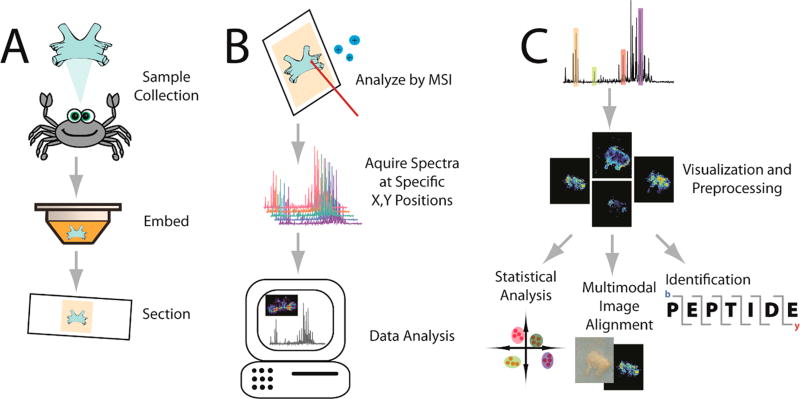
Mass spectrometry imaging (MSI) is a powerful tool that enables untargeted investigations into the spatial distribution of molecular species in a variety of samples. It has the capability to image thousands of molecules, such as metabolites, lipids, peptides, proteins, and glycans, in a single experiment without labeling.1 The combination of information gained from mass spectrometry (MS) and visualization of spatial distributions in thin sample sections makes this a valuable chemical analysis tool for biological specimen characterization. A summary workflow is depicted in Figure 1. After minimal but careful sample preparation, the general setup of an MSI experiment involves defining an (x, y) grid over the surface of the sample, with the grid area chosen by the user. The mass spectrometer then ionizes the molecules on the surface of the sample and collects a mass spectrum at each pixel on the section with the resulting spatial resolution defined by the pixel size. After collecting the spectra, computational software can be used to select an individual mass-to-charge (m/z) value, and the intensity of the m/z is extracted from each pixel’s spectrum. These intensities are then combined into a heat map image depicting the relative distribution of that m/z value throughout the sample’s surface. In order to determine the identity of a specific m/z value, tandem MS (MS/MS) fragmentation can be performed on ions from each pixel, and the fragments can be used to piece together the structure of the unknown molecule. Otherwise, the molecule can be identified based on its intact mass by accurate mass matching to databases of known molecules within a certain mass error range.2,3
Figure 1.
Visual workflow for the MSI analysis. A crustacean’s brain is used as an example tissue for this workflow. (A) Sample preparation. After collection from the animal, the sample is embedded in a supporting medium for sectioning onto slides. Other sample processing, such as applications of enzymes, matrix, or derivatization agents, may be performed depending on the molecular species of interest or the instrument being used. (B) Sample analysis. After acquiring a spectrum at each (x,y) grid point on the tissue, sophisticated software tools are used to process and visualize the data. A laser is used to ionize molecules as depicted, although several nonlaser based methods are also used. (C) Data processing. After preprocessing the data (e.g., baseline correction), the distribution of selected molecules can be visualized. From there, identification of the m/z values and statistical analysis between different images or image coregistration with other image modalities can occur.
With the numerous technological advances in recent years, MSI is becoming a more established tool in clinical practice and the pharmaceutical industry.4–6 Advances include improvements in reproducible sample preparation to ensure reliable interpretation of data and instrumentation that allows for high acquisition speeds and enhanced spatial resolution improving throughput and depth. The credibility of MSI experiments has further been enhanced by the development of methods for absolute quantitation of detected molecules. To help with large computational endeavors, statistical workflows and machine learning algorithms have been implemented to handle the large imaging data sets being produced with modern instrumentation. MSI can also be combined with other complementary imaging modalities, such as microscopy, Raman spectroscopy, and MRI, to strengthen any biological conclusions. With both hardware and software improvements, 3-dimensional (3D) renderings and even single-cell resolution using MSI are emerging as future frontiers. With all the advances in this field, MSI is rapidly evolving and requires continuous development to match the current demand.
