Abstract
The repertoire of miRNAs has considerably expanded during metazoan evolution, and duplication is an important mechanism for generating new functional miRNAs. However, relatively little is known about the functional divergence between paralogous miRNAs and the possible coevolution between duplicated miRNAs and the genomic contexts. By systematically examining small RNA expression profiles across various human tissues and interrogating the publicly available miRNA:mRNA pairing chimeras, we found that changes in expression patterns and targeting preferences are widespread for duplicated miRNAs in vertebrates. Both the empirical interactions and target predictions suggest that evolutionarily conserved homo-seed duplicated miRNAs pair with significantly higher numbers of target sites compared to the single-copy miRNAs. Our birth-and-death evolutionary analysis revealed that the new target sites of miRNAs experienced frequent gains and losses during function development. Our results suggest that a newly emerged target site has a higher probability to be functional and maintained by natural selection if it is paired to a seed shared by multiple paralogous miRNAs rather than being paired to a single-copy miRNA. We experimentally verified the divergence in target repression between two paralogous miRNAs by transfecting let-7a and let-7b mimics into kidney-derived cell lines of four mammalian species and measuring the resulting transcriptome alterations by extensive high-throughput sequencing. Our results also suggest that the gains and losses of let-7 target sites might be associated with the evolution of repressiveness of let-7 across mammalian species.
Keywords: microRNA duplication, expression divergence, target repression, target site evolution, birth-and-death, let-7 family
INTRODUCTION
Gene duplication is an important mechanism that generates genetic and phenotypic novelties (Hughes 1994; Lynch and Conery 2000, 2003; Gu et al. 2002, 2003; Zhang 2003; Taylor and Raes 2004; Kaessmann et al. 2009; Innan and Kondrashov 2010; Qian and Zhang 2014). It is well established that the duplicated copies of protein-coding genes can have distinct fates over long-term evolution, including nonfunctionalization (Albalat and Cañestro 2016), dosage effects (Hughes et al. 2007), neofunctionalization (Zhang et al. 2002; Assis and Bachtrog 2013), and subfunctionalization (Lynch and Force 2000). However, much less is known about the functional divergence between paralogous noncoding RNAs and the underlying evolutionary principles. Of the various categories of noncoding RNAs encoded in the mammalian genomes, microRNAs (miRNAs) serve as a paradigm for studying functional divergence between the paralogs and the possible coevolutionary processes between the duplicated miRNAs and the genomic contexts.
Mature miRNAs are small noncoding RNAs (20–24 nt in length) produced from precursors (60–120 nt) that form stable stem–loop structures (Kim and Nam 2006; Bartel 2009; Carthew and Sontheimer 2009). miRNAs down-regulate target genes through mRNA destabilization or translational repression (Kim and Nam 2006; Bartel 2009; Carthew and Sontheimer 2009). The miRNA:mRNA interaction in metazoans is initiated by perfect pairing between the seed sequence (position 2–8) of a miRNA and its target site that is typically located in the 3′ UTR of a protein-coding gene (Bartel 2009). Since perfect pairing between the seed and a target site is crucial for target recognition (Bartel 2009), most studies have assumed that miRNAs with the same seeds target the same set of genes (Enright et al. 2003; Stark et al. 2003; John et al. 2004; Rajewsky and Socci 2004; Brennecke et al. 2005; Grün et al. 2005; Lewis et al. 2005; Friedman et al. 2009; Li et al. 2009). Metazoan miRNAs are evolutionarily dynamic (Berezikov et al. 2006; Fahlgren et al. 2007; Zhang et al. 2007, 2008; Lu et al. 2008a,b; Liang and Li 2009; Berezikov 2011; Mohammed et al. 2013, 2014a,b; Lyu et al. 2014; Wang et al. 2016), and the repertoire of functional miRNAs has gradually expanded during metazoan evolution (Hertel et al. 2006; Wheeler et al. 2009; Berezikov 2011). Duplication is an important mechanism for miRNA expansions. A large number of functionally essential miRNAs are duplicated and highly conserved across metazoan species (Kim and Nam 2006; Carthew and Sontheimer 2009; Berezikov 2011).
The paralogous miRNAs can diversify their functions by changing the nucleotides in the seeds or shifting the seeds in the mature miRNAs (Berezikov 2011; Sun et al. 2013; Mohammed et al. 2014a; Ninova et al. 2014). Since seeds are crucial for target repression, the change or shift of seeds would direct the duplicated miRNAs to regulate different sets of target genes (Berezikov 2011). Many paralogous miRNAs maintained the same seeds (or even the whole mature sequences) after duplication (Lu et al. 2008a,b; Berezikov 2011; Ninova et al. 2016; Wang et al. 2016). Since mature miRNA sequences outside the seed regions can also affect the specificity of miRNA targeting (Grimson et al. 2007; Bartel 2009; Shin et al. 2010; Wolter et al. 2014; Moore et al. 2015), the paralogous miRNAs could also regulate different sets of target genes even if they share the same seeds (Wolter et al. 2017). Therefore, miRNA duplication followed by the possible neo/subfunctionalization of the paralogs might play crucial roles in the post-transcriptional regulation of gene expression.
miRNA regulation has a significant impact on the evolution of metazoan 3′ UTRs (Farh et al. 2005; Stark et al. 2005; Bartel 2009). Although a large number of mutations in the 3′ UTRs potentially create or destroy miRNA target sites (Chen and Rajewsky 2006; Clop et al. 2006; Saunders et al. 2007; Sethupathy and Collins 2008; Chen et al. 2009; Gong et al. 2012; Li et al. 2012; Lu and Clark 2012; Moszyńska et al. 2017), only a small fraction of the target sites are evolutionarily conserved and likely to be functional (Enright et al. 2003; Stark et al. 2003; John et al. 2004; Rajewsky and Socci 2004; Brennecke et al. 2005; Grün et al. 2005; Lewis et al. 2005; Bartel 2009; Friedman et al. 2009; Li et al. 2009). Previous studies suggest that the functional development of miRNA target sites might be evolutionarily dynamic, with frequent gains and losses of target sites during this evolutionary process (Xu et al. 2013; Liu et al. 2015). The functional targets might be maintained by purifying selection, whereas the neutral or deleterious ones would degenerate by random mutations before or even after they are fixed in the populations. Many factors might affect the evolutionary patterns of miRNA target sites. In particular, it remains unclear whether the evolution and functional development of miRNA target sites are affected by miRNA duplication. Compared to a single-copy miRNA, the duplicated copies of miRNAs might have more flexibility to fine-tune the miRNA:target pairing (Grimson et al. 2007; Bartel 2009; Shin et al. 2010; Wolter et al. 2014, 2017; Moore et al. 2015). Moreover, the duplicated miRNAs might have divergent expression patterns across tissues (Aboobaker et al. 2005; Ason et al. 2006; Landgraf et al. 2007; Ruby et al. 2007; Liu et al. 2008; Roush and Slack 2008; Berezikov 2011; Wolter et al. 2017), which allows them to coevolve with mRNAs in more temporal or spatial environments. Hence, it would be interesting to examine the relationship between miRNA duplication and target site evolution.
In this study, we first present evidence that changes in expression patterns and targeting preferences are not uncommon for duplicated miRNAs in vertebrates. Then we show that the newly emerged target sites have higher chances to become functional during evolution if they are targeted by homo-seed duplicated miRNAs than by single-copy miRNAs. Finally, we verify our results by transfecting two paralogous miRNAs (let-7a and let-7b) into kidney-derived cell lines of four mammalian species, and quantifying the transcriptome alterations with extensive high-throughput sequencing.
