Abstract
Over thirty hereditary diseases are caused by the expansion of microsatellite repeats. The length of the expandable repeat is the main hereditary determinant of these disorders. They are also affected by numerous genomic variants that are nearby (cis) and physically separated from (trans) the repetitive locus, which we review here. These genetic variants have largely been elucidated in model systems using gene knockouts, while a few have been directly observed as single nucleotide polymorphisms (SNPs) in patients. There is a notable disconnect between these two bodies of knowledge: knockouts poorly approximate the SNP-level variation in human populations that gives rise to medically-relevant cis- and trans-modifiers, while the rarity of these diseases limits the statistical power of SNP-based analysis in humans. We propose that high-throughput SNP-based screening in model systems could become a useful approach to quickly identify and characterize modifiers with clinical relevance for patients.
Microsatellite repeats and DNA secondary structures
Microsatellites (see Glossary) consist of tandem repeated units of 1-to-9 DNA base pairs that can extend from a few repeats to thousands. Trinucleotide repeats in particular are linked to a number of human genetic disorders [1–3], including (CAG)n repeats in Huntington’s disease (HD) and various spinocerebellar ataxias, (CTG)n repeats in myotonic dystrophy type 1 (DM1), (CGG)n repeats in fragile-X syndrome (FXS), (GAA)n repeats in Friedrich’s ataxia (FRDA) and many others. Repetitive sequences are subject to expansions and contractions, a unique class of mutations arising from a variety of distinct mechanisms. In each case, disease occurs in individuals who have inherited a repeat tract that has expanded beyond a certain length. Nearly all microsatellite expansion diseases are neurological or neurodegenerative, with progressive symptoms coinciding with continued somatic expansion throughout life [4–6]. Understanding the nature of repeat expansion should therefore help to explain how individuals inherit and develop microsatellite expansion diseases.
For a subset of micro- and minisatellites (slightly longer 10–100 bp repeat units), the repetition of complementary base pairs leads to stable intra-strand base-pairing, resulting in non-B-form DNA secondary structures (Fig. 1). For (AT)n dinucleotide repeats, A-T base pairs on each strand can nucleate into stable hairpin (one strand) or cruciform (both strands) structures upon unwinding [7,8]. (CAG/CTG)n repeats can form imperfect hairpins, with the strong C-G base pairs stabilizing the structure [9]. Hairpins also contain short unpaired caps at the point of symmetry, due to the limited bending angle of DNA strands. In the case of (GAA)n repeats, one strand consists entirely of purines, while the other contains only pyrimidines. This creates conditions favorable for the formation of triple-helical (triplex) H-DNA, in which the third strand is bound to the duplex via Hoogsteen (H-y) or reverse Hoogsteen (H-r) base-pairing, rather than Watson-Crick base-pairing [10,11]. (CGG)n repeats and human telomeric (TTAGGG)n repeats contains regularly phased guanines in one strand, promoting the formation of four-stranded G-quadruplex DNA (although the importance of G-quadruplex formation in (CGG)n repeats remains a subject of debate) [12–14]. All microsatellite repeats also have the potential to form slipped-strand DNA, where the DNA has unwound and reannealed out of register, leaving an unpaired or hairpin-stabilized loop on each strand [15,16]. In most cases, these structures form transiently during processes involving DNA strand separation, such as replication, repair, recombination and transcription. Longer repetitive tracts and high levels of DNA negative supercoiling lead to more energetically stable non-B DNA structures [17–19].These dynamic structure-forming properties of repetitive DNA can be linked to many of their functional and detrimental consequences.
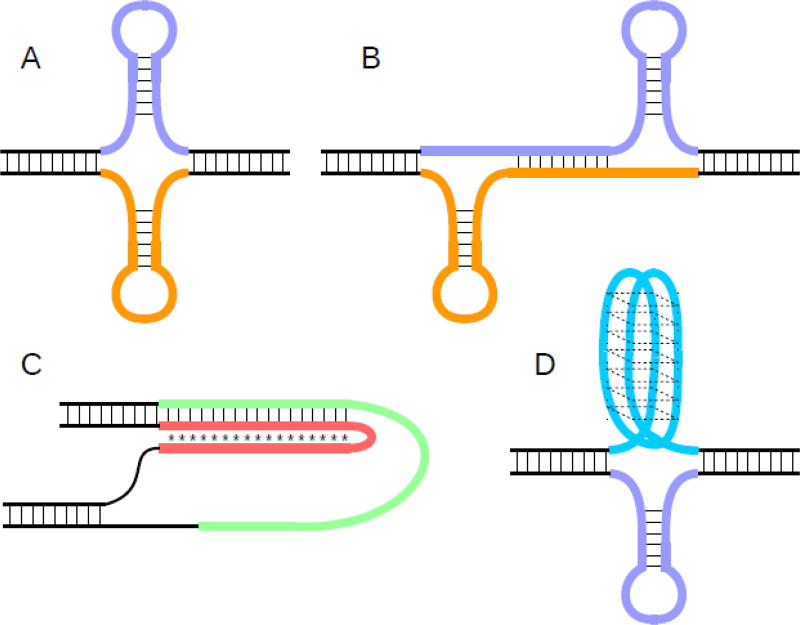
Fig. 1. DNA secondary structures.
All panels: repetitive DNA portions pictured in color, non-repetitive DNA pictured in black. A) Cruciform structure, consisting of hairpin structures on the top and bottom strands. B) Slipped-strand DNA, here shown with loop-outs on either end stabilized by hairpin structures. The loop-outs can also remain unpaired, or can be stabilized by a different secondary structure. C) H-DNA, one of several potential secondary structures involving triplex or triple-helical DNA. The triplex is stabilized by Hoogsteen or Reverse-Hoogsteen basepairs (illustrated as *). The fourth strand can remain unpaired, as shown, or can potentially incorporate into further secondary structures. D) G-quadruplex DNA (top strand), also known as G4 or tetrahelical DNA. Several different folding patterns are possible, in addition to the one shown here, involving different arrangements of parallel or anti-parallel strand orientations. Typically, only one of the two strands will contain the regularly-spaced Gs that permit G4 folding, while the other strand can remain unpaired or fold into a different secondary structure, such as a hairpin (bottom strand as pictured).
Repeats may be added or lost a few at a time, or in large jumps [20]. The rate of this instability increases exponentially with repeat length, in accordance with structure-forming potential, with long repeat tracts expanding and contracting at rates that are orders-of-magnitude higher than the rate of point mutations [21–24]. Large repetitive tracts are also frequent sources of double-strand breaks. In particular, fragile-X syndrome is named for the tendency of expanded (CGG)n repeats to break, and long (CAG)n and (GAA)n tracts also serve as fragile sites [25–27]. In addition, (AT)n-rich repeats appear to play a role in the fragility of common fragile sites, and are frequently associated with translocation breakpoints in cancer [28–30]. Repeat-containing broken DNA ends can promote homologous recombination (HR) into non-allelic genomic loci that also contain repeats, illustrated by the frequency of microsatellites appearing at the sites of complex genomic rearrangements (CGRs) in cancer, as well by repeat-mediated CGRs in model organisms [26,30–34].
In this review, we discuss the complex progression of human microsatellite diseases through the lens of cis- and trans-acting modifiers of microsatellite instability, as well as strategies for identifying new modifiers. Because microsatellite instability is intrinsically linked to basic properties of DNA, the genetic modifiers of instability include core molecular machineries that have been conserved throughout evolution. The molecular mechanisms of microsatellite instability are largely understood due to efforts in model systems, including the eukaryotic baker’s yeast, mice and cultured human cells [2,20,35–37]. Studies in human patients have uncovered a few modifiers of somatic instability that align with results from model systems. Due to the rarity of each microsatellite disease, the latter population-based studies are limited in the power to detect all but the most common and powerful modifiers. Sequencing of patient genomes could potentially reveal rare mutations in the same genes implicated in model systems, suggesting which individuals might be most subject to high rates of somatic expansion. However, human genetic variation within genes rarely resembles the most common tool used to investigate gene function in simple model systems, namely gene knockouts. While extensive structural variation exists in humans, the vast majority of large-scale deletions and insertions appears within introns and intergenic regions [38]. Variation in protein-coding regions is more likely to take the form of single nucleotide variants / polymorphisms (SNVs, SNPs) and short, non-frame-shift indels, as these less-severe variants have been tolerated through evolution. Unfortunately, it is not always straightforward to predict the effect of a SNV on a gene’s function, even when the gene knockout is well characterized. Furthermore, many gene variants may only become important in the context of a particular genetic background. We discuss here a particular strategy to aid in translating patient genome sequences into actionable medical information, namely the use of model systems for the large-scale identification of SNVs affecting conserved genes involved in microsatellite instability.
