Abstract
Unbiased allele transmission into progeny is a fundamental genetic concept canonized as Mendel’s Law of Segregation. Not all alleles, however, abide by the law. Killer meiotic drivers are ultra-selfish DNA sequences that are transmitted into more than half (sometimes all) of the meiotic products generated by a heterozygote. As their name implies, these loci gain a transmission advantage in heterozygotes by destroying otherwise viable meiotic products that do not inherit the driver. We review and classify killer meiotic drive genes across a wide spectrum of eukaryotes. We discuss how analyses of these ultra-selfish genes can lead to greater insight into the mechanisms of gametogenesis and the causes of infertility.
Keywords: Meiosis, meiotic drive, selfish genes, killer meiotic drivers, segregation distortion
Meiotic Drivers Break Mendel’s Law
The visionary monk Gregor Mendel was the first to describe the rules governing the transmission of alleles into offspring [1]. Two of Mendel’s ‘Laws’ (Dominance and Independent Assortment) have long since been downgraded and are now better described as ‘context dependent guidelines.’ His Law of Segregation, however, has endured. This law stipulates that each of the two alleles carried by a heterozygote will be transmitted equally into meiotic products (e.g. gametes, like sperm). Unbiased segregation of homologous chromosomes during meiosis is now known to underlie Mendelian (equal) allele transmission.
Meiosis and gametogenesis are, however, vulnerable to exploitation by meiotic drivers. These selfish loci can bias allele transmission in their favor so that they are found in more than half of the functional meiotic products generated by a heterozygote [2]. The transmission advantage enjoyed by these cheaters can allow them to spread in a population, even if they are deleterious to fitness, which they often are [3–6]. Meiotic drive is thus a powerful evolutionary force that has likely shaped the structure of eukaryotic chromosomes and the genes that act during meiosis and gametogenesis [7–9].
There are two general classes of meiotic drivers [9]. Class one drivers, sometimes called ‘true meiotic drivers’ (see glossary), act during the meiotic chromosome divisions. For example, loci called ‘knobs’ in maize can promote their preferential segregation during meiosis to the position that will become the egg [10]. The second class of drivers are the killer meiotic drivers that constitute the focus of this review. These drivers are sometimes called ‘ultra-selfish’ genes because of their exceptional methods [11]. These ultra-selfish alleles do not achieve a transmission bias by increasing the absolute number of meiotic products carrying them. Instead, killers gain an advantage by sabotaging otherwise viable meiotic products that do not inherit the selfish allele. In other words, they can spread through a population by actively destroying competitors. Killers often act on the products of male (symmetric) meiosis in which all meiotic products are viable [9]. However, there are also killer meiotic drivers that act on gametes produced by female (asymmetric) meiosis in which only one meiotic product becomes a gamete [12]. Some killers even destroy both male and female gametes that do not inherit them [13, 14].
Several genes used in killer meiotic drive systems have been identified. Here, we review these exciting discoveries and examine their implications. We omitted discussion of meiotic drivers that either act during the meiotic divisions or have an unknown mode of altering allele transmission frequencies [15–21].
Cryptic Criminals: How Common are Killers?
The theoretical potential of killer meiotic drivers to shape the evolution of populations is clear from years of analyses [8, 9]. The actual impact of these ultra-selfish elements on biology, however, depends on their prevalence. If killers are genetic oddities and seen in relatively few populations, their impact would be minimal. Conversely, if these selfish alleles are lurking (or previously existed) in the genomes of many populations, they would have a large impact. Indeed, it has been argued that these parasites are likely widespread and that they may explain important aspects of eukaryote biology and evolution [7, 22]. The prevalence of currently active killer meiotic drivers is not clear, even in intensely studied organisms. This ignorance of extant killer frequencies is due to the significant challenges of detecting meiotic drive (Figure 1).