RESULTS
The majority of the evolutionarily conserved human miRNAs are duplicated
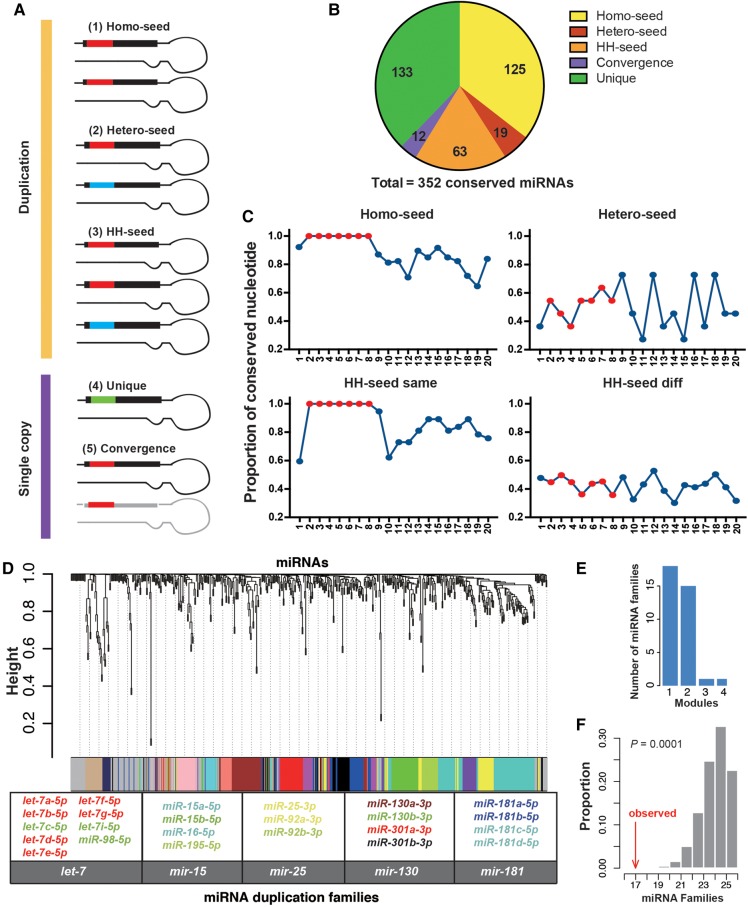
Since most nonconserved miRNAs are lowly expressed and nonfunctional (Lu et al. 2008b; Liang and Li 2009; Berezikov et al. 2010; Kozomara and Griffiths-Jones 2014; Lyu et al. 2014; Patel and Capra 2017), we primarily focused on the human miRNAs that are evolutionarily conserved throughout in this study. Briefly, we downloaded the annotations and duplication information (based on the homology of the precursor sequences) of the miRNAs from miRbaseV21 (Kozomara and Griffiths-Jones 2014). The conservation patterns of the mature miRNAs of humans were examined in the UCSC 100-way vertebrate genome alignments. We followed previous studies (Friedman et al. 2009; Agarwal et al. 2015) and defined the “mammalian conserved” and “broadly conserved” miRNAs based on the conservation patterns of the seed sequences in the mature miRNAs (Materials and Methods). In total, we identified 352 miRNA precursors that encode evolutionarily conserved mature miRNAs in vertebrates (Supplemental Table S1), and 207 (58.8%) of these miRNA precursors were duplicated based on the homology of precursor sequences. Based on the seed sequences of the paralogous miRNAs, we divided the duplicated miRNAs (DmiRs) into three categories (Fig. 1A,B; see Supplemental Fig. S1 for examples): (i) the homo-seed families, in which all the paralogous mature miRNAs in a family share the same seeds (125 precursors, 42 families); (ii) the hetero-seed families, in which each paralog of a family has a distinct seed (19 precursors, nine families); and (iii) the homo-hetero-seed (HH-seed) families (63 precursors, 11 families). The sequence comparisons suggest that nucleotides 13–17 in the mature miRNAs are generally more conserved (>80% identity) between two paralogous miRNAs that share the same seeds (Fig. 1C, left panels), which is consistent with previous observations (Baek et al. 2008; Wolter et al. 2017). Notably, between paralogous miRNAs that have different seeds, the sequence similarities are lower in both the 5′ and 3′ ends of the mature miRNAs (Fig. 1C, right panels). These results suggest (i) that maintaining the same seed between two paralogous miRNAs might also constrain the evolution of nucleotides outside the seed regions, and (ii) that considerable nucleotide substitutions have accumulated between the paralogous miRNAs after duplications, which might lead to functional diversifications. We found 133 evolutionarily conserved single-copy miRNAs, each of which has a unique seed (SCUmiRs). Interestingly, we found another 12 miRNAs that are single-copy and have seeds identical to other conserved miRNAs, although their precursors do not have sequence similarities (Supplemental Fig. S1D). Since the seeds are very short and new miRNAs often de novo originate, the most parsimonious explanation is that these miRNAs are under convergent evolution toward the same seeds (Convergence), as previously shown (Ninova et al. 2016).
FIGURE 1.
The divergence in sequences and expression patterns between duplicated miRNAs. (A) The classification of human miRNAs that are evolutionarily conserved. Based on the seed sequences of the paralogous miRNAs, the DmiRs are divided into three categories: (1) the homo-seed families; (2) the hetero-seed families; and (3) the homo–hetero-seed (HH-seed) families. For the single-copy mRNAs, they can have seeds identical to other miRNAs that do not have sequence similarity in the precursors due to convergent evolution (Convergence), or they have unique seeds (SCUmiRs). (B) The numbers of miRNA precursors in each of the five categories as described in A. (C) The proportion of nucleotides that are identical between two paralogous miRNAs (y-axis) along the position (x-axis) of the mature miRNAs. On each position, the proportion of the pairwise comparisons that have the same nucleotides out of the total number of pairwise comparisons (y-axis) is given. “HH-seed same,” the paralogous miRNAs that share the same seeds in the HH-seed families; “HH-seed diff,” the paralogous miRNAs that have different seeds in the HH-seed families. Positions 2–8 are shown in red. (D) Hierarchical clustering of 574 miRNAs from 181 nonredundant human tissues/cell lines using WGCNA. The color row below the dendrogram shows the module assignment for each miRNA. The histogram shows the number of miRNA families (y-axis) that had paralogous miRNAs assigned to a single (1, x-axis) module or multiple (2, 3, or 4, x-axis) modules. Five representative miRNA families that had paralogs assigned to at least two different modules are given, with each miRNA member labeled in the same color as the module containing that miRNA. (E) The number of miRNA families (y-axis) that had broadly conserved paralogs assigned to different numbers of modules (x-axis). (F) The observed number of miRNA families that had paralogous copies assigned to at least two different expression modules (the red arrow) and the distribution of the simulated numbers (x-axis) obtained by randomly permuting the miRNA: module assignments for 10,000 replicates (the mean is 25, and 95% CI is [22, 26]).
In summary, the majority of the evolutionarily conserved human miRNAs are duplicated, suggesting that duplication is an important mechanism to expand the repertoire of functional miRNAs. The extensive nucleotide substitutions inside and outside the seed regions might cause the paralogous miRNAs to gain targeting specificity. Convergent evolution causes miRNAs of different origins to have the same seeds, which might enable them to target overlapping genes.
Expression divergence between duplicated miRNAs across human samples
The duplicated copies of protein-coding genes are often different in expression patterns (Gu et al. 2003, 2004; Zhang 2003). Similarly, temporal or spatial expression divergence was observed between paralogs for a few miRNA families (Aboobaker et al. 2005; Ason et al. 2006; Landgraf et al. 2007; Ruby et al. 2007; Liu et al. 2008; Roush and Slack 2008; Berezikov 2011; Wolter et al. 2017). To test whether such a pattern exists for most DmiRs, we retrieved the expression profiles of mature miRNAs (precursors that encode the same mature miRNAs were collapsed; Supplemental Table S2) in 181 nonredundant small RNA libraries (Supplemental Table S3). We conducted the weighted gene coexpression network analysis (WGCNA) (Langfelder and Horvath 2008; Zhao et al. 2010) to identify the coexpressed miRNA modules (Materials and Methods). Among the 35 miRNA families that had at least two broadly conserved paralogs expressed in the examined libraries (in total, 107 miRNAs), 17 families (63 miRNAs) had paralogs assigned to at least two different expression modules (Fig. 1D,E). This result suggests that expression divergence between paralogous miRNAs is widespread in humans. For example, the paralogs of many miRNA families, such as let-7, mir-15, mir-25, mir-130, and mir-181, showed high levels of divergence in expression patterns across the human samples (Fig. 1D). As the miRNA precursors in a cluster are usually transcribed as a single unit (Baskerville and Bartel 2005; Saini et al. 2007; Ozsolak et al. 2008; Wang et al. 2009; Ryazansky et al. 2011), the expression levels of the mature miRNAs in the same cluster tend to be similar (Marco et al. 2013; Wang et al. 2016). To test whether the expression divergence between paralogous miRNAs is overall under selective constraints, we randomly permuted the miRNA:module assignments while maintaining the clustering structure of microRNAs (Materials and Methods). Occasionally, one mature miRNA was located in multiple genomic locations. We first assigned such a miRNA to the largest cluster and conducted the simulations. With this simulation approach, we found the observed number of miRNA families (17) that had paralogous copies assigned to at least two different expression modules was significantly lower (P = 0.0001) than that obtained under the assumption of randomness (the mean is 25, and 95% CI is [22, 26]; Fig. 1F). Similar results were obtained when we only considered the miRNAs that were uniquely mapped on the human genomes (the observed number is 13 versus 16 with 95% CI [14, 17] in the simulations, P = 0.0143, Materials and Methods). Thus, although many paralogous miRNAs are divergent in expression patterns, the general trend is that the expression divergence between paralogous miRNAs is under selective constraints.
