Abstract
A substantial fraction of the proteome is intrinsically disordered, and even well-folded proteins adopt non-native geometries during synthesis, folding, transport, and turnover. Characterization of intrinsically disordered proteins (IDPs) is challenging, in part because of a lack of accurate physical models and the difficulty of interpreting experimental results. We have developed a general method to extract the dimensions and solvent quality (self-interactions) of IDPs from a single small-angle x-ray scattering measurement. We applied this procedure to a variety of IDPs and found that even IDPs with low net charge and high hydrophobicity remain highly expanded in water, contrary to the general expectation that protein-like sequences collapse in water. Our results suggest that the unfolded state of most foldable sequences is expanded; we conjecture that this property was selected by evolution to minimize misfolding and aggregation.
In contrast to well-folded proteins, intrinsically disordered proteins (IDPs) sample a broad ensemble of rapidly interconverting conformations. An ongoing issue is whether IDPs and denatured state ensembles (DSEs) of foldable proteins undergo compaction under physiological conditions. Whereas IDPs and DSEs are highly expanded in high concentrations of denaturant (1), numerous Förster resonance energy transfer (FRET) studies and computational studies have indicated that they are collapsed in water (2–9). In contrast, many small-angle x-ray scattering (SAXS) studies have not detected statistically significant chain collapse during the earliest steps in folding (10–18). Establishing whether collapse in water is a general feature would have implications for our understanding of protein folding, protein stability, and the functional role of IDPs, as well as the improvement of simulations (13, 19).
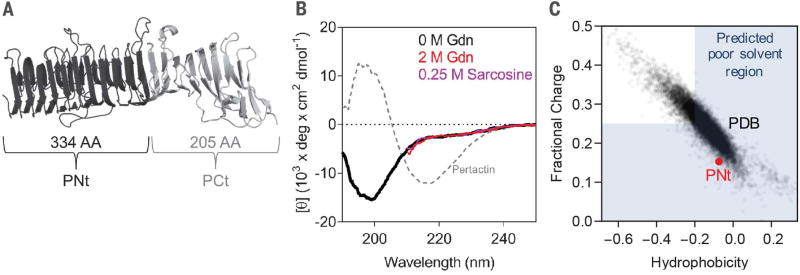
We developed a general method to extract the conformational biases of IDPs from a single SAXS measurement and applied it to DSEs having sequences typical of well-folded proteins, with low charge and high hydrophobicity. DSEs for a stably folded protein can be examined under equilibrium conditions through truncation. We applied this strategy to pertactin, a 539-residue, 16-rung parallel β helix from Bordetella pertussis (Fig. 1A). The 205-residue C-terminal truncation is independently foldable and has a far-ultraviolet circular dichroism (CD) spectrum similar to the full-length pertactin β helix (20). In contrast, the 334-residue N-terminal portion “PNt” has a CD spectrum with a near-zero θ222 value, indicative of a polypeptide lacking α-helical or β-sheet structure (Fig. 1B). Moreover, its CD spectrum changes minimally upon addition of denaturant or osmolyte [2 M guanidinium chloride (Gdn) or 0.25 M sarcosine, respectively]. The poor peak dispersion in the 15N-1H nuclear magnetic resonance (NMR) heteronuclear single-quantum coherence spectrum also is consistent with an unstructured chain (fig. S1A) (21). The disorder occurs despite PNt’s long and rather hydrophobic sequence with a low fraction of charged residues (Fig. 1C). The intrinsic disorder of PNt, along with its sequence composition, makes it an ideal model system to probe the extent of collapse expected for the DSE of a foldable protein in water.
Fig. 1. PNt is an IDP.
(A) Native pertactin consists of N-terminal (PNt) and C-terminal (PCt) domains. (B) Relative to native pertactin, isolated PNt is disordered, as shown by far-ultraviolet CD and NMR (fig. S1A). (C) PNt sequence is relatively hydrophobic with low charge, even by comparison to other proteins in the PDB (data points). In the shaded region, water is predicted to be a poor solvent according to single-molecule FRET studies (7) and simulations (26).