Overall, the aim of this review is to provide an informative resource for those in the MSI community who are interested in improving MSI data quality and analysis or using MSI for novel applications. Particularly, we discuss advances from the last 2 years in sample preparation, instrumentation, quantitation, statistics, and multimodal imaging that have allowed MSI to emerge as a powerful technique in various biomedical applications including clinical settings. Also, several novel biological applications are highlighted to demonstrate the potential for the future of the MSI field.
SAMPLE PREPARATION
The Basics
As with any methodology, one of the most crucial steps for analytical success is proper sample preparation. This is particularly true for MS, as even subtle differences in sample integrity or molecular density can have profound effects on the signal intensity, types of molecules being ionized and detected, or localizations. For example, one of the greatest challenges is ensuring that the spatial mapping of molecules in an MSI experiment is consistent with the distribution in in vivo conditions. This relies heavily on proper sample preparation strategies. Researchers have even developed a new statistical scoring system to ensure sample preparation quality.7
After any necessary dissection or collection, biological tissue samples require a step to halt enzyme activity to reduce degradation and delocalization (e.g., diffusion across the tissue) of the molecules. This is typically done by flash-freezing the sample for MSI since many other preparations (e.g., formalin fixation (FF)) are not MS compatible for most molecular species due to being cross-linked (e.g., bound) in the sample, making them unavailable for ionization. This is not the case though for some lipids, and FF can be used to preserve sample integrity for their analysis.8 New method developments have made many FF paraffin embedded (FFPE) samples more MSI accessible (see discussion below). Prior to sectioning, one unique preparation step is the decellularization (i.e., removal cells from the extracellular matrix scaffold) of the tissues, allowing for the improved signal of extracellular matrix.9 Next, these samples are thinly sectioned (6–20 µm thickness), thaw-mounted onto the appropriate surface (e.g., microscope slides), and placed into a drying system (e.g., desiccator box). In many cases, tissues are fragile and do not section well without support, thus many researchers embed samples prior to sectioning. These embedding media include materials such as gelatin,8,10 but, as always, MS-compatibility is a concern. Optimal cutting temperature compound (OCT), for example, is popular among histologists but tends to contaminate MS spectra and is thus not recommended. Because of samples flaking or washing off the slide, O’Rourke et al. recommend coating the slide in nitrocellulose as a “glue-like” substance to aid the sections in staying on the slides.11 Here, one major assumption made is that the samples can be sectioned; however, not all samples are suitable for these general steps. For example, researchers have found ways to image analytes in imprinted plant leaves,12 plant roots,13 and even agar.14 Others have gone beyond single tissues to whole body imaging, which can have its own unique challenges.15
Several different ionization techniques are compatible with MSI, and each requires a unique process to preserve the corresponding sample. Matrix-assisted laser desorption/ionization (MALDI) is the most popular ionization technique for MSI, especially due to its ability to image a wide range of molecular weights and molecular species (e.g., metabolites and proteins).16 Its requirement of a matrix for proper ionization and production of only singly charged ions often limits its applicability to larger proteins. This has prompted the development of laserspray ionization and unique matrixes (e.g., 2-nitrophologlucinol (2-NPG)).17 Of course, no one matrix, application method, or analyte extraction process works for all molecules, so optimization is important and will be discussed later in this review. Other varieties of MALDI-MSI exist, including scanning microprobe MALDI (SMALDI),15 infrared (IR) IR-matrix-assisted laser desorption electrospray ionization (MALD-ESI),18,19 and surface-assisted laser desorption/ionization (SALDI),20 although they are not as widely implemented. Other techniques worth noting include desorption electrospray ionization (DESI), secondary ion mass spectrometry (SIMS), and more recently easy ambient sonic spray ionization (EASI),21 which require minimal sample preparation in comparison to MALDI since they do not require the presence of a matrix.2,22–25 Unfortunately, each of these is more limited in the molecules they ionize (peptides and metabolites, respectively). In the most general cases, both DESI and SIMS can be performed directly after sectioning, as they depend more on the instrument parameters for proper analyte extraction. Even with all the ionization methods available, researchers are still developing new methodology, such as laser electrospray ionization.26 Each ionization method has its own advantages and disadvantages, ranging from the molecules that can be analyzed to the spatial resolution achievable, the latter to be discussed further in this review. Finally, after proper preparation and ionization, the instrument itself (e.g., mass analyzer) is important to consider before determining a proper sample handling workflow. For example, the Bruker MALDI-time-of-flight (TOF)/TOF instrument requires ITO coated slides, while the Thermo MALDI-LTQ-Orbitrap XL can analyze samples on plain microscope slides.9,27 In general, while selecting the appropriate sample preparation, ionization source, and mass analyzer is important to allow the molecular species of interest to be analyzed, care should be taken in using instruments with MS/MS abilities or high mass accuracy whenever possible to allow confident identification of the molecules in question.
