Abstract
The fidelity of RNA splicing is regulated by a network of splicing enhancers and repressors, although the rules that govern this process are not yet fully understood. One mechanism that contributes to splicing fidelity is the repression of nonconserved cryptic exons by splicing factors that recognize dinucleotide repeats. We previously identified that TDP-43 and PTBP1/PTBP2 are capable of repressing cryptic exons utilizing UG and CU repeats, respectively. Here we demonstrate that hnRNP L (HNRNPL) also represses cryptic exons by utilizing exonic CA repeats, particularly near the 5′SS. We hypothesize that hnRNP L regulates CA repeat repression for both cryptic exon repression and developmental processes such as T cell differentiation.
Keywords: alternative splicing, cryptic exons, dinucleotide repeats, HNRNPL, HnRNP L
INTRODUCTION
RNA splicing in higher eukaryotes helps produce the transcriptional diversity that is required for cellular differentiation. This robust process is governed by RNA-binding splicing factors, many of which bind to degenerate consensus sequences (Wahl et al. 2009; Fu and Ares 2014). Given this limited specificity, how is the spliceosome able to distinguish proper exons from similar intronic sequences? Most introns contain cryptic “pseudoexon” sequences that appear to have all the appropriate splicing motifs, but are never incorporated into mature RNA transcripts (Dhir and Buratti 2010). We previously reported that certain pseudoexons are flanked by repetitive dinucleotide sequences, and that the repression of these cryptic exons is mediated by splicing factors that recognize said dinucleotide repeats (Ling et al. 2015, 2016). Repression of cryptic exons may help maintain splicing fidelity, given that the sequences of cryptic exons are not conserved between species and thus appear to have arisen stochastically.
Transactivation response element DNA-binding protein 43 (TDP-43, TARDBP), polypyrimidine tract-binding protein 1 (PTB, PTBP1), and polypyrimidine tract-binding protein 2 (nPTB, PTBP2) were identified as splicing factors that mediated cryptic exon repression. Specifically, TDP-43 binds to UG repeats (Ling et al. 2015; Jeong et al. 2017; Sun et al. 2017) while PTBP1 and PTBP2 bind to CU repeats (Ling et al. 2016). Other dinucleotide repeats exist in and around cryptic exons, however, aside from UG and CU. This led us to search for other splicing factors that might repress cryptic exons using other dinucleotide sequences. Here, we demonstrate that heterogeneous nuclear ribonuclear protein L (hnRNP L, HNRNPL) is a cryptic exon repressor that binds to CA repeats.
HnRNP L was first identified as a member of the heterogeneous nuclear ribonucleoprotein (HNRNP) family (Pinol-Roma et al. 1989) and has a homolog, hnRNP LL (HNRNPLL). Among the HNRNP family members, hnRNP L and hnRNP LL share closest homology with PTBP1 and PTBP2 (Ghetti et al. 1992; Busch and Hertel 2012; Blatter et al. 2015). HnRNP L was shown to play a role in RNA splicing (Hui et al. 2003a) soon after its discovery, and like many other splicing factors, autoregulates the levels of its own transcript (Rossbach et al. 2009). HnRNPL was a leading candidate for cryptic exon repression because strong experimental consensus has indicated that hnRNP L binds to CA dinucleotide repeats (Hui et al. 2003b, 2005; Smith et al. 2013). This preference for CA repeats was further confirmed by UV cross-linking immunoprecipitation (CLIP) (Rossbach et al. 2014; Shankarling et al. 2014) and crystal structure analysis (Zhang et al. 2013a). CA repeats are among the most common repeats in the human genome (Lander et al. 2001; Subramanian et al. 2003; Sawaya et al. 2013). Furthermore, hnRNP L has been reported to act as a splicing repressor in certain contexts (Hui et al. 2005; Rothrock et al. 2005; Hung et al. 2008; Motta-Mena et al. 2010), with several reports suggesting that hnRNP L interferes with appropriate assembly of the spliceosome at the 5′ splice site (5′SS) (Hui et al. 2005; Heiner et al. 2010; Chiou et al. 2013; Loh et al. 2017). Together, these studies provided strong rationale to study hnRNP L as a cryptic exon repressor candidate.