Cis-modifiers of repeat expansions
Cis-acting modifiers of repeat expansion are those genomic variants that are found within or in the immediate vicinity of the repetitive locus. The first discovered cis-modifiers of repeat expansions were interruptions within the repeats themselves, such as (AGG)n triplets within FXS (CGG)n runs [9], (CAT)n triplets within the SCA1 (CAG)n runs [39], or (GGA)n triplets in the FRDA (GAA)n repeats [40]. These mutations disrupt the stability of secondary structures, leading to drastic reductions in expansions [41,42]. Using newly developed methods to amplify and sequence the extremely GC-rich (CGG)n repeat, it was found that the protective effect of an (AGG)n interruption diminishes as the starting length of the repeat increases, likely because the length of the uninterrupted portion of the repeat also increases [43]. This study also observed contractions of the repeat that eliminated (AGG)n interruptions, which could increase the likelihood of expansions in the next generation, despite the shorter repeat tract.
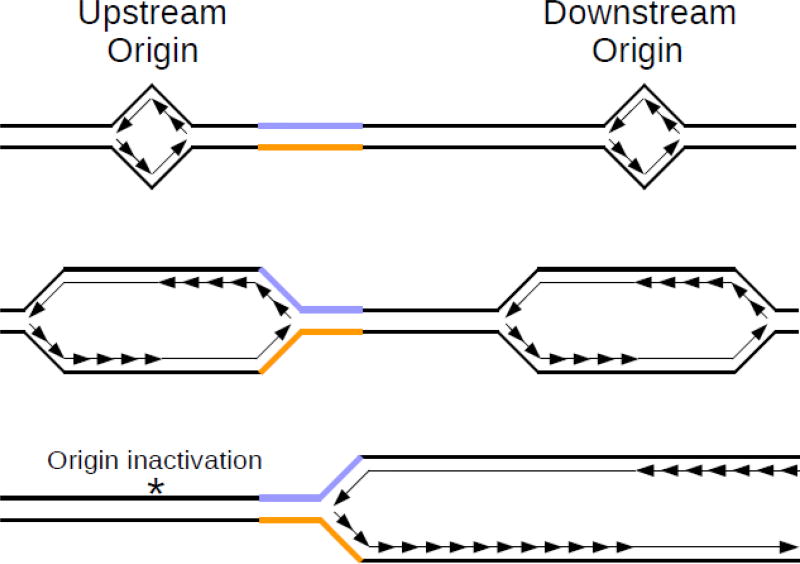
Numerous data from model systems show that microsatellite instability also depends on the orientation relative to replication origins [44–49]. This led to the “ori-switch” model, in which expansions arise frequently in the presence of a genetic or epigenetic cis-modifier that results in a switch in the direction of replication through the repeat [50] (Fig. 2). Recently, this hypothesis was confirmed by the discovery of a cis-modifier for FXS. SNP genotyping of the region surrounding FMR1 revealed a single SNP ~50 kb upstream of the (CGG)n repeats that was present in nearly all individuals with an expanded repeat, but was present in only half of normal-length individuals [51]. Furthermore, using a powerful technique known as single molecule analysis of replicating DNA (SMARD), it was shown that this SNP is correlated with the activity of an underlying replication origin [52,53]. Normally, the FMR1 locus is replicated from both directions, originating from the aforementioned upstream site and from another downstream origin. In the presence of this SNP, however, the upstream origin gets inactivated, and so replication proceeds through FMR1 in only one direction. This change places the (CGG)n repeats exclusively on the leading strand template and the nascent lagging strand of the replication fork. Formation of a secondary structure by the (CGG)n run might result in nascent strand slippage and realignment out of register with the template strand, leading to repeat expansions (Fig. 3). Note that while both (CCG)n and (CGG)n strands of the fragile X repeat can form hairpins, the stability of the two structures differ due to the C-C vs. G-G mismatches. Additionally, only the (CGG)n strand is potentially capable of forming G-quadruplex structures, though it remains unclear under what conditions this structure may be stable in vivo [13].
Fig. 2. “Ori-switch” hypothesis.
Top panel: Repetitive DNA (colored) sits between two different replication origins. Middle panel: Replication proceeds bi-directionally from each origin. In this case, the upstream origin reaches the repeats first. The top strand serves as the lagging strand template, while the bottom strand serves as the leading strand template. Bottom panel: The upstream origin is inactivated, either by a mutation of the binding site, or due to epigenetic changes such as DNA methylation. As a consequence, the downstream origin replicates through the repeats, flipping the orientation such that the top strand now serves as the leading strand template, while the bottom strand serves as the lagging strand template.
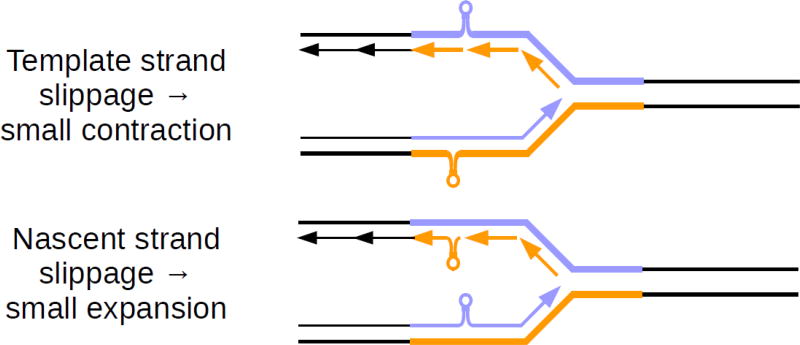
Fig. 3. Small-scale instability due to replication slippage.
Top panel: Secondary structures, here shown as hairpins, form within a repetitive region on either template strand during replication. As a result, the nascent strand skips a small portion of the template, leading to a contraction. Bottom panel: Secondary structures form on either nascent strand during replication, leading to a small expansion in the newly-generated DNA. Both panels: Lagging strand synthesis is discontinuous by nature, providing regular opportunities for slippage. Leading strand synthesis is generally continuous, but may occasionally slip while encountering DNA lesions or previously-formed secondary structures.
Similar phenomena have been observed for other repeats. (CTG)n repeats in DM1 are flanked by CTCF insulator sites, which delineate the boundaries of chromatin loops and affect heterohromatin spreading. Binding of the CTCF protein is controlled by the methylation status of the binding sites, which differ between different tissue types in DM1 patient cells and mouse models, and may affect replication direction [54]. (GAA)n repeats in cells derived from FRDA patients are replicated in a single orientation, whereas the same locus in cells from healthy individuals replicates from both directions [55]. In the orientation which places (GAA)n repeats on the leading strand template, replication fork progression was stalled more severely. Treatment of FRDA cells with a polyamide compound, which had been shown to prevent triplex formation and reduce expansions [56], here rescued replication fork stalling at the (GAA)n repeats as well. This implies that triplex formation leads to fork stalling, a process linked to expansions in model systems (see below).
While it is not known whether a genetic or epigenetic change is responsible for the observed change in replication direction in FRDA cells, these data together are indicative that “ori-switch” may be a common theme for repeat expansions in humans. Why does replication direction matter? Possible explanations include differences in the activity of the leading and lagging strand polymerases, Pol ε and Pol δ, respectively, and their differing responses to replication stress [57]. Okazaki fragment maturation on the lagging strand is another step that is vulnerable to mistakes within microsatellites (see below). In addition to changing replication direction, an “ori-switch” may also alter replication timing and/or position the repeat tract farther away from the next-closest origin. This may impact the stability of the replication fork when it reaches the repeats, or may involve changes to the chromatin environment [47,48,54].
Trans-modifiers of repeat expansions identified in model systems
Trans-modifiers of microsatellite instability may occur in any part of the genome. Most trans-modifiers have been uncovered in genetically tractable model systems including yeast, mice and cultured human cells [2,20,35–37]. A priori, these modifiers can contribute to one or more of the following broadly characterized mechanisms: secondary structure formation, processes inhibited by secondary structures, recognition and processing of secondary structures, and processes that are invoked in response to the previous categories. The misalignment of repeats represents an additional route to instability, because the repetitive sequence itself poses a problem apart from secondary-structure formation.
DNA replication is vulnerable at multiple points to each of these categories of instability mechanisms. Nearly every replication protein has been implicated in triplet repeat instability. The single-stranded binding protein complex RPA appears important for preventing expansions [58]. This strong protection may encompass multiple mechanisms, including binding to unpaired regions within secondary structures or extensive single-stranded regions which can be produced following uncoupling of the replisome [57,58]. The latter mechanism would explain the increase in expansions observed upon knocking down the replicative DNA helicase gene MCM4 [58]. Accessory DNA helicases such as the yeast Srs2 and Sgs1, and the human BLM, WRN and RTEL1, were also found to stabilize expandable repeats, likely due to their secondary-structure-unwinding capabilities [23,35,59–62]. This activity may be coordinated via PCNA [61]. Mutations in the replicative DNA polymerases delta and epsilon slow replication, which exacerbates replication blockage by secondary structures and leads to an increased rate of instability, in some cases via translesion synthesis [63]. Expansions increase in the absence the replication fork stabilizer Tof1, or Timeless in yeast and humans, respectively [23,24,64,65].