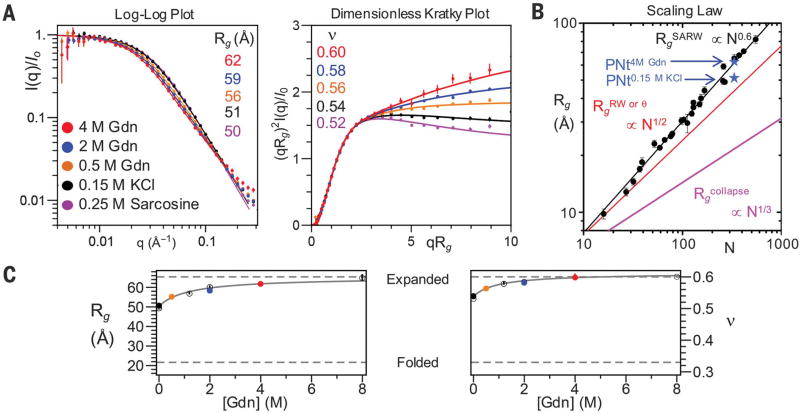
We used SAXS to probe PNt’s dimensions. In-line size exclusion chromatography eliminated oligomeric species seconds before measurement, permitting us to study the monomer in 0 to 8 M Gdn or 0.25 M sarcosine [Fig. 2, A (left) and C]. Upon shifting from aqueous buffer to 4 M Gdn, the PNt radius of gyration, Rg, increased from 51.3 ± 0.1 Å to 62.0 ± 0.4 Å (Fig. 2B), as determined using the analysis procedure presented below. The Rg value in high denaturant matched the known scaling behavior observed for other denatured proteins (1) (Fig. 2, B and C). To highlight differences at short length scales (high q), we also plotted data with the x axis scaled by Rg and the y axis multiplied by (qRg)2 (Fig. 2A, right). In this dimensionless Kratky plot, the slope at high qRg is slightly negative in water but becomes positive in high denaturant. This slope provides a quantifiable diagnostic of solvent quality (see below).
Fig. 2. Denaturant dependence of PNt SAXS.
(A) Presentations of the scattering at the solvent conditions indicated. Lines show MFF fit. (B) Rg for PNt in water and 4 M Gdn are consistent with values for chemically denatured proteins (1). Other polymer limits are shown for comparison. Most errors are smaller than data points. (C) Dependence of Rg (left) and ν (right) on Gdn [solid points are colored according to (A); open points are replicates; error bars shown are fitted error].
The degree of polypeptide chain collapse can be quantified using principles from polymer physics, where interactions and solvent quality are described in terms of the Flory exponent, ν. For polymers where intrachain interactions are less, equally, or more favorable relative to solvent-chain interactions, the solvent quality is termed good, θ, or poor, respectively. Quantitatively, ν is defined as the scaling exponent in Rg ∝ Nν, where N is the chain length and ν is greater than, equal to, or less than 0.5 for good, θ, or poor solvents, respectively. For a random walk and a self-avoiding random walk (SARW), ν = 0.5 and ~0.6, respectively. Alternatively, ν can be expressed as a function of the average intrachain pairwise distance, R|i−j| ∝ |i − j|ν.