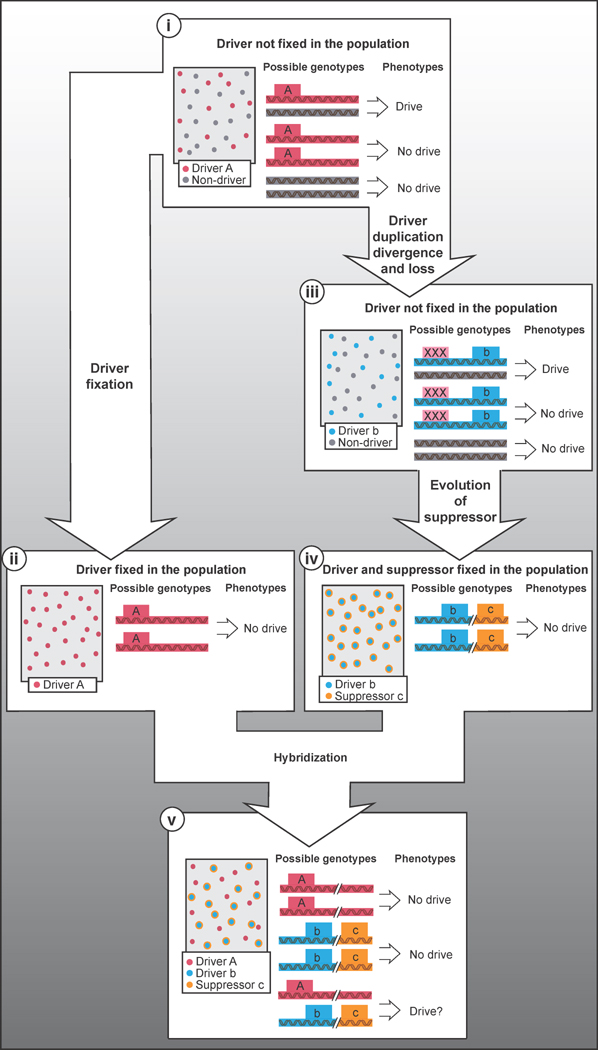
Figure 1. Hypothetical evolution of meiotic drivers.
i) A meiotic drive locus (A) has the ability to generate drive when heterozygous. Homozygotes fail to exhibit drive. ii) If a drive allele becomes fixed in a population, drive will not be observed because all individuals will be homozygous. iii) Another possible path is that the driver (A) could duplicate, creating a drive locus (b) that is free to diverge. The original drive locus could accumulate mutations and be lost in the population. iv) As drivers are generally deleterious for fitness, unlinked suppressors (c) are likely to evolve. v) Over time, populations (represented in ii and iv) evolve distinct landscapes of meiotic drivers and suppressors. If these two populations were to merge (v), the phenotypes of cryptic drivers could emerge in hybrid individuals. In this way, meiotic drivers can contribute to hybrid infertility and speciation.
Perhaps the biggest contributor inhibiting the discovery of meiotic drive loci is that geneticists often follow Mendel’s example and choose pure-breeding (i.e. homozygous) highly inter-fertile organisms for their experiments [1]. This choice of experimental systems has facilitated innumerable discoveries, but the choice has also given a biased view of how sexual reproduction happens. In the wild, extreme inbreeding is generally not a winning evolutionary strategy and gametogenesis often occurs in heterozygotes. In addition, heterozygotes generated by crossing natural isolates are not always highly fertile [23–25]. These factors are important to consider as there can be differences between gametogenesis in homozygotes versus heterozygotes [26]. For example, meiotic drivers can only be observed in heterozygotes, where a given drive allele has a competitor. Furthermore, meiotic drivers can cause infertility. Eschewing individuals with low fertility from genetic analyses could preclude discovery of meiotic drivers, not to mention other factors that cause infertility in natural populations.
An additional challenge to detecting drive alleles is that they are predicted to be transient and evolutionarily labile [9, 27]. The transmission advantage enjoyed by drivers can allow them to become fixed in a population. After fixation, all individuals will be homozygous for the driver and exhibit no drive phenotype. Alternatively, a potentially active driver may be segregating within a population, but be phenotypically invisible due to the actions of suppressors. Both fixed and successfully suppressed drive alleles can accumulate inactivating mutations and lose the ability to drive. Because non-functional drivers receive no selfish advantages and are not maintained by selection, they are likely to become extinct. Due to these factors, meiotic drivers are often discovered by chance in hybrids generated by crossing genetically diverse individuals or even (largely) reproductively isolated species [8, 24, 26, 28–30]. In these hybrids, the meiotic drive alleles are more likely to be heterozygous and can become uncoupled from suppressors. A hypothetical evolutionary scenario of a drive system illustrating some of these ideas is presented in Figure 1.
In the past, discovery of allele transmission biases associated with drive depended on genetic linkage between the drive locus and a heterozygous phenotypic marker. Many identified meiotic drivers, for example, are on sex chromosomes and lead to biased sex ratios (Box 1). Similarly, the driving t-haplotype of mouse was discovered because the haplotype carries an allele that causes taillessness [8]. In recent years, however, technological advances have bypassed this requirement and biased allele transmission can be assayed directly via whole genome sequencing of gametes or cross progeny [17, 31–33].
Box 1. Killer meiotic drivers on sex chromosomes.
Various drive systems have been identified on sex chromosomes due to their easily detectable phenotype: a sex-ratio bias in the offspring [57]. Drive of one of the sex chromosomes leads to an unequal transmission of the X or Y chromosome, resulting in a female or male-biased progeny, respectively. For example, in Drosophila simulans, the Distorter on the X (Dox) driver is thought to disrupt the maturation of Y-bearing sperm. The first apparent defect caused by Dox occurs after the meiotic divisions: however, the mechanism is unknown and it is possible Dox acts during meiosis [58, 59].