RESULTS AND DISCUSSION
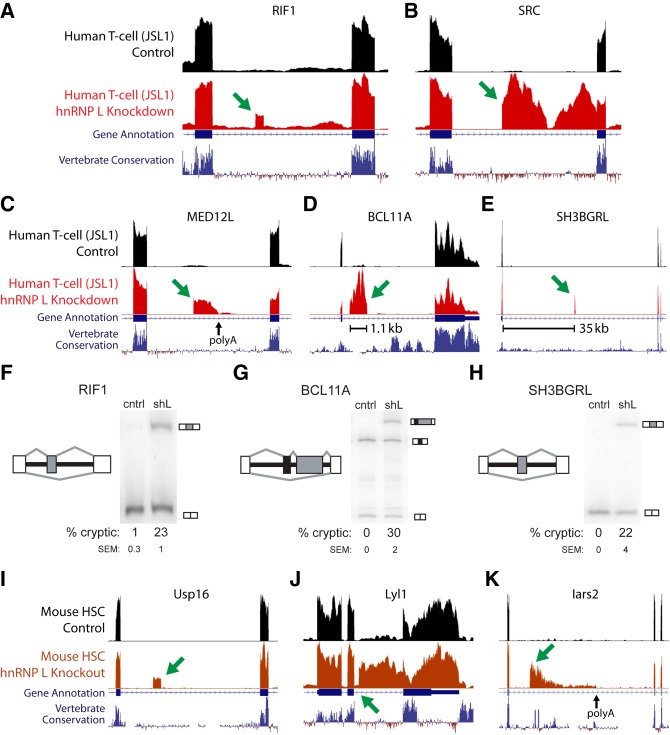
In order to determine whether hnRNP L is a cryptic exon repressor, we analyzed RNA-seq data sets from hnRNP L knockdown in three human cell lines: JSL1, HepG2, and K562 (Cole et al. 2015; Sloan et al. 2016; Sundararaman et al. 2016), as well as RNA-seq data sets from genetic knockouts of Hnrnpl in mouse fetal liver hematopoietic stem cells (HSC) (Gaudreau et al. 2016). Applying the bioinformatics pipeline previously used to identify TDP-43 and PTBP1/PTBP2 cryptic exons (Ling et al. 2015, 2016), we analyzed the above data sets for novel splice sites and found many cryptic cassette exons, cryptic alternative splice site usage (exon extensions), cryptic terminal exons and cryptic polyadenylation sites (3′ end exons) that were only observed after hnRNP L depletion (Fig. 1A–E; Supplemental Fig. S1; Supplemental Excel File). Importantly, we validated by RT-PCR use of many of the cryptic exons in JSL1 cells depleted of hnRNP L protein (Fig. 1F–H; Supplemental Fig. S2). We were also able to identify cryptic cassette exons, cryptic exon extensions, and cryptic 3′ end exons in the mouse knockout data sets (Fig. 1I–K). As expected, there was no overlap in cryptic exons between mouse and human, consistent with the lack of conservation of these elements.
FIGURE 1.
Human and mouse hnRNP L cryptic exons viewed in the UCSC Genome Browser (hg38, mm10). Human cryptic cassette exon (A), cryptic exon extension (B), and cryptic 3′ end exon (C). Examples of variability: a large cassette exon (D) and a cassette exon flanked by long introns (E). (F–H) Representative RTPCR gels showing splicing of hnRNP L repressed cryptic exons for both control JSL1 cells (cntrl) and JSL1 cells transfected with short-hairpin RNA against hnRNP L (shL). Splice graphs of each splicing event are shown to the left of each gel with the cryptic exon in gray, constitutive (upstream and downstream) exons in white, and intersecting alternative exons in black. The average percent inclusion of the cryptic exon (% cryptic) and standard error of the mean (SEM) for at least two replicates is shown (see Supplemental Fig. 1 for other examples). Mouse cryptic cassette exon (I), cryptic exon extension (J), and cryptic 3′ end exon (K). Genes displayed in UCSC views: RIF1 (A), SRC (B), MED12L (C), BCL11A (D), SH3BGRL (E), Usp16 (I), Lyl1 (J), and Iars2 (K).