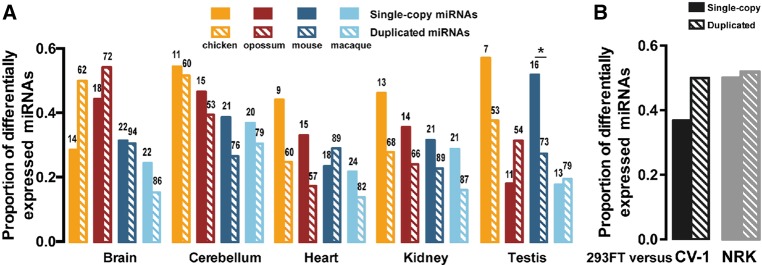
The duplicated protein-coding genes tend to be more differentially expressed across strains/species than the single-copy ones (Gu et al. 2004). Here, we asked whether such a pattern can be observed for miRNAs. We retrieved miRNA expression profiles from five tissues (brain, cerebellum, heart, kidney, and testis) of five vertebrate species (human, macaque, mouse, opossum, and chicken) quantified in a previous study (Meunier et al. 2013) (Materials and Methods; Supplemental Table S4). On average, ∼30% of the broadly conserved miRNAs were differentially expressed (greater than or equal to fourfold change) between humans and other vertebrate species (Fig. 2A). Interestingly, we did not find that the DmiRs were more differentially expressed between humans and other species than the SCUmiRs (Fig. 2A). Similar observations were made when we considered all the conserved miRNAs (Supplemental Fig. S2). To independently verify this pattern, we also deep-sequenced the small RNAs in three kidney-derived cell lines: 293FT (human), CV-1 (green monkey), and NRK (rat) (Materials and Methods). We still found that the level of differential expression between humans and other species is similar for the SCUmiRs and DmiRs (Fig. 2B). Thus, the duplicated miRNAs exhibit distinct features from duplicated protein-coding genes in expression evolution across species.
FIGURE 2.
The proportions of duplicated and single-copy miRNAs that are differentially expressed by at least fourfold change (y-axis) between humans and other vertebrate species. (A) The comparisons in five tissues between human and macaque, mouse, opossum, or chicken. (B) The comparisons between human (293FT) and green monkey (CV-1) or between human and rat (NRK) cell lines. The number above each box represents the number of single-copy or duplicated miRNAs that are broadly conserved in vertebrates and expressed in both humans and the other species.
Divergence in target pairing between paralogous miRNAs
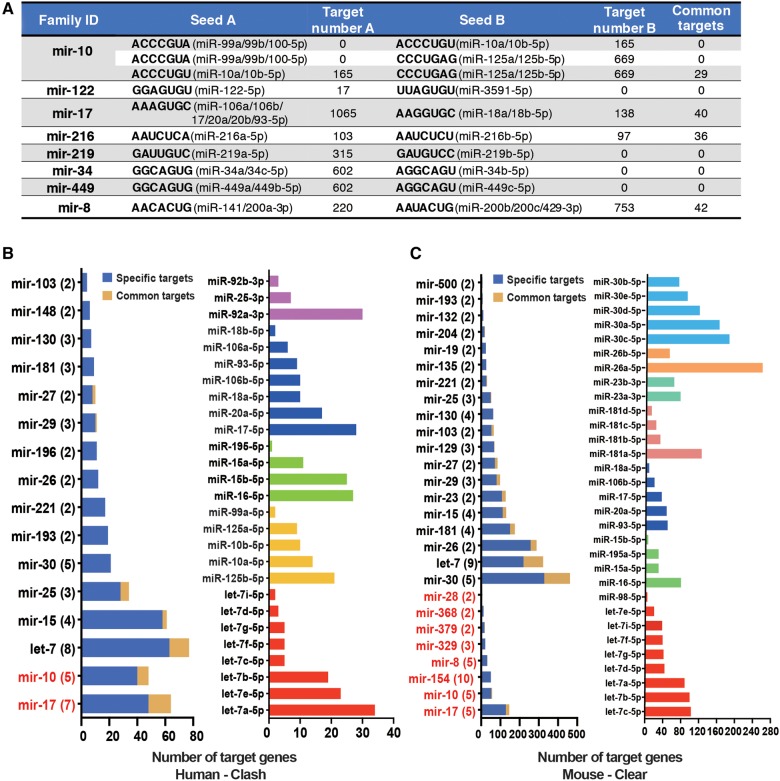
The paralogous miRNAs could change their nucleotides to pair with different target sites (Berezikov 2011). Occasionally, two paralogous miRNAs with different seeds might still target different sites of the same gene. For the broadly conserved hetero-seed or HH-seed miRNA families that have at least one evolutionarily conserved target site (PCT > 0.5 in TargetScan [Agarwal et al. 2015]), only a few target genes were shared between two paralogous miRNAs that have different seeds (Fig. 3A). For example, miR-34a/c-5p (the seed is GGCAGUG) has 602 conserved target sites (PCT > 0.5), while one nucleotide shift of the seed sequence in miR-34b-5p (AGGCAGU) causes this miRNA to have no conserved target site (Supplemental Fig. S3). Alternatively, when we examined the target sites with context++ score < −0.3 (Grimson et al. 2007; Agarwal et al. 2015), which are located in optimized genomic context for targeting but not necessarily conserved, we also found that ∼2% of the target genes were shared between two paralogous miRNAs that do not share the same seeds (Supplemental Fig. S4). Hence, changing or shifting of seeds after miRNA duplication has diversified miRNA target repertoire.
FIGURE 3.
Divergence in target pairing between the paralogous miRNAs. (A) The numbers of target sites shared between two seeds (common targets) in the hetero-seed and HH-seed miRNA families (target sites were predicted with TargetScan PCT > 0.5). The broadly conserved miRNAs that had the specified seeds are shown. (B) The number of canonical target sites in 3′ UTRs (seed pairing) bound by at least one member of a miRNA family in the CLASH data set (x-axis). “Specific targets”: the target is only bound by one paralogous miRNA; “Common targets”: the target genes that were targeted by at least two paralogous miRNAs that were expressed. (Left panel) The HH-seed family is indicated in red; the number of paralogous miRNAs in each family that were expressed in the CLASH data set is given in parentheses. (Right panel) The number of canonical target sites bound by the paralogs in five representative miRNA families. (C) The number of canonical target sites in 3′ UTRs bound by at least one member of a miRNA family in the CLEAR data set (x-axis). The figure setting is the same as in B.
Mature miRNA sequences outside the seed regions also affect the specificity of miRNA targeting (Grimson et al. 2007; Bartel 2009; Shin et al. 2010; Wolter et al. 2014; Moore et al. 2015). Hence, paralogous miRNAs might regulate different sets of target genes even if they share the same seeds (Wolter et al. 2017). Since it is challenging to in silico predict the targeting preferences for paralogous miRNAs that share the same seeds, we examined the miRNA:mRNA pairing chimeras identified in previously published AGO HITS-CLIP studies (Helwak et al. 2013; Moore et al. 2015). In the first data set, thousands of miRNA:mRNA chimeras were identified using CLASH (cross-link and sequencing of hybrids) in human HEK-293T cells (Helwak et al. 2013). A salient observation of this data set is that only a small fraction of the canonical target genes was shared between paralogs in a DmiR family (Fig. 3B, left panel; detailed information for the top five miRNA families that have the highest numbers of experimentally detected targets is given in the right panel). We also found that paralogous miRNAs tend to pair with different sets of canonical target genes in the second data set (Fig. 3C), which captured 130,000 endogenous miRNA:mRNA chimeras using a modified AGO HITS-CLIP strategy termed CLEAR (covalent ligation of endogenous Argonaute-bound RNAs)-CLIP in mouse brains (Moore et al. 2015). Altogether, these AGO HITS-CLIP data sets provide experimental evidence for the existence of widespread divergence in target site recognition between paralogous miRNAs.
The evolutionary dynamics are different for targets of SCUmiRs versus homo-seed DmiRs
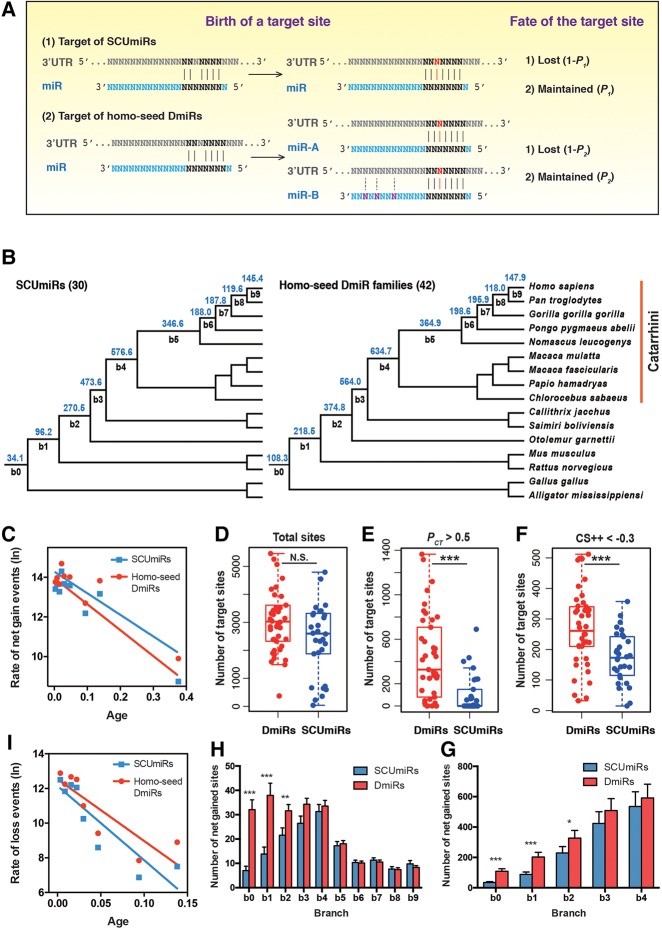
Since the DmiRs are often divergent in target recognition, we postulate that the evolutionary dynamics are different for targets of SCUmiRs versus those of homo-seed DmiRs (Fig. 4A). Under this model, a newly emerged target site has a higher probability to be functional and maintained by natural selection if it is paired to a seed shared by multiple paralogous miRNAs (P2) than being paired to a SCUmiR (P1). On one hand, the nucleotide changes outside the seed regions of the DmiRs might provide more optimized contexts for the miRNA: target pairing (Fig. 4A). Moreover, expression divergence would cause the DmiRs to be exposed to more mRNAs in more spatial and temporal environments, which might also increase the chance for a new target site to develop function (Fig. 4A). As a consequence, this model predicts that the broadly conserved homo-seed DmiRs would have higher numbers of functional target sites than the SCUmiRs.