The Slx and and Sly multigene families in mouse represent competing killer meiotic drive systems on the X and Y chromosomes, respectively. Slx promotes expression of X and Y-linked genes in spermatids, whereas Sly represses expression of X and Y-linked genes. An imbalance of Slx and Sly copy number in hybrids or knockdown of either gene family causes reduced fertility and sex-ratio bias. Slx deficiency leads to male-biased litters while Sly deficiency causes female-biased progeny. The mechanism(s) used by these genes are unclear [60–62]. Analogous amplified gene families, VCX and VCY, are present on the human X and Y chromosomes and may also be involved in meiotic drive. VCX and VCY are found only in simian primates and are expressed only in testes [62, 63].
After observing allele transmission biases, it can be difficult to distinguish meiotic drive from other causes of biased allele transmission. For example, if an allele contributes to a lethal genetic incompatibility, it will be underrepresented in the progeny of a heterozygote, but that does not make the alternative allele a meiotic driver. Careful genetic analyses must be used to demonstrate that drivers cause viable genotypes (like a parental genotype) to be underrepresented in the functional meiotic products.
A final barrier to detecting and gauging the prevalence of killer meiotic drivers in a population is their diversity. There are apparently many ways to build a killer. Of all genes known to be required for killer meiotic drive in diverse organisms, none are homologous. The filamentous fungus Podospora anserina, for example, contains at least two totally distinct types of killer meiotic drive systems [34, 35]. In addition, the genes comprising a killer drive system can be rapidly evolving [32, 36]. These factors limit the usefulness of simple bioinformatic searches (e.g. BLAST) for querying genomes for the presence of meiotic drivers. Thorough genetic analyses and a degree of luck are currently required to unambiguously detect drivers in any system.
How to Kill the Competition?
We currently know very little about the molecular mechanisms used by killer meiotic drivers. The few identified genes suggest the mechanisms are remarkably diverse. However, some general themes have emerged. To execute killing, a locus must be able to do two things. First, the driving allele must confer the ability to distinguish meiotic products that inherit the allele from those that do not. Second, the driving allele must confer the ability to kill meiotic products. There are alternative approaches to accomplishing these requirements and killing meiotic drivers can be broken down into two broad types (Figure 2, Key Figure).
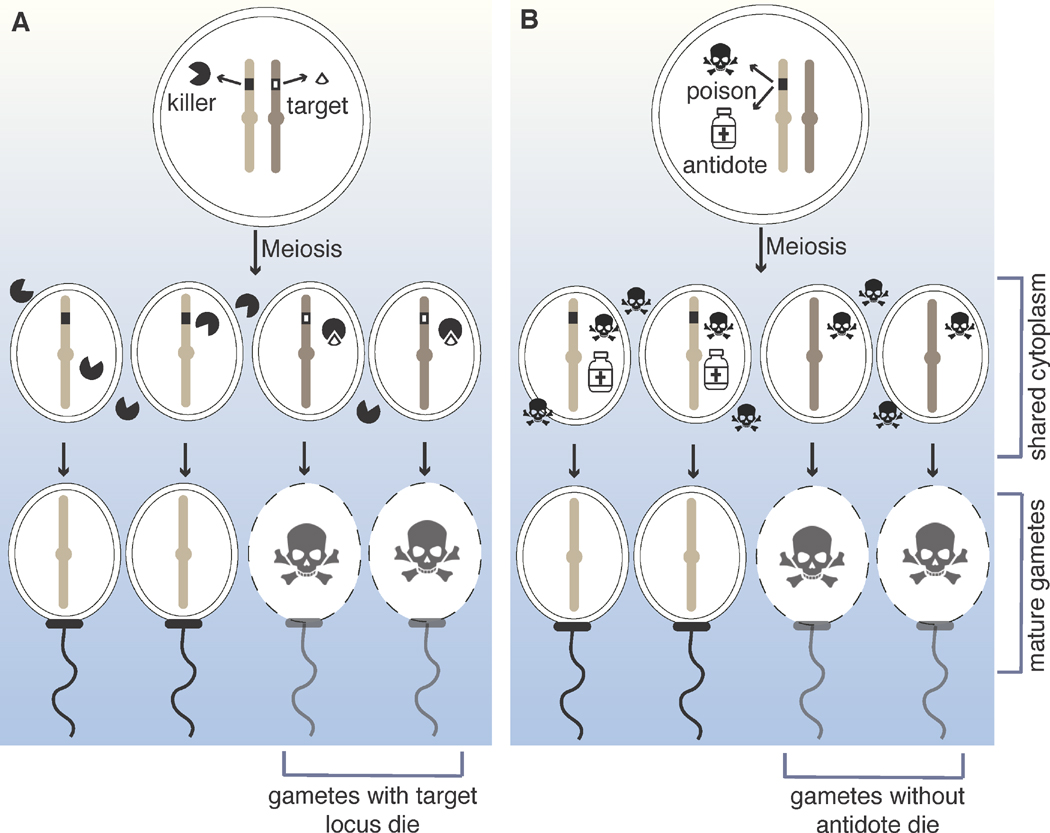
Figure 2 (Key Figure). General mechanisms of meiotic drivers.
