Abstract
Congenital disorders of glycosylation (CDG) are a rapidly expanding group of metabolic disorders that result from abnormal protein or lipid glycosylation. They are often difficult to clinically diagnose because they broadly affect many organs and functions and lack clinical uniformity. However, recent technological advances in next generation sequencing have revealed a treasure trove of new genetic disorders, expanded the knowledge of known disorders, and showed a critical role in infectious diseases. More comprehensive genetic tools specifically tailored for mammalian cell-based models have revealed a critical role for glycosylation in pathogen-host interactions, while also identifying new CDG susceptibility genes. We highlight recent advancements that have resulted in a better understanding of human glycosylation disorders, perspectives for potential future therapies and mysteries that continue to seek new insights for their solution.
Keywords: Glycosylation, Next generation sequencing
What 30 years of genetics has taught us about glycosylation
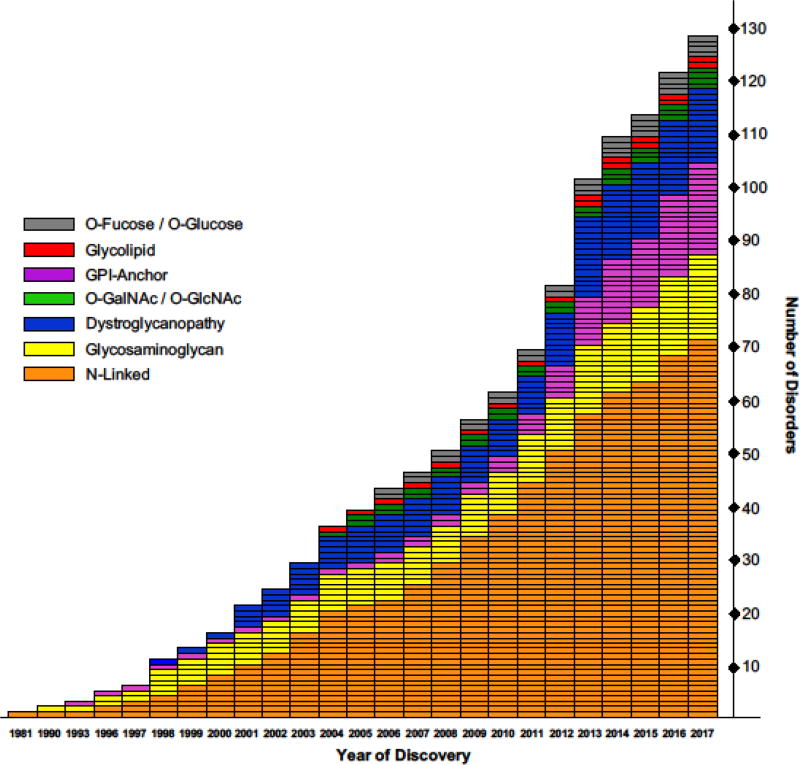
Glycosylation (see glossary) is an essential cellular process where individual monosaccharides are uniquely arranged to form the most expansive and diverse type of protein or lipid modifications across all species [1]. It is conservatively estimated that ~2% of all genes in the human genome encode for proteins involved in various aspects of glycosylation and that half of all cellular proteins are directly glycosylated in some form [2]. Thus, it is not surprising that more than 125 genetic disorders have been identified across several glycosylation pathways including N-linked, O-linked (O-Mannose, O-Glucose, O-Fucose, O-GlcNAc, O-GalNAc), glycosaminoglycan (GAG), glycosylphosphatidylinositol (GPI) and glycolipids [3,4] (Figure 1, Table 1). Adding further complexity are disorders with unanticipated roles in glycosylation, such as ER signal peptide recognition, vesicular transport, Golgi architecture and homeostasis to name a few [3,5] (Figure 2).
Figure 1. Discovery of glycosylation disorders.
The distribution of glycosylation-related disorders by the year they were identified. They are grouped according to the glycosylation pathway which they affect and while most fall within a clear specific pathway, there are those that can affect multiple glycosylation disorders. This describes disorders identified up to the end of 2017. This figure is an updated version adapted from Freeze, H.H. et al. (2014) [10].
Table 1.
Disorders Identified by WES from 2010 – 2017
| Glycosylation Pathway | Disorders identified from 2010 – 2017 |
Disorders identified by WES from 2010 – 2017 |
Total Number of Disorders |
|---|---|---|---|
| N-Linked or Multiple | 33 | 25 | 71 |
| Glycosaminoglycan | 8 | 5 | 16 |
| Dystroglycanopathy | 7 | 4 | 14 |
| O-GalNAc / O-GlcNAc | 2 | 2 | 4 |
| GPI-Anchor | 14 | 12 | 17 |
| Glycolipid | 1 | 1 | 2 |
| O-Fucose / O-Glucose | 2 | 2 | 4 |
| Totals | 67 | 51 | 128 |
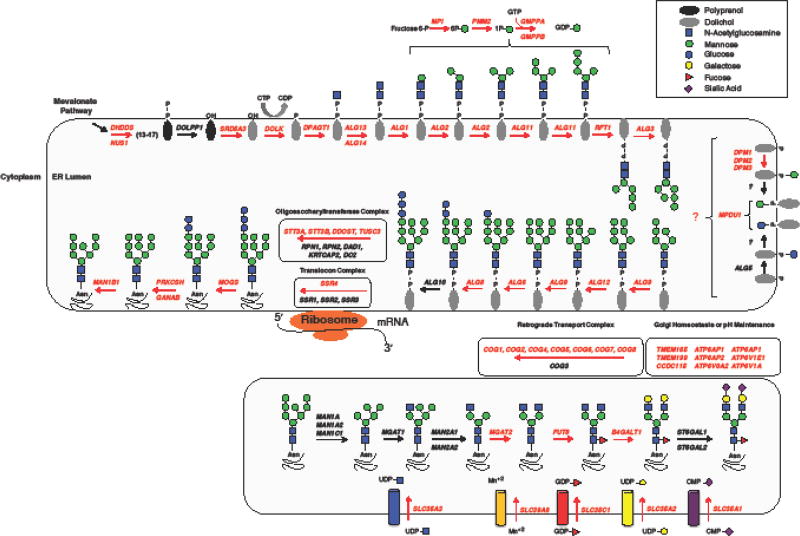
Figure 2, Key Figure. N-linked glycosylation pathway.