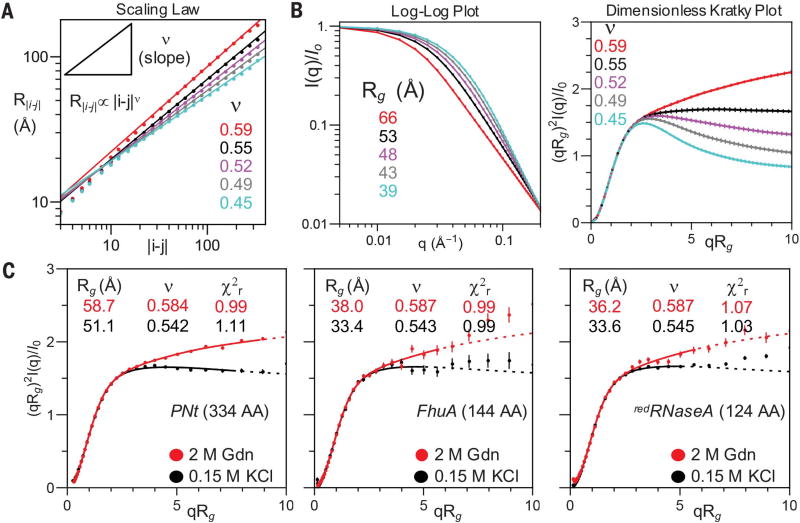
For polymers, no analytic form exists to describe scattering as a function of solvent quality ν (see supplementary text and fig. S2A). We approached this problem by developing a molecular form factor (MFF) for disordered polymers. MFFs are size-invariant functions used to describe the scattering of common shapes; for example, the MFF of an ellipsoid has a distinctive ringing pattern associated with Bessel functions (movie S1). To generate the MFF, we first ran molecular dynamics simulations using a Cβ-level polypeptide chain model in implicit solvent. Thirty different solvent conditions were obtained by varying the strength of Cβ-Cβ interactions (fig. S3A). For each resulting ensemble, the R|i−j| values were calculated as a function of sequence separation, |i − j|, and fit to the relationship R|i−j| ∝ |i − j|ν to obtain ν ranging from 0.35 to 0.6 (Fig. 3A, lines). Notably, each PNt experimental scattering pattern could be closely matched to one of the 30 simulated ensembles (Fig. 3C and fig. S3B), without resorting to the current common practice of reweighting or selection of a sub-ensemble of conformations.
Fig. 3. SAXS simulations and data fitting to MFF.
(A and B) Simulation analysis for five of the 30 simulations, with different Cβ-Cβ interaction potentials (fig. S3A). (A) ν is obtained from a fit to the slope of the dependence of the intrachain distance, R|i−j|, on sequence separation, |i − j|. (B) Presentations of simulated scattering data. Error bars are standard replicate error of five simulations. (C) Dimensionless Kratky plots for PNt, FhuA (plug domain), and redRNase A in conditions as indicated, fit to MFF. Dotted lines represent regions not fit (q > 0.15) to avoid issues related to water and denaturant scattering.
We combined the scattering profiles of the simulations using splines to generate a MFF (ν, Rg) (movie S2). To examine the robustness of this MFF to simulation parameters, we also generated five additional MFFs for models with different backbone (ϕ, ψ) Ramachandran maps (fig. S4), polypeptide chain lengths, and an alternative model where only the hydrophobic residues were attractive. Each MFF was fit to the scattering of our simulated ensembles to produce Rg and ν values that could be compared to true values obtained directly from the ensembles (Fig. 3B and table S1). For our first MFF, the fitted values of Rg and ν are within 0.3 Å and 0.002 of their true values, respectively. This accuracy is not surprising, given that the MFF was generated from the same ensemble; nonetheless, this result supports our overall procedure for generating a MFF(ν, Rg). In addition, the five other MFFs generated with the different simulation protocols produced similar values, having an average deviation of 1 Å in Rg and 0.01 in ν.
Having demonstrated the applicability of the MFF to simulated data, we next applied each of the six MFFs to the five PNt experimental data sets in Fig. 2A, where Rg and ν are unknown (table S2). Within each data set, the fits using the six MFFs produced very similar values of Rg, ν, and χ2r, with average standard deviations between the MFFs across the different conditions of 0.6 Å, 0.01, and 0.05, respectively. These small deviations indicate that the determination of Rg and ν from a scattering profile is robust to the details of the simulations used to create the MFF. Overall, these results indicate that the scattering of IDPs can be described with a general MFF, and that most of the information content of the scattering profile is contained in two parameters, Rg and ν.