Restarting replication can proceed by several mechanisms. One of these is template switch, in which replication temporarily switches to use the sister chromatid as a template, then switches back to bypass the secondary structure (Fig. 4). Work in yeast has shown that template switch uses DNA polymerase alpha to synthesize an Okazaki fragment-length segment, and that modifiers in DNA polymerase alpha can result in a larger step size of repeat expansions [63]. Deletion of yeast Rad5, which promotes template switching, results in fewer large-scale (GAA)n repeat expansions [23]. However, the opposite is true for expansions of short (CAG)n tracts, where Rad5 acts in a separate pathway [20,66,67]. The flap endonuclease Fen1/Rad27 also appears to protect against large-scale (GAA)n expansions via template switching, in addition to its role in the strand-slippage that leads to small-scale repeat expansions [68–70]. This may occur even in the absence of fork stalling during post-replicative repair, as Fen1 cleaves repeat-containing flaps left by the lagging strand synthesis. These flaps may otherwise fold back into a triplex and require bypass by template-switch [37].
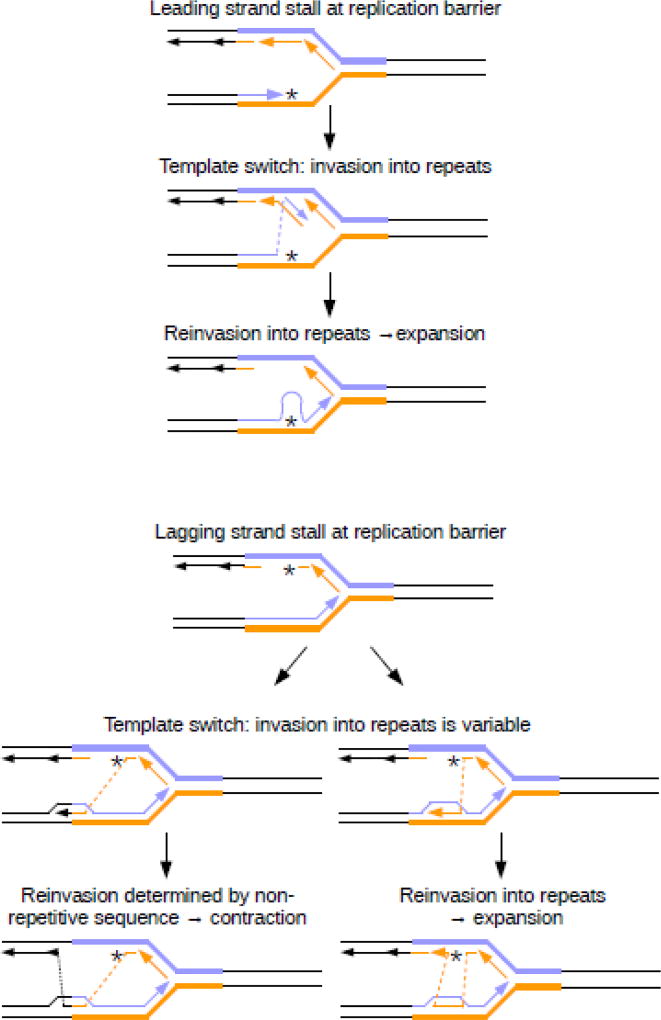
Fig. 4. Instability due to template-switch events.
Top panel: During replication, the leading strand may stall after encountering a barrier, including DNA lesions, secondary structures or bound proteins. To bypass the barrier, replication may temporarily switch to use the nascent lagging strand as a template. After reaching the end of the Okazaki fragment, replication re-invades the leading strand template ahead of the lesion. However, within a repetitive region, this re-invasion can occur out-of-register, potentially leading to a large-scale expansion. Bottom panel: The lagging strand can also encounter a barrier to replication, leading to use of the nascent leading strand as a template. Within a repetitive region, this invasion step can be variable. If it occurs close to the border of the repeat (left panel) the Okazaki fragment will contain non-repetitive sequence, leading to a contraction after re-invasion. If the Okazaki fragment contains only repetitive DNA, reinvasion can occur at any point within the repeat, potentially leading to a large-scale expansion.
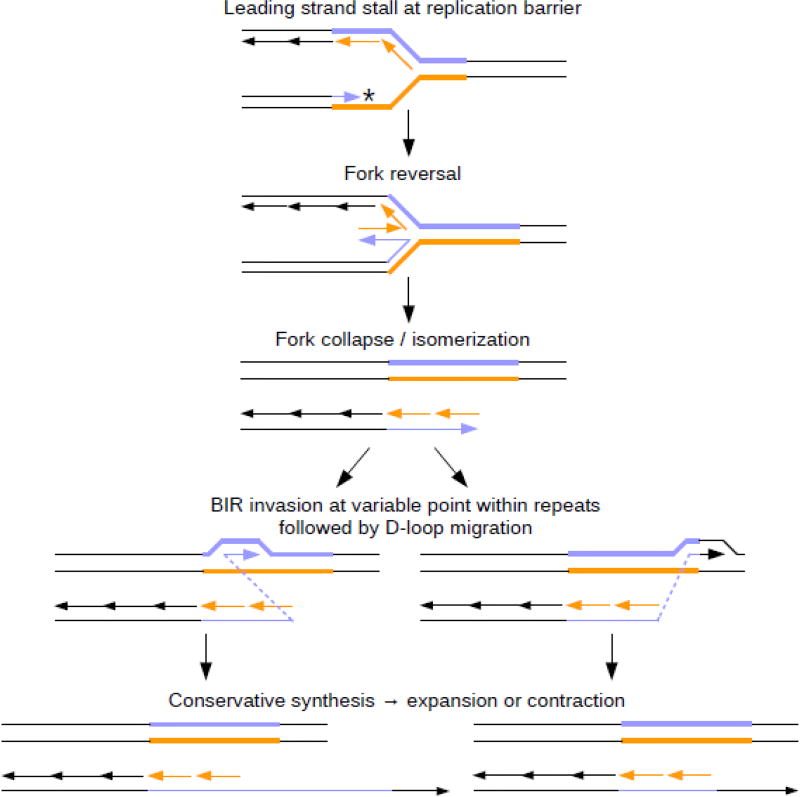
Replication can also restart by fork reversal, which can generate one-ended DSBs, the repair of which can proceed via break-induced replication (BIR). BIR uses HR to restart replication from the sister chromatid, and conservative DNA synthesis proceeds until interrupted, potentially only when reaching the end of the chromosome. [37,71,72]. Large-scale (CAG)n repeat expansions in yeast have been shown to require the Pol32 subunit of DNA polymerase delta, as well as Pif1 helicase, two proteins central to BIR [72]. BIR was also recently implicated in expansions of (CGG)n repeats in cultured mammalian cells [73]. If BIR initiates within a microsatellite tract, repeat instability is possible (Fig. 5). Surprisingly, nuclear pore components have been found to affect (CAG)n instability in yeast, through a mechanism involving relocalization of stalled replication forks to the pore for efficient fork restart [74]. This relocalization may be important for restricting the homology search during BIR initiation, helping to prevent translocations [75–77].
Fig. 5. Instability due to break-induced replication (BIR).
A stalled replication fork (top panel) that cannot be restarted by other means may lead to fork reversal, resulting in a chicken-foot structure (second panel). Together, the two template strands are intact, while the two nascent strands make up a one-ended double strand break (third panel). The nascent leading strand can then invade the template via homologous recombination to initiate BIR. If the invading end consists of repeats, invasion can occur anywhere within the repetitive tract. This can lead to a large-scale expansion – as much as a doubling of the repeat tract – if invasion occurs near the beginning of the repeats (left). In the opposite case (right), invasion can occur towards the end of the repetive tract, skipping a large portion of the repeat and leading to a large-scale contraction. Synthesis of this strand continues (bottom panel), potentially until reaching the end of the chromosome, before the remaining strand is filled in, resulting in conservative DNA replication.