A schematic of the N-linked pathway highlighting those genes required for the both the initial steps of lipid linked oligosaccharide synthesis as well as several key components for glycan processing within the Golgi. These genes highlighted in red represent known loci for glycosylation disorders. This figure is an updated version adapted from Freeze, H.H. et al. (2015) [4]
Because glycosylation occurs in every cell in every organism, it is not surprising that patients with glycosylation defects show highly diverse clinical presentations that seldom generate genetic identity. This, plus the complexity of glycosylation itself, may account for the longevity and hesitation of working on glycobiology; what some have termed as “Glycophobia”. The recent arrival of Next Generation Sequencing (NGS) has rapidly expanded both the discovery of novel glycosylation-related disorders and the unraveling of many unsolved cases. Ultimately, the likelihood of a clinician encountering a patient with a glycosylation disorder has increased propelling glycobiology to a more pronounced position. Because of the rapid expansion of disorders across several unique pathways, we often use both congenital disorders of glycosylation (CDG) and glycosylation related disorders to describe a genetic disorder whose primary defect impacts one or more glycosylation pathways. Here we will explore new developments in the field of glycosylation disorders, perspectives on therapies and attempt to highlight unanswered questions that remain in this expanding field.
How Next-Generation sequencing propelled the CDG field
The first genetic disorder definitively linked to a specific gene via Whole Exome Sequencing (WES) occurred in early 2010 with the finding that Miller syndrome was caused by mutations in DHODH, a key enzyme in the pyrimidine de novo biosynthesis pathway [6]. Since then, broader accessibility and higher rates of conclusive diagnosis have made WES a preferred method for solving undiagnosed genetic disorders [7,8]. Prior to expansion by WES, there were approximately sixty glycosylation disorders with most having been solved by painstaking biochemical trial and error or individual gene sequencing (Figure 1). However, shortly thereafter, the first glycosylation-related disorder (PIGV-CDG) was solved using WES as the cause of Hyperphosphatasia with mental retardation syndrome 1 [9]. Fast forward to today, nearly half of the 128 glycosylation-related disorders discovered since 2010, have been identified by exome sequencing [10] (Table 1).
The emergence of pathogenic de novo mutations in CDG
Most glycosylation disorders follow an autosomal recessive inheritance pattern; however, exome sequencing using family based trios has helped to shed light on how de novo mutations can also result in various forms of CDG. The first gene to be identified with confirmed pathogenic de novo mutations was the X-linked ALG13, which is primarily associated with epilepsy [11]. Since the initial single case report, nearly all subsequently identified cases have been affected females with the same specific recurrent de novo mutation, c.320A>G (p.Asn107Ser) [12–18]. However, an affected male who was hemizygous for the c.320A>G (p.Asn107Ser) was recently identified with a clinical phenotype similar to previously reported affected females [19]. Since the Asn107 is critical for the binding of ALG13 to its substrate, UDP-GlcNAc, this mutation was thought to result in a complete loss of function [20], however the presence of an affected hemizygous male suggests that the p.Asn107Ser likely has some residual activity. Is it possible that this or other individuals with ALG13-CDG have varying degrees of mosaicism or chimerism and this accounts for why glycosylation is “normal” within these cases? This has yet to be addressed.
Surprisingly, when the abundant serum glycoprotein transferrin (Box 1), a biomarker for many types of CDG was tested in p.Asn107Ser ALG13-CDG females, it revealed normal glycosylation and only minor glycosylation abnormalities were seen in the affected male [15,19]. This is curious since ALG13 encodes an essential glycosyltransferase required for the second step of lipid linked oligosaccharide (LLO) synthesis and has long been associated with N-linked glycosylation [21,22] (Figure 2). One explanation could be that in higher eukaryotes, ALG13 may have functionally evolved since it differs significantly from lower eukaryotic organisms such as the yeast strain Saccharomyces cerevisiae. In S. cerevisiae, the ALG13 gene encodes a small molecular weight protein of approximately 200 amino acids, however at some point in evolution ALG13 underwent a fusion event to form an approximately 1100 amino acid protein that now contains several other functional domains outside the highly-conserved glycosyltransferase domain [23]. Could it be that in higher eukaryotes ALG13 has evolved additional functions outside its annotated role in glycosylation? Or could the several different isoforms expressed in humans possess vastly different functions? These are questions that remain a mystery. Interestingly, in zebrafish this fusion event did not occur since there are two separate genes that align to the single human gene (https://zfin.org/). This could make zebrafish a useful model for studying the function of ALG13.
Box 1. Serum transferrin, a biomarker for most CDG types.
The abundant serum glycoprotein transferrin is primarily synthesized in the liver and normally contains two N-glycan molecules. Glycosylation status can be analyzed using a number of methods including isoelectric focusing (IEF) and electrospray ionization mass spectrometry (ESI-MS) to name a few [105].
It can often be used to identify many, but not all types of N-linked glycosylation disorders [5,10] and while useful in identifying affected individuals, an abnormal Tf profile very rarely implicates a specific gene defect.
Abnormalities in Tf glycosylation can be described as either a type I or II profile [106]. A type I profile involves a Tf molecule that has an absence of either one or both of its N-glycan chains and is most commonly associated with defects in the synthesis or transfer of LLO molecules [106]. On the other hand, a type II is most often associated with processing and remodeling of the protein bound N-glycans and can result in a large degree of N-glycan structural diversity [106].