Next, we analyzed hnRNP L knockdown in the HepG2 and K562 cell lines utilizing RNA-seq data sets from the ENCODE shRNA library. Although we found that 58% (49/84) of cryptic exons were shared between at least two cell types, 42% (35/84) of cryptic exons were unique to a single cell type (Supplemental Fig. S3). Notably, much of this cell-type specificity appears to be due to the cell-type–specific expression of the gene (Supplemental Excel File). These results are analogous to our previous observations with tissue-specific cryptic exons associated with TDP-43 loss of function (Jeong et al. 2017).
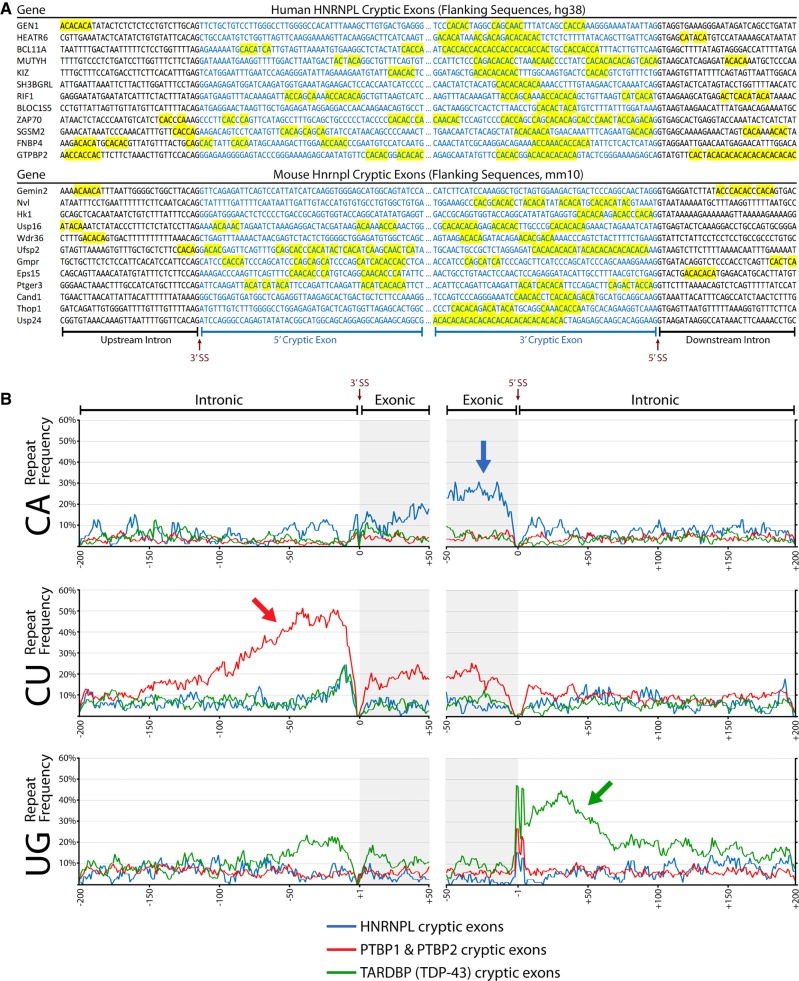
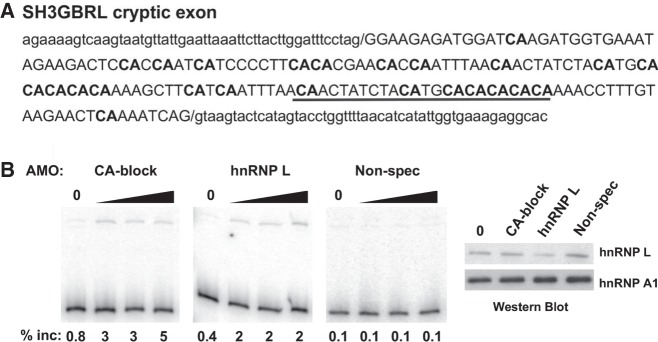
Further inspection of cryptic exons and their flanking sequences confirmed the presence of numerous CA repeat clusters. Interestingly, aligning cryptic cassette exons to the 3′SS and 5′SS revealed that most CA repeats are exonic and proximal to the 5′SS (Fig. 2A,B). We confirmed the functionality of these exonic CAs in at least one case in which they were well clustered by transfection of JSL1 cells with an antisense morpholino oligo (AMO) complementary to seven CA dinucleotides located upstream of the cryptic 5′ss in SH3BGRL (Fig. 3A). Blocking of these seven CAs by AMO resulted in a level of exon inclusion that mirrors that of hnRNP L depletion by translation blocking AMO (Fig. 3B). In contrast to the pattern of CA dinucleotides in the hnRNP L cryptic exons, analysis for TDP-43 and PTBP1/2 cryptic exons revealed that each dinucleotide repeat had different frequency distributions. TDP-43 cryptic exons had primarily intronic UG repeats that were most commonly downstream from the 5′SS, with some repeats upstream of the 3′SS. In contrast, PTBP1 and PTBP2 cryptic exons had mostly intronic CU repeats upstream of the 3′SS but also some exonic repeats as well (Fig. 2B).