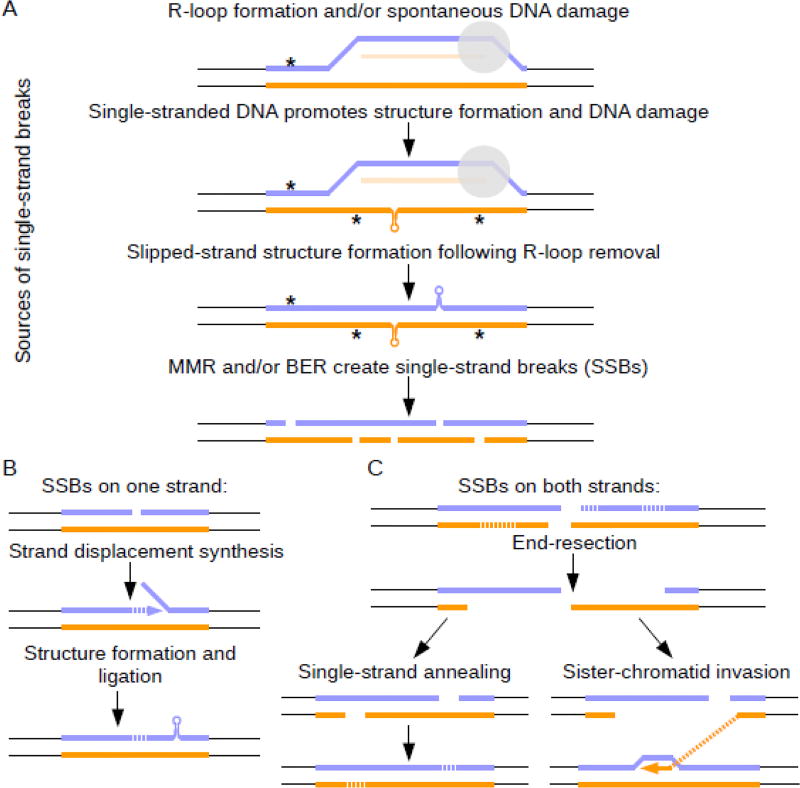
Nearly every form of DNA repair, in addition to post-replicative repair and BIR (see above), has been implicated in microsatellite instability. Secondary structures, which contain regions of single-stranded DNA, may be particularly vulnerable to DNA damage, including cytosine deamination and oxidative damage. This damage is then repaired by base excision repair (BER), wherein the damaged base is removed to create an abasic site that is then cleaved by AP endonuclease, leading to a single-strand nick. The nick can be processed into a single strand gap, and the resulting fill-in synthesis is vulnerable to strand slippage, leading to small-scale instability, as well as further secondary structure formation (Fig. 6). This process has been shown to involve Fcy1 and Ung1 in yeast, as well as OGG1 and NEIL1 in mouse models [78–83]. Mismatch repair (MMR) has also been found to promote expansions in numerous systems [83–86], as well as in human genetic studies (see below). It is thought that MMR components mistakenly recognize hairpins as mismatches, either at the capped ends or at actual mismatches within imperfect hairpins, and either stabilize the secondary structure or unnecessarily initiate repair [87–90]. As with BER, the resulting repair generates a nick, which can lead to strand slippage during fill-in synthesis (Fig. 6). Inappropriate MMR also appears to act on (GAA)n repeats in yeast, perhaps at the single-stranded portion of a triplex or at the unpaired regions of slipped-strand structures [26]. Single-strand nicks occurring nearby on opposite strands ultimately lead to DSBs. In such cases, repair can occur by non-homologous end joining (NHEJ), resulting in a contraction or larger deletion, or by one or more branches of HR (Fig. 6). Importantly, DNA repair can occur in both proliferating and non-proliferating cells. Thus, these modifiers may be particularly important in diseases originating from non-dividing cells.
Fig. 6. Instability due to DNA damage and repair.
A) Sources of single strand breaks (SSBs): In addition to spontaneous DNA damage (*), R-loops (extended RNA-DNA hybrids) can expose long stretches of single-stranded DNA, which can increase the rate of DNA damage, including oxidative damage and cytosine deamination. DNA damage undergoes base excision repair, leading to single-strand breaks. Secondary structure formation in repetitive tracts occurs in ssDNA exposed on the non-template strand while the R-loop is present, and can lead to slipped-strand structures when the R-loop is removed. Secondary structures can be recognized and cleaved by various enzymes, potentially leading to contractions, and also leading to SSBs. B) If SSBs occur only on one strand, repair can occur via strand displacement synthesis, creating a flap that can form secondary structures. The flap can be stabilized by mismatch repair enzymes and incorporated into the repaired DNA strand, causing a repeat expansion. C) If SSBs occur on both strands, this results in a double strand break. DSB repair can occur by a number of mechanisms, including non-homologous end joining (not shown), BIR (see Fig. 5), single-strand annealing between repetitive tracts, which can result in contractions, and sister chromatid invasion and recombination, which can potentially result in expansions or contractions.
Homologous recombination genes implicated in repeat instability include recombinases Rad52 and Rad51, the end-resection complex MRX/Sae2, as well as Mus81/Yen1 resolvases [31,58,72,91–94]. The process of HR is intrinsically vulnerable to destabilizing repetitive DNA. Realignment of broken repeat ends can occur between any two segments of the repeat, assuring that a length change will frequently occur. This can potentially happen during BIR, as mentioned above, as well as during single-strand annealing (SSA) or synthesis-dependent strand annealing (SDSA) [95–100]. That being said, sister chromatid exchange may have a stabilizing role in repeat maintenance [93,101,102]. Broken ends within repeats may be resected beyond the repetitive tract, allowing the homology search to take advantage of non-repetitive sequence.
Transcription: cis- and trans-modifiers of repeat expansions
Genes involved in various stages of the transcription process have also been shown to affect microsatellite instability, and may be very important in accounting for expansions that occur in non-dividing cells [35,36,103,104]. This could be due to several mechanistic reasons. Transcription necessarily involves accessing single-stranded DNA to generate the complementary RNA. DNA unwinding generates negative supercoils behind the RNA polymerase, creating favorable conditions for secondary structure formation [17–19]. Nucleosome remodeling and/or removal also accompanies active transcription, leading to secondary structure formation and thus instability [105]. Secondary structure can then inhibit the movement of RNA polymerase in further rounds of transcription [106,107]. These mechanisms likely explain the involvement of chromatin modifiers and transcription initiation factors in repeat expansion observed in yeast, flies and human cells [58,102,108–110]. This also suggests that mutations within the promoter or enhancer of a repeat-containing gene might serve as cis-modifiers by increasing or decreasing transcription levels. Trans-modifiers of genome stability affecting other stages of transcription have also been uncovered, namely a polyadenylation and 3’-end processing factor [94] (see below), as well as multiple mRNA packaging and export factors in yeast and human cells [111–117]. The former appears to lead to instability by preventing transcript cleavage and detachment of RNA polymerase, thus promoting collisions between RNA and DNA polymerases that can lead to DSBs. The latter appears to lead to increased R-loop formation (extended RNA/DNA hybrids). R-loops may lead to instability by stalling RNA polymerase while also leaving the non-template strand unpaired. This can allow access by DNA damaging agents, leading to BER (Fig. 6), or transcription-coupled nucleotide excision repair (NER), which has separately been implicated in instability [83,110,118–121]. Secondary structure formation on the unpaired strand may further stabilize the R-loop [118]. (GAA)n repeats may be particularly susceptible to R-loop formation, as purine-pyrimidine DNA-RNA bonds are stronger than purine-pyrimidine DNA-DNA bonds [120].
Transcription and fragility in FRDA and FXS: consequences and considerations
Considering the above ways in which transcription of unstable microsatellites is a risky proposition, it may ultimately be beneficial for a cell to inhibit transcription of repeat-containing genes. In fact, FMR1 and FXN genes in FXS and FRDA patients, respectively, typically show chromatin and/or DNA methylation changes that reduce transcription, and the resulting insufficient protein levels lead to disease phenotypes. While this is disastrous for the individual, from the perspective of the cell, this may be the best option for maintaining the stability of the repetitive tract. The alternative is to risk repeat expansion, exponentially increasing the risk of subsequent instability. Note that expansions are not the most detrimental possible outcome when cellular machinery encounters long microsatellite tracts. Chromosomal fragility has been observed for nearly all structure-prone repeats that reach a certain length [29,35]. Interestingly, significant overlap has been observed between genes affecting fragility and instability of repeats [58,122], and repair of DSBs, which can lead to further repeat expansions (see above). Unrepaired DSBs can lead to the loss of entire chromosome arms and numerous essential genes, likely leading to cell death. Misrepaired DSBs can lead to deletions of various sizes, as well as chromosomal translocations and copy number changes, all potential drivers of cancer. Triplex-forming repeats as well as AT-rich repeats have been found to be prevalent at the breakpoints of translocation in cancer [30]. As mentioned above, DSB repair involving repeats is highly prone to errors, which can also include non-allelic recombination due to the multitude of microsatellites throughout the genome [26,33,34,99,122]. BIR is also prone to generating frequent point mutations, which may account for the phenomenon of repeat-induced mutagenesis (RIM) [99,123–125].
Thus, a somewhat paradoxical situation arises, in that repeat-containing genes must be re-activated to prevent disease symptoms, but higher transcription of the repeats may increase the likelihood of further genomic instability. Treatments designed to reactivate FMR1 or FXN, such as histone deacetylase (HDAC) inhibitors, have not yet been successful in clinical trials, however more approaches along this line are being developed [126,127]. One might speculate that such treatments may be limited in long-term effectiveness by the side effect of promoting further somatic expansions. In this light, two other proposed therapies for FRDA may have greater success. The first involves using (UUC)n synthetic oligonucleotides to bind (GAA)n transcripts, thus preventing R-loop formation [128]. This has the intended effect of promoting efficient FXN transcription, but may also help to prevent further R-loop-dependent somatic expansions. A second therapy involves a synthetic peptide targeted to the mitochondria that promotes efficient translation of FXN transcripts [126]. This has the benefit of increasing FXN protein expression without increasing risky FXN transcription. However, the lasting effectiveness of this therapy may still be limited by ongoing somatic expansions. Perhaps this therapy will prove effective in combination with treatments designed to disrupt secondary structures[129], slow somatic expansions [81] or promote somatic contractions [130,131].
