within the intergenic spacer region separating rDNA repeat units (Fig. 6B–D). The centromere-proximal junction sites comprise DNA from the wild-type STE regions to which the rDNA jumps. Such junctions are unique to HAATI; they are absent from wild-type genomes. Third, the polarity of the rDNA repeat unit—with RNA Pol I-mediated transcription occurring toward the chromosome end—is preserved following rDNA translocation. Fourth, the sequences directly abutting the junctions comprise one of two sequence units, GAAAG or AGGGGGA, that occur as dimers within the intergenic spacer units of the rDNA on wild-type chromosome III (Fig. 6D); these dimer units are severed upon translocation. In the case of AGGGGGA, the STE region (Fig. 6E) accommodating the translocation harbors a single AGGGGG unit; for the other translocation junctions, we failed to detect any obvious homology between STE and rDNA sequences surrounding the junction site. Moreover, while some S. pombe strains are reported to have STE regions on wild-type chromosome III (Ohno et al. 2016; Tashiro et al. 2017), our starting wild-type strain showed no evidence of such STE repeats. Fifth, the SNP variability among wild-type isolates is equivalent to the SNP variability between HAATI isolates or between wild-type and HAATI cells, indicating that the HAATIrDNA genome is not characterized by internal mutations. Hence, HAATIrDNA formation involves translocation of rDNA tracts onto exposed STE regions in a manner that preserves their original polarity. The uniqueness of the translocations identified in HAATIrDNA demonstrate that RNAi factors trigger a newly recognized illegitimate translocation pathway.
Figure 6.
Whole-genome sequencing of HAATIrDNA isolates identifies junctions derived from rDNA translocation. (A) Screenshot of Integrated Genomic Viewer browser depicting paired-end sequencing reads mapped to the subtelomeric (subTELO) contig, which harbors 7848 bp of STE DNA and 202 bp of telomere repeats (see the Materials and Methods). Three replicates of wild-type cells show robust mapping of reads to the telomeric repeats; the near-complete absence of corresponding reads in HAATIrDNA isolates confirms the absence of canonical telomeres. The few sequencing reads culminating in telomeric repeats in HAATIrDNA correspond to the four telomere repeat units found within the STE region centromere-proximal to the translocation site in a subset of HAATI isolates. The scale for bar height in both wild-type and HAATIrDNA samples is 0–400 and indicates the relative level of coverage of each base in a DNA string. (B, left) Wild-type genome organization. (Right) Sequences just centromere-proximal to the telomeric repeats on each chromosome, as identified by whole-genome sequencing from three wild-type isolates, are shown. Red asterisks denote the junctions between wild-type telomeric and STE (top sequence) or rDNA (bottom sequence) regions. (C, left) HAATIrDNA genome organization. (Right) Unique junctions (J1 and J2) between the centromere-proximal STE regions and rDNA in HAATIrDNA genomes. Green asterisks denote the junctions, which are unique to HAATIrDNA strains. (D) Schematic of the rDNA region. Each 10.9-kb rDNA repeat is composed of the 35S rDNA transcriptional unit (pink boxes) and an intergenic spacer that includes one origin of replication (ars3001) and four closely spaced polar replication fork barriers (RFBs). Green asterisks within the intergenic spacer indicate the sites of junctions between the STE regions (always on the centromere-proximal side) and the rDNA repeats; these junctions are unique to HAATIrDNA cells. (E) Schematic of the STE regions on chromosome I and chromosome II, which comprise ∼20 kb of imperfect tandem repeats (light-blue boxes) whose sequences modulate from the distal to the proximal ends of the region. Restriction sites (gray arrows) in the STEs are shown. Green asterisks within the STE region indicate the sites of junctions between STEs and rDNA (as indicated by an asterisk in C) in HAATIrDNA.
Discussion
RNAi determines repair dynamics at unshielded chromosome ends
First described mechanistically in Caenorhabditis elegans, RNAi is a highly conserved pathway that has been implicated in modulation of gene expression and an increasing number of cognate cellular processes (Napoli et al. 1990; Guo and Kemphues 1995; Fire et al. 1998). A growing body of evidence implicates RNAi in not only HC formation but also genome surveillance, the DDR, and eviction of RNA Pol II to avert collisions between transcription and replication (Peng and Karpen 2006; Zaratiegui et al. 2011; Francia et al. 2012; Castel et al. 2014). Here we demonstrate an unexpected and seemingly paradoxical role for the RNAi pathway in propelling illegitimate translocation events that place rDNA repeats at the ends of unprotected chromosomes, forming HAATIrDNA.