Abnormalities in sugar nucleotide metabolism, Golgi homeostasis and architecture, even pH maintenance within the different Golgi compartments can all contribute to type II disorders.
It is of interest that a few reports have described individuals with a classical X-linked form of ALG13-CDG [24,25], however upon closer inspection these variants are present in the hemizygous state in several individuals from the gnomAD-ExAC database (http://gnomad.broadinstitute.org/gene/ENSG00000101901). Since the gnomAD-ExAC database has “removed individuals affected by severe pediatric disease” it seems unlikely that these variants are truly pathogenic. This finding underscores the importance of having an independent functional assay to assess pathogenicity.
De novo mutations in SLC35A2, which in humans encodes the major uridine diphosphate (UDP)-galactose transporter in the Golgi, are associated with another CDG originally described in three unrelated individuals (2 males and 1 female) [26] (Figure 2). What makes SLC35A2-CDG unusual is that, SLC35A2 is essential for transporting UDP-galactose into the Golgi and ultimately affects several types of glycosylation, yet nearly all patients who were tested show normal glycosylation [27,28]. A few affected individuals have shown severely abnormal galactosylation early in life that later became completely normal [26]. Once again, the question of why the loss of a key glycosylation transporter results in such varied glycosylation status remains a mystery. It is possible that the UDP-GlcNAc transporter, SLC35A3, could have a broader specificity and potentially compensate for the loss of SLC35A2 in certain tissue types. This substrate recognition and compensation by SLC35A2-SLC35A3 chimera proteins has been seen in cell based models [29]. Alternatively, the normalization of transferrin and serum glycoproteins from the liver in some affected individuals could be explained by the positive selection for the wild-type SLC35A2 allele [26].
Identification of CDG genes that cause divergent disorders
It is not unheard of for different mutations within a specific gene to cause distinct clinical disorders that can also vary in their inheritance patterns. Until recently, this was seldom seen for CDG. Perhaps the best example involves the PIGA gene, which encodes a protein required for the synthesis of N-acetylglucosaminyl phosphatidylinositol (GlcNAc-PI), the first step in GPI anchor biosynthesis [30]. Somatic mutations in PIGA can cause paroxysmal nocturnal hemoglobinuria, an acquired hematologic disorder characterized by hemolytic anemia, thrombosis and expansion of hematopoietic stem cells expressing the mutant PIGA. However, X-linked forms that present with severe neurological and frequent lethality also exist [31–35].
It was recently reported that recurrent de novo mutations within DHDDS and NUS1 can cause developmental and epileptic encephalopathies [36]. DHDDS and NUS1, interact and play pivotal roles in the earliest part of the LLO pathway (Figure 2, Key Figure), and have already been shown to cause multiple recessive disorders [37–40]. In fact, DHDDS has now been shown to cause three clinically distinct disorders. The first, is due to a founder mutation c.124A>G (p.Lys42Glu) within the Ashkenazi Jewish population that causes an autosomal recessive form of retinitis pigmentosa [37,38], the second is a lethal autosomal recessive form that presents with a more traditional multisystem involvement CDG phenotype [39] and the third is due to de novo mutations causing a developmental and epileptic encephalopathy disorder [36]. These highly divergent presentations are unlikely to be isolated occurrences and more examples of this will likely appear as more affected individuals and trios undergo in-depth sequencing analysis.
In fact, we are already seeing this occur in other LLO pathway genes (DPAGT1, ALG14, ALG2) that can cause a severe form of CDG, but also a milder form of congenital myasthenic syndrome [41–45]. It remains unclear why mutations in this set of genes cause these two specific and very different disorders. It is known the early LLO biosynthesis enzymes do form specific protein complexes and we speculate that it may possible certain mutations can dramatically affect enzyme activity while other less severe mutations, alter complex formation.
Expanding the molecular toolbox for glycobiology
For decades, most genetic analysis of glycosylation was done in Saccharomyces cerevisiae using phenotypic mutants [46–48]. However, this work focused primarily on the early N-linked and Golgi processing pathways [46,49]. The major downside to using Saccharomyces cerevisiae is that it does not have machinery for N-glycan branching, galactosylation, sialylation or fucosylation pathways to name a few [46]. Therefore, a mammalian cell equivalent to S.cerevisiae was needed. Chinese hamster ovary (CHO) mutant cell lines have provided an immense wealth of knowledge about specific glycosylation pathways and their proteins, but isolating and identifying the mutated genes can often be painstaking [50].
Recently, technological advances have been made in developing comprehensive whole genome shRNA and CRISPR/CAS9 libraries for such screens. As a proof of principle, several independent groups employed multiple screening methods (RNAi, Gene-Trap and CRISPR) to identify genes required for cellular resistance to the toxicity of the lectin ricin [51–54]. Importantly, while different methods were used, each study identified similar factors.
Since then, several genetic screens have been applied to identifying key host factors for several infectious agents. One example of such a genetic screen involved taking advantage of the pathogen, Lassa virus (LASV), which requires properly glycosylated α-dystroglycan (α-DG) as a ligand for viral entry [55]. α-DG is an extensively glycosylated extracellular peripheral protein that contains unique O-Mannose based glycans, which are critical for linking the extracellular matrix with the cytoskeleton [56]. An inability to properly assemble these O-Mannose glycans on α-DG results in a group of 17 glycosylation-related disorders termed “Dystroglycanopathies” [57–59]. Prior to the LASV screen several key steps in the assembly of the O-Mannose glycan had not been identified and in fact the complete structure of the glycan was still a mystery. Yet the screen identified, what is believed to be, every component required to make the functional O-Mannose glycans. Importantly, the screen identified established dystroglycanopathy genes (DAG1, LARGE, FKRP, FKTN, POMT1, POMT2, POMGNT1, B3GALNT2) but also revealed several potential candidate genes, that would later be found to cause various types of dystroglycanopathy (ISPD, TMEM5, POMGNT2, B4GAT1, POMK) [60–65] (Table 2). Since dolichol phosphate mannose (Dol-P Man) is required to generate O-Mannose glycans, it is not surprising that the several established CDG genes were identified (i.e. PMM2, MPDU1, DPM1,3) [56] (Table 2). Ultimately the identification of these novel genes led to the unraveling of these unique O-mannose glycans, using both traditional and state-of-the-art glycobiology tools [56,66–68].