Complex cis- and trans-modifier interactions
Although it may appear that “everything but the kitchen sink” in DNA-related processes affects microsatellite instability, the difficulty in translating this knowledge to a disease treatment is that each of these factors interacts in a complex network, which can change drastically based on the presence of particular modifiers and the particular microsatellite sequence in question. A few examples of this have been worked out in yeast: The Rad5 helicase/ubiquitin ligase protects against small-scale (CAG)n expansions during replication slippage, but promotes (GAA)n expansions via template switching at a stalled replication fork [20]. Another example is the role of HR factor Rad52. In studies of very short (CAG)n repeat tracts, knocking out Rad52 had little to no effect [20]. However, Rad52 has been shown to have a role in instability of longer (CAG)n repeat tracts, particularly in conditions of elevated DNA fragility [93,132]. Large-scale (CAG)n expansions in yeast were also found to occur through a Rad52- and Pol32-dependent BIR mechanism, rather than by replication slippage [72]. For (GAA)n repeats in yeast, knockout of Rad52 normally does not greatly affect the rate of expansions, which occur by template switch during replication [23]. However, deletion of Rad52 reduces expansions when in the presence of a mutation in the RNA 3’ end-processing gene YSH1. In this genetic background, (GAA)n expansions are frequently generated via a Rad52-dependent response to DSBs [94]. Overall, a pattern emerges where the role of HR components in microsatellite instability becomes evident in situations where DNA breaks occur frequently, which may itself be dependent upon any number of cis- and trans-modifiers [122]. In humans, the picture is further complicated by the relative prominence of HR compared with non-homologous end-joining in particular cell types. Similarly, it has been shown in mouse models that tissue-specific differences in (CAG)n instability can be linked to differing expression levels of various replication and repair genes [133,134].
The action of various trans-modifiers also appears to depend on the cis-conditions of the repeat, including the direction of replication through the repeats, the status of the chromatin and whether the region is actively transcribed [65,94,135]. In multi-cellular organisms, different cell- and tissue-types present an array of cis- and trans-conditions that are known to result in striking differences in instability rates within the same individuals [5,136–139]. The challenge, therefore, is how to fully assay the complexity of the human disease, which will require the integration of knowledge generated in simple, often-times unicellular model systems, as well as more closely-related mouse and human cell models, into actionable clinical genetics.
Trans-modifiers of microsatellite disorders in humans
Inherited repeat length is the greatest determinant of disease onset and severity, but does not tell the complete picture. The progressive, late-onset characteristics of most microsatellite diseases were initially attributed to a low toxicity of the repetitive RNA or polyQ proteins expressed from expanded microsatellites [140,141]. However, it is becoming increasingly clear that this may also be due to somatic expansions that occur throughout life [81,142]. Indeed, due to somatic instability, it is difficult to accurately determine the germline repeat length that an individual has inherited. PCR or Southern blot analysis of a typical genomic DNA preparation typically produces a smear, rather than a clear band. A number of studies have used small-pool PCR, in which the input DNA is diluted to a single copy per reaction, and a large number of reactions are performed per individual. In a study of DM1 pioneering this analysis, a sharp lower limit was found for (CTG)n length in each individual, interpreted as the inherited repeat length [4]. Contractions below this length were rare, while expansions beyond it were common and highly variable. This technique was also used to measure somatic instability in FRDA, which showed that contractions of (GAA)n repeats were more frequent than expansions in most tissue types. However, expansions predominated in dorsal root ganglia, consistent with the phenotypic degeneration [5]. FRDA has also been seen to develop with a late age of onset in individuals carrying one expanded allele and one pre-mutation-length allele. Small-pool PCR revealed many somatic expansions in the shorter allele that reached disease length, suggesting an explanation for the eventual development of symptoms [6]. FXS mouse models display somatic variability in repeat length, with clear tissue-specific differences [138]. In Huntington’s disease, higher levels of somatic instability measured in the brain cortex was found to be associated with earlier age of onset [143]. This reflects additional work from mouse models [84,86]. Furthermore, in mouse models, both the knockout of Msh2, as well as chemical compounds that suppressed somatic expansions, were able to substantially delay the onset of symptoms of neurodegeneration [81,135,144]. Thus, it follows that for the various microsatellite disorders, the rate at which somatic expansions occur will critically impact disease progression. As discussed above, this appears to be a highly complex characteristic affected by numerous genetic modifiers.
In a pivotal study of DM1 patients, the small-pool PCR approach was used along with extensive statistical analysis to examine the relationship between somatic (CTG)n instability and age of onset [145]. While confirming that the inherited repeat length is the largest contributor to age of onset, this study also found that the amount of instability observed in each individual also accounted for some of the variation in age of onset. Furthermore, it was shown that the amount of instability was a heritable trait within families, demonstrating that cis- and/or trans-modifiers in the genome were altering expansion rates and meaningfully contributing to progression of DM1. Evidence of the existence of familial risk factors for increased repeat instability has also been found for fragile X [42].
Following up with a larger DM1 cohort, polymorphisms within several candidate genes were examined, leading to the observation that a non-synonymous SNP in the DNA mismatch repair gene MSH3 was correlated with increased (CTG)n instability [146]. Another variant in MSH3 was also identified via genome-wide association study (GWAS) as contributing to the progression of Huntington’s disease [142]. This reflects earlier work in mouse models, showing that variations in MSH3 occurring naturally between various strain backgrounds contributed to differences in somatic (CAG)n instability [147]. However, neither of these MSH3 variants were found to affect the rate of (CAG)n expansions in a human cell-based reporter system [148]. A likely possibility is that these SNPs do not affect MSH3 function themselves, but rather serve as markers in the genotyping analysis for one or more other SNPs that are present in the same haplotype block [146]. A larger GWAS involving HD patients resulted in two variants reaching statistical significance, implicating the DNA repair-related genes FAN1 and RRM2B, as well as a variant in the mismatch repair gene MLH1 that reached significance upon incorporating data from an additional patient cohort [149,150]. Pathway analysis of this data set, which aggregates the effects of genes that fit into various categories, also implicated DNA repair. These studies confirm a link, long established in model systems (see above), between DNA repair and repeat instability.
The rarity of microsatellite disorders is a significant obstacle in identifying modifier genes directly from human genetic data. The above-mentioned studies were limited to only a few hundred to a few thousand individuals. In contrast, the most successful GWAS discoveries have involved cohorts numbering in the tens to hundreds of thousands, focused on common diseases such as type II diabetes [151]. These massive studies have greater statistical power to detect rare and/or low-impact variants. It has been suggested that more statistical power can be gained by combining cohorts from different microsatellite disorders [152]. This was demonstrated in a study testing a panel of candidate variants among patients of various polyglutamine (protein-coding (CAG)n repeat) disorders, including HD and multiple forms of spinocerebellar ataxia (SCA) [153]. FAN1 and RRM2B, found previously in the above-mentioned study, were significantly associated with age of onset in the collective polyglutamine cohort, along with an additional DNA repair factor, PMS2. However, it is not a given that this approach can be extended to the non-polyglutamine microsatellite disorders. In comparing the above-mentioned studies, we can see consistencies between HD and DM1, such as in MSH3, but also differences, as in PMS2 and MLH1, in which no associations were found in DM1 patients [146]. While these particular examples could possibly be due to statistical power or differences in the underlying populations, i.e. whether these SNPs actually appear in both patient populations, it is also quite clear from work in model systems that not all repeats behave alike [20]. Differences between HD and DM1 may be explained by the orientation of the repeats, their placement in the carrier genes, the chromatin environment surrounding the repeats, as well as a number of other factors. Grouping together of other diseases, such as FRDA and FXS, may be even more likely to turn up differences rather than similarities, as the repeats form different types of secondary structures that may involve different molecular processes, or may even respond in different directions to the same trans-modifiers. Thus, more inclusive combinatorial studies should be approached with some caution. Certainly, some modifiers of repeat instability will be specific to certain diseases. In such cases, it may not be possible to uncover modifiers purely through population genetics. Thus, much of what has been found in model systems is likely to be important in understanding rare microsatellite diseases, predicting progression and pointing toward therapies.
Strategies for the characterization of modifiers of microsatellite instability
In the era of personal genomics, one might envision the sequencing of patient genomes to reveal risk factors for high levels of somatic instability, or to reveal a particular pathway that may be therapeutically targetable. There are several obstacles to this goal. The complexity described above suggests that the interpretation of patient genomes will benefit from knowing the status of numerous modifier genes. It is highly likely that we still do not know all of the genes involved in microsatellite instability. Furthermore, it is difficult to know whether or how a particular SNP or other mutation affects each gene. And finally, patient genomes contain combinations of SNPs that may behave in unexpected ways.