While HAATIrDNA dominates the telomerase-negative survival spectrum under competitive growth conditions, it is exceedingly rare under noncompetitive conditions, occurring in only ∼1.7% of telomerase-negative survivors. Here we found that this rarity stems exclusively from the rDNA jumping step; when telomerase is lost in a prerearranged genome, HAATIrDNA arises readily under all growth conditions. This was an initially surprising result, as we expected the engagement of Pot1 by nontelomeric repeats, an event anathema to the concept of telomeric identity, to be a major stumbling block for HAATI formation. On the contrary, it appears that when high-affinity Pot1-binding sequences (single-stranded telomeric DNA) are absent, the affinities involved in the rDNA-to-Pot1 linkage are sufficient to ensure quantitative Pot1 recruitment to chromosome termini. Here we used a series of fusion proteins combined with mutations in shelterin and SHREC to delineate the pathway from nontelomeric HC to Pot1. rDNA-bound SHREC recruits Ccq1, which in turn binds Tpz1 and Poz1, both of which are required for Pot1 binding; the SHREC can be replaced experimentally with a Reb1–Ccq1 fusion, and the Tpz1–Poz1 linkage can be replaced by a Ccq1–Pot1 fusion. The HAATIrDNA requirement for Poz1 is notable, as Poz1 is dispensable for Pot1 recruitment in wild-type cells; we surmise that high-affinity sequence-specific binding by Pot1/Tpz1 in wild-type cells obviates the need for stabilization by Poz1. Intriguingly, Pot1 recruitment in the HAATI setting, but not wild type, appears to require Rad51 and Rad52 (Fig. 5E,F). We suggest that this requirement again reflects the challenge of recruiting Pot1 without the reinforcement of high-affinity single-stranded telomere binding and propose the following HAATIrDNA maintenance scenario: The stalled forks that arise during replication of the intergenic spacers between rDNA repeats lead to local ssDNA formation. With assistance from Rad52, Rad51 forms filaments on this ssDNA and channels subsequent binding away from RPA and toward Pot1. These ideas are currently being tested.
Our observation that HAATIrDNA formation upon telomere attrition is limited solely by the improbability of rDNA jumping throws the spotlight on this pivotal event. This event shows an absolute dependence on the RNAi pathway, requiring catalytically active Dcr1 and Ago1 (and Rdp1) (data not shown), none of which are required for HAATI maintenance or formation of HAATI from a genome in which rDNA has been pretransferred to all subtelomeres. We note that the RNAi dependence of the jumping event was unexpected, as RNAi is generally associated with HC formation, which protects underlying repetitive DNA sequences from undergoing recombination (Ellermeier et al. 2010; Aygun et al. 2013). Thus, rDNA jumping appears to be governed by a heretofore undiscovered pathway of RNAi-dependent translocation.
RNA molecules have been implicated in promoting recombination by templating the repair of homologous DSB-containing DNA fragments in the absence of RNase H function (Keskin et al. 2014). This runs contrary to our observations, in which RNA is clearly involved but there is no pre-existing template for the rearrangements that lead to HAATIrDNA formation; moreover, the RNAi pathway has not been implicated in transcript RNA-templated DNA repair. However, it is conceivable that the “repair” reaction conferring rDNA translocation is templated by a transcript arising from an RNA polymerase that has switched templates from the subterminal region of one chromosome to that of another chromosome (see below).
Our sequencing data lend some hints as to the mechanism of illegitimate rDNA transfer. The sites at which native rDNA sequences become severed upon transfer to heterologous (STE) chromosome termini during HAATIrDNA formation lie in the intergenic spacer region between rDNA repeat units, a region of intense regulatory activity. The intergenic spacer of budding yeast has been shown to accommodate RNA Pol II-mediated transcription, the levels of which vary in response to rDNA copy number; increased transcription leads to local cohesin eviction and, in turn, enhanced rDNA recombination and copy number changes (Kobayashi 2008). RNA Pol II-mediated transcription through the rDNA intergenic spacer has also been observed in fission yeast (Castel et al. 2014). While rDNA replication is directionally controlled to prevent collisions between the replisome and the RNA Pol I transcription machinery, RNA Pol II runs antisense to RNA Pol I, on a collision course with the replication machinery (Zhao et al. 1997). Dcr1 has been shown to limit these collisions by evicting RNA Pol II from the rDNA, averting collision-associated replication fork stalling and recombination (Castel et al. 2014). Unlike the RNAi-mediated rDNA jumping described here, Dicer-mediated transcription termination was shown to rely on neither the catalytic activity of Dicer nor its RNAi companion, Ago1, although Ago1 localizes to the rDNA (Cam et al. 2005).