Improving the Basics
Applying an Internal Standard
To qualitatively evaluate different tissues or different analytes within a tissue, appropriate normalization and internal standards are expected if semiquantitative comparisons are to be made. These standards could be included as early in the workflow as dosing the animals/cells up to right before the ions enter the instrument. 2,28,29 For MALDI, the standards are typically applied prior to matrix application using the same automatic sprayer systems described below.30–32 Chumbley et al. has done a comprehensive study to determine the proper inclusion of the standard (e.g., with matrix, under the tissue section, or sandwiching the section with matrix), and it was found that depositing the standards followed by matrix to be optimal for MSI mapping of spatial distribution of the drug rifampicin.33 This sample protocol can also be applied to sections used in DESI experiments (applying prior to analysis), or standards can be added directly to the DESI extraction solvent for inclusion in sample analysis.2
Matrix Choice and Application (MALDI Only)
For MALDI ionization, a matrix is required to allow proper ionization of the molecules of interest. As the matrix crystallizes, analytes are extracted from the tissue section and cocrystallized. If analytes are not in this crystal structure, it is unlikely that they will be ionized. Thus, the availability of the molecule, the matrix application, and the matrix itself can all have an effect on this process. For the case of some proteins, a fixation wash is necessary to make the molecules available for cocrystallization. 9,11 The Carnoy’s solution (i.e., 6:3:1 ethanol:chloroform:-glacial acetic acid) is a common wash used for protein MSI.11 Other washes, such as ammonium citrate, have also been utilized to analyze low molecular weight species. Besides washing, prespraying with solvents can also aid in the extraction of peptides. The combination of ammonium citrate washes and prespraying with cyclohexane proved to be effective in extracting clozapine from rat brain sections.34 Vapor chambers have also been found to be effective, specifically trifluoroacetic acid vapors for SIMS imaging of lipids.23 It should be noted that all of the preparations described here may be applicable for other ionization methodology if appropriate, for example matrix-enhanced nanostructure initiator MS.35
Several matrixes have found popularity for their “universal analysis” including 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA), especially for metabolites and peptides in positive mode. A 1:1 mixture of these matrixes is also commonly used.36 Another matrix, sinapinic acid, has been well vetted for proteins in positive mode. On the other hand, negative mode has been found useful for metabolites, for which 1,5-diaminonaphthalene (DAN) and 9-aminoacridine (9-AA) are among the most commonly used matrixes.12 Interestingly, the use of water as a “matrix” in MALDESI has been explored recently.19 Furthermore, nano-materials have been utilized as an alternative matrix, though oftentimes these situations are considered as a different ionization technique (e.g., SALDI).20 Matrix has also been used to enhance SIMS signals.25 It is expected that alternative new matrixes possessing similar properties to 2-NPG (i.e., multiply-charged ion production) are likely to be developed and applied to matrix-based or matrix-enhanced MSI techniques.