As discussed above, human population genetics has revealed a handful of modifiers, and model system studies have revealed many more. Much of the work described above was done via the candidate-gene approach: knocking out or knocking down genes suspected to be involved in some aspect of instability. However, gene knockouts or knock-downs can behave differently than mutations in the same gene. A mutation may alter or eliminate only one of a gene’s multiple functions, and this may further affect how the protein behaves as a part of a complex. Conversely, the appearance of a mutation in a gene known to affect instability is not a guarantee that the mutation is biologically significant. This problem is common to many fields, where patient genome sequences reveal numerous variants of unknown significance (VUS) [154]. In each case, further work is required in order to know whether or not these mutations may be medically relevant, and this can be difficult to accomplish in a time frame that benefits the patient. Far better would be to characterize numerous mutations ahead of time using model systems.
The gene candidate approach to modifier discovery also suffers from issues of scope and bias. A favorite unbiased tool of yeast geneticists is deletion library screening. Each individual knockout of a non-essential gene is represented in the library, and an accompanying library alters the expression of each essential gene. A similar approach involves the random insertion of a plasmid to disrupt genes. This type of screening has been applied to microsatellite instability, as well as other related phenomenon, leading to several unexpected discoveries [58,64,132,155–158]. However, there are several gaps in this screening method. In addition to the above-mentioned issues with using gene knockouts and knock-downs, epistatic interactions are not assayed. Many modifiers may only appear when a redundant pathway cannot rescue their effects. An impressive study generated more than 23 million yeast double knockouts to uncover genetic interactions affecting overall fitness [159], though it would not be feasible to apply this approach to more specific questions like microsatellite instability, or to interactions of more than two genes.
Recently, a novel high-throughput screening method was developed to begin to address some of these shortcomings [94]. Yeast strains containing (GAA)n repeats were mutated with UV, generating mostly single nucleotide variants in an otherwise uniform genetic background. Strains with elevated rates of repeat expansion underwent whole-genome sequencing and bioinformatic approaches to identify the causal variants. This approach identified a new gene, YSH1, which affects repeat expansion in an unexpected manner (see above). This initial success demonstrated several key points: Deletion library screening was not comprehensive in finding all modifiers of (GAA)n expansion, and many more genes may remain to be found. Not only is YSH1 conserved from yeast to humans, where it is known as CPSF-73, but even the affected amino acids themselves are conserved. This suggests that individual mutations characterized in a yeast model system may be directly applicable to patient genomes. Such high levels of conservation have been used to predict the severity of mutations [160]. Due to this relationship, it is likely that further screening will produce many more variants at locations conserved in humans. We suggest that this approach can be carried out to the point of saturation, in order to collect a comprehensive list of conserved variants that affect microsatellite instability. In addition, it is feasible to conduct additional screening in various mutant backgrounds, in order to begin to address combinatorial effects.
This study took advantage of the low cost and dense genome of S. cerevisiae, but future approaches may take advantage of CRISPR-Cas9-based techniques to perform screens directly in human cells, although at greater expense [161]. Finally, after a modifier has been identified, it is valuable to understand how that modifier leads to microsatellite instability, including whether it reveals a new mechanism or contributes indirectly to a known pathway. Here too, genetic manipulation in model systems, including CRISPR-based approaches in human cell culture systems, will be a key tool for characterizing trans-modifiers, as has already been demonstrated in studies of MSH3 variants affecting (CAG)n instability [148]. This approach also has the advantage of directly measuring the contribution of individual SNPs to a functional consequence, without having to disentangle the multitude of variants present in a human haplotype block. Given that human population genetics does not have the same power to uncover and characterize modifiers of rare microsatellite diseases, such approaches may be the key to understanding and treating these diseases.
Concluding remarks
Spurred by the discovery that expanded microsatellite repeats are at the root of numerous hereditary disorders, much work has been done to elucidate the mechanisms of repeat instability. Using model systems as key experimental tool, we and others have gained a broad understanding of how the core cellular machineries that replicate, repair and transcribe DNA are implicated at nearly every level of microsatellite instability. Several challenges and larger questions remain (see Outstanding Questions box). Further study will bring clarity to the finer points of each mechanism and reveal unanticipated new mechanisms. Future research is also needed to determine the universality of each mechanism, and conversely, the idiosynchrasies associated with different types of repeats and their settings within genes and chromatin regions. Characterizing these complexities will be a challenge perhaps best approached with the power of model systems. Thus, the integration of model system research and clinical genetics is a valuable goal in the study of microsatellite repeat disorders.
Highlights.
Repeat expansions leading to disease are affected by cis- and trans-acting genomic variants
Modifiers of repeat expansions are extensively studied in model systems using gene knockouts
Some modifiers of repeat expansions have been identified as single nucleotide polymorphisms (SNPs) in patients
High-throughput SNP-based screening in yeast could be used to bridge the gap between model systems and human studies
Outstanding Questions.
Model system studies have uncovered numerous modifiers of microsatellite instability, which have differing effects on small- and large-scale expansions in various experimental systems. To what extent can these numerous observations be unified?
To what extent will SNP-based detection of expansion modifiers in humans be capable of elucidating the complexity of the process, given the rarity of the diseases?
Can SNP-based expansion modifiers identified in model systems provide a quick way to verify the role of ambiguous genetic variants that come from patient genome sequencing?
Acknowledgments
We are exceptionally grateful to Catherine Freudenreich, Bob Lahue, Michael Leffak, Mitch McVey, Marek Napierala, Shamil Sunyaev and Karen Usdin for their comments and suggestions. Research in the Mirkin lab is supported by the grants from National Institute of General Medical Sciences (R01GM60987 and P01GM105473) and the generous contribution from the White family.
Glossary
- microsatellite
tandemly repeated unit of 1-to-9 DNA base pairs
- expansion
a mutation that increases the length of a microsatellite
- contraction
a mutation that decreases the length of a microsatellite
- SNV
(single nucleotide variant) a mutation altering a single DNA base pair
- SNP
(single nucleotide polymorphism) a mutation altering a single DNA base pair, shared by at least some fraction of the population
- Non-B DNA
DNA structures that differ from the most prevalent form, B-DNA - a right-handed double-helix joined by Watson-Crick base pairs. Several aspects of B-form DNA can be altered, including the handedness, the number of strands, and the presence of intrastrand base pairs.
- Triplex H-DNA
a three stranded DNA structure which forms at homopurine/homopyrimidine mirror repeats. In this structure, two Watson-Crick paired DNA strands join with a third DNA strand via Hoogsteen or reverse Hoogsteen base pairing, while the complement of the third strand remains unpaired.
- G-quadruplex DNA
a four-stranded DNA secondary-structure which is formed by sequences containing regularly spaced (G)n blocks. The main element of this structure is stacked guanine quartets.
- DNA supercoiling
a topological property of DNA describing the writhing of the DNA double helix around itself. Its sign, positive or negative, results from the over- or under-winding, respectively, of the two DNA strands relative to each other.
- helicase
an enzyme that unwinds double-stranded nucleotide polymers into single strands
- polymerase
an enzyme that attaches nucleotides to generate or elongate a polymer, using an existing strand as a template
- endonuclease
an enzyme that cleaves the phosphodiester backbone within a nucleotide polymer
- BIR
(break-induced replication) a form of DNA double-strand break repair, in which a single broken end initiates a long tract of replication from a single strand of a homologous template. The newly synthesized strand then serves as the template for the second strand synthesis, resulting in conservative DNA replication.