(A) Killer-target meiotic drivers create a killer element that acts only on the meiotic products that inherit the target. The target can be a DNA locus, or a gene product encoded by the locus. (B) Poison-antidote meiotic drivers create a transacting poison that acts indiscriminately on all meiotic products. Only meiotic products that inherit the meiotic drive locus, and thus the antidote, live. A clear way to distinguish the two types is to consider the phenotypes of mutants in which the gamete (or spore)-specific component is absent. In killer-target systems, individuals containing at least one copy of the killer allele, but no target, exhibit no killing of meiotic products. In contrast, in poison-antidote systems, all the meiotic products are destroyed in individuals that encode the poison but lack the antidote.
Killer-Target Drive Systems
The first type of killer meiotic drive system is the killer-target drivers. These loci encode something that can be considered a ‘killer element.’ The killer element must be trans-acting so that it can interact with all the meiotic products generated by an individual heterozygous for the meiotic drive locus. The killer element could gain access to all meiotic products via expression in the cell prior to meiosis, or via the cytoplasmic connections commonly observed between developing meiotic products. The killer only becomes deadly in meiotic products that contain a second ‘target’ element. The target is most commonly a protein but can also be a genomic locus. Importantly, localization of the target must be restricted to the meiotic products that do not inherit the drive locus. This ensures that only meiotic products that do not carry the drive locus are destroyed. Finally, it is important that the loci encoding the killer element and the target are tightly linked to each other, but on different haplotypes. This arrangement protects the drive locus from self-destruction (Figure 2A). The three killer-target systems with known components (described below) utilize a remarkably wide range of molecular approaches.
Drosophila melanogaster SD
One of the first and most intensively studied meiotic drive systems is Segregation Distorter (SD), a killer-target system found in the fruit fly Drosophila melanogaster. The driving SD allele can be transmitted to >90% of the progeny of SD/SD+ heterozygous males. The killer of the SD system is the Sd protein, a truncated duplicate copy of the RanGAP protein involved in nuclear transport [37]. Sd kills developing spermatids that inherit sensitive alleles of the linked target DNA sequence, a large satellite repeat known as Responder (Rsp) [38]. The SD system has been extensively reviewed elsewhere, but how Sd causes death of Rsp-bearing spermatids is still unknown [6].
Oryza sativa Sa
The Sa drive locus of cultivated rice (Oryza sativa) also has two key loci and acts via a killer-target mechanism, but is quite different from SD. The driving Sa allele found in the indica subspecies encodes SaM+, a SUMO E3 ligase-like protein, and SaF+, an F-box protein. The Sa locus in the japonica subspecies encodes alternate alleles of the genes denoted SaM− and SaF−. The SaM− allele is a truncated (40 amino acids shorter) version of SaM+. SaF− differs from SaF+ by a single amino acid substitution. In indica/japonica hybrids, the SaM+ and SaF+ proteins together form a killer that targets pollen that inherit the SaM− locus. This results in transmission of the indica allele to almost all (99.5%) of the viable pollen. The mechanism of Sa drive is unknown, although both SaF+ and SaF− physically interact with the SaM− protein, but neither F protein showed an interaction with SaM+ [39].
Podospora anserina het-s
The het-s allele in P. anserina represents a killer-target meiotic driver that has only one key gene [35]. The molecular mechanism of this driver is the best understood of any due to experiments characterizing how het-s contributes to cell death in an asexual fusion process known as post-fusion heterokaryon incompatibility [40, 41]. The het-s allele encodes the HET-s protein, which can form a prion. The alternate allele, het-S, encodes the HET-S protein, which is not a prion. Interestingly, the prions themselves are not toxic to the cells. Rather, maternally inherited cytoplasmic HET-s prions (the killers) are transmitted to developing meiotic products (spores). Spores that inherit the het-S allele also contain the HET-S protein (the target) [35]. The HET-s prions convert HET-S into an oligomeric integral membrane protein that perforates the cell membrane, thus killing the het-S cell [40]. Because it depends on maternally inherited prions, the transmission advantage enjoyed by the het-s allele in heterozygotes is variable; drive only occurs in ~25% of meiosis at the optimal temperature [35].