FIGURE 2.
Cryptic exon repressors demonstrate differential preference in repeat location. (A) Examples of sequences flanking hnRNP L cassette cryptic exons found in human (top) and mouse (bottom) data sets. Sequences are aligned to the 3′SS and 5′SS and CA repeat clusters are highlighted in yellow. (B) Frequency of CA, CU, and UG dinucleotide repeats (top, middle, bottom) for cassette cryptic exons associated with hnRNP L, PTBP1/PTBP2, and TDP-43 (blue, red, green). hnRNP L cryptic cassette exons have a high frequency of exonic CA repeats near the 5′SS. PTBP1 and PTBP2 cryptic cassette exons exhibit CU repeats mostly upstream of the 3′SS, with some also located in the exon. TDP-43 cryptic cassette exons primarily have UG repeats downstream from the 5′SS, although some repeats can also be upstream of the 3′SS.
FIGURE 3.
Blocking CA repeats mimics depletion of hnRNP L in inclusion of the SH3BGRL cryptic exon. (A) Sequence of the SH3BGRL cryptic exon (uppercase) and flanking intron (lowercase) with sequence complementary to CA blocking AMO (underlined). CA dinucleotides are in bold. (B) RT-PCR gel showing inclusion of SH3BGRL cryptic exon upon transfection of CA blocking AMO, hnRNP L-depleting AMO or an unrelated GT-rich AMO. Western blot confirming depletion of hnRNP L upon treatment with the hnRNP L AMO, but not others, shown on right. HnRNP L depletion by AMO was performed as described previously (Cole et al. 2015). Translation blocking AMO for hnRNP L was used at 2, 5, and 10 nmol. CA and nonspecific AMO were used at 5, 10, and 20 nmol.
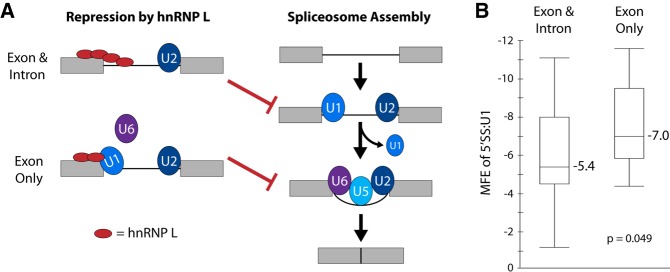
In general, recognition and splicing of exons involves the stepwise assembly of various U snRNP components of the spliceosome with the pre-mRNA substrate (Fig. 4A, right). Regulation of any of these steps can alter the efficiency of splicing (Wahl et al. 2009; Fu and Ares 2014). Interestingly, previous work has shown that hnRNP L represses exon inclusion through several distinct mechanisms. Binding of hnRNP L in the intron immediately proximal to the 5′ SS results in repression, presumably through steric hindrance of U1 recruitment (Hui et al. 2005; Hung et al. 2008). In contrast, in the case of CD45 exon 4, binding of hnRNP L within the exon functions to recruit hnRNP A1, remodel the U1 snRNP association with the 5′SS, and thereby inhibit association of U6 snRNP (Chiou et al. 2013). Importantly, we showed that a hallmark of the U1-remodeling mechanism of hnRNP L repression is a relatively strong association of the snRNA component of the U1 snRNP with the 5′SS. Specifically, exons for which the 5′SS:U1 snRNA base paring had a minimal free energy (MFE) of −7 or lower (more negative) were prone to repression when hnRNP L bound to the exon (Chiou et al. 2013).