- somatic mutation
a mutation occurring in a non-germline cell
- genotyping
the determination of DNA sequence limited to specific SNPs
- haplotype block
a contiguous group of SNPs that are typically inherited together
- essential gene
a gene required for the survival of the cell/organism
- epistatic interaction
when the phenotypic effect of a gene or mutation is masked by the effect of another gene or mutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pearson CE, et al. Repeat instability: Mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 4.Monckton DG, et al. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 5.De Biase I, et al. Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Ann. Neurol. 2007;61:55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- 6.Sharma R, et al. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum. Mol. Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 7.McClellan JA, et al. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayn A, et al. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquis Gacy A, et al. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 10.Dayn A, et al. Intramolecular DNA triplexes: unusual sequence requirements and influence on DNA polymerization. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11406–11410. doi: 10.1073/pnas.89.23.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto N, et al. Sticky DNA: Self-association properties of long GAA·TTC repeats in R·R·Y triplex structures from Friedreich’s ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 12.Fry M, Loeb La. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renciuk D, et al. Quadruplex-forming properties of FRAXA (CGG) repeats interrupted by (AGG) triplets. Biochimie. 2009;91:416–422. doi: 10.1016/j.biochi.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Kwok CK, Merrick CJ. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017;35:997–1013. doi: 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel Ta. Nucleotide repeats. Slippery DNA and diseases. Nature. 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 16.Pearson CE, et al. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–47. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirkin SM, Frank-Kamenetskii MD. H-DNA and Related Structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 18.Grabczyk E. The GAATTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napierala M, et al. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 20.Kim JC, Mirkin SM. The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 2013;23:280–288. doi: 10.1016/j.gde.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolfsmeier ML, et al. Cis-elements governing trinucleotide repeat instability in Saccharomyces cerevisiae. Genetics. 2001;157:1569–1579. doi: 10.1093/genetics/157.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon MJ, Lahue RS. DNA elements important for CAG??CTG repeat thresholds in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:1289–1297. doi: 10.1093/nar/gkh292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shishkin AA, et al. Large-Scale Expansions of Friedreich’s Ataxia GAA Repeats in Yeast. Mol. Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherng N, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freudenreich CH. Expansion and Length-Dependent Fragility of CTG Repeats in Yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 26.Kim HM, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumari D, et al. Evidence for chromosome fragility at the frataxin locus in Friedreich ataxia. Mutat. Res. 2015;781:14–21. doi: 10.1016/j.mrfmmm.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Freudenreich CH. An AT-Rich Sequence in Human Common Fragile Site FRA16D Causes Fork Stalling and Chromosome Breakage in S. cerevisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thys RG, et al. DNA Secondary Structure at Chromosomal Fragile Sites in Human Disease DNA Secondary Structure in Human Disease. Curr. Genomics. 2015;16:60–70. doi: 10.2174/1389202916666150114223205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacolla A, et al. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res. 2016;44:5673–5688. doi: 10.1093/nar/gkw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meservy JL, et al. Long CTG tracts from the myotonic dystrophy gene induce deletions and rearrangements during recombination at the APRT locus in CHO cells. Mol. Cell. Biol. 2003;23:3152–3162. doi: 10.1128/MCB.23.9.3152-3162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacolla A, et al. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair. 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Aksenova AY, et al. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19866–19871. doi: 10.1073/pnas.1319313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGinty RJ, et al. Nanopore sequencing of complex genomic rearrangements in yeast reveals mechanisms of repeat-mediated double-strand break repair. Genome Res. 2017;27:2072–2082. doi: 10.1101/gr.228148.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usdin K, et al. Repeat instability during DNA repair: Insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015;50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyzos AA, McMurray CT. Close encounters: Moving along bumps, breaks, and bubbles on expanded trinucleotide tracts. DNA Repair. 2017;56:144–155. doi: 10.1016/j.dnarep.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neil AJ, et al. Precarious maintenance of simple DNA repeats in eukaryotes. BioEssays. 2017;39:1–10. doi: 10.1002/bies.201700077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huddleston J, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–685. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menon RP, et al. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9:e 1003648. doi: 10.1371/journal.pgen.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto N, et al. GGA·TCC-interrupted Triplets in Long GAA·TTC Repeats Inhibit the Formation of Triplex and Sticky DNA Structures, Alleviate Transcription Inhibition, and Reduce Genetic Instabilities. J. Biol. Chem. 2001;276:27178–27187. doi: 10.1074/jbc.M101852200. [DOI] [PubMed] [Google Scholar]
- 41.Yrigollen CM, et al. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet. Med. 2012;14:729–736. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolin SL, et al. Fragile X AGG analysis provides new risk predictions for 45–69 repeat alleles. Am. J. Med. Genet. 2013;161:771–778. doi: 10.1002/ajmg.a.35833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nolin SL, et al. Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med. 2015;17:358–364. doi: 10.1038/gim.2014.106. [DOI] [PubMed] [Google Scholar]
- 44.Kang S, et al. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 45.Freudenreich CH, et al. Stability of a CTG/CAG Trinucleotide Repeat in Yeast Is Dependent on Its Orientation in the Genome. Mol. Cell. Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miret JJ, et al. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cleary JD, et al. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 48.Rindler PM, et al. Replication in mammalian cells recapitulates the locus-specific differences in somatic instability of genomic GAA triplet-repeats. Nucleic Acids Res. 2006;34:6352–6361. doi: 10.1093/nar/gkl846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G, et al. Replication-dependent instability at (CTG)·(CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirkin SM, Smirnova EV. Positioned to expand. Nat. Genet. 2002;31:5–6. doi: 10.1038/ng0502-5. [DOI] [PubMed] [Google Scholar]
- 51.Ennis S, et al. Closely linked cis-acting modifier of expansion of the CGG repeat in high risk FMR1 haplotypes. Hum. Mutat. 2007;28:1216–1224. doi: 10.1002/humu.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerhardt J, et al. Cis-acting DNA sequence at a replication origin promotes repeat expansion to fragile X full mutation. J. Cell Biol. 2014;206:599–607. doi: 10.1083/jcb.201404157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerhardt J, et al. The DNA Replication Program Is Altered at the FMR1 Locus in Fragile X Embryonic Stem Cells. Mol. Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cleary JD, et al. Tissue-and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat. Struct. Mol. Biol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- 55.Gerhardt J, et al. Stalled DNA Replication Forks at the Endogenous GAA Repeats Drive Repeat Expansion in Friedreich’s Ataxia Cells. Cell Rep. 2016;16:1218–1227. doi: 10.1016/j.celrep.2016.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du J, et al. Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in friedreich ataxia induced pluripotent stem cells. J. Biol. Chem. 2012;287:29861–29872. doi: 10.1074/jbc.M112.391961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gan H, et al. Checkpoint Kinase Rad53 Couples Leading- and Lagging-Strand DNA Synthesis under Replication Short Article Checkpoint Kinase Rad53 Couples DNA Synthesis under Replication Stress. Mol. Cell. 2017;68:446–455. doi: 10.1016/j.molcel.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell. 2012;48:254–265. doi: 10.1016/j.molcel.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharyya S, Lahue RS. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frizzell A, et al. RTEL1 inhibits trinucleotide repeat expansions and fragility. Cell Rep. 2014;6:827–835. doi: 10.1016/j.celrep.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen JHG, et al. Differential requirement of Srs2 helicase and Rad51 displacement activities in replication of hairpin-forming CAG/CTG repeats. Nucleic Acids Res. 2017;45:4519–4531. doi: 10.1093/nar/gkx088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anand RP, et al. Overcoming natural replication barriers: Differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah KA, et al. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2012;2:1088–1095. doi: 10.1016/j.celrep.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG·CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 2008;7:633–640. doi: 10.1016/j.dnarep.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu G, et al. Altered Replication in Human Cells Promotes DMPK (CTG)n {middle dot} (CAG)n Repeat Instability. Mol. Cell. Biol. 2012;32:1618–1632. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daee DL, et al. Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:102–110. doi: 10.1128/MCB.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye Y, et al. The Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex promotes trinucleotide repeat expansions independently of homologous recombination. DNA Repair. 2016;43:1–8. doi: 10.1016/j.dnarep.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 68.Spiro C, et al. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y, et al. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsutakawa SE, et al. Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 2017;8:1–14. doi: 10.1038/ncomms15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fouché N, et al. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim JC, et al. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat. Struct. Mol. Biol. 2017;24:55–60. doi: 10.1038/nsmb.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebersole T, et al. Mechanisms of genetic instabilities caused by the (CGG)n repeats in an experimental mammalian system. (under submission) doi: 10.1038/s41594-018-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su XA, et al. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 2015;29:1006–1017. doi: 10.1101/gad.256404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miné-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 2012;14:510–517. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]
- 76.Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 77.Becker A, et al. ATM alters the otherwise robust chromatin mobility at sites of DNA Double-Strand Breaks (DSBs) in human cells. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0092640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 79.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Møllersen L, et al. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum. Mol. Genet. 2017;21:4939–4947. doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Budworth H, et al. Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease. PLoS Genet. 2015;11:1–22. doi: 10.1371/journal.pgen.1005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lokanga RA, et al. Heterozygosity for a Hypomorphic Pol?? Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders. PLoS Genet. 2015;11:1–16. doi: 10.1371/journal.pgen.1005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su XA, Freudenreich CH. Cytosine deamination and base excision repair cause R-loop– induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E8392–E8401. doi: 10.1073/pnas.1711283114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kovalenko M, et al. Msh2 acts in medium-spiny striatal neurons as an enhancer of CAG instability and mutant huntingtin phenotypes in Huntington’s disease knock-in mice. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0044273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pinto RM, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: Genome-wide and candidate approaches. PLoS Genet. 2013;9:e 1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iyer RR, et al. DNA Triplet Repeat Expansion and Mismatch Repair. Annu. Rev. Biochem. 2015;84:199–226. doi: 10.1146/annurev-biochem-060614-034010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manley K, et al. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 88.van den Broek WJAA, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 89.Owen BAL, et al. (CAG)n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 90.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pluciennik A, et al. Long CTG·CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 2002;277:34074–34086. doi: 10.1074/jbc.M202127200. [DOI] [PubMed] [Google Scholar]