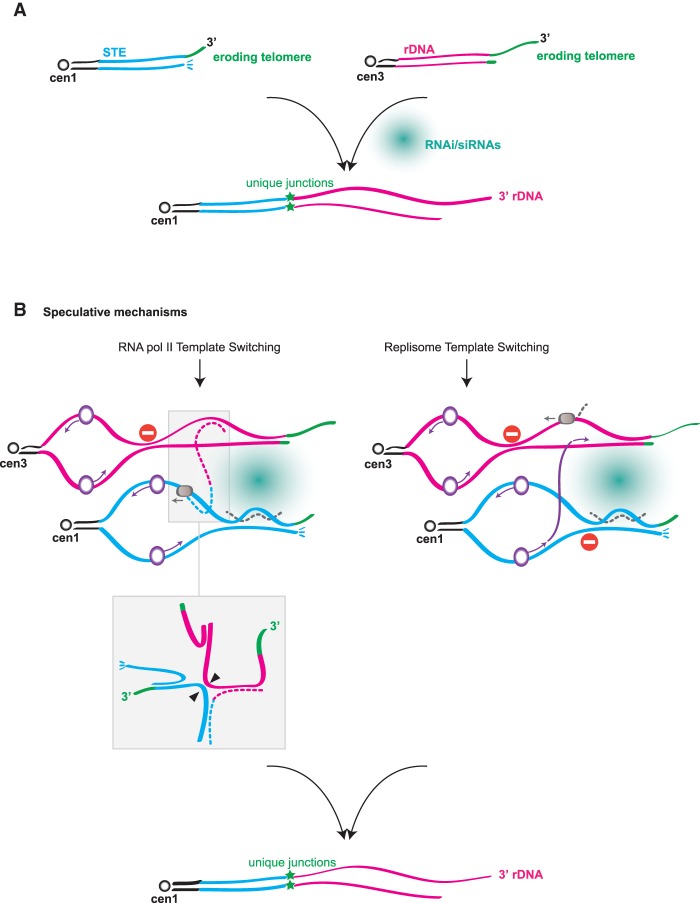
Collectively, our data paint a picture of the local conditions that promote HAATIrDNA formation (Fig. 7A). As telomeres erode following loss of telomerase, telomere-mediated inhibition of local transcription is lost (Greenwood and Cooper 2011; Bah et al. 2012; Maestroni et al. 2017; J Greenwood and JP Cooper, unpubl.), leading to the accumulation of subtelomeric RNA Pol II transcripts as well as RNAi pathway components; enhanced transcription will occur at both STE and rDNA termini. Simultaneously, the eroding telomeres and subtelomeres are likely to harbor stalled replication forks due to both the repetitiveness of these regions and enhanced levels of replication/transcription collisions as local silencing is lost. Furthermore, telomere loss abolishes the DDR prohibition at chromosome termini, rendering them vulnerable to activities that degrade and mobilize DNA ends. We propose that this scenario can result in physical interactions, perhaps RNA-mediated, between stressed STE and rDNA regions (Fig. 7).
Figure 7.
Potential routes to rDNA jumping. (A) Telomere erosion triggers the RNAi-dependent translocation of subtelomeric rDNA to STE chromosome ends. (B) Two possibilities for the mechanism of RNAi-dependent translocation are diagrammed. In both scenarios, local instability is engendered by telomere erosion, which leads to increased local transcription and DDR activation. (Left) RNA Pol II (gray circle) switches template from the subterminal region of chromosome III to that of chromosome I (or II), perhaps due to nicks, DSBs, or stalled forks at eroding chromosome ends. (Insets) A variant branched structure (the bottom inset represents an isomer of the top inset) harboring a hybrid (STE- and rDNA-containing) RNA strand forms and prompts illegitimate translocation. Arrowheads represent potential cleavage sites for an enzyme that resolves branched DNA structures. (Right) The replisome (purple circle) undergoes template switching, with DNA replication proceeding from a difficult-to-replicate STE region to the similarly compromised rDNA intergenic spacer region.