Poison-Antidote Drive Systems
The second type of killer meiotic driver is the poison-antidote drive systems. These drivers produce two factors: a poison and an antidote. Like the killers described above, the poisons of these drive systems are trans-acting to spread to all the developing meiotic products. Unlike the killers described above, these poisons kill indiscriminately. Not all meiotic products are destroyed, however, because an antidote selectively rescues those that inherit the drive locus. How specificity is ensured is generally unclear, but may be due to transcription of the antidote within the developing meiotic product itself. Like the killer-target systems, it is essential that the sequences encoding the poison and antidote of a driver are tightly linked to each other. In this case, however, the two components must be linked on the same haplotype (Figure 2B).
Mus musculus t-haplotype
The mouse t-haplotype carries the most complex characterized poison-antidote killer meiotic drive system. The locus encompasses roughly 1% of the mouse genome, yet it is inherited as a unit due to four inversions that suppress recombination. t-haplotype heterozygotes transmit the driving locus to almost all (99%) of the functional sperm [42]. Multiple proteins encoded in the t-haplotype act as poisons to additively disrupt sperm flagellar function. All three of the identified poison proteins affect Rho G-proteins. The first identified poison is a hypermorphic allele of Tagap1, which encodes a G-protein inhibitor. The Tagap1 signaling pathway is hypothesized to activate an inhibitor of the Sperm Motility Kinase (SMOK1) [43]. The second identified poison, encoded by a hypermorphic allele of Fgd2, is also hypothesized to cause over activation of SMOK1. Fgd2 encodes a G-protein activator that is hypothesized to act via a distinct signaling pathway from Tagap1 [44]. The final poison is encoded by a hypomorphic allele of Nme3, which phosphorylates GDP to GTP. It is unclear how NME3 fits into the putative TAGAP1 and FGD2 signaling pathways [45]. The antidote that rescues flagellar function in sperm that inherit the t-haplotype is a dominant negative version of SMOK1. Smok1 is expressed in sperm and is gamete-specific, because neither the mRNA nor the protein is shared with other sperm, despite their cytoplasmic connections [46].
Oryza sativa S5
The S5 locus found in Oryza sativa represents a less complex poison-antidote meiotic drive system. In indica/japonica hybrids, female gametes (megaspores) that inherit the japonica S5 haplotype are preferentially destroyed. The indica allele is thus transmitted to ~90% of the viable megaspores. The S5 drive system requires three genes: ORF3, ORF4 and ORF5. Isolates of the indica subspecies have the genotype ORF3+, ORF4−, ORF5+, whereas isolates of the japonica subspecies are ORF3−, ORF4+, ORF5−. ORF3 encodes a protein that shares homology with Hsp70 and the ORF3− allele found in japonica has a 13 bp deletion near the C-terminus. ORF4 has no homology to characterized proteins, but it contains a predicted transmembrane domain. The ORF4− allele found in indica has an 11 bp deletion in the middle of the protein that results in the loss of the transmembrane domain [12]. ORF5 encodes an aspartic protease and the indica and japonica alleles differ by only two amino acids [47].
In indica/japonica hybrids, the products of the ORF4+ and ORF5+ alleles combine to form a poison. Megaspores that inherit ORF3+ (the antidote) are rescued, whereas those that inherit ORF3− are destroyed. The ancestral genotype (ORF3+ ORF4+ and ORF5+) of indica and japonica was thus likely an intact meiotic driver that has decayed within the two subspecies [48]. It is hypothesized that ORF5+ might interact with ORF4+ protein on the plasma membrane of developing megaspores. This interaction is predicted to initiate a signal that leads to endoplasmic reticulum (ER) stress and ultimately programmed cell death. The ORF3+ protein is hypothesized to prevent gamete death by suppressing ER stress [12].
Neurospora intermedia Sk-2 and Sk-3
The Sk spore killers were the first recognized meiotic drive alleles in fungi. Sk-2 and Sk-3 were both isolated from the filamentous fungus Neurospora intermedia. These loci act in heterozygotes and can kill almost all of the spores (>96%) that do not inherit them when the loci are introgressed into N. crassa [49]. Distinct alleles of the rsk (resistant to spore killer) gene serve as antidotes for both drivers [50]. The Sk-2 rsk allele does not protect against the Sk-3 poison and vice versa [49]. rsk encodes a 486 amino acid protein with no recognized motifs or homology to proteins outside of fungi closely related to Neurospora. It is unclear how rsk prevents spore destruction and the poison(s) used by the Sk drivers are unknown [51].