FIGURE 4.
Model for different mechanisms of hnRNP L repression of weak and strong 5′ splice sites. (A) Schematic of normal splicesome assembly (right) and mechanisms by which hnRNP L represses normal assembly. Exons are designated as gray boxes, introns are black lines. The U snRNP components of the spliceosome are indicated by blue and purple ovals. HnRNP L is indicated by a red oval. HnRNP L can either block accessibility of the 5′SS to U1 (exon and intron), or inhibit exchange of U1 for U6 (exon only). In either case, assembly of the final spliceosomal complex is prevented. (B) Minimal free energy (MFE) of the association of the 5′SS with the U1 snRNA for hnRNP L-repressed exons that have CA dinucleotides in the exon and/or intron flanking the 5′SS. See Supplemental Table 1 for details. As described in the text, binding of hnRNP L to the exon only is predicted to repress U1–U6 exchange and is typically marked by a strong 5′SS:U1 interaction. P-value is a two-tailed t-test.
Given our observation that hnRNP L cryptic exons have a high frequency of CA repeats near the 5′SS (Fig. 2), we sought to determine if the U1 remodeling mechanism may account for some of the repression of cryptic exons by assessing the MFE of 5′SS:U1 snRNA interaction as done in the previous study (Chiou et al. 2013). We first identified the cryptic exons with a strong presence of CA repeats (three or more dinucleotide repeats) in the final 50 nucleotides (nt) of the exon. We then separated these into exons which had additional CA dinucleotide repeats in the intronic 50 nt downstream from the 5′SS, which we reasoned would result in multimerization of hnRNP L across the 5′SS (Fig. 4A, left). Notably, the 5′SS flanked only by exonic CA repeats had a median MFE of −7.0, while those flanked by both intronic and exonic repeats had a median MFE of −5.4 (Fig. 4B; Supplemental Table S1). These results are consistent with a model whereby hnRNP L mediates repression of these cryptic exons by mechanisms described previously; namely, binding of hnRNP L across a 5′SS can sterically block recruitment of the U1 snRNP (especially when this binding is already weak), while binding of hnRNP L to the exon hyperstabilizes an already strong interaction of the U1 snRNP with the 5′SS to prevent association of the U6 snRNP (Fig. 4A). Likewise, the demonstration that TDP-43 and PTBP1/PTBP2 repress cryptic exons from upstream and downstream intronic positions is consistent with the known activities of these proteins (Ashiya and Grabowski 1997; Pérez et al. 1997; Ayala et al. 2005; Oberstrass et al. 2005; Boutz et al. 2007; D'Ambrogio et al. 2009; Kuo et al. 2009; Xue et al. 2009; Chiang et al. 2010; Polymenidou et al. 2011; Tollervey et al. 2011; Kafasla et al. 2012; Keppetipola et al. 2012; Licatalosi et al. 2012; Lukavsky et al. 2013; Zhang et al. 2013b; Humphrey et al. 2017). Therefore, our results suggest that cryptic pseudoexons are repressed by mechanisms similar to standard exons, and that a widespread activity of hnRNP L is repression of strong 5′SSs, presumably through inhibiting replacement of U1 by U6.
Finally, in order to investigate the potential functional importance of cryptic exon repression by hnRNP L, we looked at consequences of cryptic exon inclusion on protein production. Given that cryptic exons are not conserved across species, we predict that their inclusion is likely to disrupt reading frame and protein expression. Almost half (244/550) of annotated exons repressed by hnRNP L in human JSL1 cells maintain reading frame (Cole et al. 2015). In contrast, 97% (60/62) of human cryptic exons and 86% (25/29) of mouse cryptic exons alter reading frame and are predicted to induce premature termination codons (Supplemental Excel File). For example, the gene encoding ZAP70, a critical signaling protein downstream from the T-cell receptor, contains a cryptic exon between exons 13 and 14 that introduces several stop codons. Upon knockdown of hnRNP L, this ZAP70 cryptic exon is included in almost half of the ZAP70 transcripts (Supplemental Fig. S2). Importantly, inclusion of the ZAP70 cryptic exon correlates with a marked decrease in ZAP70 protein levels (Supplemental Fig. S2, bottom right). This result indicates that repression of cryptic exons by hnRNP L serves to protect the fidelity of the proteome.
