Abstract
PccA and SenC are periplasmic copper chaperones required for the biogenesis of cbb3-type cytochrome c oxidase (cbb3-Cox) in Rhodobacter capsulatus at physiological Cu concentrations. However, both proteins are dispensable for cbb3-Cox assembly when the external Cu concentration is high. PccA and SenC bind Cu using Met and His residues and Cys and His residues as ligands, respectively, and both proteins form a complex during cbb3-Cox biogenesis. SenC also interacts directly with cbb3-Cox, as shown by chemical cross-linking. Here we determined the periplasmic concentrations of both proteins in vivo and analyzed their Cu binding stoichiometries and their Cu(I) and Cu(II) binding affinity constants (KD) in vitro. Our data show that both proteins bind a single Cu atom with high affinity. In vitro Cu transfer assays demonstrate Cu transfer both from PccA to SenC and from SenC to PccA at similar levels. We conclude that PccA and SenC constitute a Cu relay system that facilitates Cu delivery to cbb3-Cox.
Graphical Abstract
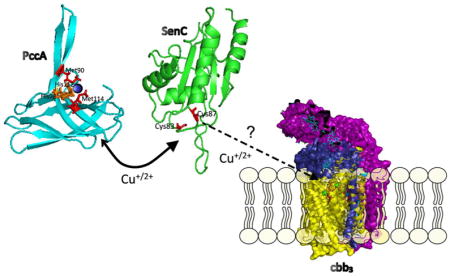
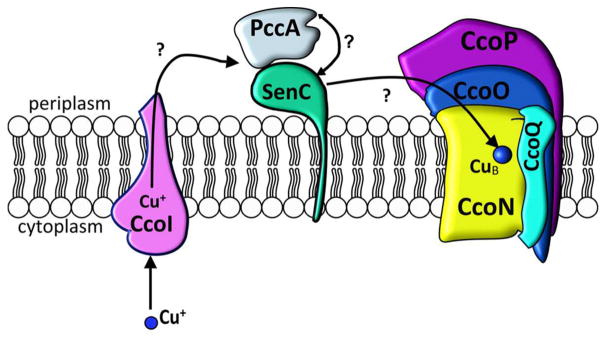
The essential microelement copper (Cu) is required for the activity of multiple enzymes such as cytochrome (cyt) c oxidases (Cox), superoxide dismutases, and tyrosinases.1,2 Because of its electrochemical properties, Cu is an excellent catalyst for electron transfer reactions, but its undesirable reactions with oxygen pose a risk for oxidative cellular damage.3,4 Cu also reacts readily with thiol groups, thereby inactivating enzymes.4 Consequently, Cu trafficking in cells is highly coordinated for preventing the occurrence of free Cu. Central to this strategy are Cu chaperones, a heterogeneous group of proteins that bind Cu and facilitate its controlled delivery to target enzymes. This is exemplified for cbb3-type Cox (cbb3-Cox) assembly in bacteria, which depends on two periplasmic Cu chaperones, PccA and SenC5,6 (Figure 1). SenC belongs to the Sco family of chaperones, and its Cu binding site is composed of two Cys residues in a CXXXCP motif and a conserved distant His residue.7 Sco-like chaperones are tethered to the inner mitochondrial membrane in eukaryotes or to the cytoplasmic membrane in bacteria, with their Cu binding motif facing the intermembrane space or the periplasm, respectively.1,8 In addition to Cu binding, a thioredoxin-like function of Sco-like chaperones has also been suggested.9 Initially these chaperones were implicated in the assembly of the binuclear CuA center of subunit II of aa3-type Cox (aa3-Cox),10 but increasing evidence shows that they are also involved in CuB center formation in cbb3- and aa3-Cox, in particular at low Cu concentrations.5,11,12 In contrast to aa3-Cox and some ba3-type Cox, cbb3-Cox lacks a CuA center; instead, it contains two membrane-anchored c-type cyt subunits, CcoO and CcoP, that transfer electrons to the binuclear CuB-heme b3 center within the catalytic subunit CcoN.13,14
Figure 1.
Putative copper delivery pathway for cbb3-Cox in Rhodobacter capsulatus. Cu is exported to the periplasm by the P1B-type ATPase CcoI.16,17 Two periplasmic chaperones, PccA and SenC, are required for transfer of the exported Cu to the catalytic subunit CcoN of cbb3-Cox.5 Whether this is a sequential transfer and the directionality of the transfer are unknown. In addition to CcoN, cbb3-Cox in R. capsulatus contains two membrane-bound c-type cyt subunits (CcoO and CcoP) and a small subunit that is required for stability (CcoQ).45 In addition, the assembly factor CcoH is stably associated with the cbb3-Cox complex in Rhodobacter membranes.46
During the assembly of cbb3-Cox, SenC cooperates with PccA, a periplasmic Cu chaperone of the PCuAC family.5 Homologues of PccA are present only in bacteria, and they bind Cu via two conserved Met and His residues.15 In the absence of either SenC or PccA, cbb3-Cox assembly is diminished.5 Importantly, Cu supplementation (>1 μM) of the growth medium rescues the cbb3-Cox assembly defect in the absence of SenC or PccA or both.5,6 However, even at high Cu concentrations, cbb3-Cox assembly still depends on the Cu-exporting P1B-type ATPase CcoI,16,17 which likely excludes the possibility of unassisted Cu insertion into CcoN at high Cu concentrations. CcoI exports Cu from a cytosolic Cu pool maintained by the Cu-uptake activity of the major facilitator superfamily protein CcoA, in concert with CopA, which is another P1B-type ATPase that exports superfluous Cu.18,19 Although CcoI and CopA are both Cu-exporting ATPases, only CcoI provides Cu for cbb3-Cox assembly,16 suggesting a specific interaction with SenC or PccA under Cu limitation.