Podospora anserina Spok1 and Spok2
The Spok1 and Spok2 poison-antidote meiotic drivers recently mapped in P. anserina greatly expanded the field’s view of the requirements for meiotic drive and how these selfish elements can spread through populations. The Spok genes were the first examples of single genes constituting autonomous drive systems. A single Spok gene can kill nearly all the spores that do not inherit the gene from a heterozygous diploid [34]. Separation of function alleles revealed that these genes each encode both poison and antidote functions. Interestingly, the antidote of Spok1 protects against killing by Spok2, but not vice versa. It is not clear, however, how the poison and antidote functions are partitioned within the gene or how the spore killing is executed [34].
An additional remarkable feature of the Spok genes is their evolutionary mobility between and within genomes. Spok1 and Spok2 are members of a multigene family found in up to 11 copies in a wide range of filamentous fungi. Phylogenies suggest that functionally intact Spok genes may have moved into some lineages via horizontal gene transfer. Consistent with this idea, Spok1 driven by its endogenous promotor can cause drive in Sordaria macrospora, even though the fungi are roughly as diverged as mammals are from fishes. Similarly, a Spok gene cloned from Nectria haematococca, an even more distantly related fungus, was also able to cause spore death in P. anserina. The Spok genes do not share apparent homology to genes outside of fungi [34].
Schizosaccharomyces pombe wtf genes
The fission yeast wtf poison-antidote meiotic drivers expand upon themes discovered in the Spok genes. The wtf genes are also autonomous one-gene drivers and they constitute a multigene family with as many as 32 genes (including likely pseudogenes) per isolate. The compact structure of the Spok and wtf drivers has likely facilitated their spread to multiple copies within a genome [26, 32, 36].
Driving wtf genes produce both a poison and an antidote using different transcriptional and translational start sites. The poison is encoded on a transcript with five exons, whereas the antidote is encoded on a longer transcript that includes an upstream exon in addition to the same five that encode the poison protein. The poison protein is expressed prior to spore individualization and the poison is found within all four developing spores. The antidote, on the other hand, is produced after spore individualization and remains within the spores that encode the wtf allele, biasing wtf transmission into > 70% of the viable spores [32, 36]. Like the Spok genes, the wtf genes do not share apparent homology to genes found outside of fungi. It is also unknown how a given Wtf poison protein kills or how the antidote neutralizes the poison [32, 34, 36].
Concluding Remarks and Future Perspectives
The meiotic drive field is 60 years old, but is still quite young in terms of molecular understanding [2]. The handful of cloned genes described in this review constitute the bulk of our molecular understanding about how killer meiotic drivers work. This is, however, an exciting time to study meiotic drive. The field is poised to greatly expand in the coming years as next generation sequencing facilitates detection of meiotic drivers [17, 19, 31, 32]. The widening implementation of CRISPR genome editing technology will also likely hasten identification of the responsible genes, even in model systems that were previously genetically intractable. These advances are necessary to address the many questions that remain unanswered (See Outstanding Questions).
We currently know the molecular mechanisms of too few drivers from too few organisms to determine if non-homologous killer meiotic drivers have converged on any common mechanistic themes. It is possible there are conserved aspects of gametogenesis that have been repeatedly targeted by these killers. If so, studying killer meiotic drivers could represent a useful approach to understanding the biology of those targeted features. There are, however, a plethora of ways to destroy a cell and killer meiotic drivers may exploit numerous diverse paths. If so, then studying killer meiotic drivers could provide an approach to learn about many critical areas of gametogenesis. For example, studies of the mechanism of the t-haplotype have provided novel insights into gamete-specific gene products and sperm motility in mammals [45, 46].
We labeled killer meiotic drivers as genetic villains in this review, because they are expected to generally decrease the fitness of populations that carry them [3, 4]. Indeed, these selfish loci may be major drivers of reproductive isolation in natural populations [8, 26, 35, 52, 53]. Killer meiotic drivers are also widespread in important crop species, including rice and wheat, where they create formidable barriers to crop improvement via crosses with divergent populations [13, 24].
Finally, it may be possible to use molecular biology to exploit meiotic drivers for a beneficial purpose. Killer meiotic drivers could be used to generate synthetic gene drives that will spread in natural populations [9, 54–56]. For example, the single-gene drivers could be modified for expression in Anopheles mosquitoes. These killers could then be used to spread linked variants that decrease the insect’s ability to transmit disease or to eliminate a population entirely. In this way, we can manipulate the ‘genetic villains’ to improve the lives of humans.
Killer meiotic drivers are ultra-selfish alleles that bias their transmission by destroying meiotic products that do not inherit them.
Meiotic drivers have been found in a wide range of eukaryotes, but are notoriously hard to detect. Killer meiotic drivers contribute to reproductive isolation in natural populations.
There are two broadly defined mechanisms killer meiotic drivers commonly use: poison-antidote and killer-target.
Cloned killer meiotic drive systems are beginning to provide mechanistic insight into how these selfish loci bias their transmission.