We note that our results raise the question of the role of hnRNP L's homolog hnRNP LL, given that hnRNP L is alone sufficient to repress cryptic exons. In contrast to the ubiquitous expression of hnRNP L, hnRNP LL is highly expressed only under certain conditions such as following T-cell activation, where it regulates the alternative splicing of genes such as CD45 (Hung et al. 2008; Oberdoerffer et al. 2008; Topp et al. 2008; Wu et al. 2008; Preußner et al. 2012). Therefore, we assume that hnRNP LL is not expressed at sufficient levels to functionally compensate for loss of hnRNP L cryptic exon repression in most cell types and conditions, including those analyzed in this study.
In summary, we have demonstrated that hnRNP L plays an important role in repressing nonconserved cryptic exons by utilizing exonic CA repeats. Together with our earlier studies, these results suggest that at least three out of six possible dinucleotide repeats are utilized to repress cryptic exon splicing (Supplemental Table S2). Of the remaining repeats, AG repeats may not be utilized because exonic splicing enhancers are often purine rich; a broad splicing repressor with an AG repeat consensus motif may interfere with normal splicing. Alternatively, GC repeats may not be useful for a cryptic exon repressor because GC repeats are extremely rare within the genome due to cytosine methylation and spontaneous deamination. However, other homopolymers and tri/tetranucleotide repeats may also be used to repress cryptic exons. For example, hnRNP C binds to continuous uridine tracks to repress cryptic exons derived from Alu elements (Zarnack et al. 2013). Future work will be required to identify additional splicing factors and their cognate sequences that may function to guard the transcriptome against cryptic splicing.
MATERIALS AND METHODS
RNA-seq data sets analyzed
To identify cryptic exons, we accessed RNA-seq data sets from the following cell types: human JSL1 Jurkat T cells—wild-type and hnRNP L knockdown (SRP059357, PMID: 26437669), human HepG2 cells—wildtype and hnRNP L knockdown wild-type and hnRNP L knockdown (ENCODE Project Accession: ENCSR155BMF), human K562 cells—wild-type and hnRNP L knockdown (ENCODE Project Accession: ENCSR563YIS), mouse embryo—wild-type and hnRNP L knockout (SRP042164, PMID: 27271479).
Bioinformatics analysis of cryptic exons
Fastq files were aligned to mouse and human genomes using HISAT2 (Kim et al. 2015), transcript assemblies were generated using StringTie (Pertea et al. 2015, 2016), and RPKM values were calculated using feature Counts (Liao et al. 2014) on Galaxy (Afgan et al. 2016). Cryptic exons were identified as previously described (Ling et al. 2015, 2016). Briefly, relative read coverage and split read counts were calculated between control and experimental condition for each exon in StringTie generated transcript assemblies. Data were visualized on the UCSC Genome Browser and false positives were removed following manual curation.
Calculation of repeat frequency around hnRNP L-repressed cryptic exons in Figure 2B was done by (i) Masking all “CA” and “AC” as “YY”; (ii) replacing all “A,” “C,” “T,” “G” as “N”; (iii) identifying all pentamer and longer repeat sequences allowing for a single N insertion (i.e., “YYYYY,” “YYYYYY,” “YYYYYYY,” “YNYYYY,” “YYNYYY,” “YYYNYY,” “YYYYNY,” …); (iv) assigning the “Y”s in sequences from step 3 a value of 1 and all other sequences a value of 0; (v) aligning all sequences to the 3′SS and 5′SS and calculating the repeat frequency for each base pair position (i.e., sum vertically and divide by the total number of sequences). Repeat frequencies for UG repeats (TDP-43) and CU repeats (PTBP1/PTBP2) were derived in a similar fashion.