Table 2.
Glycosylation genes identified as critical host factors for various toxins or pathogens.
| Toxin or Pathogen |
Genes identified | Glycosylation pathway utilized |
Reference |
|---|---|---|---|
| Ricin Toxicity | FUT9, SLC35C1, GALNT2, B4GALT1, B4GALNT3, ST3GAL4 | Fucosylation and galactosylation | [51,102] |
| Dengue Virus | MAGT1, STT3A, STT3B, DDOST, RPN1, RPN2, OSTC, OST4, KRTCAP2, DAD1, B3GALT6, EXT1, SLC35B2, SSR1, SSR2, SSR3, SRP9, SRP14, HM13, SPCS3, SEC61A1 | OST and translocon complexes | [69,70] |
| Lassa Virus | LARGE, ISPD, B3GALNT2, TMEM5, B4GAT1, POMK, DAG, POMT1, POMT2, FKTN, FKRP, ST3GAL4, SLC35A1, GNE, CMAS, SLC35A2, PTAR1, COG5, 7, 8, MAN1B1, MGAT1, MAN1A1, ALG5, ALG6, ALG8, MPDU1, DPM1, DPM3 | O-Mannose | [64] |
| Cholera Toxin | ST3GAL5, B3GALT4, SLC35A2 | Galactosylation | [77] |
| Enterovirus D68 | SLC35A1, GNE, NANS, ST6GAL1, MGAT5, B4GALT1, ST3GAL4 | Sialic Acid | [78] |
| Chikungunya Virus | EXT1, EXT2, PTAR1, TMEM165, COG 1– 8, NDST1, EXTL3, B3GAT3 | GAG synthesis | [79] |
| Rift Valley fever virus | SLC35B2, EXT1, EXT2, EXTL3, XYLT2 UXS1, UGDH, NDST1, B3GAT3, B3GALT6, B4GALT7, COG1, COG2, COG3, COG4, COG5, COG7, COG8, PTAR1 | GAG synthesis | [103] |
| Monkeypox Virus | EXT1, B3GAT3, B4GALT7, B3GALT6, SLC35B2, XYLT2, COG3, COG4, COG7, COG8, PTAR1, TRAPPC13, PIGL, DPM3, MPDU1 | GAG synthesis | [104] |
Another genetic screen highlighting the importance of N-glycosylation in pathogen susceptibility was done by two independent groups studying host factors for Flaviviridae viruses (i.e. dengue virus (DENV) [69,70]. The screen specifically performed by Carette et al., used either a Gene-Trap mutagenized population of haploid cells or a stably infected HUH7 population of lentiviral CRISPR cells and took advantage of the cytopathic effects of DENV to identify critical host factors [69]. From this screen two critical pathways for DENV toxicity were identified, first was the oligosaccharide transferase complex (OST) (STT3A, STT3B, RPN1, RPN2, OSTc, KRTCAP2, MAGT1, DAD1, OST4) and second were factors in the translocon complex (SSR1, SSR2 and SSR3), which has been shown to bind directly to and influence substrate specificity of the OST complex [71] (Table 2). As with the LASV screen, several of the identified factors from the DENV screen have been established as CDG genes including STT3A, STT3B and MAGT1 [72,73] (Table 2). Interestingly, SSR4 another core component of the TRAP complex was not identified in the screen, but does cause a form of CDG termed SSR4-CDG [74,75]. It is possible that unlike SSR1–3, SSR4 could be an essential gene whose knock out phenotype is lethal to the cell type used in the screen.
These are just two examples of nearly a dozen screens performed to date, other screens have identified glycosylation factors critical for Influenza A virus, Enterovirus D68, cholera toxin and chikungunya virus to name a few [76–79] (Table 2). These findings reiterate the importance of glycosylation genes in infectious diseases.
The importance of the Flaviviridae viral screen is highlighted by a recent paper in which a small molecule inhibitor to the OST subunit, STT3b, was shown to have potent antiviral activity and a potentially high barrier of resistance [80]. This potentially opens the door to more glycosylation specific targeted therapies.
Perspective therapies
Over one-hundred glycosylation disorders exist, but there are relatively few treatment options for CDG’s or glycosylation-related disorders in general. The first CDG to be efficiently treated was MPI-CDG, which simply required dietary supplementation with D-mannose [81]. However, MPI-CDG is unlike most CDG types since it lacks the severe neurological presentation [4]. Another disorder benefiting from “monosaccharide therapy” is SLC35C1-CDG which is a defect in the transport of GDP-fucose into the Golgi [82, 83]. Here individuals present with leukocyte adhesion deficiency (LADII) which, in some cases, can be quickly corrected by providing dietary L-fucose [83]. Interestingly, not all reported cases of LADII respond to fucose therapy, suggesting that certain patient mutations my act through an inability to directly bind GDP-fucose [84].