To date there is no clear consensus as to the directionality of Cu transfer between SenC and PccA. PCuAC-like chaperones were considered to represent functional homologues of the mitochondrial Cox17 chaperones, which deliver Cu to Sco1 in eukaryotes.2 A function of PCuAC upstream of Sco1 was also proposed during Cox assembly in Rhodobacter sphaeroides and Bradyrhizobium japonicum.11,12 Alternatively, Sco proteins have been suggested to reduce Cys residues in the CuA binding motif of subunit II of aa3-Cox, thereby facilitating Cu insertion via PCuAC.10 In this model, PCuAC would work downstream of Sco1. It is noteworthy that several organisms contain more than one Sco paralogue, which seem to cooperate during CuA assembly.20 Finally, the deletion of PCuAC-like chaperones had no or only a weak effect on aa3-Cox assembly in Paracoccus dentrificans21 and R. sphaeroides,11 suggesting that Sco proteins do not necessarily depend on PCuAC-like chaperones for CuA assembly. These different models for Sco- and PCuAC-dependent Cox assembly probably reflect differences in (1) the strategies for CuA and CuB assembly, (2) the CuB assembly processes in different heme copper oxygen reductases (e.g., aa3- vs cbb3-Cox), (3) the orthologues present in the studied model organisms, and (4) the cellular concentrations of these Cu chaperones and their Cu affinities.
In this work, we analyzed the cellular concentrations of SenC and PccA chaperones in the model organism Rhodobacter capsulatus, which is a perfect model organism for studying cbb3-Cox assembly because it is the sole Cox present in this organism. We determined the Cu stoichiometries of PccA and SenC and their Cu(I) and Cu(II) affinities and directly monitored Cu transfer between these two proteins in order to provide further insight into their roles in cbb3-Cox assembly.
RESULTS AND DISCUSSION
Cellular Concentrations of SenC and PccA in R. capsulatus Cells
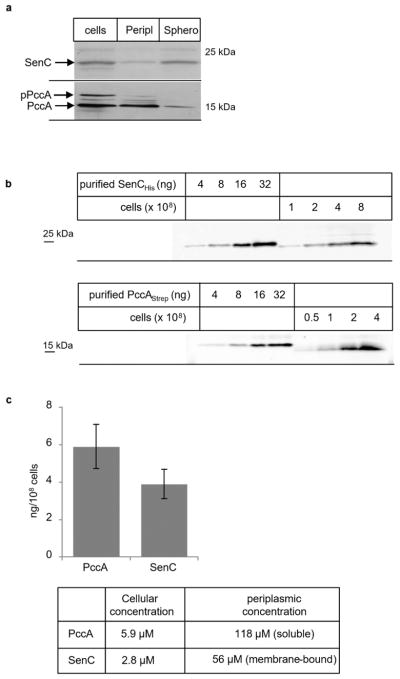
For determination of the cellular concentrations of PccA and SenC in wild-type R. capsulatus cells, the localization of both proteins was first determined by immune detection after cell fractionation. The periplasmic Cu binding domain of SenC is tethered to the cytoplasmic membrane via a single transmembrane domain,6 and the majority of SenC was detected in the membrane fraction after spheroplast preparation (Figure 2a). A small amount was also seen in the periplasmic fraction, which likely reflects some membrane contamination of this fraction. PccA is a periplasmic protein that is synthesized with a cleavable signal sequence, and it is predominantly detected as two bands in whole cells. The mature form of PccA (i.e., after its signal sequence cleavage) was detected mainly in the periplasmic fraction and only weakly in the spheroplast fraction. The small amount of PccA present in the spheroplast fraction probably reflects the portion of PccA that is associated with membrane-bound proteins, such as SenC.5 Thus, in whole cells the majority of each protein is present in the periplasm as either soluble (PccA) or membrane-tethered (SenC) protein.
Figure 2.
Quantification of SenC and PccA in R. capsulatus cells. (a) Rhodobacter cells were directly TCA-precipitated, and cell pellets were separated by SDS-PAGE followed by Western blotting and immune detection using α-SenC and α-PccA antibodies. From the same cell culture, spheroplasts (Sphero) were generated and a periplasmic fraction (Peripl) was prepared as described.35 These samples were separated by SDS-PAGE and analyzed by immune detection. pPccA corresponds to the signal sequence containing premature PccA. (b) Different numbers of Rhodobacter cells were directly TCA-precipitated. As references, different amounts of SenCHis or PccAStrep purified from Escherichia coli were loaded on the same gel and subjected to immune detection. (c) The chemiluminescent signal intensities of the immune blots in (b) were quantified using ImageJ. Quantification was based on at least three biological replicate samples with two technical replicates for each sample. The mean values are shown, and the error bars indicate the standard deviation (SD). The mean values for PccA (5.9 ng/108 cells) and SenC (3.9 ng/108 cells) were used to calculate their cellular concentrations assuming a total volume of 0.6 × 10−15 L for a single Rhodobacter cell22 and the predicted molecular weights of PccA (16.4 kDa) and SenC (23 kDa). The periplasmic concentrations were calculated on the basis of the assumption that the periplasmic volume corresponds to approximately 5% of the total cellular volume in Gram-negative bacteria.23
Different amounts of cells grown under semiaerobic conditions on enriched (MPYE) medium were precipitated using trichloroacetic acid (TCA) and separated by SDS-PAGE, together with defined amounts of purified proteins for quantitative Western blotting (Figure 2b). The concentrations of mature PccA and SenC were determined to be 5.9 ± 1.1 ng/108 cells and 3.9 ± 0.8 ng/108 cells, respectively (Figure 2c). The total volume of a single Rhodobacter cell was reported to correspond to 0.6 × 10−15 L.22 On the basis of these values, the total cellular concentrations of PccA (MW = 16.4 kDa) and SenC (MW = 23 kDa) in R. capsulatus were calculated to be ~5.9 and ~2.8 μM, respectively. As these proteins are primarily located in the periplasm, their periplasmic concentration was also calculated. The periplasmic volume corresponds to ~5% of the total cell volume in Gram-negative bacteria,23 i.e., 3 × 10−17 L for Rhodobacter, resulting in periplasmic concentrations of ~118 μM for PccA and ~56 μM for SenC (Figure 2c). The total cellular concentrations of SenC and PccA are comparable to those of yeast mitochondrial Sco1 (2 μM), Sco2 (3 μM), and Cox17 (10 μM).24 As these yeast proteins are mainly located in the intermembrane space of mitochondria,1 their local concentrations would be significantly higher and probably also similar to the periplasmic concentrations of SenC and PccA. The high periplasmic concentrations of SenC and PccA correlate with the facts that most bacterial cuproenzymes reside in the periplasm and that bacteria generally have few cytosolic Cu-binding proteins.25 For membrane-integral Cu-dependent enzymes like Cox, metalation presumably occurs from the periplasmic side of the membrane,11,26–28 also requiring a periplasmic Cu delivery system.