Killer meiotic drivers may guide construction of synthetic gene drives which could be used to modify or eliminate pest populations.
Are meiotic drivers a genetic oddity or is it just hard to detect them? Do meiotic drivers generally cause a strong transmission bias, or do drivers with weaker phenotypes tend to go undetected?
Will new genetic tools facilitate discovery of novel drivers? Are there killer meiotic drivers present in humans?
What aspects of gametogenesis do killer meiotic drivers exploit? What are the molecular mechanisms by which uncharacterized drive systems act? Are there mechanistic themes commonly used by killer meiotic drivers?
Are there evolutionary signatures that tend to be shared by killer meiotic drivers? If so, could these features be used to guide discovery of new drive systems?
Acknowledgments
We would like to thank members of the Zanders lab and three anonymous reviewers for feedback that improved the manuscript and Mark Miller for assistance with the figures. This work was supported by the Stowers Institute for Medical Research and the National Institutes of Health under Award Number R00GM114436. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Glossary
- True meiotic driver
a locus that biases allele transmission via altered chromosome segregation during meiosis.
- Killer meiotic driver
a locus that biases allele transmission by disabling meiotic products that do not inherit the drive locus.
- RanGAP
activating protein for the Ran GTPase involved in transport of molecules between the cytoplasm and nucleus.
- Satellite
large, repetitive arrays of non-coding DNA sequences.
- SUMO E3 ligase
enzyme that can modify other proteins by attaching SUMO (small ubiquitin-like modifier) proteins.
- F-box protein
contain an F-box domain that mediates protein-protein interactions.
- Post-fusion heterokaryon incompatibility
a process in which filamentous fungi with incompatible genotypes cannot form viable heterokaryons after asexual fusion.
- Prion
aggregating conformation of a protein that can induce other proteins with the same amino acid sequence to refold into the prion conformation.
- Rho G-proteins
GTPase proteins involved in cellular signaling.
- Hsp70
chaperone protein originally identified in fruit flies that helps to fold nascent proteins and to refold misfolded proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abbott S, Fairbanks DJ. Experiments on Plant Hybrids by Gregor Mendel. Genetics. 2016;204(2):407–422. doi: 10.1534/genetics.116.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandler L, Novitski E. Meiotic Drive as an Evolutionary Force. The American Naturalist. 1957;91(857):105–110. [Google Scholar]
- 3.Crow JF. Why is Mendelian segregation so exact? Bioessays. 1991;13(6):305–12. doi: 10.1002/bies.950130609. [DOI] [PubMed] [Google Scholar]
- 4.Price TA, Wedell N. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica. 2008;134(1):99–111. doi: 10.1007/s10709-008-9253-y. [DOI] [PubMed] [Google Scholar]
- 5.Sutter A, Lindholm AK. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc Biol Sci. 2015;282(1811) doi: 10.1098/rspb.2015.0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larracuente AM, Presgraves DC. The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics. 2012;192(1):33–53. doi: 10.1534/genetics.112.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice WR. Nothing in Genetics Makes Sense Except in Light of Genomic Conflict. The Annual Review of Ecology, Evolution, and Systematics. 2013;44:217–237. [Google Scholar]
- 8.Burt A, Trivers R. Genes in conflict: the biology of selfish genetic elements. Cambridge, Mass: Belknap Press of Harvard University Press; 2006. p. viii.p. 602. 8 p. of plates. [Google Scholar]
- 9.Lindholm AK, et al. The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol Evol. 2016;31(4):315–26. doi: 10.1016/j.tree.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Rhoades MM. Preferential Segregation in Maize. Genetics. 1942;27(4):395–407. doi: 10.1093/genetics/27.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow JF. The ultraselfish gene. Genetics. 1988;118(3):389–91. doi: 10.1093/genetics/118.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science. 2012;337(6100):1336–40. doi: 10.1126/science.1223702. [DOI] [PubMed] [Google Scholar]
- 13.Endo TR. Gametocidal chromosomes and their induction of chromosome mutations in wheat. The Japanese Journal of Genetics. 1990;65(3):135–152. [Google Scholar]
- 14.Rick CM. Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics. 1966;53(1):85–96. doi: 10.1093/genetics/53.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323(5912):376–9. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helleu Q, et al. Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc Natl Acad Sci U S A. 2016;113(15):4110–5. doi: 10.1073/pnas.1519332113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottolini CS, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47(7):727–35. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322(5907):1559–62. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 19.Didion JP, et al. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 2015;11(2):e1004850. doi: 10.1371/journal.pgen.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chmatal L, et al. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24(19):2295–300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akera T, et al. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358(6363):668–672. doi: 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2(8):597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 23.Jeffares DC, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8:14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang Y, Liu YG, Zhang Q. Hybrid sterility in plant: stories from rice. Curr Opin Plant Biol. 2010;13(2):186–92. doi: 10.1016/j.pbi.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Prakash S. Origin of reproductive isolation in the absence of apparent genic differentiation in a geographic isolate of Drosophila pseudoobscura. Genetics. 