Against this backdrop, we envision several possible mechanisms for rDNA jumping. Two such possibilities comprise template-switching events (Fig. 7B). (1) RNA Pol II may switch templates from the subterminal region of one chromosome to that of another. Template switching by mammalian RNA Pol II has been observed not only in vitro but also in vivo (Nudler et al. 1996; Kandel and Nudler 2002) using a system in which template switching allows a retroviral vector to be converted into an integrated provirus; in the latter case, switching showed no requirement for obvious homology between sequences flanking the translocation. In cells forming HAATIrDNA, we could envision RNA Pol II switching from a nick, DSB, stalled fork, or R loop on one eroding chromosome end to that of another; the resulting STE/rDNA hybrid transcript would serve as a template for DNA synthesis. (2) The replisome may undergo template switching, with DNA replication proceeding from a difficult-to-replicate STE region to a similarly compromised rDNA region, thus leading to translocation. (3) A microhomology-mediated end-joining (MMEJ) pathway may comprise the translocation event in some instances, as an AGGGGGA microhomology is seen at some of the translocation junctions; in those cases, MMEJ would occur between an acentric rDNA fragment and a broken STE end. We view MMEJ as the least likely possibility, as our sequencing data have thus far revealed no instance in which translocation resulted in a switched orientation of the rDNA unit with respect to the chromosome terminus; given that perfect homology surrounding translocation points is unlikely, we would expect the rDNA to fuse occasionally in the “head-on” orientation with respect to the STE if MMEJ were frequent. We suggest that rather than conferring MMEJ, microhomologies assist in aligning disparate chromosome ends for translocation, which can be accomplished through template switching by RNA or DNA polymerases.
What explains the absolute dependence of the rDNA jumping reaction on RNAi? In the RNA Pol II switching model, the RNAi pathway might be envisioned to act after template switching has occurred, cleaving an RNA-containing translocation intermediate (Fig. 7B). For any model, the RNAi dependence of rDNA jumping could be attributed to a role for RNAi pathway components and/or noncoding RNAs (ncRNAs) in creating a platform for the switching event. This idea recalls studies of the mammalian (Francia et al. 2012) and plant (Wei et al. 2012) DDR, in which siRNAs corresponding to the sequences surrounding a DSB are generated upon breakage; these siRNAs, along with Dicer and Drosha, are thought to provide a platform for assembly of DDR foci. Moreover, as RNAs can promote the formation of phase-separated liquid droplets (Shin and Brangwynne 2017), we might postulate that upon telomere erosion, the subtelomeric (STE and rDNA) RNAi products promote the nucleation of droplets that subsequently fuse, bringing together disparate chromosome ends and facilitating translocation. We note that while the RNAi pathway has been implicated in promoting interphase telomere clustering (Hall et al. 2003), the modest level of disruption of this clustering upon loss of RNAi components (Supplemental Fig. S2) seems incommensurate with the 100% loss of HAATI formation in RNAi mutants. However, under the conditions of subtelomeric turmoil brought on by telomere loss, the enhanced presence of RNAi factors and siRNAs might significantly change the dynamics of dysfunctional chromosome ends to a degree not seen at unperturbed chromosome ends. Alternatively or in addition, R loops may be crucial for either RNA Pol II or replisome template switching and may be promoted by siRNAs.
Dicer quashes STE mobility
Even more rare than HAATIrDNA formation is that of HAATISTE. Here we show that this severe rarity is due to the presence of Dicer, as, in a dcr1Δ setting, HAATISTE formation rates increase (by 10-fold under noncompetitive growth conditions and much greater-fold in competitive conditions). In marked contrast to Dcr1, Ago1 (and Rdp1) (data not shown) fails to share Dcr1's role in prohibiting HAATISTE formation. Hence, beyond conferring translocation of rDNA repeats to unshielded chromosomes ends as part of the nuclear RNAi pathway, Dcr1 alone actively inhibits the rearrangement of STE arrays. As terminal transcripts accumulate upon telomere loss, we presume that Dicer normally prevents HAATISTE formation by cleaving a STE transcript that has the potential to confer dramatic genomic invasion by the STE. While the mechanism underlying this ncRNA-mediated invasion is far from clear, we can attest that it is clearly distinct from the mechanism underlying rDNA jumping and likely involves RNA–DNA hybridization at internal genomic sites. We speculate that telomere loss in a dcr1Δ setting leads to mobilization of STE transcripts as transposon-like elements.
New avenues to genome plasticity and telomerase escape
We reiterate that the translocation events placing rDNA or STE at all chromosome ends have never been seen in a trt1+ setting; hence, this translocation reaction can be added to the list of events, such as nonhomologous end-joining (NHEJ)-mediated chromosome end fusions, that are prohibited by functioning telomeres. Whether this RNAi-mediated translocation might occur at other genomic loci is a crucial question that we are approaching: Do DSBs at non-telomere-adjacent sites occasionally lead to RNAi-dependent sequence jumping? If rDNA were located away from telomeres, would it be more mobile? The implications of such reactions for genome organization are interesting questions for the future.