Cu Stoichiometry and Affinity to SenC and PccA
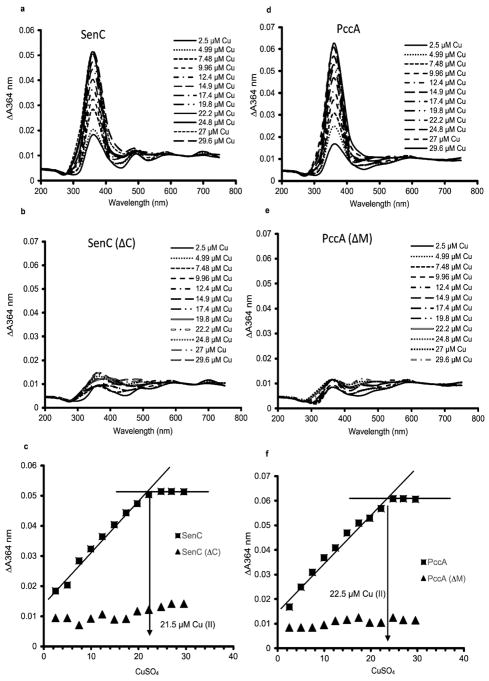
For determination of the stoichiometry of Cu binding, the formation of the thiolate–Cu(II) charge transfer band6 was monitored using a defined concentration of SenC (23 μM) and increasing CuSO4 concentrations (Figure 3). The visible spectrum revealed a major Cu-dependent peak at 364 nm, which is diagnostic for the thiolate–Cu(II) charge transfer complex (Figure 3a). As a control, for a SenC mutant lacking its conserved Cys residues (Cys83 and Cys87, Rhodobacter numbering), no charge transfer complex was observed (Figures 3c and S1). These data further established that Cu binding involves the two conserved Cys residues of SenC, supporting previous studies.6 Plotting the absorbance at 364 nm (A364) against the Cu(II) concentration revealed saturation at 21.5 μM (Figure 3c), indicating a 1:1 Cu(II):SenC stoichiometry, in line with the results obtained for other Sco homologues.7
Figure 3.
Determination of the Cu binding stoichiometries of SenC and PccA. (a) Purified SenC (23 μM) was incubated with increasing concentrations of CuSO4 (2.5 to 30 μM), and the absorbance spectra were recorded between 200 and 750 nm. Cu-free SenC (as purified) was used to record the baseline. (b) The same experiment as in (a) was repeated with 23 μM SenC(ΔC), a purified mutant variant of SenC lacking the conserved Cys residues of its Cu binding motif. (c) The absorbances at 364 nm (indicative of a Cu(II)–thiolate charge transfer complex) measured in (a) and (b) were plotted against the CuSO4 concentrations used for determining the Cu(II) binding stoichiometry of SenC. Binding saturation was reached at ~21.5 μM Cu(II), indicating a 1:1 Cu(II):SenC stoichiometry. No significant Cu binding was detected with SenC(ΔC). (d) As in (a) except that purified PccA (23 μM) was used. (e) As in (d) except that 23 μM PccA(ΔM), a purified PccA variant lacking the two conserved Met residues of its Cu binding motif, was used. (f) As in (c). Cu(II) binding saturation was reached at ~22.5 μM Cu(II), indicating a 1:1 Cu(II):PccA stoichiometry. No significant Cu binding was detected with PccA(ΔM).
The same approach was also employed for PccA, which binds Cu via a conserved His-Met motif.5 A Cu-concentration-dependent peak at 364 nm was observed (Figure 3d), which was not observed for a PccA variant in which its two conserved Met residues were replaced by Leu (Figures 3e and S1). Plotting the absorption as a function of the Cu(II) concentration also revealed a 1:1 Cu(II):PccA stoichiometry (Figure 3f).
As both SenC and PccA bind a single Cu(II) ion, competition experiments with the Cu(II) chelator EDTA were also performed. At pH 7.0, EDTA binds Cu(II) with an apparent of 3.2 × 10−15 M and provides suitable competition to SenC and PccA.29 SenC or PccA (20 μM in 50 mM HEPES, pH 7.5, containing 150 mM NaCl) was incubated with increasing Cu(II) concentrations (0–30 μM) in the absence or presence of 20 μM EDTA. Relative absorbances for SenC and PccA, A364/Amax (absorbance at 364 nm, baseline-corrected and normalized to the maximum value) were plotted against [Cu(II)/protein] (Figure 4a). For both proteins, the absorbance arising from Cu(II)–protein complexes shifted downward when EDTA was present, indicating that SenC or PccA competed with EDTA for the available Cu(II). The absorbance shift was more pronounced for PccA than for SenC, indicating that SenC has a higher affinity for Cu(II) than PccA. On the basis of the apparent competition, the KD values for Cu(II) were calculated according to published protocols29 and found to be 3.7 × 10−17 M for SenC and 1.4 × 10−16 M for PccA (Figure 4a).
Figure 4.