FIGURE 4.
The evolutionary dynamics of canonic target sites for the SCUmiRs and homo-seed DmiRs. (A) A scheme describing how the evolutionary dynamics are different for targets of SCUmiRs versus homo-seed DmiRs. A point mutation (red) in the 3′ UTR generates a new target site that is perfectly paired to the seed of a miRNA. The probability that the target site becomes functional and maintained by natural selection during evolution is P1 if it is paired to the seed of SCUmiR, and P2 if it is paired to the seed of homo-seed DmiRs. Accordingly, the probability that the target site is lost during evolution is 1 − P1 and 1 − P2, respectively. We postulate that P2 would be higher than P1 based on the following considerations: (1) The nucleotides outside the seed regions of the homo-seed DmiRs might have more flexibility to optimize the miRNA: target pairing; and (2) the expression divergence would cause the duplicated copies of the homo-seed DmiRs to be exposed to more mRNAs in more spatial and temporal environments, which might also increase the chance for a new target site to develop function. Based on this model, during long-term evolution, the seeds of the homo-seed DmiRs would have higher numbers of functional target sites than those of the SCUmiRs. (B) The average number of net-gained target sites per seed in the branches leading to extant humans for the broadly conserved miRNAs that originated before the split of birds and mammals (30 SCUmiRs, left; and 42 homo-seed DmiR families, right). The target sites were predicted with TargetScan with the requirement of perfect seed pairing (7mer-m8 and 8mer). b0–b9 represents the branches leading to extant humans. The number of target sites in b0 is the target sites (per seed) that are ancient and conserved in all 16 species. Branch length is not scaled. (C) Correlation between the age of a branch (x-axis) and the rate of net gained target sites in that branch (y-axis). The age of each branch is calculated based on the middle point of the branch length and the offspring branches, if applicable. The rate is defined as the number of target sites gained per evolutionary unit (mutations per nucleotide per generation). (D) The boxplot of canonical target sites of the homo-seed DmiRs and SCUmiRs (N.S., not significant). (E) The seed of a homo-seed DmiR family pairs a significantly higher number of conserved target sites (PCT > 0.5) than that of a SCUmiR; (***) P < 0.001. (F) The seed of a homo-seed DmiR family pairs a significantly higher number of optimized target sites (context++ score < −0.3) than that of a SCUmiR; (***) P < 0.001. (G) The average numbers of target sites that originated in the ancient branches (b1, b2, b3, and b4) and conserved in extant humans and at least seven other Catarrhini species (hominoids and old world monkeys, see Fig. 4B). For each branch, a WMW test was used to test whether there is a significant difference in the numbers of target sites for a homo-seed DmiR family and a SCUmiR (*) P < 0.05; (***) P < 0.001. The number of target sites in b0 is the target sites (per seed) that are ancient and conserved in all 16 species. The bars represent the standard errors. (H) The number of the net-gained target sites in each branch (b1–b9) leading to extant human and that are located in the optimized genomic contexts for miRNA targeting (context++ score < −0.3). For each branch, a WMW test was used to test whether there is a significant difference in the numbers of target sites for a homo-seed DmiR family and a SCUmiR; (**) P < 0.01, (***) P < 0.001. The number of target sites in b0 is the target sites (per seed) that are ancient and conserved in all 16 species. The bars represent the standard errors. (I) Correlation between the age of a branch (x-axis) and the loss rate of putative target sites lost in that branch.
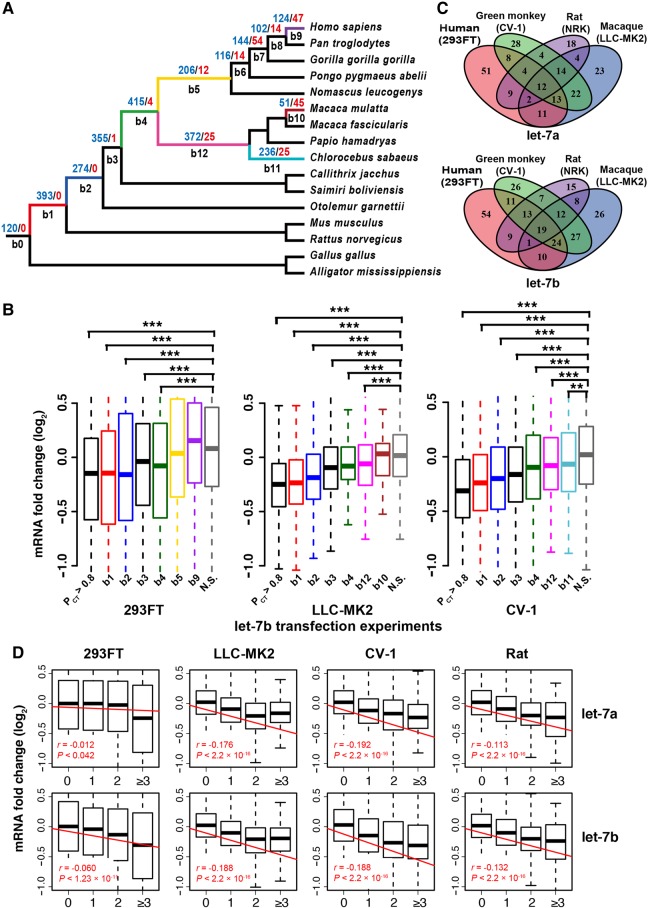
Since the functional target sites are usually evolutionarily conserved (Bartel 2009), we first examined the conservation patterns of the target sites for the SCUmiRs and homo-seed DmiRs. Based on the 3′ UTR alignments of 16 vertebrate species (12 primates, mouse, rat, chicken, and American alligator), we traced the births and deaths of the canonical target sites (perfect seed matching) for each broadly conserved miRNA (42 homo-seed DmiR families and 30 SCUmiRs). For both classes of miRNAs, the target sites evolved in a birth-and-death manner (Supplemental Fig. S5A; the detailed gain-and-loss information for each miRNA/family was given in Supplemental Fig. S5B,C). We primarily focused on the target sites that are maintained in extant humans, with the assumption that a functional target site would not be lost after emergence. We inferred the length of each branch in the phylogenetic tree regarding evolutionary units (number of mutations per nucleotide site per generation) and summarized the net gains of putative target sites in that branch (Fig. 4B). We uncovered a significantly negative correlation between the age of a branch (the middle point of the branch to extant humans) and the number of target sites net-gained per evolutionary unit in that branch (Pearson's r = −0.94 and −0.92 for targets of the SCUmiRs and homo-seed DmiR families, respectively, P < 0.01 in each analysis; Fig. 4C). This pattern suggests (i) that the newly emerged miRNA target sites experience births and deaths at high rates, and (ii) that the majority of the newly emerged target sites are evolutionarily transient, and have not had time to degenerate. This pattern holds for both the SCUmiRs and homo-seed DmiR families (Fig. 4C).