Over the last five years the field has seen increased success in finding treatments that either resolve most clinical issues or at least provide some measurable clinical benefits. Three such disorders (PGM1-CDG, SLC35A2-CDG and TMEM165-CDG) have been reported to show clinical benefit by simply providing the monosaccharide, D-galactose [85–88]. Another disorder, SLC39A8-CDG which is a defect in manganese transport can be effectively treated with both D-galactose and manganese supplementation [89–91]. In a few individuals with CAD-CDG, a defect in de novo pyrimidine biosynthesis, uridine supplementation has been shown to virtually reverse clinical symptoms in a surprisingly short period of time. One affected individual went from a minimal state of responsiveness to communicating and assisted walking within nine weeks of treatment [92]. While these therapies may not apply to all affected individuals, manipulating metabolic flux does appear to be a possible and a realistic option.
Still, a treatment for PMM2-CDG continues to garner the most attention given that it is the most common CDG type with > 900 reported cases [5,93]. Phosphomannomutase 2 (PMM2) is a 246-amino acid protein that is required for converting mannose-6 phosphate (Man6p) to mannose-1 phosphate (Man1p) within the cytoplasm. To date ~115 disease causing PMM2 mutations have been identified and listed in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=PMM2) [94]. The primary glycosylation defect in PMM2-CDG arises because mannose-1 phosphate (Man1p) functions as the intermediate for making GDP-mannose, which then serves as a high-energy nucleotide sugar donor to various mannosyltransferases [95] (Figure 2).
Initially, promising results utilizing primary fibroblast from various CDG cases (including PMM2-CDG) showed that when cells were incubated with mannose, temporary improvements to glycosylation could be achieved [96,97]. However, attempts to replicate this success in PMM2-CDG patients showed no significant benefit of oral mannose [96]. More recently, pharmacological chaperons have been used to stabilize specific patient mutations [98]. However, one challenge that has plagued many therapy efforts is how to target such a large array of patient-relevant mutations. Since more than half of PMM2-CDG patients carry the common p.R141H mutation, efforts have focused on how to target this one specific mutation, which has a carrier frequency of ~1/90, but is lethal when homozygous [99,100]. However, none has been successful thus far.
Attempts have been made to directly provide hydrophobic Man1p derivatives that cross the plasma membrane into the cytoplasm of cultured fibroblasts. Penetration occurred, but compound toxicity related to the removal of the substituents covering the charged phosphate group was unacceptable [101]. Technological and chemical advancements have improved shuttling charged molecules, such as Man1P, across the plasma membrane and thus efforts have resurfaced at trying to facilitate Man1p penetration of the membrane [5].
Concluding Remarks
Over the last ten years the number of genetic disorders related to abnormal glycosylation has more than tripled from ~40 to over 125 today. This has been driven largely by advancements in next generation sequencing, specifically whole exome sequencing. (See Outstanding Questions Box) While WES has been incredibly successful in identifying causal candidate genes, the need for discriminating between benign and pathogenic variants is still an absolute necessity. Especially as an increased number of individuals from both healthy and diseased populations are sequenced revealing more rare variants. For many years, the complex world of glycosylation may have had a dampening effect on its embrace by the larger community, but the emergence of glycosylation pathways in genetic disorders, infectious diseases, cancer and metabolism has propelled it to the forefront of science requiring both new and continued collaborations between the medical and research communities.
Outstanding Questions Box.
There are unanswered questions regarding glycosylation disorders on both the basic research and medical fronts. However, with the recent rise of next generation sequencing, more genetic screening methods and development of more sensitive biochemical instrumentation, specifically in mass spectrometry, the field of glycosylation has propelled into prominence. As these tools become increasingly sophisticated, mysteries will be answered and dogmas may be challenged.
Will glycosylation-related disorders that are due to de novo mutations or those that display multiple inheritance patterns and diverse phenotypes become more frequent as more affected families are sequenced?
Can monosaccharides alone or in combination with other means be used as a therapeutic strategy for additional glycosylation-related disorders or even in complex disorders like cancer?
Knowing the critical roles of glycosylation in infectious diseases, will additional screens of viruses or other pathogens reveal additional glycosylation targets? Perhaps small molecules will be developed targeting those pathways.
Highlights.
Over 125 Congenital disorders of glycosylation (CDG) are clinically diverse and cover all major glycosylation pathways.
Next Generation Sequencing (NGS) enabled discovery of over 51 novel glycosylation disorders and identified de novo mutations in several disorders.
Complex genetic screening of human cells shows glycosylation is critical for many pathogens, especially viruses. Targeting glycosylation pathways could become short-term treatments.
Simple monosaccharide therapies and novel chemical approaches offer treatments for several glycosylation disorders.
Acknowledgments
We would like to thank all the families, medical and research collaborators who have contributed to furthering glycosylation disorder research. The Rocket Fund and National Institutes of Health (NIH) grant R01DK099551 supported this work.
Glossary
- Dystroglycanopathies
a group of genetic disorders that most often involve abnormal glycosylation of α-dystroglycan.
- Glycosylation
a metabolic enzyme dependent process where sugar molecules (monosaccharides) are attached to an acceptor molecule, most often proteins or lipids.
- Gnome-ExAC database
a free public database resource of unrelated individuals that excludes individuals with severe pediatric disease, as well as their first-degree relatives. It numbers 123,136 exomes and 15,496 genomes sequences.
- Lipid linked oligosaccharide
also known as LLO refers to the glycolipid structure which is the typical precursor for N-glycans prior to transfer of the glycan to a nascent proteins.
- Pharmacological chaperons
a group of molecules that are used to aid in the proper folding of proteins that are misfolded.
- Next Generation sequencing
a general term for high throughput DNA sequencing.