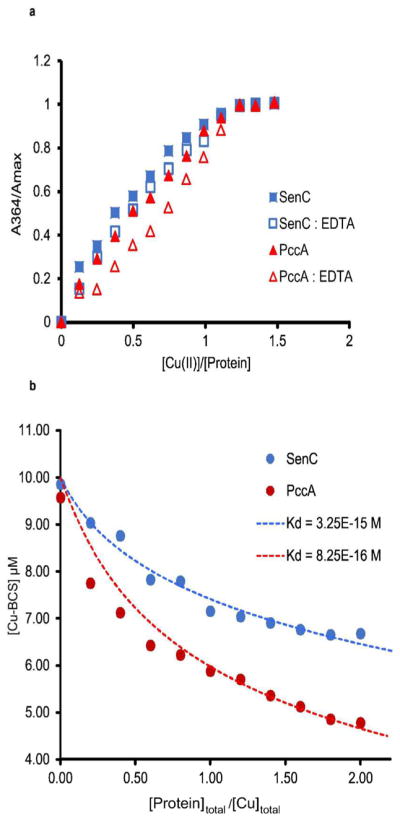
Determination of Cu(II) and Cu(I) affinities of SenC and PccA. (a) Competition between PccA or SenC and EDTA for Cu(II) binding. Purified PccA or SenC (20 μM in 50 mM HEPES, pH 7.5, containing 150 mM NaCl) was mixed with varying concentrations of CuSO4 (0 to 30 μM) in the absence (solid blue squares for SenC and solid red triangles for PccA) or presence (open blue squares for SenC and open red triangles for PccA) of 20 μM EDTA, and the binding equilibrium was followed by monitoring the absorbance at 364 nm. For ease of comparison, A364 was normalized to the maximum value (Amax) and plotted vs the ratio of the Cu(II) and SenC or PccA concentrations. The KD values for Cu(II) were determined using the plots of A364 vs [Cu(II)] following the method of Xiao and Wedd (eq 14 in ref 29; see the Supporting Information for details). (b) Determination of dissociation constants KD for binding of Cu(I) to SenC or PccA using Cu-BCS as a competitive probe. The [Cu-BCS] concentration was plotted as a function of [SenC]total/[Cu]total (blue circles) or [PccA]total/[Cu]total (red circles). Dashed lines represent the best fits of the experimental data to eq 1 in the Supporting Information, yielding KD values of 3.25 × 10−15 and 8.25 × 10−16 M for SenC and PccA, respectively. The extinction constant of Cu-BCS at 483 nm (ε483) is 13 000 cm−1 M−1; [Cu]total = 10 μM; [BCS]total = 25 μM; [SenC]total or [PccA]total = 0 to 20 μM; β2 (formation constant for Cu-BCS) = 1019.8 M−2; Cu-BCS stands for the [CuI(BCS)2]3− complex.
Sco-like proteins have been shown to bind both Cu(II) and Cu(I), but their respective affinities appear to be species-dependent.10,30,31 The affinities of R. capsulatus SenC and PccA for Cu(I) were determined using a competition assay with bathocuproine disulfonate (BCS) as described previously29,32 (Figure 4b). Briefly, 10 μM Cu(II) was reduced with 10 μM ascorbic acid and incubated with 25 μM BCS, and the [CuI(BCS)2]3− complex (Cu-BCS) thus formed was titrated with different SenC or PccA concentrations varying from 0 to 20 μM. The decrease in Cu-BCS with increasing total protein concentration was monitored at 483 nm, and the competition values were determined as described in the Supporting Information. This yielded values of 3 × 10−15 M for SenC and 8 × 10−16 M for PccA (Figure 4b).
The R. capsulatus SenC and PccA KD values for Cu(I) and Cu(II) are in the same ranges as those reported for PCuAC-like10,11,15 and other Cu chaperones.29,33 An exception is the Bacillus subtilis ScoI homologue, which seems to have approximately picomolar affinity for Cu(II), but only approximately micromolar affinity for Cu(I).31,34 The basis of this difference is unclear, although the Gram-positive bacterium B. subtilis lacks a PccA homologue that could transfer Cu(I) to ScoI. Perhaps preferential binding of Cu(II) by B. subtilis ScoI in an oxidizing environment, such as the extracellular face of the membrane, might be beneficial in this case.
Cu Transfer between PccA and SenC
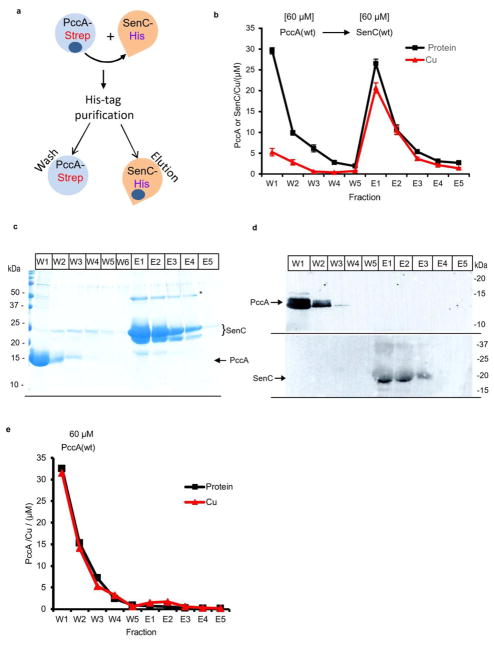
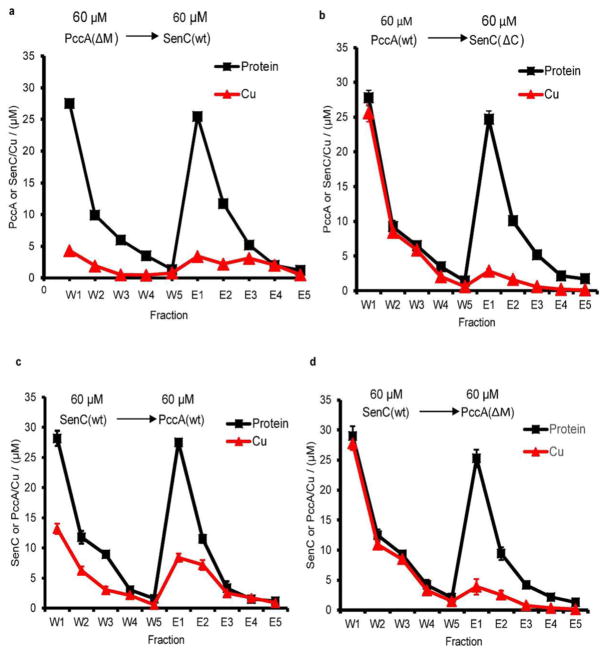
Although both PccA and SenC are involved in Cu delivery to cbb3-Cox, it is unknown in which order both proteins link Cu export via CcoI with Cu insertion into cbb3-Cox.5,6 Thus, an in vitro assay based on affinity chromatography using differentially epitope-tagged purified proteins was developed to probe the Cu transfer between PccA and SenC (Figure 5a). As both proteins bind Cu(II) and Cu(I) tightly, this assay was first performed with Cu(II) under aerobic conditions. Purified PccAStrep or SenCHis was incubated with a 1.5-fold excess of Cu(II), and unbound Cu(II) was removed by size-exclusion chromatography, after which the Cu contents of the Cu-loaded and Cu-free forms of these proteins were verified by atomic absorption spectroscopy (AAS) (Figure S2).35 These experiments also showed that the His and Strep tags used did not interfere with Cu binding, as no appreciable amount of Cu was detectable in mutants of tagged SenC and PccA devoid of their liganding residues. In this assay (Figure 5a), a Cu-loaded protein (e.g., PccAStrep) was incubated with another Cu-free protein (e.g., SenCHis) for 10 min at room temperature, and then the protein mixture was loaded onto an appropriate affinity column (e.g., Talon). The unbound protein (e.g., PccAStrep) was washed off (fractions W1–W6), and subsequently, the bound protein (e.g., SenCHis) was eluted (fractions E1–E5) by increasing the concentration of the appropriate affinity ligand (e.g., imidazole). The protein and Cu contents of each fraction were respectively determined by the Lowry assay and by measuring the amount of Cu-BCS obtained after release of the protein-bound Cu by SDS denaturation and its reduction by ascorbate to allow Cu-BCS formation (Figure 5b). All of the fractions were also analyzed by SDS-PAGE and immune detection to identify the proteins present in these fractions (Figure 5c,d). As a further control, Cu-loaded PccAStrep was loaded onto the Talon column in the absence of the acceptor protein SenC. No significant amounts of Cu were detected in the elution fraction, demonstrating that no Cu was retained on the column (Figure 5e).