To examine the generality of the PNt results as well as the robustness of the MFF for fitting different protein sizes, we performed SAXS measurements on two other disordered proteins: (i) the 144-residue “plug” domain from a TonB-dependent receptor, FhuA, that unfolds once outside of its β barrel (22) (fig. S1B), and (ii) reduced ribonuclease A (redRNase A), a 124-residue model DSE (14, 23) (Fig. 3C). The quality of the fits obtained using the MFF is similar to the fit obtained for PNt. Upon addition of 2 M Gdn to the FhuA plug, Rg increased from 33.4 ± 0.2 Å to 38.0 ± 0.3 Å while ν increased from 0.543 ± 0.009 to 0.587 ± 0.009. Similarly for redRNase A, Rg increased from 33.6 ± 0.1 Å to 36.2 ± 0.2 Å while ν increased from 0.545 ± 0.002 to 0.587 ± 0.008. The value of ν in the absence of denaturant was very similar for all three proteins (ν ~ 0.54) (Fig. 3C), as was its denaturant dependence. These findings indicate that water is a good solvent (ν > 0.5) for all three disordered proteins.
The experimentally determined Rg and ν pairs obtained for PNt at the five solvent conditions are very close to corresponding pairs obtained from the simulations (fig. S5A). This similarity further supports our modeling procedure and demonstrates that PNt is behaving near the SARW limit. To compare the PNt results with results for the shorter proteins, we calculated the prefactor Ro in the relationship Rg = RoNν as a function of ν for the three proteins. All three proteins followed the same Ro:ν trend observed in the simulations, which suggests that they all behave near the SARW limit (fig. S5B). Conversely, a deviation from this trend (e.g., smaller Rg than expected for a given value of ν) is a useful diagnostic that a protein deviates from the limit (e.g., as a result of residual structure).
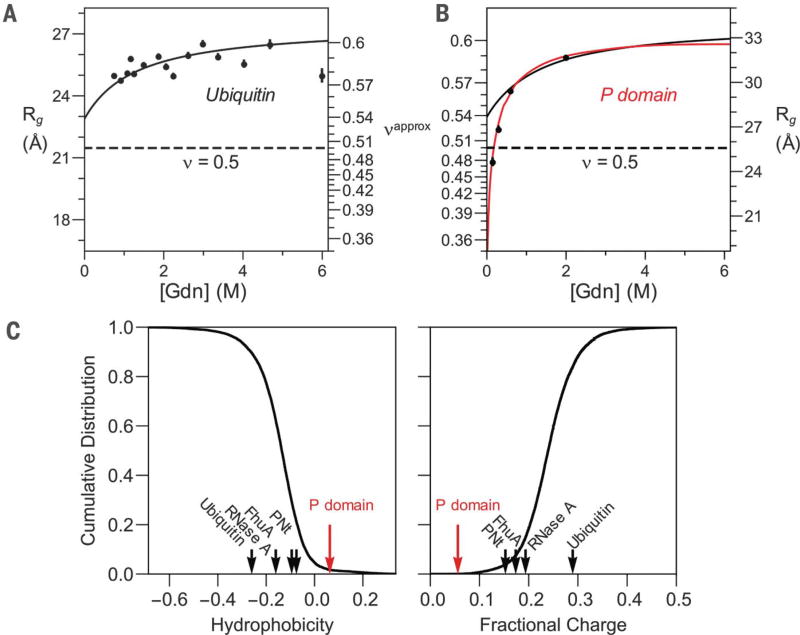
Upon transfer from high denaturant to aqueous conditions for the three IDPs, about half of the observed contraction occurred below 1 M (Fig. 2C). In previous SAXS studies of DSEs, the denaturant concentration remained above 0.5 M (10–18), likely explaining why little if any contraction was previously observed for DSEs by SAXS. For example, our prior SAXS study of the DSE for ubiquitin found no measurable contraction for denaturant jumps from 6 M down to 0.7 M Gdn (12). On the basis of our current results, we expect that the ubiquitin Rg should have contracted by 2.2 Å in this measurement—a value consistent with the noise level in the older data (Fig. 4A).
Fig. 4. SAXS data yield consistent results.