- Whole exome sequencing
a form of next generation sequencing that specifically targets the protein-coding regions of the genome termed “exons” that make up ~ 1% of the genome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertozzi CR, Rabuka D. Essentials of Glycobiology (nd et al. eds) Chapter 2. Cold Spring Harbor Laboratory Press; 2009. Structural Basis of Glycan Diversity; pp. 23–36. [PubMed] [Google Scholar]
- 2.Apweiler R, et al. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 3.Jaeken J, Peanne R. What is new in CDG? J. Inherit. Metab. Dis. 2017;40:569–586. doi: 10.1007/s10545-017-0050-6. [DOI] [PubMed] [Google Scholar]
- 4.Freeze HH, et al. Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 2015;38:105–125. doi: 10.1146/annurev-neuro-071714-034019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peanne R, et al. Congenital disorders of glycosylation (CDG): Quo vadis? Eur. J. Med. Genet. 2017 doi: 10.1016/j.ejmg.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers L, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet. Med. 2017;19:1055–1063. doi: 10.1038/gim.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monroe GR, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet. Med. 2016;18:949–956. doi: 10.1038/gim.2015.200. [DOI] [PubMed] [Google Scholar]
- 9.Krawitz PM, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 10.Freeze HH, et al. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timal S, et al. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum. Mol. Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 12.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 13.Epi KC, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaud JL, et al. The genetic landscape of infantile spasms. Hum. Mol. Genet. 2014;23:4846–4858. doi: 10.1093/hmg/ddu199. [DOI] [PubMed] [Google Scholar]
- 15.Smith-Packard B, et al. Girls with Seizures Due to the c.320A>G Variant in ALG13 Do Not Show Abnormal Glycosylation Pattern on Standard Testing. JIMD Rep. 2015;22:95–98. doi: 10.1007/8904_2015_416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimassi S, et al. Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin. Genet. 2016;89:198–204. doi: 10.1111/cge.12636. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi Y, et al. High prevalence of genetic alterations in early-onset epileptic encephalopathies associated with infantile movement disorders. Brain Dev. 2016;38:285–292. doi: 10.1016/j.braindev.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Ortega-Moreno L, et al. Molecular diagnosis of patients with epilepsy and developmental delay using a customized panel of epilepsy genes. PLoS One. 2017;12:e0188978. doi: 10.1371/journal.pone.0188978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galama WH, et al. ALG13-CDG with Infantile Spasms in a Male Patient Due to a De Novo ALG13 Gene Mutation. JIMD Rep. 2017 doi: 10.1007/8904_2017_53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase. Structure. 2008;16:965–975. doi: 10.1016/j.str.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao XD, et al. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J. Biol. Chem. 2005;280:36254–36262. doi: 10.1074/jbc.M507569200. [DOI] [PubMed] [Google Scholar]
- 22.Bickel T, et al. Biosynthesis of lipid-linked oligosaccharides in Saccharomyces cerevisiae: Alg13p and Alg14p form a complex required for the formation of GlcNAc(2)-PP-dolichol. J. Biol. Chem. 2005;280:34500–34506. doi: 10.1074/jbc.M506358200. [DOI] [PubMed] [Google Scholar]
- 23.Esposito T, et al. Dysregulation of the Expression of Asparagine-Linked Glycosylation 13 Short Isoform 2 Affects Nephrin Function by Altering Its N-Linked Glycosylation. Nephron. 2017;136:143–150. doi: 10.1159/000455129. [DOI] [PubMed] [Google Scholar]
- 24.Bissar-Tadmouri N, et al. X chromosome exome sequencing reveals a novel ALG13 mutation in a nonsyndromic intellectual disability family with multiple affected male siblings. Am. J. Med. Genet. A. 2014;164A:164–169. doi: 10.1002/ajmg.a.36233. [DOI] [PubMed] [Google Scholar]
- 25.Gadomski TE, et al. ALG13-CDG in a male with seizures, normal cognitive development, and normal transferrin isoelectric focusing. Am. J. Med. Genet. A. 2017;173:2772–2775. doi: 10.1002/ajmg.a.38377. [DOI] [PubMed] [Google Scholar]
- 26.Ng BG, et al. Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 2013;92:632–636. doi: 10.1016/j.ajhg.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodera H, et al. De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum. Mutat. 2013;34:1708–1714. doi: 10.1002/humu.22446. [DOI] [PubMed] [Google Scholar]
- 28.Kimizu T, et al. A case of early onset epileptic encephalopathy with de novo mutation in SLC35A2: Clinical features and treatment for epilepsy. Brain Dev. 2017;39:256–260. doi: 10.1016/j.braindev.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Olczak M, et al. UDP-Gal/UDP-GlcNAc chimeric transporter complements mutation defect in mammalian cells deficient in UDP-Gal transporter. Biochem. Biophys. Res. Commun. 2013;434:473–478. doi: 10.1016/j.bbrc.2013.03.098. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson MAJ, et al. Essentials of Glycobiology (rd et al. eds) Chapter 12. Cold Spring Harbor Laboratory Press; 2015. Glycosylphosphatidylinositol Anchors; pp. 137–150. [Google Scholar]
- 31.Johnston JJ, et al. The phenotype of a germline mutation in PIGA: the gene somatically mutated in paroxysmal nocturnal hemoglobinuria. Am. J. Hum. Genet. 2012;90:295–300. doi: 10.1016/j.ajhg.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Crabben SN, et al. Expanding the spectrum of phenotypes associated with germline PIGA mutations: a child with developmental delay, accelerated linear growth, facial dysmorphisms, elevated alkaline phosphatase, and progressive CNS abnormalities. Am. J. Med. Genet. A. 2014;164A:29–35. doi: 10.1002/ajmg.a.36184. [DOI] [PubMed] [Google Scholar]
- 33.Belet S, et al. Early frameshift mutation in PIGA identified in a large XLID family without neonatal lethality. Hum. Mutat. 2014;35:350–355. doi: 10.1002/humu.22498. [DOI] [PubMed] [Google Scholar]