Another fascinating set of questions surrounds the uniqueness of rDNA as a proxy for telomeres. The association with SHREC is clearly essential for recruiting Pot1; whether a distinct (non-rDNA) repeat that also associates with SHREC, if located near chromosome termini, could confer a HAATIrDNA-like mechanism is a current focus. The special properties of the rDNA intergenic spacer regions, which attract RNA polymerases, RNAi factors, and fork-stalling proteins, may make them optimal for this type of telomerase-negative survival. Hence, the proximity of rDNA to chromosome termini in human cells, Allium species (Fajkus et al. 2016), Brachidontes (Pérez-García et al. 2010), and other species is intriguing; not only the principle of HAATIrDNA using nontelomeric HC to recruit Pot1 might be conserved but also the specific ability of rDNA to translocate and supersede telomeres. The unusual and striking similarities between such widely divergent organisms as flies and fission yeast argue that sequence-independent strategies may provide alternative mechanisms of telomerase escape in some human cancers.
In conclusion, our results show that both growth environment and prior genomic rearrangements play a fundamental role in favoring one or the other type of telomerase-minus survival. Similarly, we can envision that the specific environment in which human tumor cells arise—the local nutritional state and the extent to which genome-destabilizing processes have occurred before the cells undergo crisis—could favor or disfavor HAATI mechanisms.
Materials and methods
Strains and media
S. pombe strains used in this study were derivatives of the standard laboratory strain 972 and are listed in Supplemental Table S1. Strains were grown at 32°C in standard rich medium (yeast extract with supplements [YE5S]) unless indicated otherwise (Moreno et al. 1991). Plasmid-containing strains were grown under conditions selecting for the appropriate marker. All strains were constructed by one-step gene replacement (Bahler et al. 1998), starting by constructing heterozygous diploids. For HAATI maintenance experiments, strains were constructed by one-step gene replacement in already formed HAATI survivors. For reintroduction of telomerase, strains were transformed with p-kanMX-trt1+-myc (Haering et al. 2000).
Generating trt1Δ survivors
For competitive culturing in liquid, 20-mL liquid cultures were inoculated at 104 cells per milliliter and propagated for ∼28 d. Every ∼24 h, cell density was measured by optical density, and fresh 20-mL cultures were inoculated at 104 cells per milliliter. For competitive culturing in patches, equal volumes of trt1Δ cells were patched onto plates and propagated for ∼28 d by repatching equal volumes onto fresh plates every ∼24 h. For noncompetitive culturing, trt1Δ offspring were streaked to single colonies iteratively for ∼28 d.
Dilution assays
Cells were grown in liquid culture to log phase, and their density was measured using a haemocytometer. Cultures were adjusted to a final concentration of 1 × 107 cells per milliliter, and fivefold serial dilutions were made in a 96-well microtiter plate with repeated agitation. Diluted cells were stamped onto plates using a metal stamper (which transfers ∼5 µL).
Cytological analysis
Cells were grown to log phase (∼0.4–0.6 OD) in rich medium at 32°C, adhered to a glass culture dish (MatTek) precoated with 0.2 mg/mL soyabean lectin (Calbiochem), immersed in YE5S liquid, and visualized by fluorescence microscopy on a DeltaVision Spectris or DeltaVision OMX conventional mode (Applied Precision) using Soft-WoRx software (Applied Precision) to capture images and quantify foci. Equal exposures were used across all compared genetic backgrounds. Using two to three independent biological replicates, 50–80 cells per genotype were imaged and analyzed for each experiment. Images were deconvolved, and all Z-stacks were projected into a single-plane image.
DNA isolation and Southern blotting
DNA isolation and Southern blot analysis were performed as described previously (Supplemental Table S2; Miller et al. 2006).
PFGE of whole chromosomes
PFGE of whole chromosomes was performed as described previously (Jain et al. 2010) with the following modifications: Agarose plugs were loaded onto 0.5% agarose gels in 1× TAE (40 mM Trisacetate buffer, 2 mM Na2EDTA at pH 8.3). PFGE was performed on a Bio-Rad CHEF DR-III system in 1× TAE at 14°C using the following program: step 1, 30 h at 2 V/cm, 96° angle, and 1200-sec switch time; step 2, 30 h at 2 V/cm, 100° angle, and 1500-sec switch time; and step 3, 30 h at 2 V/cm, 106° angle, and 1800-sec switch time. After electrophoresis, DNA was visualized by ethidium bromide staining, and gels were processed for Southern blot analysis using the STE1 probe (Nakamura et al. 1998) or the rDNA probe (Supplemental Table S2; Toda et al. 1984).