Calculation of minimal free energy of the 5′SS region of hnRNP L-repressed cryptic exons was done using RNAcofold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAcofold.cgi) to assess the free energy of the interaction of the 5′SS interacting nucleotides of U1: (5′ ACUUACCUG) with the 9 nt encompassing positions +3 to −6 around the 5′SS of the cryptic exons (cleavage bond is designated as 0).
hnRNP L knockdown, AMO transfection, RT-PCR validation, and western blots
RNA from JSL1 Jurkat cells (Lynch and Weiss 2000) was generated as previously described (Cole et al. 2015) using RNA-Bee (Tel-Test). HnRNP L depletion was achieved using a stably integrated, doxycycline-inducible short hairpin RNA against the hnRNP L mRNA as described in Cole et al. (2015). The CA-blocking or hnRNP L-depletion antisense morpholino oligos (AMO) were purchased from GeneTools and transfected at the indicated concentrations by electroporation as described previously (Cole et al. 2015). CA-blocking AMO is TTGTGTGTGTGCATGTAGATAGTTG. Nonspecific GT AMO is CATGTAGTGTGCAGGTTAAGAGTGA. hnRNP L-depleting AMO was described previously in Cole et al. (2015) (CGCCCGCCGCCGCCATCTTCACCAT). Cells were incubated following transfection for 24 h and RNA was harvested using RNA-Bee reagent. Low cycle RT-PCR was performed as previously described (Lynch and Weiss 2000), using radiolabeled primers. For genes where the cryptic exon was >1000 nt, two reverse primers were used. Following resolution of products on a 5% denaturing polyacrylamide gel, results were imaged using a Typhoon 9200 (GE) and analyzed using ImageQuant (GE). Percent spliced in of the cryptic exon was determined as the ratio of the signal from PCR product or products containing the cryptic exon sequence to the total PCR product signal. Western blots were performed as described previously (Table 1; Lynch and Weiss 2000).
TABLE 1.
Primer sequences used in this study
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a National Institute of General Medical Sciences, National Institutes of Health (NIGMS, NIH) grant to K.W.L. (R35GM118048). J.P.L. is a recipient of a Johns Hopkins Kavli Neuroscience Discovery Institute post-doctoral fellowship award.
Author contributions: All authors contributed equally to this work.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065508.117.
REFERENCES
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44: W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya M, Grabowski PJ. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3: 996–1015. [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. 2005. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 348: 575–588. [DOI] [PubMed] [Google Scholar]
- Blatter M, Dunin-Horkawicz S, Grishina I, Maris C, Thore S, Maier T, Bindereif A, Bujnicki JM, Allain FHT. 2015. The signature of the five-stranded vRRM fold defined by functional, structural and computational analysis of the hnRNP L protein. J Mol Biol 427: 3001–3022. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Hertel KJ. 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, Wong PC. 2010. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci 107: 16320–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou NT, Shankarling G, Lynch KW. 2013. HnRNP L and HnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell 49: 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole BS, Tapescu I, Allon SJ, Mallory MJ, Qiu J, Lake RJ, Fan H, Fu X, Lynch KW. 2015. Global analysis of physical and functional RNA targets of hnRNP L reveals distinct sequence and epigenetic features of repressed and enhanced exons. RNA 21: 2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, Baralle FE. 2009. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res 37: 4116–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Buratti E. 2010. Alternative splicing: role of pseudoexons in human disease and potential therapeutic strategies. FEBS J 277: 841–855. [DOI] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau MC, Grapton D, Helness A, Vadnais C, Fraszczak J, Shooshtarizadeh P, Wilhelm B, Robert F, Heyd F, Möröy T. 2016. Heterogeneous Nuclear Ribonucleoprotein L is required for the survival and functional integrity of murine hematopoietic stem cells. Sci Rep 6: 27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Piñol-Roma S, Michael WM, Morandi C, Dreyfuss G. 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 20: 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. 2010. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol 7: 56–64. [DOI] [PubMed] [Google Scholar]
- Hui J, Reither G, Bindereif A. 2003a. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA 9: 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, Stangl K, Lane WS, Bindereif A. 2003b. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol 10: 33–37. [DOI] [PubMed] [Google Scholar]