Figure 5.
PccA transfers Cu(II) to SenC. (a) Schematic view of the Cu transfer assay used in these experiments. In brief, Strep-tagged PccA was loaded with Cu and incubated with Cu-free His-tagged SenC. The mixture was purified via Talon, resulting in wash fractions containing PccA and elution fractions containing SenC. The protein and Cu contents of each fraction were then determined. (b) Cu(II)-loaded PccAStrep (60 μM) was incubated with Cu-free SenCHis (60 μM) for 10 min at room temperature, and the mixture was loaded onto a Talon affinity column. PccA was recovered in the flow-through and wash fractions (W1–W5) and SenC in the elution fractions (E1–E5) (see the Supporting Information). For each fraction, the protein content was determined by the Lowry method and converted to molarity using the molecular weight of PccA for fractions W1–W5 and that of SenC for E1–E5. The Cu content of each fraction was determined by the BCS method after SDS denaturation and reduction to Cu(I) by ascorbate, as described in the Supporting Information. (c) Coomassie staining of the wash and elution fractions and (d) immune detection using antibodies against PccA or SenC. Cu transfer experiments were repeated three times. (e) A control experiment as in (b) but in the absence of the acceptor protein SenC indicated that a negligible amount of Cu was retained by the column matrix used.
When Cu-loaded PccA and Cu-free SenC were used, the data showed that the flow-through and wash fractions (W1–W5) contained low amounts and the elution fractions (E1–E5) high amounts of Cu per protein content (Figure 5b and Table 1). In addition, the corresponding SDS-PAGE (Figure 5c) and immune blots (Figure 5d) indicated that fractions W1–W5 and E1–E5 contained primarily PccA and SenC proteins, respectively. Thus, a large portion (~80%) of the Cu initially present in PccA was transferred to SenC (Table 1). Control experiments showed that Cu transfer from a PccA variant with a mutated Cu binding motif (PccA(ΔM)) to wild-type SenC (Figure 6a) and Cu transfer from wild-type PccA to a SenC variant lacking its Cu-binding Cys residues (SenC(ΔC)) (Figure 6b) were low (~10–20%), while the protein elution profiles remained comparable to those seen with native PccA and SenC (Figure 5b). Thus, the mutated PccA was unable to transfer Cu to SenC, and similarly, the Cu-loaded native PccA was unable to transfer Cu to the SenC mutant. Overall, the data therefore established that efficient Cu transfer from PccA to SenC requires functional Cu binding motifs in both proteins.
Table 1.
Quantification of the Cu Transfer Efficiencya
| transfer | Cu(I)
|
Cu(II)
|
||||
|---|---|---|---|---|---|---|
| unbound/bound to donor protein | coelution with acceptor protein | recovery rate | unbound/bound to donor protein | coelution with acceptor protein | recovery rate | |
| PccA → SenC | 66% | 29% | 81% | 19% | 79% | 81% |
| PccA(ΔM) → SenC | n.a. | n.a. | n.a. | 16% | 21% | 80% |
| PccA → SenC(ΔC) | 90% | 10% | 96% | 88% | 12% | 83% |
| SenC → PccA | 67% | 32% | 98% | 47% | 46% | 79% |
| SenC → PccA(ΔM) | 91% | 5% | 97% | 81% | 18% | 95% |
Quantification of the Cu transfer reactions from PccA to SenC and from SenC to PccA based on the data shown in Figures 6 and 7. The Cu and protein contents in the individual fractions were determined and calculated in μM. The Cu occupancy was then calculated as (ΣCu(W1–W5)[μM)]/Σprotein(W1–W5)[μM]) × 100% to determine the unbound Cu and the Cu still bound to the donor protein after the transfer reaction (wash fractions). The Cu occupancy of the acceptor protein after the transfer reaction was calculated accordingly as (ΣCu(E1–E5)[μM]/Σprotein(E1–E5)[μM]) × 100% (elution fractions). PccA(ΔM) and SenC(ΔC) refer to protein variants with a mutated Cu binding motif. The Cu recovery rate refers to the total amount of Cu that was recovered in the wash and elution fractions. It was calculated as (ΣCu(W1–E5)[μM]/60 μM) × 100%. It should be noted that the absolute values of the recovery rate for the transfer PccA(ΔM) → SenC are significantly lower than for the other transfer pairs.