Although the numbers of target sites of the broadly conserved homo-seed DmiR families were slightly higher than those of the SCUmiRs, the difference was not statistically significant (P = 0.088, Wilcoxon–Mann–Whitney [WMW] test, Fig. 4D; similar results were obtained when we separately examined the 7mer-m8 and 8mer types of target sites, Supplemental Fig. S6A,B). Nevertheless, among the conserved target sites predicted by TargetScan (PCT > 0.5), the seeds of the homo-seed DmiR families paired to significantly higher numbers of target sites than those of the SCUmiRs (P = 1.48 × 10−5, WMW test, Fig. 4E; see Supplemental Table S5 for details). Moreover, the homo-seed DmiR families had a significantly higher number of target sites that were located in the optimized genomic contexts for miRNA targeting (context++ score < −0.3) than the SCUmiRs (P = 6.93 × 10−4, WMW test, Fig. 4F). Analogous results were obtained when we examined the CLASH and CLEAR experimental data sets (Supplemental Fig. S6C,D). All these results support the notion that the homo-seed DmiR families have acquired higher numbers of putative functional target sites than the SCUmiRs, as we illustrate in Figure 4A. Next, we stratified the net-gained target sites into different ages on the phylogenetic tree. In each branch, we examined the difference in numbers of acquired target sites for the SCUmiRs versus the homo-seed DmiR families (Supplemental Fig. S6E). Interestingly, the numbers of target sites that were net-gained in the ancient branches (b1, b2, b3, and b4) and conserved in humans and at least seven other Catarrhini species (hominoids and the old world monkeys, see Fig. 4B) were higher for the homo-seed DmiR families than the SCUmiRs (P < 0.05 for the comparisons in b1 and b2, WMW test, Fig. 4G). Similarly, at the cutoff of context++ score < −0.3, the numbers of target sites acquired in branches b1 and b2 and preserved in extant humans were also significantly higher for the homo-seed DmiR families than the SCUmiRs (P < 0.01 in each comparison, WMW test, Fig. 4H). Therefore, our results suggest that a newly emerged target site is more likely to be functional and preserved during evolution if it is targeted by the homo-seed DmiRs. It is known that two rounds of whole-genome duplications (2rWGDs) occurred at the base of the vertebrate lineage, which preceded the most recent common ancestor of the 16 vertebrate species we examined (Ohno et al. 1968; Dehal and Boore 2005; Panopoulou and Poustka 2005). Since all the broadly conserved homo-seed DmiR families and SCUmiRs we examined also formed before the radiation of these 16 species (Fig. 4B), the significant differences in the numbers of target sites gained in the ancient mammalian branches (b1 and b2) between the two classes of miRNAs are unlikely to be directly caused by the 2rWGDs. In summary, all these observations support the notion that the putatively functional target sites are recruited at higher rates if they are paired to homo-seed DmiRs than the SCUmiRs.
Notably, target sites were frequently lost during evolution (red in Supplemental Fig. S5A), and such loss events were significantly enriched in the younger branches that lead to extant humans (Pearson's coefficient between the rate and age of a branch is r = −0.86 and −0.89 for sites pairing to the seeds of the broadly conserved homo-seed families and SCUmiRs, respectively; P < 0.05 in each analysis; Fig. 4I). To test whether the loss events are different for the target sites of the homo-seed DmiR families versus the SCUmiRs, for each branch we calculated the proportion of the target sites that initially originated in that branch but were eventually lost in extant humans (Supplemental Fig. S7A). In general, ∼10% of the target sites that originated in the ancient branches (b1–b6) are finally lost in extant humans. Interestingly, compared to those of SCUmiRs, the target sites of the homo-seed DmiR families overall experienced marginally lower loss probabilities during evolution (P = 0.05, WMW test, Supplemental Fig. S7B). Similar patterns were observed when we individually examined the target sites that originated in branches b1, b3, and b5 (Supplemental Fig. S7A). Thus, the putative functional target sites are also under stronger selective constraints from being lost if they are paired to the homo-seed DmiRs than by SCUmiRs.
Stronger repressive effects of let-7b than let-7a in cellular transfection experiments
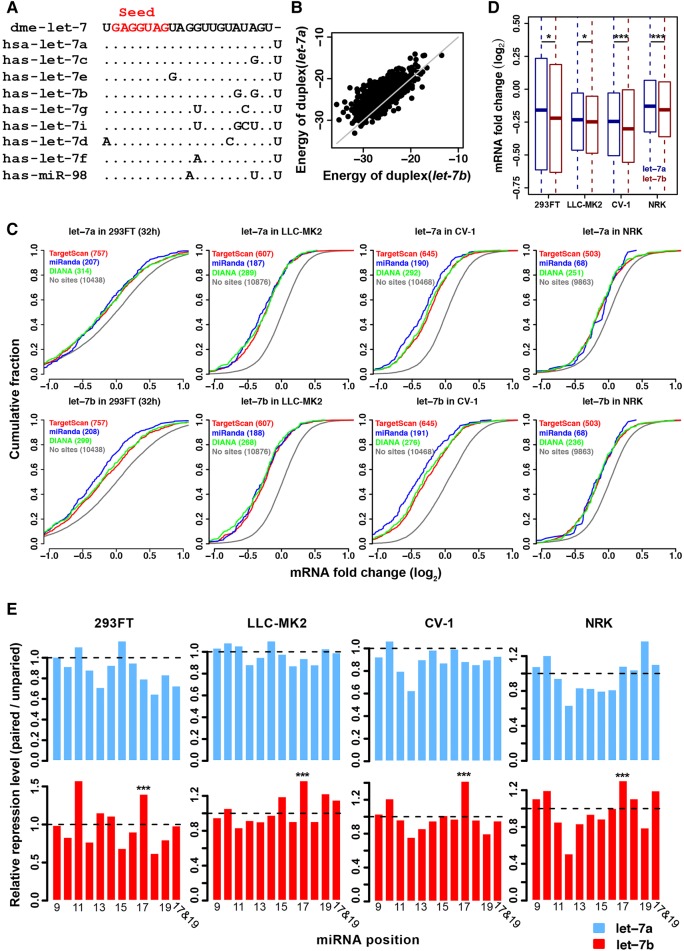
Our results suggest that paralogous miRNAs can target different sets of target genes even if they share the same seeds. Remarkably, the evolutionary dynamics of target sites are different for SCUmiRs and homo-seed DmiRs. To experimentally verify these results, we transfected let-7a and let-7b mimics into kidney-derived cell lines of four mammalian species (Human, 293FT; Macaque, LLC-MK2; Green Monkey, CV-1; and Rat, NRK) and measured the resulting transcriptome alterations using high-throughput sequencing (Materials and Methods). These four cell lines were chosen because they provide experimentally tractable systems to study the evolution and conservation of gene regulation mediated by miRNAs. Moreover, these cell lines were derived from the same tissue, which minimized the tissue-specific effects in comparing miRNA mediated regulation across species. The let-7 family, whose paralogous members share the same seed, is one of the first miRNA families discovered in animals (Pasquinelli et al. 2000; Reinhart et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Roush and Slack 2008; Hertel et al. 2012). let-7a, which is the ancestral copy in this family, differs from its paralogs in vertebrates by 1–4 nucleotides (nt) outside the seed regions (Fig. 5A). Previous studies suggest a substitution at position 12 of let-7f directed it to repress novel targets (Wolter et al. 2017). Notably, the mature sequences of let-7a and let-7b differ by 2 nt (positions 17 and 19), and both miRNAs are highly conserved in the vertebrates (Supplemental Fig. S8). Our bioinformatics analysis suggests the A > G substitutions in let-7b lead to higher binding affinities of the target sites (Fig. 5B). Furthermore, the relative expression of the two miRNAs differed dramatically across species: The ratio of let-7a/let-7b abundance was 17.7, 2.47, and 0.55 in the 293FT, CV-1, and NRK cell lines (Supplemental Fig. S9A), and similar patterns were observed in the kidney tissues across species (Supplemental Fig. S9B). Thus, the divergence in sequences and expression patterns might cause divergence in target recognition between let-7a and let-7b.
FIGURE 5.
The repressive effect mediated by let-7a and let-7b in transfection experiments. (A) Sequence alignment of human let-7 family miRNAs. The seed region (positions 2–8) is shown in red. let-7 of D. melanogaster was used as an outgroup. (B) The two A > G substitutions in let-7b enable let-7b to exert stronger binding affinities (lower energy, kcal/mol) to the target sites than let-7a. (C) Cumulative distribution of log2 fold changes in mRNA levels (x-axis) after transfecting let-7a or let-7b into human, macaque, green monkey, or rat cell lines. The target genes (with canonical sites in the 3′ UTRs) of let-7a and let-7b were predicted by TargetScan with a PCT > 0.8 (red), miRanda with a mirSVR score < −0.8 (blue), or DIANA with a threshold score > 0.5 (green). The mRNAs without any site complementary to the seed sequence of let-7a/b (controls, gray) were used as controls. The log2 fold change of the predicted target sites is significantly lower compared to the control mRNAs in each comparison (P < 0.0001 in each comparison, Kolmogorov–Smirnov test). (D) let-7b (purple) exerted stronger repressive effects on the canonical target genes than let-7a (blue) in the cellular transfection experiments ([*] P < 0.05; [***] P < 0.001; paired t-tests). The target genes from different sources in C were pooled. (E) The ratio of median log2 fold change for target genes (y-axis) that have canonical sites paired at the indicated position of let-7a (upper panel) or let-7b (lower panel) relative to the target genes not paired at that position. Pairing at position 17 of let-7b is associated with significantly stronger repressive efficiencies compared to the other conserved canonical sites, but such a pattern does not exist for let-7a; ([***] P < 0.001, Fisher's exact test).