(A) Ubiquitin stopped-flow SAXS data (12). (B) P domain equilibrium SAXS data (27). For comparison, the predicted trend line based on PNt data is shown as a black curve in both (A) and (B). (C) Global hydrophobicity (left) and fractional charge (right) trends are shown as the cumulative distribution of proteins in the PDB (black curve) compared to the respective property for ubiquitin, RNase A, FhuA (plug domain), PNt, and the P domain.
Our SAXS-based identification of a relatively small amount of chain contraction upon removal of denaturant is in apparent contradiction to a variety of FRET measurements (2–8). Although improved FRET analysis procedures have narrowed the inconsistency (24), the Flory exponent of ν ~ 0.54 determined here remains well above the FRET-determined range of ν = 0.45 ± 0.05 for foldable sequences (7). Further, as measured by SAXS, the denaturant dependence of Rg is nearly saturated by 2 M Gdn (Fig. 2C), whereas FRET signals often continue to exhibit changes at higher denaturant concentrations (2, 4–8, 25). A recent study using dye-labeled polyethylene glycol, a reported SARW, observed a denaturant-dependent FRET signal change of the same magnitude as seen for unfolded proteins, but no corresponding change in the Rg was observed in small-angle scattering measurements of dye-free versions (25). Taken together, these findings suggest that the addition of fluorophores with hydrophobic character may lead to chain compaction and may contribute to FRET signal changes. This possibility, combined with the mild chain contraction observed here by SAXS, appears sufficient to resolve the discrepancy between the two techniques.
The charge and hydrophobicity of a sequence have been used to infer the extent of collapse in the absence of denaturant. Typically, sequences having less than 25% charged residues have been predicted to collapse into globules (7, 26). Such a view suggests that the majority of DSEs of foldable proteins should be collapsed in water (Fig. 1C). Yet we find that redRNase A, the FhuA plug, and PNt behave as polymers in a good solvent even under physiological conditions. It is noteworthy that RNase A, FhuA, and PNt are more hydrophobic than 40%, 70%, and 80%, respectively, of the sequences in the Protein Data Bank (PDB) (Fig. 4C). These results suggest that water will be a good solvent for the DSE of a majority of well-folded proteins.
In contrast to well-folded proteins, many IDPs never adopt a folded structure and have distinct amino acid composition. Previously, we showed that the isolated proline-rich, low-charge P domain of Pab1 contracts more in water (27) than did the three proteins studied here (ν ~ 0.4 in water, Fig. 4B). The P domain Rg is sensitive to net hydrophobicity, indicating that P domain hydrophobicity is near a threshold necessary for chain collapse. The hydrophobicity of the P domain is higher and total fractional charge is lower than 98% of proteins in the PDB (Fig. 4C), providing a reference point for the level of hydrophobicity necessary for polypeptide chain collapse.
We have shown that SAXS data from three disordered proteins of various lengths and composition can be accurately modeled using a MFF obtained from simulations near the SARW limit. Crucially, this MFF is robust to features such as the backbone conformational preferences and whether the chain is modeled as a hetero- or homopolymer. Accurate values of Rg and ν are obtained in part because the MFF is fit to the entire scattering profile, including data above qRg ~ 1. For disordered proteins, this feature is a major advantage over typical procedures that rely on data below qRg ~ 1 to 1.5, which often is challenging to acquire for unfolded proteins (see supplementary text and fig. S2 for more details). The agreement of our MFF across the scattering profile out to qRg ~ 6 to 8 suggests that for disordered proteins, the majority of the information content in SAXS profiles is contained in just two parameters, ν and Rg.
The approach presented here should be broadly useful for future studies of DSEs and IDPs. The molecular form factor MFF(ν, Rg) can be used to fit disordered IDPs without additional simulations (http://sosnick.uchicago.edu/SAXSonIDPs). Our results indicate that the DSEs of most proteins should be expanded in water and that early collapse is not an obligatory initial step in protein folding. In fact, the behavior of water as a good solvent may assist folding by enabling the polypeptide chain to avoid stable misfolded conformations. Good solvent quality may help proteins in the cell avoid non-native protein-protein associations (28) and prevent large-scale, deleterious aggregation. It is therefore possible that polypeptide chains constructed of α-amino acids were selected by evolution in part because water acts as a good solvent for this class of biomolecules.