TSA assay
TSA was dissolved in 2 mg/mL ethanol and stored at −20°C. Cells were precultured overnight in rich medium to reach log phase. TSA was added to cultures of 5 × 105 cells per milliliter for a final drug concentration of 35 µg/mL. Control samples were treated with 96% ethanol. After ∼24 h, cell density was estimated by OD, cultures were adjusted to a final concentration of 5 × 105 or 5 × 106 cells per milliliter, and 87.5 µL of fresh TSA/ethanol was added. After another 24 h, cells were pelleted, washed extensively in ddH2O, adjusted to a final concentration of 1 × 107 cells per milliliter in YE5S, and stamped onto plates in fivefold serial dilution.
ChIP
ChIP was performed as described previously (Jain et al. 2010). Quantification of DNA by PCR was realized using SYBR Green mix. Each sample was amplified in triplicate, and enrichment was expressed as input/whole-cell extract (Supplemental Tables S2, S3). For each experiment, a standard curve was calculated from serial dilution of whole-cell extract.
PFGE of NotI-digested chromosomes
PFGE of NotI-digested chromosomes was performed as described previously (Jain et al. 2010) with the following modifications: NotI-digested agarose plugs were loaded onto 1% agarose gels in 0.5× TBE (1× TBE: 89 mM Tris-borate, 2 mM EDTA). PFGE was performed on a Bio-Rad CHEF DR-III system in 0.5× TBE at 14°C using the following program: 28 h at 6 V/cm, 120° angle, and 60- to 120-sec switch time. After electrophoresis, DNA was visualized by ethidium bromide staining, and gels were subjected to Southern blot analysis using LMIC probes (Ferreira and Cooper 2001) or the STE1 probe (Supplemental Tables S2, S3).
Preparation of genomic DNA libraries
The genomic DNA libraries were made using Illumina TruSeq Nano DNA library preparation kits. The S. pombe genomic DNA was fragmented to a 400-bp insert size on the Covaris, which generates dsDNA fragments with 3′ or 5′ overhangs. The sheared DNA was blunt-ended, and library size selection was performed using sample purification beads. A single “A” nucleotide was added to the 3′ ends of the blunt fragments to prevent them from ligating to each other during the adapter ligation reaction. A corresponding single “T” nucleotide on the 3′ end of the adapter provided a complementary overhang for ligating the adapter to the fragment. The indexed adapters were ligated to the ends of the DNA fragments and then PCR-amplified to enrich for fragments that had adapters on both ends. The final purified product was then quantitated by quantitative PCR (qPCR) before cluster generation, and paired-end sequencing was done on the MiSeq sequencer (Supplemental Table S3).
Paired-end Illumina sequencing and SNP analysis
The genomes of three wild-type and three HAATIrDNA strains were sequenced using the Illumina MiSeq sequencing platform to generate between 5 million and 5.5 million 150-bp paired-end reads per strain. The reads were trimmed to remove adapter sequence using Trimmomatic (Bolger et al. 2014). Duplicate reads were removed using Picard Tools Markduplicates (Picard Tools, http://broadinstitute.github.io/picard). The trimmed deduplicated reads mapped to the S. pombe reference (ASM294v2.30), to which was added a contig (termed the subtelomeric contig) containing an additional 8050 bases of subtelomeric sequences (from pNSU21 harboring the chromosome IIR sequence) (Sugawara 1988) and telomeric repeats not present in the reference genome. This contig was assembled by performing telo PCR using wild-type strains in our laboratory and adding the existing sequencing information from pNSU21 chromosome IIR clones to the sequenced region. Read alignment was performed with the BWA aligner (Wood et al. 2002). Fast and accurate short read alignment was performed with Burrows-Wheeler Transform. (Li and Durbin 2009). Alignment quality was assessed using Qualimap (Okonechnikov et al. 2016). For assessment of telomere structure, BWA was run using the “-a” flag to output all alignments regardless of uniqueness. For the analysis of variants, BWA was run using default parameters. The resulting alignments covered the reference at a mean depth of >50× with 90% of the reference covered at >35×. Mean alignment quality was >57 for all strains. Coverage per chromosome showed differences between wild-type and HAATI strains. Typical coverage for chromosomes I and II was 50×–57× for wild-type strains but lower, at only 40×–45×, for the HAATI strains. Conversely, chromosome III coverage was higher in the HAATI strains at ∼80× compared with ∼67× for the wild type. Coverage of the subtelomeric contig was much higher in the wild-type strains, at 90×–160× as opposed to 35×–80× in the HAATI strains; note that the residual coverage reflects the subterminal regions of the subtelomeric contig.