1972;72(1):143–55. doi: 10.1093/genetics/72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanders SE, et al. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife. 2014;3:e02630. doi: 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin RN, Jr, Malik HS. Genetic conflicts: the usual suspects and beyond. J Exp Biol. 2017;220(Pt 1):6–17. doi: 10.1242/jeb.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orr HA, Irving S. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics. 2005;169(2):671–82. doi: 10.1534/genetics.104.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98(23):13183–8. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dermitzakis ET, et al. Non-Mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics. 2000;154(2):687–94. doi: 10.1093/genetics/154.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei KH, et al. A Pooled Sequencing Approach Identifies a Candidate Meiotic Driver in Drosophila. Genetics. 2017;206(1):451–465. doi: 10.1534/genetics.116.197335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu W, et al. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. Elife. 2017:6. doi: 10.7554/eLife.26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbett-Detig R, et al. Direct Gamete Sequencing Reveals No Evidence for Segregation Distortion in House Mouse Hybrids. PLoS One. 2015;10(6):e0131933. doi: 10.1371/journal.pone.0131933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grognet P, et al. Genes that bias Mendelian segregation. PLoS Genet. 2014;10(5):e1004387. doi: 10.1371/journal.pgen.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalstra HJ, et al. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A. 2003;100(11):6616–21. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuckolls NL, et al. wtf genes are prolific dual poison-antidote meiotic drivers. Elife. 2017:6. doi: 10.7554/eLife.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrill C, et al. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283(5408):1742–5. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 38.Wu CI, et al. Association between a satellite DNA sequence and the Responder of Segregation Distorter in D. melanogaster. Cell. 1988;54(2):179–89. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- 39.Long Y, et al. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci U S A. 2008;105(48):18871–6. doi: 10.1073/pnas.0810108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seuring C, et al. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol. 2012;10(12):e1001451. doi: 10.1371/journal.pbio.1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riek R, Saupe SJ. The HET-S/s Prion Motif in the Control of Programmed Cell Death. Cold Spring Harb Perspect Biol. 2016;8(9) doi: 10.1101/cshperspect.a023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schimenti J. Segregation distortion of mouse t haplotypes the molecular basis emerges. Trends Genet. 2000;16(6):240–3. doi: 10.1016/s0168-9525(00)02020-5. [DOI] [PubMed] [Google Scholar]
- 43.Bauer H, et al. The t complex-encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nat Genet. 2005;37(9):969–73. doi: 10.1038/ng1617. [DOI] [PubMed] [Google Scholar]
- 44.Bauer H, et al. The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes Dev. 2007;21(2):143–7. doi: 10.1101/gad.414807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauer H, et al. The nucleoside diphosphate kinase gene Nme3 acts as quantitative trait locus promoting non-Mendelian inheritance. PLoS Genet. 2012;8(3):e1002567. doi: 10.1371/journal.pgen.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrmann BG, et al. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature. 1999;402(6758):141–6. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci U S A. 2008;105(32):11436–41. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouyang Y, et al. Origination and Establishment of a Trigenic Reproductive Isolation System in Rice. Mol Plant. 2016;9(11):1542–1545. doi: 10.1016/j.molp.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Turner BC, Perkins DD. Spore killer, a chromosomal factor in neurospora that kills meiotic products not containing it. Genetics. 1979;93(3):587–606. doi: 10.1093/genetics/93.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammond TM, et al. Molecular dissection of Neurospora Spore killer meiotic drive elements. Proc Natl Acad Sci U S A. 2012;109(30):12093–8. doi: 10.1073/pnas.1203267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harvey AM, et al. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics. 2014;197(4):1165–74. doi: 10.1534/genetics.114.167007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank SA. Divergence of Meiotic Drive-Suppression Systems as an Explanation for Sex-Biased Hybrid Sterility and Inviability. Evolution. 1991;45(2):262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 53.Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics. 1991;128(4):841–58. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1645):20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112(49):E6736–43. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esvelt KM, et al. Concerning RNA-guided gene drives for the alteration of wild populations. Elife. 2014:3. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaenike J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics. 2001;32:25–49. [Google Scholar]
- 58.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 2007;5(11):e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 2007;5(11):e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cocquet J, et al. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8(9):e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cocquet J, et al. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 2009;7(11):e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soh YQ, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159(4):800–13. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lahn BT, Page DC. A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum Mol Genet. 2000;9(2):311–9. doi: 10.1093/hmg/9.2.311. [DOI] [PubMed] [Google Scholar]