In general, most of the focus for sample preparation has been on the matrix application process. When applying matrix, the best method would provide appropriate analyte extraction, small crystal size, and homogeneous application. Unfortunately, no universal method exists. Classically, researchers would spray matrix over the tissue sample using a painter’s airbrush. While this can be reproducible between applications by the same individual, person-to-person variability is high, and there is little adjustability. For example, the “wetness” of the surface of the tissue during application defines the appropriate analyte extraction. An appropriate balance needs to be found, as a too “wet” application can cause molecular diffusion while a too “dry” method may not effectively extract the molecules. “Wet” vs “dry” methods also have an effect on the crystal size, the wetter methods yielding larger crystal sizes. Substrate (i.e., where the sample is placed) versus its surrounding temperatures can also affect heterogeneity, but this has been only applied to MALDI spots.37 Automated sprayers have allowed reproducible application methods across individuals and laboratories, and as such, their popularity has grown in the past few years.38 Several application notes from different vendors exist, but researchers should take time to optimize their application methods to their specific systems. This will likely increase lab-to-lab reproducibility, but it is expected that similar methodologies will be utilized. To increase clarity, all developed methods and their parameters should be included in publications. Interestingly, alternative ionization methods (e.g., SIMS) have been used to characterize the analyte incorporation into spots and, although difficult to implement, similar imaging-based studies would be interesting.25 Homogenous application has also been a major focus, and researchers have utilized alternative application methods to improve this facet in the past few years. One example is electrospray deposition, for which units tend to be home-built. This dry application method usually requires an additional “incorporation spray” after the matrix has been applied.39 Some electrospray devices have allowed for control of the crystal size, which can directly relate to the spatial resolution achievable.40 Other methods have also benefited from the inclusion of an electric field, decreasing crystal size and thereby increasing spatial resolution.41 Finally, the “driest” method used is sublimation, which is popular for its low-cost, small crystal size, and high homogeneity. Commercial and partially modified apparatuses are highly published.11,20,42 When individuals want to use several matrixes on a tissue section or staining, they will tend to wash off the original and apply the new matrix, but this unsurprisingly produces signal loss and diffusion. As an alternative, using a commercial sprayer, Urbanek et al. have developed a multigrid MALDI (mMALDI) methodology, where different matrixes are “printed” into predefined dots on a grid. By targeting these specific matrix dots during the imaging run, a researcher is able to gather multiple data sets (e.g., metabolites, peptides, and proteins) from a single tissue section without washing.43 Finally, with all of the variations in equipment and methodology, an emphasis should be placed on sharing automated matrix application methods and cross-lab communication to allow for reproducible results. The use of open-source software and easily fabricated instrumentation is an example of this, although the ease of commercial instrumentation will continually compete with this notion.44
Specific Molecular Considerations
On-Tissue Digestion
Molecular imaging of proteins has been of major interest, but high-mass resolution analysis of proteins has been out of reach due to the mass range limitations of current mass analyzers (e.g., Orbitraps), especially for MALDI. This has been alleviated for extract analysis by the inclusion of an initial protein digestion step (i.e., bottom-up proteomics), so in some cases trypsin on-tissue digestion protocols have been employed for MSI.9,30,45 However, as with every method developed, the steps should be optimized specifically for each tissue type.30,46 For example, Heijs et al. has shown the appearance of different myelin basic protein fragments over longer trypsin incubation times.30 With the recent surge of interest in mapping glycans in tissue sections, PNGase F, which cleaves N-glycans, has found application into in situ digestion and sequential enzyme application allowing the imaging of both glycans and protein fragments in a single MS imaging run.47 Overall, while immunostaining/labeling approaches are very effective, they can be nonspecific due to possible cross-reactivity, and MALDI MSI provides an orthogonal yet highly specific cross-validation of the labeling-based strategies. The most challenging part of in situ digestion is appropriately identifying the protein fragments. In some cases, on-tissue MS/MS is difficult depending on the instrumentation, and a complementary liquid chromatography coupled with tandem MS experiment may need to be performed.9,47 It is worth noting that other ionization techniques (nanoDESI) allow for intact protein imaging up to 15 kDa on Orbitrap systems.48
Formalin-Fixed Paraffin Embedded (FFPE) Samples
While there is preference in obtaining freshly excised samples for MSI analysis, sometimes that is not possible for hard-to-obtain biological samples, especially rare, human specimens. With the wide availability of FFPE tissues, which are not typically compatible with MS, researchers have been motivated to develop methods to release the analytes of interest to image these tissues.49 As stated previously, optimization for specific tissue types is important, and Oetjen et al. has provided a comprehensive, guided study to do this for other researchers.46 Unfortunately, not all molecular species can be extracted from these tissues, although Pietrowska et al. reported that lipids can be analyzed by avoiding paraffin embedding after fixing the tissue with formalin.50 Originally, most studies targeted proteins and peptides in the FFPE tissue sections, mainly using the in situ digestion methods described above.46,50 More recently, researchers have been able to extract metabolites and glycans.47,51 With more standardized protocols, the extensive FFPE samples available will be utilized more readily for MSI workflow, allowing for exciting possibilities to examine many clinical specimens and a flood of new information to help guide researchers in future endeavors.