Junctions between subtelomeric sequence on the subtelomeric contig and rDNA sequence on chromosome III in HAATI were detected on the basis of “split reads,” in which one segment of a sequencing read aligned to the subtelomeric contig, while the remaining segment aligned to the chromosome III contig. Such split reads show that subtelomeric sequences are contiguous with rDNA sequences in the sequenced genome and allow the junctions between these sequences to be mapped with single-base precision. Simultaneous consideration of the map of the paired ends of the split read allows the determination of an ∼300-bp region that includes the junction. Split reads were identified on the basis of the supplementary alignment flag set by the aligner that marks individual reads whose alignments are split into two separate chromosomal segments. The reads so flagged were scanned using a script to collect reads in which the first segment of alignment was to the subtelomeric contig, while the second segment (supplementary alignment) was to chromosome III or vice versa. The paired ends of the reads meeting these criteria were also collected. The sets of split reads and their paired ends were then precisely remapped to the reference genome using the BLAST sequence alignment program (Altschul et al. 1997). The sequences of the reference genome in the region covered by the telomeric and chromosome III segments, respectively, of the split read in the BLAST alignment were extracted. The two segments of sequence were then joined to recreate the genomic sequence corresponding to the original sequencing read, with the point of the join corresponding to the subtelomere:chromosome III junction. The genomic sequence extending from the end of the split read alignment to the distal end of the BLAST alignment of its paired end was then appended to that of the reconstructed sequence. In this way, a sequence context in the area of the subtelomere:chromosome III junction was obtained. Mutations were called from aligned reads using SAMtools Mpileup (Li et al. 2009) followed by conversion from BCL to VCF format with bcltools (http://github.com/samtools/bcftools). Variants were annotated using Snpeff (Cingolani et al. 2012) with Pombase feature annotations (Wood et al. 2011).
Data and software availability
The sequencing data reported in this study were deposited in the BioSample database (BioProject-NCBI Portal) with accession numbers SAMN07964785, SAMN07964786, SAMN07964787, SAMN07964788, SAMN07964789, and SAMN07964790. The six accession numbers correspond to three wild-type and three HAATIrDNA isolates. Software and algorithms used for the analysis are listed in Supplemental Table S4.
Supplementary Material
Acknowledgments
We thank all members of the J.P.C. laboratory for useful discussion and experimental help; special thanks for advice go to our former laboratory member, Jessica Greenwood. We thank the Center for Cancer Research Sequencing Facility at National Cancer Institute Frederick for help with whole-genome sequencing. We thank Marc Buhler, Toru Nakamura, and Paul Nurse for kind gifts of reagents and advice, and Alex Kelly for discussion and comments on the manuscript. This work was supported by Cancer Research UK and the National Cancer Institute.
Author contributions: M.B. and J.P.C. designed and interpreted this study with contributions from M.S.A. and D.J. M.B. performed the experiments in Figures 1, 2A, and 3–5 and Supplemental Figures S1, S2A, S3, and S4. M.S.A. performed the experiments in Figures 2, B–D, and 6. D.L.W. analyzed sequencing experiments in Figure 6. D.J. performed experiments in Figure 1, A and D. H.M. performed the experiments in Supplemental Figure S2, B and C. M.B. and J.P.C. wrote the paper, and all authors edited it. J.P.C. obtained funding for this study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.311712.118.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte MS, Cooper JP. 2017. Life and cancer without telomerase: ALT and other strategies for making sure ends (don't) meet. Crit Rev Biochem Mol Biol 52: 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O, Mehta S, Grewal SI. 2013. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol 20: 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Wischnewski H, Shchepachev V, Azzalin CM. 2012. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res 40: 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886. [DOI] [PubMed] [Google Scholar]
- Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. 2013. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J 32: 1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, Fitzgerald PC, Grewal SI. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang AY, Sanchez M, Valbuena A, Antequera F, Martienssen RA. 2014. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 159: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. 2003. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5: 82–84. [DOI] [PubMed] [Google Scholar]
- Choi ES, Shin JA, Kim HS, Jang YK. 2005. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res 33: 7102–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- Costa A, Daidone MG, Daprai L, Villa R, Cantù S, Pilotti S, Mariani L, Gronchi A, Henson JD, Reddel RR, et al. 2006. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res 66: 8918–8924. [DOI] [PubMed] [Google Scholar]
- Cowieson NP, Partridge JF, Allshire RC, Mclaughlin PJ. 2000. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol 10: 517–525. [DOI] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. 1990. Structure and variability of human chromosome ends. Mol Cell Biol 10: 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. 2010. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci 107: 8701–8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M. 2010. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113. [DOI] [PubMed] [Google Scholar]