Figure 6.
Cu(II) transfer between PccA and SenC is bidirectional and depends on the functional Cu binding motifs. (a) Cu(II) transfer was determined as in Figure 5, except that a mutant variant of PccA lacking its Cu-binding Met residues (PccA(ΔM)) was used as the Cu donor instead of native PccA. (b) Cu(II) transfer between native PccA and a mutant variant of SenC lacking its Cys residues (SenC(ΔC)). (c) Transfer of Cu(II) from SenC to PccA was determined as described in Figure 5, but in the reverse direction, using Cu-loaded SenCHis and Cu-free PccAStrep. After incubation, the mixture was loaded onto a Strep-Tactin affinity column, and SenC was recovered in the flow-through and wash fractions (W1–W5) and PccA in the elution fractions (E1–E5). The protein and Cu contents of each fraction were determined as before. (d) As in (c) except that PccA(ΔM) was used as the Cu acceptor instead of native PccA as a control for Cu transfer.
The reverse Cu transfer from SenC to PccA was also tested. Cu-loaded SenC was incubated with Cu-free PccA and loaded onto a Strep-Tactin column. Analyses of the W1–W5 and E1–E5 fractions indicated that a smaller amount of Cu (46%) was transferred from SenC to PccA. About half of the SenC molecules still retained their bound Cu, and only about half of the PccA molecules received Cu from SenC (Figure 6c). Again, a control experiment in which Cu-loaded native SenC was incubated with PccA(ΔM) showed that almost all of the Cu (~90%) remained associated with SenC (Figure 6d). The data further indicated that the less efficient Cu transfer was not due to low Cu occupancy of SenC as a Cu donor (Figure 6d).
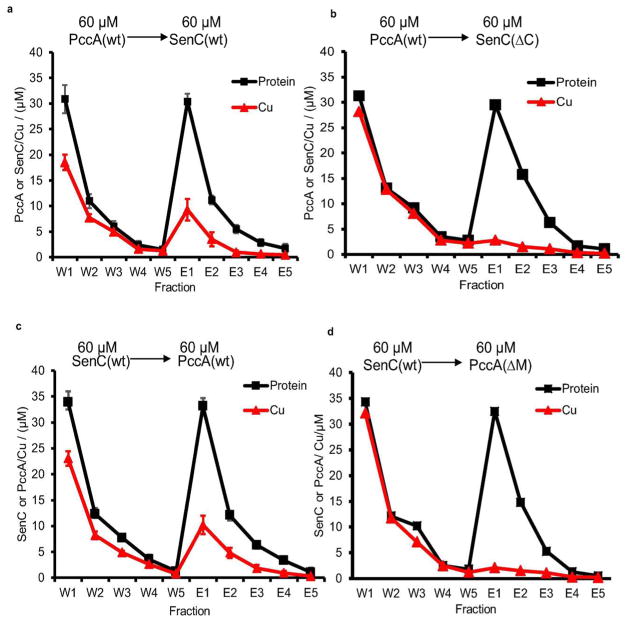
A similar experimental approach was then tested for Cu(I) under anaerobic conditions in the presence of 10 mM ascorbate. About 30% of the initially PccA-bound Cu(I) was transferred to wild-type SenC (Figure 7a), but only about 10% was transferred to SenC(ΔC) (Figure 7b). The opposite transfer from SenC to PccA was equally efficient (Figure 7c) and compromised when PccA(ΔM) was used as the acceptor (Figure 7d). The data showed that Cu(I) was less efficiently transferred than Cu(II) (Figure 7 and Table 1). This was not related to a generally lower Cu recovery rate in these experiments, because 80–100% of the initially present Cu was recovered in both the Cu(I) and Cu(II) experiments (Table 1).
Figure 7.
PccA and SenC also exchange Cu(I) bidirectionally. Cu(I) loading and transfer between PccA and SenC were performed in the presence of 10 mM ascorbate under anaerobic conditions. Cu(I) transfer between the donor and acceptor proteins was analyzed as described in Figures 5 and 6 (see the Supporting Information for details).
In summary, in vitro Cu transfer between PccA and SenC works in both directions for both Cu(I) and Cu(II). For Cu(II), the transfer from PccA to SenC appears to be more efficient than that in the opposite direction, which is in line with their calculated KD values (Figure 4a). Furthermore, Cu transfer from PccA to SenC would also be in line with the observation that SenC but not PccA directly interacts with cbb3-Cox.6 However, it is important to emphasize that the coordination ligands His, Cys, and Met are highly sensitive to the environment36,37 and therefore the in vitro-determined KD values and that the directionality of Cu transfer is likely further fine-tuned under in vivo conditions. Vectorial Cu transfer in the bacterial periplasm is probably further influenced by a complex interplay with environmental factors and small-molecule reductants like glutathione or bacillithiol38 as well as by protein–protein interactions (e.g., with disulfide oxidoreductases like DsbA39 or TlpA,40 with the Cu exporter CcoI,16 or with the target protein CcoN6).