Chemical Derivatization/On-Tissue Labeling
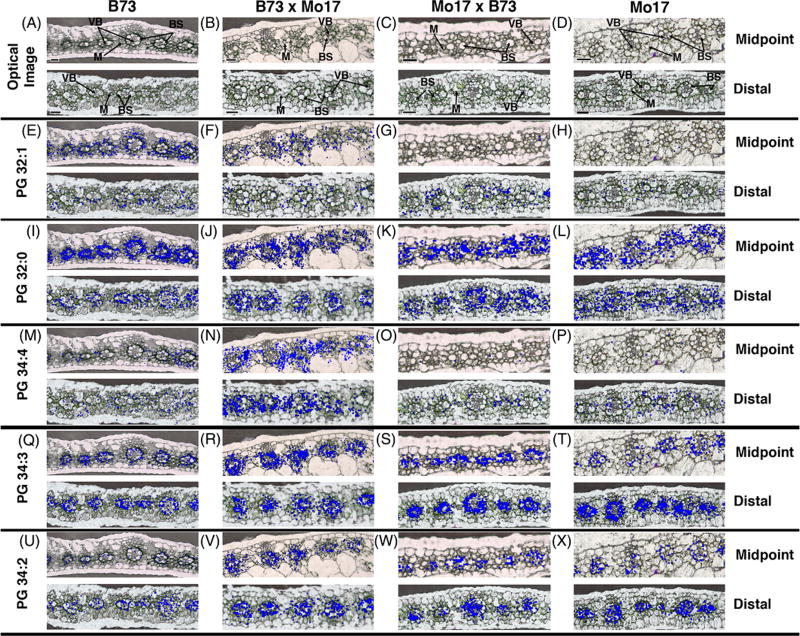
MS is often touted as a universal technique for all molecular species, but there are several classes of molecules that are difficult to ionize and thus analyzed directly by MS. Most targets thus far have been small molecules, such as metabolites, but the inclusion of derivatizing other molecules, such as peptides and glycans, is expected.52 The overall goal of derivatizing molecules is to change their physicochemical properties and to aid in ionization for MS analysis. For example, the Girard T (GirT) reagent has been applied successfully to several steroids, including testosterone and triamcinolone acetonide.31,53 Other steroids (e.g., tetrahydrocannabinal) have also been targeted using 2-fluoro-1-methylpyridinium p-tolunesolfonate as a derivatization agent.29 N-glycans (Figure 2), fatty acids, and neurotransmitters have all been targets through other, unique on-tissue assays.28,42,52 Compared to the traditional spraying of reagent, which usually produces poor spatial resolutions (>100 µm), electrospray deposition has been successfully utilized to derivatize fatty acids while achieving a high spatial resolution (20 µm).42
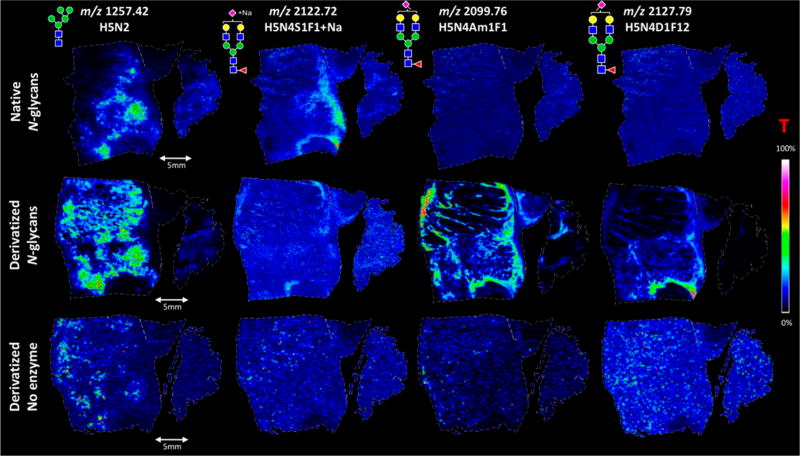
Figure 2.