- 92.Napierala M, et al. Long CTG·CAG repeat sequences markedly stimulate intramolecular recombination. J. Biol. Chem. 2002;277:34087–34100. doi: 10.1074/jbc.M202128200. [DOI] [PubMed] [Google Scholar]
- 93.Sundararajan R, et al. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McGinty RJ, et al. A Defective mRNA Cleavage and Polyadenylation Complex Facilitates Expansions of Transcribed (GAA)nRepeats Associated with Friedreich’s Ataxia. Cell Rep. 2017;20:2490–2500. doi: 10.1016/j.celrep.2017.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pollard LM, et al. Repair of DNA double-strand breaks within the (GAA·TTC)n sequence results in frequent deletion of the triplet-repeat sequence. Nucleic Acids Res. 2008;36:489–500. doi: 10.1093/nar/gkm1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mott C, Symington LS. RAD51-independent inverted-repeat recombination by a strand-annealing mechanism. DNA Repair. 2011;10:408–415. doi: 10.1016/j.dnarep.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crespan E, et al. Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res. 2012;40:5577–5590. doi: 10.1093/nar/gks186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Finn KJ, Li JJ. Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination. PLoS Genet. 2013;9:e 1003192. doi: 10.1371/journal.pgen.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tang W, et al. Genomic deletions and point mutations induced in Saccharomyces cerevisiae by the trinucleotide repeats (GAA/TTC) associated with Friedreich’s ataxia. DNA Repair. 2013;12:10–17. doi: 10.1016/j.dnarep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chumki SA, et al. Remarkably long-tract gene conversion induced by fragile site instability in Saccharomyces cerevisiae. Genetics. 2016;204:115–128. doi: 10.1534/genetics.116.191205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nag DK, et al. Both CAG repeats and inverted DNA repeats stimulate spontaneous unequal sister-chromatid exchange in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5677–5684. doi: 10.1093/nar/gkh901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.House NCM, et al. NuA4 Initiates Dynamic Histone H4 Acetylation to Promote High-Fidelity Sister Chromatid Recombination at Postreplication Gaps. Mol. Cell. 2014;55:818–828. doi: 10.1016/j.molcel.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin Y, et al. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 104.Groh M, et al. R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome. PLoS Genet. 2014;10:e 1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shah KA, et al. Coupling Transcriptional State to Large-Scale Repeat Expansions in Yeast. Cell Rep. 2014;9:1594–1603. doi: 10.1016/j.celrep.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Belotserkovskii BP, et al. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 107.Pandey S, et al. Transcription blockage by stable H-DNA analogs in vitro. Nucleic Acids Res. 2015;43:6994–7004. doi: 10.1093/nar/gkv622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for PolyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 109.Debacker K, et al. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e 1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koch MR, et al. The Isw1 chromatin remodeler prevents excision repair-induced CAG repeat expansions during transcription in Saccharomyces cerevisiae. Genetics. 2018 doi: 10.1534/genetics.117.300529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prado F, et al. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 1997;16:2826–35. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dominguez-Sanchez MS, et al. Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO / TREX. PLoS Genet. 2011;7:19–22. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gómez-González B, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gavaldá S, et al. R-Loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0065541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santos-Pereira JM, et al. Npl3, a new link between RNA-binding proteins and the maintenance of genome integrity. Cell Cycle. 2014;13:1524–1529. doi: 10.4161/cc.28708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gavaldá S, et al. Excess of Yra1 RNA-binding factor causes transcription-dependent genome instability, replication impairment and telomere shortening. PLoS Genet. 2016;12:e1005966. doi: 10.1371/journal.pgen.1005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Salas-Armenteros I, et al. Human THO – Sin 3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 2017;36:3532–3547. doi: 10.15252/embj.201797208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duquette ML, et al. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin Y, Wilson JH. Transcription-Induced CAG Repeat Contraction in Human Cells Is Mediated in Part by Transcription-Coupled Nucleotide Excision Repair. Mol. Cell. Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Belotserkovskii BP, et al. Transcription blockage by homopurine DNA sequences: Role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013;41:1817–1828. doi: 10.1093/nar/gks1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Polleys EJ, et al. Role of recombination and replication fork restart in repeat instability. DNA Repair (Amst) 2017;56:156–165. doi: 10.1016/j.dnarep.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saini N, et al. Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in Saccharomyces cerevisiae. PLoS Genet. 2013;9:1–12. doi: 10.1371/journal.pgen.1003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shah KA, Mirkin SM. The hidden side of unstable DNA repeats: Mutagenesis at a distance. DNA Repair (Amst) 2015;32:106–112. doi: 10.1016/j.dnarep.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sakofsky CJ, Malkova A. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 2017;52:395–413. doi: 10.1080/10409238.2017.1314444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao H, et al. Peptide SS-31 upregulates frataxin expression and improves the quality of mitochondria: Implications in the treatment of Friedreich ataxia. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-10320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Erwin G, et al. Synthetic transcription elongation factors license transcription across repressive chromatin. Science. 2017;6414(358):1617–1622. doi: 10.1126/science.aan6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li L, et al. Activating frataxin expression by repeat-targeted nucleic acids. Nat. Commun. 2016;7:1–8. doi: 10.1038/ncomms10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bergquist H, et al. Disruption of higher order DNA structures in friedreich’s ataxia (GAA)n Repeats by PNA or LNA Targeting. PLoS One. 2016;11:1–24. doi: 10.1371/journal.pone.0165788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Richard G. Shortening trinucleotide repeats using highly specific endonucleases: a possible approach to gene therapy? Trends Genet. 2015;31:177–186. doi: 10.1016/j.tig.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 131.Cinesi C, et al. Contracting CAG/CTG repeats using the CRISPR-Cas9 nickase. Nat. Commun. 2016;7:e 13272. doi: 10.1038/ncomms13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gellon L, et al. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genet. 2011;7:e 1001298. doi: 10.1371/journal.pgen.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goula A, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mason AG, et al. Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Hum. Mol. Genet. 2014;23:1606–1618. doi: 10.1093/hmg/ddt551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wheeler VC, et al. Mismatch repair gene Msh2 modifies the timing of early disease in HdhQ111 striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 136.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goula AV, et al. Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet. 2012;8:e1003051. doi: 10.1371/journal.pgen.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lokanga RA, et al. Somatic Expansion in Mouse and Human Carriers of Fragile X Premutation Alleles. Hum. Mutat. 2013;34:157–166. doi: 10.1002/humu.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dion V. Tissue specificity in DNA repair: Lessons from trinucleotide repeat instability. Trends Genet. 2014;30:220–229. doi: 10.1016/j.tig.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 140.Amiri K, et al. Fragile X–Associated Tremor/Ataxia Syndrome. Neurol. Rev. 2008;65:19–25. doi: 10.1001/archneurol.2007.30. [DOI] [PubMed] [Google Scholar]
- 141.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, Spinocerebellar ataxia type 1. J. Biol. Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Moss DJH, et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 2017;16:701–711. doi: 10.1016/S1474-4422(17)30161-8. [DOI] [PubMed] [Google Scholar]
- 143.Swami M, et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suelves N, et al. A selective inhibitor of histone deacetylase 3 prevents cognitive deficits and suppresses striatal CAG repeat expansions in Huntington’s disease mice. Nat. Sci. Reports. 2017;7:1–15. doi: 10.1038/s41598-017-05125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morales F, et al. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum. Mol. Genet. 2012;21:3558–3567. doi: 10.1093/hmg/dds185. [DOI] [PubMed] [Google Scholar]
- 146.Morales F, et al. A polymorphism in the MSH3 mismatch repair gene is associated with the levels of somatic instability of the expanded CTG repeat in the blood DNA of myotonic dystrophy type 1 patients. DNA Repair. 2016;40:57–66. doi: 10.1016/j.dnarep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 147.Tome S, et al. MSH3 Polymorphisms and protein levels affect CAG repeat instability in Huntington’s disease mice. PLoS Genet. 2013;9:e1003280. doi: 10.1371/journal.pgen.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keogh N, et al. MutSBeta abundance and Msh3 ATP hydrolysis activity are important drivers of CTG•CAG repeat expansions. Nucleic Acids Res. 2017;45:10068–10078. doi: 10.1093/nar/gkx650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee JM, et al. Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell. 2015;162:516–526. doi: 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee J, et al. A modifier of Huntington’s disease onset at the MLH1 locus. 2017;26:3859–3867. doi: 10.1093/hmg/ddx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Holmans PA, et al. Genetic modifiers of Mendelian disease: Huntington’s disease and the trinucleotide repeat disorders. Hum. Mol. Genet. 2017;26:R83–R90. doi: 10.1093/hmg/ddx261. [DOI] [PubMed] [Google Scholar]
- 153.Bettencourt C, et al. DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 2016;79:983–990. doi: 10.1002/ana.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bhattacharyya S, et al. Identification of RTG2 as a Modifier Gene for CTG • CAG Repeat Instability in Saccharomyces cerevisiae. Genetics. 2002;589:579–589. doi: 10.1093/genetics/162.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y, et al. Genome-wide screen reveals replication pathway for quasi-palindrome fragility dependent on homologous recombination. PLoS Genet. 2013;9:e1003979. doi: 10.1371/journal.pgen.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pan X, et al. Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library. BMC Genomics. 2012;13:e662. doi: 10.1186/1471-2164-13-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saka K, et al. More than 10% of yeast genes are related to genome stability and influence cellular senescence via rDNA maintenance. Nucleic Acids Res. 2016;44:4211–4221. doi: 10.1093/nar/gkw110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Adzhubei I, et al. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7: Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang T, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;80:80–85. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]