We performed two biological replicates in the transfection and sequencing experiments (see Table 1 and Supplemental Table S6 for statistics of all the sequencing libraries) and obtained high correlations between the biological replicates in each experiment (Supplemental Fig. S10). We identified hundreds of genes that were up- or down-regulated in the let-7a and let-7b transfection experiments in human 293FT cells (Supplemental Fig. S11) and the cell lines from the other species (Supplemental Fig. S12). In our transfection experiments, the canonical target genes of let-7a/b that were predicted with TargetScan v7.0 (Agarwal et al. 2015), DIANA (Reczko et al. 2012; Paraskevopoulou et al. 2013), or miRanda (Betel et al. 2010) were significantly down-regulated compared to the transcripts without any predicted site of let-7a/b (P < 0.0001 in each comparison, KS test, Fig. 5C). We performed qRT-PCR to verify the down-regulation of a subset of target genes in human 293FT cells (EIF4G2, TIMM17B, MAPK6, USP38, IGDCC4, and SLC25A4; Supplemental Fig. S13A) or in macaque and green monkey cells (DCR1, TMEM8A, TARBP2, TTLL4, and PBX2; Supplemental Fig. S13B). Furthermore, we also conducted luciferase reporter assays to verify all the six human target genes in 293FT cells (Supplemental Fig. S13C).
TABLE 1.
Expression profiles of mRNA transcripts in let-7a and let-7b transfection experiments
Remarkably, let-7b exerted stronger repressive effects on the canonical target genes than let-7a in the transfection experiments for all four cell lines (Fig. 5D). For example, in human 293FT cells at 32 h post-transfection, the median log2 fold-change (LFC) for the conserved target genes was −0.157 in cells transfected with let-7a versus −0.220 in cells transfected with let-7b (P = 0.018; paired Student's t-test). Furthermore, among all the conserved target sites (perfect seed pairing), the sites paired at position 17 of let-7b were associated with significantly stronger mRNA destabilization than the sites not paired at that position, whereas such a pattern was not observed for let-7a (Fig. 5E; Supplemental Fig. S14). Altogether, these results suggest that pairing between the target sites and let-7b at position 17 might be associated with higher binding affinities and thus stronger repressive effects, and the A > G change at that position might change the targeting specificity of let-7b and let-7a.
The newly emerged target sites of let-7 affect mammalian gene expression evolution
Our results suggest the target sites of miRNAs are continuously gained during vertebrate evolution, but the extent to which such target sites affect gene expression evolution remains elusive. To explore the relationship between evolutionary patterns and repressive effects of the target sites, we predicted the target sites of let-7a/b based on perfect seed pairing (8mer and 7mer-m8 in TargetScan) and stratified them into different ages on the phylogenetic tree (Fig. 6A; see Supplemental Fig. S15 for the results that considered the 7mer-A1 sites). It should be noted that here we considered all the target sites in the 3′ UTRs that are perfectly paired to the seed of let-7 since we could not accurately separate the sites specifically targeted by let-7a from those by let-7b. As expected (Bartel 2009), we observed stronger repression on genes containing target sites gained in the older branches (originating in b1, b2, or older branches) than genes harboring younger target sites (b3–b9) in both let-7a (Supplemental Fig. S16) and let-7b (Fig. 6B) transfection experiments. Specifically, genes containing target sites highly conserved in vertebrates (defined with TargetScan PCT > 0.8) or conserved target sites that originated prior to the radiation of mammals (b1 branch, red) were the most severely repressed; genes containing target sites that were conserved only in primates (b2, blue) had the second highest level of repression; and genes harboring species-specific target sites (b9, purple; b10, brown; or b11, cyan) were generally weakly suppressed (Fig. 6B). Although the evolutionarily young target sites were associated with weak repressive effects (e.g., the sites originating in b3, b4, or b5, Fig. 6B; Supplemental Fig. S16), some might also contribute to the divergence of transcriptomes across species. For example, among the 77 human-specific let-7 target genes expressed in 293FT cells, 12 and 12 were significantly down-regulated in the let-7a and let-7b experiments, respectively (five of them were significantly down-regulated in both experiments). Overall, our results suggest that the newly emerged target sites of let-7 gradually develop function as they mature, and such target sites might have made nonnegligible contributions to the evolution of target gene expression.
FIGURE 6.
Birth and death of let-7 target sites during mammalian evolution and the contributions to the evolution of transcriptomes. (A) The gains and losses of canonical let-7a/b target sites in the branches leading to extant primates. The numbers of gain and loss events are in blue and red, respectively. The number of target sites in b0 is the target sites that are ancient and conserved in all 16 species. (B) Changes in the mRNA abundance of genes with ancient or newly emerged target sites after transfecting let-7b into human, macaque, and green monkey cells. The target sites of let-7b predicted with PCT > 0.8 are also shown (these sites are not exclusive to sites in branches b1–b9 of Fig. 6A). The genes with target sites originating in a branch and maintained thereafter were used in the analysis. If a gene had multiple target sites of different ages, the gene was assigned to the most ancient class. N.S., nontarget sites. (C) Venn diagram showing only a small fraction of the evolutionarily conserved canonical target sites (TargetScan v7.0, PCT > 0.8) consistently mediated repression across the four species in the let-7a and let-7b experiments. The number of the significantly down-regulated genes after transfection is shown for each species. (D) Genes whose 3′ UTRs have higher numbers of canonical let-7a (upper) and let-7b (lower) target sites are more strongly repressed in the transfection experiments. Both the conserved and nonconserved target sites were considered. The raw Pearson's correlation coefficient r is shown for each experiment.
Next, we asked whether the newly emerged target sites might cooperate with the ancient and conserved sites that were located in the same genes in regulating gene expression. We identified 368 genes that were expressed in all the four cell lines we examined and had at least one evolutionarily conserved target site of let-7a/b in the 3′ UTRs (PCT > 0.8). In both the let-7a or let-7b experiments, the observed numbers of target genes that were down-regulated in multiple species were significantly higher than the number obtained with the assumption of randomness (P < 0.05, Supplemental Fig. S17). However, most of these conserved target genes were down-regulated in a species-specific manner, and only a small fraction of them was down-regulated in all four species (Fig. 6C). For example, only 22%–37% of these target genes were significantly down-regulated with LFC <−0.2 in the let-7b experiments (141, 127, 139, and 84 targets were down-regulated in 293FT, LLC-MK2, CV-1, and NRK cells, respectively, Fig. 6C), and very few of them were overlapped. We observed similar patterns when we examined the extent of mRNA down-regulation for the conserved orthologous target genes. For example, in the let-7b experiments, Pearson's r was 0.17, 0.28, and 0.18 for human versus macaque, green monkey, and rat, respectively (Supplemental Fig. S18). Similar results were obtained in the let-7a experiments (Fig. 6C and Supplemental Fig. S18). Notably, besides these highly conserved target sites, these target genes also had numerous births and deaths of let-7 target sites in other locations of the 3′ UTRs (Supplemental Fig. S19). Importantly, we found that multiple target sites of let-7 (either conserved or nonconserved) are associated with stronger repressive effects in all the let-7a and let-7b transfection experiments (Fig. 6D). Overall, our in vitro cellular transfection experiments suggest that the newly emerged target sites of let-7 affect mammalian gene expression evolution, although further in vivo studies are needed to physiologically verify this pattern in the physiological conditions.
DISCUSSION
Duplication is an important mechanism to expand the repertoire of metazoan miRNAs. This present study has unveiled widespread divergence in expression patterns and targeting preferences between paralogous miRNAs. Our results suggest the broadly conserved homo-seed DmiR families pair with significantly higher numbers of conserved (putatively functional) target sites than those of the SCUmiRs, which supports the notion that a newly emerged target site would have a higher probability to become functionally preserved during evolution if it is targeted by the homo-seed DmiRs than by the SCUmiRs (Fig. 4A). Our results also suggest that the divergence or convergence of miRNA seeds have different effects on the evolution of the target sites. Specifically, for the hetero-seed DmiR families, where the paralogous miRNAs have different seeds from each other, we did not find that the seeds pair with significantly more target sites than the SCUmiRs. Nevertheless, the broadly conserved single-copy miRNAs that experienced convergent evolution (Convergence) also had significantly higher numbers of conserved target sites than the SCUmiRs (P < 0.01, WMW test; Supplemental Fig. S20). Furthermore, after grouping miRNAs based on the seed sequences, we found the seeds shared by multiple broadly conserved miRNAs overall pair with significantly higher numbers (and higher fractions) of target sites than those of the single miRNAs (P < 10−9, WMW test, Supplemental Fig. S21). Altogether, these results suggest that the modes of functional diversification and convergence of miRNAs have had the different impact on the evolution patterns of the target sites. Although purifying selection is important in maintaining the functional target sites, further studies are needed to dissect the roles of positive selection and genetic drift in shaping the landscapes of miRNA target sites.