Supplementary Material
Acknowledgments
We thank S. Chakravarthy for assistance with the SAXS measurements and J. Peng for assistance with the pertactin NMR measurements. Supported by NIH grants GM055694 (T.R.S., K.F.F.), GM097573 (P.L.C.), GM103622 and 1S10OD018090-01 (T. C. Irving), T32 EB009412 (T.R.S.), T32 GM007183 (B. Glick), and T32 GM008720 (J. Picirrilli) and by NSF grants GRF DGE-1144082 (J.A.R.) and MCB 1516959 (C. R. Matthews). Use of the Advanced Photon Source was supported by the U.S. Department of Energy under contract DE-AC02-06CH11357. All the observational data analyzed, simulation code, and other relevant files used in this paper are available from https://github.com/sosnicklab/SAXSonIDPs.
Footnotes
REFERENCES AND NOTES
- 1.Kohn JE, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziv G, Thirumalai D, Haran G. Phys. Chem. Chem. Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dasgupta A, Udgaonkar JB. J. Mol. Biol. 2010;403:430–445. doi: 10.1016/j.jmb.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 5.Haran G. Curr. Opin. Struct. Biol. 2012;22:14–20. doi: 10.1016/j.sbi.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgia A, et al. J. Am. Chem. Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann H, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voelz VA, et al. J. Am. Chem. Soc. 2012;134:12565–12577. doi: 10.1021/ja302528z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy G, Thirumalai D. J. Phys. Chem. B. 2017;121:995–1009. doi: 10.1021/acs.jpcb.6b13100. [DOI] [PubMed] [Google Scholar]
- 10.Plaxco KW, Millett IS, Segel DJ, Doniach S, Baker D. Nat. Struct. Biol. 1999;6:554–556. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
- 11.Yoo TY, et al. J. Mol. Biol. 2012;418:226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob J, Krantz B, Dothager RS, Thiyagarajan P, Sosnick TR. J. Mol. Biol. 2004;338:369–382. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 13.Skinner JJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15975–15980. doi: 10.1073/pnas.1404213111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Trewhella J, Goldenberg DP. J. Mol. Biol. 2008;377:1576–1592. doi: 10.1016/j.jmb.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathuria SV, et al. J. Mol. Biol. 2014;426:1980–1994. doi: 10.1016/j.jmb.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob J, Dothager RS, Thiyagarajan P, Sosnick TR. J. Mol. Biol. 2007;367:609–615. doi: 10.1016/j.jmb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Konuma T, et al. J. Mol. Biol. 2011;405:1284–1294. doi: 10.1016/j.jmb.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 18.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. J. Mol. Biol. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Piana S, Klepeis JL, Shaw DE. Curr. Opin. Struct. Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Junker M, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renn JP, Junker M, Besingi RN, Braselmann E, Clark PL. Chem. Biol. 2012;19:287–296. doi: 10.1016/j.chembiol.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udho E, Jakes KS, Finkelstein A. Biochemistry. 2012;51:6753–6759. doi: 10.1021/bi300493u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi PX, Sosnick TR, Englander SW. Nat. Struct. Biol. 1998;5:882–884. doi: 10.1038/2321. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Gomes GN, Gradinaru CC, Chan HS. J. Phys. Chem. B. 2015;119:15191–15202. doi: 10.1021/acs.jpcb.5b09133. [DOI] [PubMed] [Google Scholar]
- 25.Watkins HM, et al. Proc. Natl. Acad. Sci. U.S.A. 2015;112:6631–6636. doi: 10.1073/pnas.1418673112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das RK, Ruff KM, Pappu RV. Curr. Opin. Struct. Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riback JA, et al. Cell. 2017;168:1028–1040. e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tompa P, Rose GD. Protein Sci. 2011;20:2074–2079. doi: 10.1002/pro.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.