- Fajkus P, Peška V, Sitová Z, Fulnečková J, Dvořáčková M, Gogela R, Sýkorová E, Hapala J, Fajkus J. 2016. Allium telomeres unmasked: the unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J 85: 337–347. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538. [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell 7: 55–63. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, De Hoon M, Anelli V, Mione M, Carninci P, D'adda Di Fagagna F. 2012. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488: 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J 29: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J, Cooper JP. 2011. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res 40: 2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues K. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. [DOI] [PubMed] [Google Scholar]
- Haering CH, Nakamura TM, Baumann P, Cech TR. 2000. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc Natl Acad Sci 97: 6367–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci 100: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland JL, Chang YT, Moser BA, Nakamura TM. 2014. Tpz1-Ccq1 and Tpz1-Poz1 interactions within fission yeast shelterin modulate Ccq1 Thr93 phosphorylation and telomerase recruitment. PLoS Genet 10: e1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. 2006. Argonaute slicing is required for heterochromatic silencing and spreading. Science 313: 1134–1137. [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP. 2010. Telomeric strategies: means to an end. Annu Rev Genet 44: 243–269. [DOI] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Gettings EJ, Schwalm J, Pei J, Testa JR, Litwin S, von Mehren M, Broccoli D. 2007. Whole-genome profiling in liposarcomas reveals genetic alterations common to specific telomere maintenance mechanisms. Cancer Res 67: 9221–9228. [DOI] [PubMed] [Google Scholar]
- Kandel ES, Nudler E. 2002. Template switching by RNA polymerase II in vivo. Evidence and implications from a retroviral system. Mol Cell 10: 1495–1502. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819. [DOI] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. 2014. Transcript-RNA-templated DNA recombination and repair. Nature 515: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khair L, Subramanian L, Moser BA, Nakamura TM. 2010. Roles of heterochromatin and telomere proteins in regulation of fission yeast telomere recombination and telomerase recruitment. J Biol Chem 285: 5327–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. 2008. A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays 30: 267–272. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestroni L, Audry J, Matmati S, Arcangioli B, Géli V, Coulon S. 2017. Eroded telomeres are rearranged in quiescent fission yeast cells through duplications of subtelomeric sequences. Nat Commun 8: 1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H. 2008. Drosophila telomeres: an exception providing new insights. Bioessays 30: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP. 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 24: 3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1–Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344. [DOI] [PubMed] [Google Scholar]
- Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cooper JP, Cech TR. 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493–496. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Avetissova E, Markovtsov V, Goldfarb A. 1996. Transcription processivity: protein–DNA interactions holding together the elongation complex. Science 273: 211–217. [DOI] [PubMed] [Google Scholar]
- Ohno Y, Ogiyama Y, Kubota Y, Kubo T, Ishii K. 2016. Acentric chromosome ends are prone to fusion with functional chromosome ends through a homology-directed rearrangement. Nucleic Acids Res 44: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Conesa A, García-Alcalde F. 2016. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32: 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. 2006. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García C, Guerra-Varela J, Morán P, Pasantes JJ. 2010. Chromosomal mapping of rRNA genes, core histone genes and telomeric sequences in Brachidontes puniceus and Brachidontes rodriguezi (Bivalvia, Mytilidae). BMC Genet 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett HA, Reddel RR. 2015. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol 22: 875–880. [DOI] [PubMed] [Google Scholar]
- Roche B, Arcandioli RA, Martienssen RA. 2016. RNA interference is essential for cellular quiescence. Science 354: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 22: 357. [DOI] [PubMed] [Google Scholar]
- Sugawara N. 1988. “DNA sequences at the telomeres of the fission yeast S. pombe.” PhD thesis, Harvard University, Cambridge, MA. [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma KI, Zofall M, Kobayashi R, Grewal SI. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504. [DOI] [PubMed] [Google Scholar]
- Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, Jia S. 2013. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev 27: 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Nishihara Y, Kugou K, Ohta K, Kanoh J. 2017. Subtelomeres constitute a safeguard for gene expression and chromosome homeostasis. Nucleic Acids Res 45: 10333–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Nakaseko Y, Niwa O, Yanagida M. 1984. Mapping of rRNA genes by integration of hybrid plasmids in Schizosaccharomyces pombe. Curr Genet 8: 93–97. [DOI] [PubMed] [Google Scholar]
- Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O. 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165: 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaecke T, Papeleu P, Elaut G, Rogiers V. 2004. Trichostatin A-like hydrox-amate histone deacetylase inhibitors as therapeutic agents: toxicological point of view. Curr Med Chem 11: 1629–1643. [DOI] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White Charles I, Rendtlew Danielsen JM, Yang YG, Qi Y. 2012. A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–878. [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, et al. 2011. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, De Castro E, Marín L, Chang AY, Goto D, et al. 2011. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Guo A, Liu Z, Pape L. 1997. Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res 25: 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.