By transfecting let-7a and let-7b into four mammalian cell lines, we presented evidence that the A > G changes (positions 17 and 19) enable let-7b to exert stronger repressive effects on canonical target genes than let-7a. We may have underestimated the divergence in target repression between the paralogous miRNAs because we did not consider noncanonical target pairing (i.e., pairing to the centers or 3′ ends of miRNAs), which might not be uncommon (Broderick et al. 2011; Moore et al. 2015). Indeed, the AGO HIST-CLIP data set revealed substantial differences in target recognition between paralogous miRNAs in the same family, particularly for the noncanonical target sites, through 3′ pairing (Supplemental Fig. S22). To test whether let-7a and let-7b repress noncanonical target sites in our transfection experiments, we predicted all the possible pairing duplexes between let-7a or let-7b and 3′ UTRs of the down-regulated genes without requiring perfect seed matching. We calculated the fraction of potential target sites that were paired at each position of let-7a or let-7b, and found that significantly higher fractions of the putative target sites were bound to the 3′ end of let-7b compared to let-7a (Supplemental Fig. S23A). Similarly, the putative target sites pairing to the 3′ end (position 17–21) of let-7a or let-7b were also associated with target repression (Supplemental Fig. S23B), suggesting that the difference in target repression between let-7a and let-7b might be mediated by these noncanonical target sites as well. Indeed, the genes with putative target sites paired with let-7b at either the 5′ end, center, or 3′ end were significantly overrepresented in the genes down-regulated by let-7b transfection in 293FT, LLC-MK2, and CV-1 cells (Supplemental Fig. S23C). In contrast, the predicted target sites that paired with the 3′ end of let-7a were not associated with mRNA destabilization in either humans or macaques (Supplemental Fig. S23C). Altogether, our results suggest that let-7a and let-7b might also be divergent in regulating the noncanonical target genes.
In summary, this present study advances our understanding of the functional divergence between paralogous miRNAs pertaining to expression patterns and target repression. Importantly, we found that the functional diversification of duplicated miRNA genes significantly expands the miRNA target sets and impacts the evolution of vertebrate 3′ UTRs. Future studies will be necessary to dissect the mechanisms and evolutionary forces underlying the coevolutionary process between miRNAs and 3′ UTRs.
MATERIALS AND METHODS
miRNA sequences and expression data
We downloaded the sequences, annotation and expression data of miRNAs from miRBaseV21 (http://www.mirbase.org). The duplication information, which is based on the homology of miRNA precursor sequences, was also obtained from miRBase V21. Also, by all-to-all miRNA sequence homology searches with FASTA36 (Pearson 2016), we found another four human miRNA precursors (hsa-mir-548o, hsa-mir-3118-4, hsa-mir-5701-3, and hsa-mir-5692b) are also duplicated (E-value < 0.0001). The orthologous sequences of miRNAs in the UCSC 100-way vertebrate genome alignments (genome.ucsc.edu) were retrieved from the Galaxy workflows at https://usegalaxy.org (Goecks et al. 2010). We followed previous procedures (Friedman et al. 2009; Agarwal et al. 2015) and defined a mature miRNA of humans to be “broadly conserved” if the seed (positions 2–8) sequences were identical in at least 60 vertebrate species, and a mature miRNA to be “mammalian conserved” if its seed sequences were identical in between 40 and 60 vertebrate species. Occasionally, some broadly conserved miRNAs might be classified as mammalian conserved due to poor genomic alignments. We manually changed such “mammalian conserved” miRNAs to “broadly conserved” ones if the miRNAs were homologous between humans and chicken, frog, or zebrafish (we required the homologous miRNAs to have the same seeds, and the sequences to be identical for at least 18 nt in the mature miRNAs), as previously described (Agarwal et al. 2015). If a miRNA precursor had both the guide and the passenger (*) strands (or the 5p and 3p) annotated in miRBase V21, we defined the guide strand (mature miRNA) based on the higher expression level or higher conservation level. For the duplicated miRNAs, we preferentially chose the mature miRNAs that had seed sequences conserved between paralogs as the guide strands. We also filtered the miRNAs that had no expression evidence supported in miRBase V21. In total, we identified 352 miRNA precursors whose mature miRNAs were evolutionarily conserved and could be unambiguously classified into duplicated or single-copy miRNAs (Supplemental Table S1).
We obtained 484 miRNA expression libraries from different human tissues and cell lines from the NCBI SRA database (www.ncbi.hlm.nih.gov/sra, last accessed May 16, 2016). The reads were mapped to the miRNA sequences (miRBase V21) using Bowtie v1.0.1 (Langmead et al. 2009). After removing technical replicates and redundant data sets as previously described (Wang et al. 2016), we obtained expression levels of 574 mature miRNAs (including conserved and nonconserved) that had mean RPM (reads per million) ≥ 5 (Supplemental Table S2) in 181 nonredundant libraries (Supplemental Table S3). The expression levels of mature miRNAs across libraries were normalized using DESeq2 (1.16.1) (Love et al. 2014). The miRNA coexpression modules were detected with WGCNA (V1.61) (Langfelder and Horvath 2008; Zhao et al. 2010).
We retrieved miRNA expression profiles from five tissues (brain, cerebellum, heart, kidney, and testis) of five vertebrate species (human, macaque, mouse, opossum, and chicken) that were quantified previously (Meunier et al. 2013). For each tissue, we normalized the expression levels of the orthologous miRNAs between human and another species using DESeq2 (1.16.1).
Permutation analysis of miRNA expression in WGCNA modules
Human miRNAs within 10 kb of each other were clustered together as previously described (Marco et al. 2013; Wang et al. 2016). We shuffled the assignment of miRNAs to the WGCNA expression modules and counted the number of miRNA families that had paralogous copies assigned to at least two different modules. miRNAs that were located in a genomic cluster and coexpressed in the same WGCNA module were treated as a single unit during shuffling. In case a miRNA was located in more than one cluster, it was assigned to the largest cluster to which it belongs. This procedure was repeated for 10,000 replicates. The confidence intervals were calculated as the 2.5% and 97.5% quantiles in the permutation analysis. The P-value was calculated by comparing the observed versus the simulated values in the permutation analysis. We also only considered the mature miRNAs that were uniquely mapped to the human genome and performed the same analytical procedures.
Evolutionary analysis of miRNA target sites
We retrieved the sequence alignments of the 3′ UTRs in 16 vertebrate species (12 primates, mouse, rat, chicken, and American alligator) from the website of TargetScan 7.0 (www.targetscan.org) (Agarwal et al. 2015). We used tree_doctor (Hubisz et al. 2011) to extract the phylogenetic information of the 16 species from the 100-way vertebrate phylogenetic tree (genome.ucsc.edu). We predicted all the conserved and nonconserved target sites perfectly matching the seed (position 2–8) of a miRNA (7mer-m8 and 8mer types) with the Perl script of TargetScan 7.0, and traced the gain and loss of each target site using the Gain Loss Mapping Engine (GLOOME) (Cohen and Pupko 2011). We also retrieved the evolutionarily conserved target sites (PCT > 0.5) or the sites in the optimized genomic context of miRNA targeting (context++ score < −0.3) from the website of TargetScan v7.0. The targets of let-7a and let-7b predicted by miRanda (mirSVR < −0.8) (Betel et al. 2010) and DIANA-microT (threshold score > 0.5) (Reczko et al. 2012; Paraskevopoulou et al. 2013) were also used in this study. Duplex structure and noncanonical target predictions for miRNAs and targets were made with RNAhybrid v2.1.2 (Rehmsmeier et al. 2004) without requiring seed matching. For duplex heat maps, base-paired (Watson–Crick or G:U) miRNA sites were assigned a score of 1, and unpaired sites were assigned a score of 0. k-means clustering of the resulting matrix was performed with Cluster 3.0 and visualized with Java TreeView (Eisen et al. 1998; Saldanha 2004).
Cell transfection, library construction, NGS analyses, and target validation
The kidney-derived 293FT (human, Homo sapiens), CV-1 (green monkey, Chlorocebus sabaeus), LLC-MK2 (macaque, Macaca mulatta), and NRK (rat, Rattus norvegicus) cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences. For transfection, the cells were incubated with synthesized double-stranded hsa-let-7a (sense: 5′-UGAGGUAGUAGGUUGUAUAG-UU-3′, antisense: 5′-CUAUACAACCUACUACCUCAUU-3′) or hsa-let-7b (sense: 5′-UGAGGUAGU-AGGUUGUGUGGUU-3′, antisense: 5′-CCACACAACCUACUACCUCAUU-3′) mimics or a negative control (NC, 5′-UUCUCCGAACGUGUCACGUUU-3′ and 5′-ACGUGACACGUUCGGAGAAUU-3′) small RNA duplex at the same final concentration of 50 nM for 12 or 32 h. The Lipofectamine 2000 transfection reagent (Thermo Fisher) was used according to the manufacturer's instructions. Mock-treated cells were treated with Lipofectamine 2000 in the same manner, except that no miRNA mimic or NC RNA was used. The small RNA and poly(A) tailed mRNA libraries were constructed according to previously described protocols (Wang et al. 2016). The deep sequencing was conducted on the Illumina HiSeq-2500 platform. The annotations and sequences of the reference genomes of human (GRCh38), macaque (MMUL1.0), green monkey (ChlSab1.1), and rat (Rnor6.0) were downloaded from Ensembl Genome Browser (genome.ensembl.org). The NGS reads were mapped on the reference genomes using STAR (2.4.2a) (Dobin et al. 2013). The gene expression levels were determined using HTSeq-count (0.9.0) (Anders et al. 2015), and differential gene expression was detected with DESeq2 (1.16.1).