- 34.Kato M, et al. PIGA mutations cause early-onset epileptic encephalopathies and distinctive features. Neurology. 2014;82:1587–1596. doi: 10.1212/WNL.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 35.Tarailo-Graovac M, et al. The genotypic and phenotypic spectrum of PIGA deficiency. Orphanet J. Rare Dis. 2015;10:23. doi: 10.1186/s13023-015-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamdan FF, et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zelinger L, et al. A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am. J. Hum. Genet. 2011;88:207–215. doi: 10.1016/j.ajhg.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuchner S, et al. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabry S, et al. A case of fatal Type I congenital disorders of glycosylation (CDG I) associated with low dehydrodolichol diphosphate synthase (DHDDS) activity. Orphanet J. Rare Dis. 2016;11:84. doi: 10.1186/s13023-016-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park EJ, et al. Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 2014;20:448–457. doi: 10.1016/j.cmet.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, et al. Deficiency of UDP-GlcNAc:Dolichol Phosphate N-Acetylglucosamine-1 Phosphate Transferase (DPAGT1) causes a novel congenital disorder of Glycosylation Type Ij. Hum. Mutat. 2003;22:144–150. doi: 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- 42.Belaya K, et al. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am. J. Hum. Genet. 2012;91:193–201. doi: 10.1016/j.ajhg.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel C, et al. A new type of congenital disorders of glycosylation (CDG-Ii) provides new insights into the early steps of dolichol-linked oligosaccharide biosynthesis. J. Biol. Chem. 2003;278:22498–22505. doi: 10.1074/jbc.M302850200. [DOI] [PubMed] [Google Scholar]
- 44.Cossins J, et al. Congenital myasthenic syndromes due to mutations in ALG2 and ALG14. Brain. 2013;136(Pt 3):944–956. doi: 10.1093/brain/awt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schorling DC, et al. Early and lethal neurodegeneration with myasthenic and myopathic features: A new ALG14-CDG. Neurology. 2017;89:657–664. doi: 10.1212/WNL.0000000000004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doering TL, et al. Essentials of Glycobiology (rd et al. eds) Chapter 23. Cold Spring Harbor Laboratory Press; 2015. Fungi; pp. 293–304. [Google Scholar]
- 47.Huffaker TC, Robbins PW. Temperature-sensitive yeast mutants deficient in asparagine-linked glycosylation. J. Biol. Chem. 1982;257:3203–3210. [PubMed] [Google Scholar]
- 48.Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. U S A. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley P, et al. Essentials of Glycobiology (rd et al. eds) Chapter 9. Cold Spring Harbor Laboratory Press; 2015. N-Glycans; pp. 99–111. [PubMed] [Google Scholar]
- 50.Patnaik SK, Stanley P. Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 51.Moreau D, et al. Genome-wide RNAi screens identify genes required for Ricin and PE intoxications. Dev. Cell. 2011;21:231–244. doi: 10.1016/j.devcel.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Elling U, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han K, et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao W, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 56.Sheikh MO, et al. Recent advancements in understanding mammalian O-mannosylation. Glycobiology. 2017;27:806–819. doi: 10.1093/glycob/cwx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanagawa M, Toda T. Muscular Dystrophy with Ribitol-Phosphate Deficiency: A Novel Post-Translational Mechanism in Dystroglycanopathy. J. Neuromuscul. Dis. 2017;4:259–267. doi: 10.3233/JND-170255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godfrey C, et al. Dystroglycanopathies: coming into focus. Curr. Opin. Genet. Dev. 2011;21:278–285. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Manya H, Endo T. Glycosylation with ribitol-phosphate in mammals: New insights into the O-mannosyl glycan. Biochim. Biophys. Acta. 2017;1861:2462–2472. doi: 10.1016/j.bbagen.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 60.Willer T, et al. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 2012;44:575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vuillaumier-Barrot S, et al. Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am. J. Hum. Genet. 2012;91:1135–1143. doi: 10.1016/j.ajhg.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manzini MC, et al. Exome sequencing and functional validation in zebrafish identify GTDC2 mutations as a cause of Walker-Warburg syndrome. Am. J. Hum. Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buysse K, et al. Missense mutations in beta-1,3-N-acetylglucosaminyltransferase 1 (B3GNT1) cause Walker-Warburg syndrome. Hum. Mol. Genet. 2013;22:1746–1754. doi: 10.1093/hmg/ddt021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jae LT, et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340:479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida-Moriguchi T, et al. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Praissman JL, et al. The functional O-mannose glycan on alpha-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 2016;5 doi: 10.7554/eLife.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerin I, et al. ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto alpha-dystroglycan. Nat. Commun. 2016;7:11534. doi: 10.1038/ncomms11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riemersma M, et al. Human ISPD Is a Cytidyltransferase Required for Dystroglycan O-Mannosylation. Chem. Biol. 2015;22:1643–1652. doi: 10.1016/j.chembiol.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 69.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang R, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfeffer S, et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017;8:14516. doi: 10.1038/ncomms14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shrimal S, et al. Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum. Mol. Genet. 2013;22:4638–4645. doi: 10.1093/hmg/ddt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li FY, et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature. 2011;475:471–476. doi: 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Losfeld ME, et al. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum. Mol. Genet. 