MSI comparison of digestion and derivatization of N-glycans in FFPE colon carcinoma sections. Different section preparations are shown in each row, while each column is a different N-glycan. Native N-glycans (e.g., digested) are shown in the first row. The second row shows the digested, derivatized samples. This in situ derivatization specifically targeted sialic acids by dimethylation and subsequent amidation. Finally, the last row shows a negative control sample where derivatization was performed but no digestion was done. Without a digestion step, N-glycans should not be available for analysis. On the basis of the results, it is clear that the derivatized, digested N-glycan (middle row) method produces the best extraction and ionization of N-glycans with sialic acids. Green circle, mannose; yellow circle, galactose; blue square, N-acetylglucosamine; yellow square, N-acetylgalactosamine; white square, N-acetylhexosamine; red triangle, fucose; purple diamond, N-acetylneuraminic acids; T, total ion current normalization. Reproduced from Holst, S.; Heijs, B.; Haan, N.; van Zeijl, R. J. M.; Briaire-de Bruijn, I. H.; van Pelt, G. W.; Mehta, A. S.; Angel, P. M.; Mesker, W. E.; Tollenaar, T. A.; Drake, R. R.; Bovee, J. V. M. G.; McDonnell, L. A.; Wuhrer, M. Analytical Chemistry 2016, 88 (11), 5904-5913 (ref 52). Copyright 2016 American Chemical Society.
DEVELOPMENTS IN INSTRUMENTATION
MSI often requires specially developed instrumentation in order to address challenges unique to image acquisition, such as spatial resolution or surface homogeneity. Numerous advancements have been made in recent years to improve the quality and reproducibility of generated images. The main distinction between MSI and liquid chromatography (LC)-MS experiments is the preservation of a spatial dimension. Thus, most instrumentation developments have focused on the ionization source, with several exceptions related to ion accumulation. The two main ionization methods for MSI are laser-based and secondary ion-based, and most of the progress in recent years has focused on these sources. As such, they will be the focus of discussion of this section.
Laser-Based Ionization
Spatial Resolution
Arguably the most sought-after improvements in MSI are related to spatial resolution, which is the area corresponding to each individual mass spectrum in an imaging acquisition. Improving the spatial resolution enables more discrete localization patterns to be observed throughout a tissue, but since improving spatial resolution decreases the area of tissue ionized, there is a trade-off between spatial resolution and sensitivity. The resolution can be changed by adjusting the optics of the ionization source or otherwise changing the instrument’s geometry to decrease the laser diameter. Sample preparation can also affect the spatial resolution, which is discussed above. Numerous groups have recently reported drastic instrumental improvements in spatial resolution. Spengler and co-workers reported a lateral spatial resolution of 1.4 µm on an atmospheric pressure MALDI source by adjusting its geometry, allowing for the visualization of subcellular distributions of lipids, metabolites, and peptides.54 The Lee group achieved a spatial resolution of 5 µm on a vacuum pressure MALDI instrument by using a simple modification to the optical instrument. The system was easily interchangeable between various laser spot sizes, allowing for greater flexibility in the trade-off between sensitivity and resolution based on each individual experiment’s needs.55 Numerous other notable advancements have also been made to improve spatial resolution recently.56–59
However, with the rapid developments being reported by researchers across the field, it was found that spatial resolution was being defined differently between groups, instruments, and samples. As this makes it difficult to form a standard of comparison between methods and instruments, developing a universal method for both defining and measuring spatial resolution is crucial to proper data reporting and comparison of images acquired on different instruments with different sample preparation methods or with different users. Typically, the limiting factor in spatial resolution is the laser, as the laser spot diameter determines the ablation area. Therefore, efforts have been devoted to characterizing the ablation pattern in imaging experiments, particularly with MALDI-MSI, the most widespread imaging technique. It was found that laser ablation patterns follow a Gaussian distribution, with incomplete ionization around the outside of the pixel. Furthermore, there is the ability to “shear” matrix crystals, scattering debris across the sample after laser ablation. This finding led to the assertion that MALDI-MSI resolution should be defined as (1) the homogeneity of the matrix crystals once they have been applied and cocrystallized with the analyte and (2) the effective ablation diameter of the laser.60 The hope is that this new definition will allow for more uniform reporting of spatial resolution between research laboratories on different instruments and with different sample preparation methodologies.