The importance of PccA- and SenC-like Cu chaperones for both aa3- and cbb3-Cox assembly has been documented in several studies, but whether Cu is indeed transferred between these proteins or each protein acts independently in Cu transfer to cbb3-Cox has not been settled.10–12,15 The absence of SenC prevents cbb3-Cox assembly under physiological Cu concentrations, while that of PccA still allows partial assembly.5 If both proteins were to act independently, the drastically greater effect of SenC on cbb3-Cox assembly could be explained by its higher efficiency due to either its higher affinity for Cu or its higher periplasmic concentration. However, PccA is more abundant than SenC, and both have very high but comparable Cu affinities. Thus, the different phenotypes of strains lacking PccA or SenC might rather point out their sequential role during cbb3-Cox assembly, with SenC likely working downstream of PccA. Cu transfer between PccA and SenC probably involves a transient PccA–SenC protein complex that was identified in the Rhodobacter periplasm.5 Transient protein complexes have been also reported for other Cu transfer reactions and appear to be the means of preventing Cu toxicity.41
This Cu delivery pathway is not exclusive, however, as in the absence of PccA, SenC can still convey some Cu to cbb3-Cox. Indeed, residual Cox assembly in the absence of PccA has also been suggested for its respective homologues during aa3-Cox assembly in B. japonicum12 and R. sphaeroides.11 Why cbb3-Cox assembly involves the cooperation between two periplasmic Cu chaperones is intriguing. Cu delivery to two target proteins has been shown for Cox17, the predicted functional homologue of PccA in mitochondria, which delivers Cu to both Sco1 and Cox11.42 In R. capsulatus a possibility might be that PccA delivers Cu not only to SenC but also to other periplasmic cuproproteins, such as multicopper oxidase43 or periplasmic nitrous oxide reductase.44
The available data indicate that in the absence of PccA, SenC can still obtain Cu that is exported by CcoI, although possibly with lower efficiency. However, it remains to be analyzed whether SenC inserts Cu directly into cbb3-Cox, as suggested by the cross-link data between SenC and cbb3-Cox,6 or transfers Cu to as-yet-unidentified protein(s) that mediate CuB center formation. Alternatively, the absence of PccA might promote another less efficient Cu delivery pathway to cbb3-Cox that still involves SenC. The existence of such a pathway is supported by the observation that the cbb3-Cox assembly defects of single and double mutants lacking PccA or SenC or both, but not a mutant lacking CcoI, can be rescued by increasing the Cu concentration in the medium.5 This observation points out that although cbb3-Cox might receive Cu from the periplasmic side of the membrane, this Cu atom inevitably needs to originate from a cytosolic pool, as it still relies on the function of CcoA, which is the cytoplasmic Cu importer in R. capsulatus.
Supplementary Material
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG RTG2202) to H.-G.K., the Motivate College of the University Freiburg Medical School to D.M. and M.U., and the Deutscher Akademischer Austausch-dienst to P.-I.T. and partly by grants to F.D. from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences, Department of Energy (DOE DE-FG02-91ER20052) (protein purifications) and the National Institutes of Health (NIH GM 38237) (various KD determinations).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.8b00293.
Methods and Figures S1–S3 (PDF)
References
- 1.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metal-loenzymes. Biochim Biophys Acta, Mol Cell Res. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: a mechanistic systems biology approach. Cell Mol Life Sci. 2010;67:2563–2589. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palumaa P. Copper chaperones. The concept of conformational control in the metabolism of copper. FEBS Lett. 2013;587:1902–1910. doi: 10.1016/j.febslet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Argüello JM, Raimunda D, Padilla-Benavides T. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol. 2013;3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trasnea PI, Utz M, Khalfaoui-Hassani B, Lagies S, Daldal F, Koch HG. Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3-type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Mol Microbiol. 2016;100:345–361. doi: 10.1111/mmi.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohmeyer E, Schroder S, Pawlik G, Trasnea PI, Peters A, Daldal F, Koch HG. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochim Biophys Acta, Bioenerg. 2012;1817:2005–2015. doi: 10.1016/j.bbabio.2012.06.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banci L, Bertini I, Calderone V, Ciofi-Baffoni S, Mangani S, Martinelli M, Palumaa P, Wang S. A hint for the function of human Sco1 from different structures. Proc Natl Acad Sci U S A. 2006;103:8595–8600. doi: 10.1073/pnas.0601375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buggy J, Bauer CE. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:6958–6965. doi: 10.1128/jb.177.23.6958-6965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badrick AC, Hamilton AJ, Bernhardt PV, Jones CE, Kappler U, Jennings MP, McEwan AG. PrrC, a Sco homologue from Rhodobacter sphaeroides, possesses thiol-disulfide oxidoreductase activity. FEBS Lett. 2007;581:4663–4667. doi: 10.1016/j.febslet.2007.08.058. [DOI] [PubMed] [Google Scholar]
- 10.Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ, Wang S. Mechanism of Cu(A) assembly. Nat Chem Biol. 2008;4:599–601. doi: 10.1038/nchembio.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson AK, Gray J, Liu A, Hosler JP. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa(3)-type and the cbb(3)-type cytochrome c oxidases. Biochim Biophys Acta, Bioenerg. 2012;1817:955–964. doi: 10.1016/j.bbabio.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serventi F, Youard ZA, Murset V, Huwiler S, Buhler D, Richter M, Luchsinger R, Fischer HM, Brogioli R, Niederer M, Hennecke H. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. J Biol Chem. 2012;287:38812–38823. doi: 10.1074/jbc.M112.406173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 14.Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- 15.Banci L, Bertini I, Ciofi-Baffoni S, Katsari E, Katsaros N, Kubicek K, Mangani S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Proc Natl Acad Sci U S A. 2005;102:3994–3999. doi: 10.1073/pnas.0406150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch HG, Winterstein C, Saribas AS, Alben JO, Daldal F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. J Mol Biol. 2000;297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- 17.Kulajta C, Thumfart JO, Haid S, Daldal F, Koch HG. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J Mol Biol. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 18.Ekici S, Turkarslan S, Pawlik G, Dancis A, Baliga NS, Koch HG, Daldal F. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio. 2014;5:e01055–13. doi: 10.1128/mBio.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekici S, Yang H, Koch HG, Daldal F. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio. 2012;3:e00293–11. doi: 10.1128/mBio.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgada MN, Abriata LA, Cefaro C, Gajda K, Banci L, Vila AJ. Loop recognition and copper-mediated disulfide reduction underpin metal site assembly of CuA in human cytochrome oxidase. Proc Natl Acad Sci U S A. 2015;112:11771–11776. doi: 10.1073/pnas.1505056112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dash BP, Alles M, Bundschuh FA, Richter OM, Ludwig B. Protein chaperones mediating copper insertion into the CuA site of the aa3-type cytochrome c oxidase of Paracoccus denitrif icans. Biochim Biophys Acta, Bioenerg. 2015;1847:202–211. doi: 10.1016/j.bbabio.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Gaju N, Guerrero R, Pedrós-Alió C. Measurement of cell volume of phototrophic bacteria in pure cultures and natural samples: phase contrast, epifluorescence and particle sizing. FEMS Microbiol Lett. 1989;62:295–302. [Google Scholar]
- 23.Sundararaj S, Guo A, Habibi-Nazhad B, Rouani M, Stothard P, Ellison M, Wishart DS. The CyberCell database (CCDB): a comprehensive, self-updated, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 2004;32:D293–D295. doi: 10.1093/nar/gkh108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 25.Rensing C, McDevitt SF. The copper metallome in prokaryotic cells. Met Ions Life Sci. 2013;12:417–450. doi: 10.1007/978-94-007-5561-1_12. [DOI] [PubMed] [Google Scholar]
- 26.Ekici S, Pawlik G, Lohmeyer E, Koch HG, Daldal F. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim Biophys Acta, Bioenerg. 2012;1817:898–910. doi: 10.1016/j.bbabio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greiner P, Hannappel A, Werner C, Ludwig B. Biogenesis of cytochrome c oxidase—in vitro approaches to study cofactor insertion into a bacterial subunit I. Biochim. Biophys Acta, Bioenerg. 2008;1777:904–911. doi: 10.1016/j.bbabio.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Buhler D, Rossmann R, Landolt S, Balsiger S, Fischer HM, Hennecke H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J Biol Chem. 2010;285:15704–15713. doi: 10.1074/jbc.M109.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Z, Wedd AG. The challenges of determining metal-protein affinities. Nat Prod Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 30.McEwan AG, Lewin A, Davy SL, Boetzel R, Leech A, Walker D, Wood T, Moore GR. PrrC from Rhodobacter sphaeroides, a homologue of eukaryotic Sco proteins, is a copper-binding protein and may have a thiol-disulfide oxidoreductase activity. FEBS Lett. 2002;518:10–16. doi: 10.1016/s0014-5793(02)02532-2. [DOI] [PubMed] [Google Scholar]
- 31.Cawthorn TR, Poulsen BE, Davidson DE, Andrews D, Hill BC. Probing the kinetics and thermodynamics of copper(II) binding to Bacillus subtilis Sco, a protein involved in the assembly of the Cu(A) center of cytochrome c oxidase. Biochemistry. 2009;48:4448–4454. doi: 10.1021/bi802288m. [DOI] [PubMed] [Google Scholar]
- 32.Quintana J, Novoa-Aponte L, Arguello JM. Copper homeostasis networks in the bacterium Pseudomonas aeruginosa. J Biol Chem. 2017;292:15691–15704. doi: 10.1074/jbc.M117.804492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 34.Hill BC, Andrews D. Differential affinity of BsSCO for Cu(II) and Cu(I) suggests a redox role in copper transfer to the Cu(A) center of cytochrome c oxidase. Biochim Biophys Acta, Bioenerg. 2012;1817:948–954. doi: 10.1016/j.bbabio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Trasnea PI, Marckmann D, Utz M, Koch HG. Measurement of cellular copper in Rhodobacter capsulatus by atomic absorption spectroscopy. Bio-Protoc. 2016;6:e1948. [Google Scholar]
- 36.Rubino JT, Franz KJ. Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J Inorg Biochem. 2012;107:129–143. doi: 10.1016/j.jinorgbio.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Badarau A, Dennison C. Copper trafficking mechanism of CXXC containing domains: Insight from the pH-dependence of their Cu(I) affinities. J Am Chem Soc. 2011;133:2983–2988. doi: 10.1021/ja1091547. [DOI] [PubMed] [Google Scholar]
- 38.Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 39.Onder O, Verissimo AF, Khalfaoui-Hassani B, Peters A, Koch HG, Daldal F. Absence of Thiol-Disulfide Oxidoreductase DsbA impairs cbb3-type cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. Front Microbiol. 2017;8:2576. doi: 10.3389/fmicb.2017.02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abicht HK, Scharer MA, Quade N, Ledermann R, Mohorko E, Capitani G, Hennecke H, Glockshuber R. How periplasmic thioredoxin TlpA reduces bacterial copper chaperone ScoI and cytochrome oxidase subunit II (CoxB) prior to metallation. J Biol Chem. 2014;289:32431–32444. doi: 10.1074/jbc.M114.607127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kay KL, Zhou L, Tenori L, Bradley JM, Singleton C, Kihlken MA, Ciofi-Baffoni S, Le Brun NE. Kinetic analysis of copper transfer from a chaperone to its target protein mediated by complex formation. Chem Commun. 2017;53:1397–1400. doi: 10.1039/c6cc08966f. [DOI] [PubMed] [Google Scholar]
- 42.Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 43.Wiethaus J, Wildner GF, Masepohl B. The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus. FEMS Microbiol Lett. 2006;256:67–74. doi: 10.1111/j.1574-6968.2005.00094.x. [DOI] [PubMed] [Google Scholar]
- 44.Richardson DJ, Bell LC, McEwan AG, Jackson JB, Ferguson SJ. Cytochrome c2 is essential for electron transfer to nitrous oxide reductase from physiological substrates in Rhodobacter capsulatus and can act as an electron donor to the reductase in vitro. Correlation with photoinhibition studies. Eur J Biochem. 1991;199:677–683. doi: 10.1111/j.1432-1033.1991.tb16170.x. [DOI] [PubMed] [Google Scholar]
- 45.Peters A, Kulajta C, Pawlik G, Daldal F, Koch HG. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J Bact. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawlik G, Kulajta C, Sachelaru I, Schröder S, Waidner B, Hellwig P, Daldal F, Koch HG. The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. J Bact. 2010;192:6378–6389. doi: 10.1128/JB.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.