Quantitative real-time PCR and luciferase reporter assay
qRT-PCR was performed with SYBR Green Master Mix (Thermo Fisher) in a 20-µL reaction volume and monitored on a StepOnePlus Real-Time PCR System (Thermo Fisher). Fragments containing the examined target sites or mutant target sites in the 3′ UTRs of EIF4G2, TIMM17B, SLC25A4, MAPK6, USP38, and IGDCC4 were cloned behind the Renilla luciferase gene in the psiCHECK2 vector (Promega). This vector also carries the firefly luciferase gene, which can be used for normalization to rule out variations in transfection efficiency. A total of 293FT cells in 24-well plates were cotransfected with 0.5 µg of each construct with or without 20 nM hsa-let-7a or has-let-7b mimic or negative control small RNA duplex using Lipofectamine 2000. The cells were harvested 48 h after transfection for dual luciferase measurements using the Dual-Glo Luciferase Assay System (Promega), according to the manufacturer's instructions. All transfection experiments were repeated at least three times. The primer sequences for qRT-PCR and luciferase reporter assays are shown in Supplemental Tables S7 and S8, respectively.
DATA DEPOSITION
The sequence data obtained in this study have been submitted to the NCBI Sequence Read Archive (SRA) under SRA accession number SRP073287.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Margarida Cardoso Moreira, Maria Warnefors, and Bin He for constructive comments regarding this manuscript. We thank the Biodynamic Optical Imaging Centre at Peking University for sequencing services. We thank Xingliang Qin and Qi Zhang for technical assistance. We thank the Henrik Kaessmann laboratory for making the large-scale miRNA sequencing data publicly available. This study was supported by grants from the National Natural Science Foundation of China (31571333, 91731301, 31771411, and 91431101), Ministry of Science and Technology of the People's Republic of China (2016YFA0500800), and the Peking-Tsinghua Center for Life Sciences to Jian Lu.
Author contributions: J. Luo and Y.W. conducted the experiments; Y.W., J.Y., Z.Z. and J. Lu analyzed the data; J. Lu wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062752.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. 2005. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci 102: 18017–18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet 17: 379–391. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeqa Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. 2006. Differences in vertebrate microRNA expression. Proc Natl Acad Sci 103: 14385–14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D. 2013. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci 110: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12: 846–860. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. 2006. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 38: 1375–1377. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. 2010. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet 42: 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. 2011. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 17: 1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. 2006. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38: 1452–1456. [DOI] [PubMed] [Google Scholar]
- Chen K, Maaskola J, Siegal ML, Rajewsky N. 2009. Reexamining microRNA site accessibility in Drosophila: a population genomics study. PLoS One 4: e5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. 2006. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38: 813–818. [DOI] [PubMed] [Google Scholar]
- Cohen O, Pupko T. 2011. Inference of gain and loss events from phyletic patterns using stochastic mapping and maximum parsimony—a simulation study. Genome Biol Evol 3: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. 2007. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KKH, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J; Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G, Sun J, Guo AY. 2012. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat 33: 254–263. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 1: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZL, Nicolae D, Lu HHS, Li WH. 2002. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18: 609–613. [DOI] [PubMed] [Google Scholar]
- Gu ZL, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66. [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li W-H. 2004. Duplicate genes increase gene expression diversity within and between species. Nat Genet 36: 577–579. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF; Students of Bioinformatics Computer Labs 2004 and 2005. 2006. The expansion of the metazoan microRNA repertoire. BMC Genomics 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Bartschat S, Wintsche A, Otto C; Students of the Bioinformatics Computer Lab, Stadler PF. 2012. Evolution of the let-7 microRNA family. RNA Biol 9: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform 12: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Hughes T, Ekman D, Ardawatia H, Elofsson A, Liberles DA. 2007. Evaluating dosage compensation as a cause of duplicate gene retention in Paramecium tetraurelia. Genome Biol 8: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. 2004. Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Nam JW. 2006. Genomics of microRNA. Trends Genet 22: 165–173. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Xin X, Kim TS, Cabeza EA, Ren J, Nielsen R, Wrana JL, Zhang Z. 2012. Evidence for positive selection on a number of microRNA regulatory interactions during recent human evolution. PLoS Genet 8: e1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. 2009. Lowly expressed human microRNA genes evolve rapidly. Mol Biol Evol 26: 1195–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. 2008. The evolution and functional diversification of animal microRNA genes. Cell Res 18: 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang R, Xu J, Wu C-I, Lu X. 2015. Functional conservation of both CDS- and 3′-UTR-located microRNA binding sites between species. Mol Biol Evol 32: 623–628. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. 2012. Impact of microRNA regulation on variation in human gene expression. Genome Res 22: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Fu Y, Kumar S, Shen Y, Zeng K, Xu A, Carthew R, Wu CI. 2008a. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol 25: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. 2008b. The birth and death of microRNA genes in Drosophila. Nat Genet 40: 351–355. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2003. The evolutionary demography of duplicate genes. J Struct Funct Genomics 3: 35–44. [PubMed] [Google Scholar]
- Lynch M, Force A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Y, Shen Y, Li H, Chen Y, Guo L, Zhao Y, Hungate E, Shi S, Wu CI, Tang T. 2014. New microRNAs in Drosophila—birth, death and cycles of adaptive evolution. PLoS Genet 10: e1004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Ninova M, Ronshaugen M, Griffiths-Jones S. 2013. Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res 41: 7745–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. 2013. Birth and expression evolution of mammalian microRNA genes. Genome Res 23: 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Flynt AS, Siepel A, Lai EC. 2013. The impact of age, biogenesis, and genomic clustering on Drosophila microRNA evolution. RNA 19: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Bortolamiol-Becet D, Flynt AS, Gronau I, Siepel A, Lai EC. 2014a. Adaptive evolution of testis-specific, recently evolved, clustered miRNAs in Drosophila. RNA 20: 1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Siepel A, Lai EC. 2014b. Diverse modes of evolutionary emergence and flux of conserved microRNA clusters. RNA 20: 1850–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TKH, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB. 2015. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6: 8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moszyńska A, Gebert M, Collawn JF, Bartoszewski R. 2017. SNPs in microRNA target sites and their potential role in human disease. Open Biol 7: 170019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S. 2014. Fast-evolving microRNAs are highly expressed in the early embryo of Drosophila virilis. RNA 20: 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S. 2016. MicroRNA evolution, expression, and function during short germband development in Tribolium castaneum. Genome Res 26: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkin NB. 1968. Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev 22: 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulou G, Poustka AJ. 2005. Timing and mechanism of ancient vertebrate genome duplications–the adventure of a hypothesis. Trends Genet 21: 559–567. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. 2013. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41: W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. [DOI] [PubMed] [Google Scholar]
- Patel VD, Capra JA. 2017. Ancient human miRNAs are more likely to have broad functions and disease associations than young miRNAs. BMC Genomics 18: 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. 2016. Finding protein and nucleotide similarities with FASTA. Curr Protoc Bioinformatics 53: 3.9.1–3.9.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhang J. 2014. Genomic evidence for adaptation by gene duplication. Genome Res 24: 1356–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N, Socci ND. 2004. Computational identification of microRNA targets. Dev Biol 267: 529–535. [DOI] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. 2012. Functional microRNA targets in protein coding sequences. Bioinformatics 28: 771–776. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. 2008. The let-7 family of microRNAs. Trends Cell Biol 18: 505–516. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazansky SS, Gvozdev VA, Berezikov E. 2011. Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 12: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. 2007. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci 104: 17719–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. 2007. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci 104: 3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Collins FS. 2008. MicroRNA target site polymorphisms and human disease. Trends Genet 24: 489–497. [DOI] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KKH, Chiang HR, Shkumatava A, Bartel DP. 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 38: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. 2003. Identification of Drosophila microRNA targets. PLoS Biol 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. 2005. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Sun YM, Lin KY, Chen YQ. 2013. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. 2004. Duplication and divergence: The evolution of new genes and old ideas. Annu Rev Genet 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Wang X, Xuan Z, Zhao X, Li Y, Zhang MQ. 2009. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res 19: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo J, Zhang H, Lu J. 2016. microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol Biol Evol 33: 2232–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. 2009. The deep evolution of metazoan microRNAs. Evol Dev 11: 50–68. [DOI] [PubMed] [Google Scholar]
- Wolter JM, Kotagama K, Pierre-Bez AC, Firago M, Mangone M. 2014. 3′LIFE: a functional assay to detect miRNA targets in high-throughput. Nucleic Acids Res 42: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter JM, Le HHT, Linse A, Godlove VA, Nguyen TD, Kotagama K, Lynch A, Rawls A, Mangone M. 2017. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res 27: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang R, Shen Y, Liu G, Lu X, Wu CI. 2013. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res 23: 1810–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ. 2003. Evolution by gene duplication: an update. Trends Ecol Evol 18: 292–298. [Google Scholar]
- Zhang JZ, Zhang YP, Rosenberg HF. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet 30: 411–415. [DOI] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B. 2007. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res 17: 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang Y-Q, Su B. 2008. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol 25: 1493–1502. [DOI] [PubMed] [Google Scholar]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. 2010. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat 20: 281–300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.