Several research groups have developed methods for measuring the actual spatial resolution achievable by an instrument, which can differ from the reported pixel size of the instrument acquisition parameters due to previously mentioned factors such as crystal size and laser beam profile. A simple way to measure effective spatial resolution of an instrument based on user-defined instrument parameters is with a standardized imaging plate. Caprioli and co-workers developed such a slide that incorporated a pattern of crystal violet using lithography in order to measure the beam diameter in MALDI-MSI experiments by visually inspecting the ablation pattern.61 Another slide for measuring spatial resolution was developed using a slightly different technique, in which a sample solution can be dragged over the slide’s surface, allowing it to be automatically retained in hydrophilic grooves of the slide. The slide can then be imaged on the instrument in order to determine the lower threshold of the instrument’s spatial resolution.62 These strategies can provide a valuable method for testing the spatial resolution when adjusting instrumental parameters or performing quality assurance on images to ensure that proper resolution is being reported.
Matrix-Free Laser-Based Ionization
Though highly beneficial in many regards, MALDI-MSI’s requirement for a matrix coating is often a major drawback in imaging experiments. Matrix application can be a limitation because it requires an additional step in sample preparation, it suffers from poor homogeneity that can affect spatial resolution, and it results in excessive noise peaks in certain mass ranges of the spectrum due to the interference of matrix ions. As a result, ionization sources are being developed to utilize laser ablation techniques without the requirement of matrix. For example, improvements in the sensitivity and coverage of laser ablation electrospray ionization (LAESI)-MS were made for metabolite analysis.63 Laser desorption post-ionization MS, though still in its early stages of development, has been demonstrated to have a promising potential as a complementary tool for in situ localization and quantitation. It has the benefit of not requiring matrix application or sample preparation, though currently its resolution and mass accuracy are 500 µm and 300 ppm, respectively, which is not competitive with commercial instruments.64 However, with further development, it may earn its place as a prominent ionization source. Another method for ionization without the application of matrix is nanophotonic laser desorption ionization, which ionizes analytes from a highly uniform silicon nanopost array.65 This method has achieved 40 µm spatial resolution for over 80 molecular species, giving it the potential to be competitive with MALDI upon further exploration.
Throughput
Another frequently cited challenge with MSI is the long analysis time typically required, which can range from several hours to several days, depending on the selected area and pixel size. These long analysis times limit the practicality of MSI for routine applications, particularly in clinical settings. As a result, developments have been made in order to increase throughput without sacrificing image quality. One notable example involved utilizing a solid state laser with a 5 kHz repetition rate to perform continuous laser raster sampling on a MALDI-TOF/TOF instrument. This method achieved an acquisition rate of up to 50 pixels per second, an 8 to 14-fold improvement over conventional lasers.66 Throughput becomes even more of a challenge when molecules in the same tissue ionize differently, thus requiring different polarities for acquisition. This is particularly the case with lipid analysis, as lipids are a diverse class with high structural variability. Methods have been developed for imaging in both positive and negative polarity while minimizing analysis time using high speed MALDI-MSI technology and precise laser control.67 The field is moving toward real-time imaging capabilities for immediate spatial analysis for guidance during surgeries. As an example, Fowble et al. have applied a laser ablation imaging approach in ambient conditions in order to obtain spatial distribution of metabolites with a range of polarities in real time without the use of any matrix or sample pretreatment.68 Another method couples a picosecond IR laser to an electrospray ionization (ESI) source in order to provide ambient MS imaging without causing thermal damage to tissue. This allows molecules to remain in their native environment until ionization, allowing better insight into the tissue’s condition.69 The iKnife has also demonstrated real-time capabilities, most recently with real-time analysis of the mucosal lipidome by Takats and co-workers.70 There have been several other developments in technologies to use MSI with surgical procedures in order to guide surgical decision-making using MALDI and nanoDESI MSI.71–73 These developments demonstrate great potential in moving MSI technology from laboratories to clinical settings for improved patient treatment.
Another approach for improving throughput is microscope mode MSI.74 Here, ions from a relatively large sample area (typically 100–300 µm in diameter) are desorbed simultaneously. Then the ion optics of the instrument project the ionized substances from this area to a position-sensitive detection system such as Medipix or MicroChannelPlate detectors.75,76 These types of detectors allow for registration of a single m/z acquired from the whole scanned area at once, while magnifying the image and retaining spatial information. Because in the microscope mode a large area is simultaneously measured, a substantial reduction in analysis time is achieved.