- Hui J, Hung LH, Heiner M, Schreiner S, Neumüller N, Reither G, Haas SA, Bindereif A. 2005. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J 24: 1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey J, Emmett W, Fratta P, Isaacs AM, Plagnol V. 2017. Quantitative analysis of cryptic splicing associated with TDP-43 depletion. BMC Med Genomics 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. 2008. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA 14: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YH, Ling JP, Lin SZ, Donde AN, Braunstein KE, Majounie E, Traynor BJ, LaClair KD, Lloyd TE, Wong PC. 2017. Tdp-43 cryptic exons are highly variable between cell types. Mol Neurodegener 12: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Mickleburgh I, Llorian M, Coelho M, Gooding C, Cherny D, Joshi A, Kotik-Kogan O, Curry S, Eperon IC, et al. 2012. Defining the roles and interactions of PTB. Biochem Soc Trans 40: 815–820. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. 2012. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol 47: 360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CKJ, Yuan HS. 2009. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. 2012. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev 26: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Pletnikova O, Troncoso JC, Wong PC. 2015. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349: 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JP, Chhabra R, Merran JD, Schaughency PM, Wheelan SJ, Corden JL, Wong PC. 2016. PTBP1 and PTBP2 repress nonconserved cryptic exons. Cell Rep 17: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh TJ, Choi N, Moon H, Jang HN, Liu Y, Zhou J, Zheng X, Shen H. 2017. Suppression of 5′ splice-sites through multiple exonic motifs by hnRNP L. Biochim Biophys Acta 1860: 363–373. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain FHT. 2013. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol 20: 1443–1449. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A. 2000. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol 20: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW. 2010. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell 37: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. 2008. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057. [DOI] [PubMed] [Google Scholar]
- Pérez I, Lin CH, McAfee JG, Patton JG. 1997. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778. [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Swanson MS, Gall JG, Dreyfuss G. 1989. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol 109: 2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preußner M, Schreiner S, Hung LH, Porstner M, Jäck HM, Benes V, Rätsch G, Bindereif A. 2012. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res 40: 5666–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. 2009. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol 29: 1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Khrameeva E, Schreiner S, König J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A. 2014. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol 11: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. 2005. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J 24: 2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, Gemmell N. 2013. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One 8: e54710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarling G, Cole BS, Mallory MJ, Lynch KW. 2014. Transcriptome-wide RNA interaction profiling reveals physical and functional targets of hnRNP L in human T cells. Mol Cell Biol 34: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. 2016. ENCODE data at the ENCODE portal. Nucleic Acids Res 44: D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Ray D, Cook KB, Mallory MJ, Hughes TR, Lynch KW. 2013. Paralogs hnRNP L and hnRNP LL exhibit overlapping but distinct RNA binding constraints. PLoS One 8: e80701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Mishra RK, Singh L. 2003. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol 4: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Bell W, LaClair KD, Ling JP, Han H, Kageyama Y, Pletnikova O, Troncoso JC, Wong PC, Chen LL. 2017. Cryptic exon incorporation occurs in Alzheimer's brain lacking TDP-43 inclusion but exhibiting nuclear clearance of TDP-43. Acta Neuropathol 133: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman B, Zhan L, Blue SM, Stanton R, Elkins K, Olson S, Wei X, Van Nostrand EL, Pratt GA, Huelga SC, et al. 2016. Resources for the comprehensive discovery of functional RNA elements. Mol Cell 61: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, et al. 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW. 2008. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wu Z, Jia X, de la Cruz L, Su XC, Marzolf B, Troisch P, Zak D, Hamilton A, Whittle B, Yu D, et al. 2008. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity 29: 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. 2013. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zeng F, Liu Y, Zhao Y, Lv H, Niu L, Teng M, Li X. 2013a. Crystal structures and RNA-binding properties of the RNA recognition motifs of heterogeneous nuclear ribonucleoprotein L: insights into its roles in alternative splicing regulation. J Biol Chem 288: 22636–22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Caulfield T, Xu YF, Gendron TF, Hubbard J, Stetler C, Sasaguri H, Whitelaw EC, Cai S, Lee WC, et al. 2013b. The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum Mol Genet 22: 3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.