2014;23:1602–1605. doi: 10.1093/hmg/ddt550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng BG, et al. Expanding the Molecular and Clinical Phenotype of SSR4-CDG. Hum. Mutat. 2015;36:1048–1051. doi: 10.1002/humu.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 77.Guimaraes CP, et al. Identification of host cell factors required for intoxication through use of modified cholera toxin. J. Cell. Biol. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baggen J, et al. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. U S A. 2016;113:1399–1404. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka A, et al. Genome-Wide Screening Uncovers the Significance of N-Sulfation of Heparan Sulfate as a Host Cell Factor for Chikungunya Virus Infection. J. Virol. 2017;91 doi: 10.1128/JVI.00432-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puschnik AS, et al. A Small-Molecule Oligosaccharyltransferase Inhibitor with Pan-flaviviral Activity. Cell Rep. 2017;21:3032–3039. doi: 10.1016/j.celrep.2017.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Niehues R, et al. Carbohydrate-deficient glycoprotein syndrome type Ib. Phosphomannose isomerase deficiency and mannose therapy. J. Clin. Invest. 1998;101:1414–1420. doi: 10.1172/JCI2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luhn K, et al. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- 83.Lubke T, et al. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- 84.van de Vijver E, et al. Hematologically important mutations: leukocyte adhesion deficiency (first update) Blood Cells Mol. Dis. 2012;48:53–61. doi: 10.1016/j.bcmd.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tegtmeyer LC, et al. Multiple phenotypes in phosphoglucomutase 1 deficiency. N. Engl. J. Med. 2014;370:533–542. doi: 10.1056/NEJMoa1206605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morava E. Galactose supplementation in phosphoglucomutase-1 deficiency; review and outlook for a novel treatable CDG. Mol. Genet. Metab. 2014;112:275–279. doi: 10.1016/j.ymgme.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nolting K, et al. Limitations of galactose therapy in phosphoglucomutase 1 deficiency. Mol. Genet. Metab. Rep. 2017;13:33–40. doi: 10.1016/j.ymgmr.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wong SY, et al. Oral D-galactose supplementation in PGM1-CDG. Genet. Med. 2017;19:1226–1235. doi: 10.1038/gim.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park JH, et al. SLC39A8 deficiency: biochemical correction and major clinical improvement by manganese therapy. Genet. Med. 2017 doi: 10.1038/gim.2017.106. [DOI] [PubMed] [Google Scholar]
- 90.Riley LG, et al. A SLC39A8 variant causes manganese deficiency, and glycosylation and mitochondrial disorders. J Inherit Metab Dis. 2017;40:261–269. doi: 10.1007/s10545-016-0010-6. [DOI] [PubMed] [Google Scholar]
- 91.Park JH, et al. SLC39A8 Deficiency: A Disorder of Manganese Transport and Glycosylation. Am. J. Hum. Genet. 2015;97:894–903. doi: 10.1016/j.ajhg.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koch J, et al. CAD mutations and uridine-responsive epileptic encephalopathy. Brain. 2017;140:279–286. doi: 10.1093/brain/aww300. [DOI] [PubMed] [Google Scholar]
- 93.Altassan R, et al. Renal involvement in PMM2-CDG, a mini-review. Mol. Genet. Metab. 2017 doi: 10.1016/j.ymgme.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 94.Vals MA, et al. The Prevalence of PMM2-CDG in Estonia Based on Population Carrier Frequencies and Diagnosed Patients. JIMD Rep. 2017 doi: 10.1007/8904_2017_41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matthijs G, et al. Mutations in PMM2, a phosphomannomutase gene on chromosome 16p13, in carbohydrate-deficient glycoprotein type I syndrome (Jaeken syndrome) Nat. Genet. 1997;16:88–92. doi: 10.1038/ng0597-88. [DOI] [PubMed] [Google Scholar]
- 96.Kjaergaard S, et al. Failure of short-term mannose therapy of patients with carbohydrate-deficient glycoprotein syndrome type 1A. Acta. Paediatr. 1998;87:884–888. doi: 10.1080/080352598750013680. [DOI] [PubMed] [Google Scholar]
- 97.Panneerselvam K, Freeze HH. Mannose corrects altered N-glycosylation in carbohydrate-deficient glycoprotein syndrome fibroblasts. J. Clin. Invest. 1996;97:1478–1487. doi: 10.1172/JCI118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yuste-Checa P, et al. Pharmacological Chaperoning: A Potential Treatment for PMM2-CDG. Hum. Mutat. 2017;38:160–168. doi: 10.1002/humu.23138. [DOI] [PubMed] [Google Scholar]
- 99.Matthijs G, et al. Lack of homozygotes for the most frequent disease allele in carbohydrate-deficient glycoprotein syndrome type 1A. Am. J. Hum. Genet. 1998;62:542–550. doi: 10.1086/301763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kjaergaard S, et al. Absence of homozygosity for predominant mutations in PMM2 in Danish patients with carbohydrate-deficient glycoprotein syndrome type 1. Eur. J. Hum. Genet. 1998;6:331–336. doi: 10.1038/sj.ejhg.5200194. [DOI] [PubMed] [Google Scholar]
- 101.Eklund EA, et al. Hydrophobic Man-1-P derivatives correct abnormal glycosylation in Type I congenital disorder of glycosylation fibroblasts. Glycobiology. 2005;15:1084–1093. doi: 10.1093/glycob/cwj006. [DOI] [PubMed] [Google Scholar]
- 102.Taubenschmid J, et al. A vital sugar code for ricin toxicity. Cell Res. 2017;27:1351–1364. doi: 10.1038/cr.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Riblett AM, et al. A Haploid Genetic Screen Identifies Heparan Sulfate Proteoglycans Supporting Rift Valley Fever Virus Infection. J. Virol. 2015;90:1414–1423. doi: 10.1128/JVI.02055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Realegeno S, et al. Monkeypox Virus Host Factor Screen Using Haploid Cells Identifies Essential Role of GARP Complex in Extracellular Virus Formation. J. Virol. 2017;91 doi: 10.1128/JVI.00011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lacey JM, et al. Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin. Chem. 2001;47:513–518. [PubMed] [Google Scholar]
- 106.Jaeken J, Matthijs G. Congenital disorders of glycosylation: a rapidly expanding disease family. Annu. Rev. Genomics Hum. Genet. 2007;8:261–278. doi: 10.1146/annurev.genom.8.080706.092327. [DOI] [PubMed] [Google Scholar]