SIMS
Resolution and Mass Accuracy
The other most common method of ionization is SIMS, which has seen notable improvements in instrumentation. In SIMS imaging, spatial resolution is often better than the other MSI counterparts but at the expense of sensitivity. This is largely a consequence of the ion beam, either due to low ionization probability or beam focusing difficulties. An argon gas cluster ion beam is typically used for TOF-SIMS, but, despite its many benefits, it suffers from poor sensitivity and mass accuracy and requires the sacrifice of either spatial or mass resolution. Delayed extraction, a method widely used for MALDI-TOF in which an initial pulse is implemented on the ions to correct for velocity distributions, is becoming more prominent in TOF-SIMS imaging and has been shown to be successful in maintaining both the high mass resolution and spatial resolution.77 By implementing external mass calibration, the mass accuracy can also be preserved.78
Methods involving delayed extraction have been explored as a means to improve mass resolution, but these methods often make mass calibration difficult, resulting in poor mass accuracy. Other groups have explored alternative primary ion sources, such as a CO2 cluster ion beam, which possesses many similarities to argon but improved the imaging resolution by more than a factor of 2 due to increased stability of the beam.79
Parallel Imaging MS/MS
With the inferior mass spectral resolution of TOF-SIMS compared to other ionization methods, the mass accuracy is usually not high enough to make confident identifications of the detected molecules by mass measurement alone. Therefore, it is usually necessary to acquire MS/MS spectra on ions of interest. However, collecting MS/MS spectra is difficult in imaging experiments because performing sequential MS/MS scans after a full-MS scan causes misalignment between spectra and spatial information. To address this, progress in parallel imaging MS/MS has been implemented, in which MS/MS spectra are collected simultaneously with MS spectra using two mass analyzers. This acquisition method differs from traditional MS/MS acquisitions, in which all ions other than the precursor ion are discarded. As a result, MS and MS/MS images are in perfect alignment with each other, allowing for more precise mapping of molecular distribution.80,81 With fully optimized parallel imaging, identification confidence can be drastically improved without sacrificing the integrity of localization information.
Ambient/Low-Vacuum TOF-SIMS
As MSI is very commonly used for the analysis of biological tissue, it is highly desirable for analyses to be conducted in near-native environments, such as in the presence of water, in order to get an accurate understanding of the chemical environment. Low-vacuum and ambient MALDI imaging have already been well-explored, but progress has recently been made with SIMS, denoted as Wet-SIMS.82 Currently, the technique is able to acquire images at 80 Pa in imaging experiments.83 With further development, this technique could be used to ionize biomolecules in their native environment, allowing for analysis in biologically relevant experimental conditions.
Separation
A significant limitation to MSI compared to LC-MS analysis is the lack of separation capabilities, as retaining spatial information typically requires ablating all ions present in a pixel of sample at the same time for a single scan. This often leads to problems such as ion suppression, but techniques that allow post-ionization separation are being developed to overcome this challenge. To separate analytes from noise or undesired compounds, a simple sample cleanup step was incorporated into MALDI-MSI by first introducing laser ablation with vacuum capture to collect the ions. The ions are then eluted by a C18 column (or other packing materials or beads) onto the MALDI target plate, effectively desalting the sample and removing background ions. The method demonstrated an improved signal from the sample and decreased background interference compared to direct MALDI-MSI, resulting in higher quality MS/MS data, cleaner spectra, and more confident identification.84 For separation of analytes, ion mobility has been a popular choice, as it can and has been seamlessly integrated into MALDI-MSI workflows, such as demonstrated by Trimpin and co-workers.85 Enhancements to the sensitivity were recently made by the McLean group using a silver-sputtered matrix coating.86 Ion mobility has also been recently demonstrated to be highly effective for coupling with ambient ionization techniques, such as LAESI, LESA, and DESI.87–89 The results showed an increase in detected molecules and the ability to select specific classes to image and offers the capability of using MSE fragmentation, in which all ions are fragmented, improving MS/MS coverage.90 An alternative, pseudoseparation method has also been employed, in which subsequent MS scans covered differing m/z windows in order to detect low-intensity ions characteristic of specific ranges, providing the effect of gas-phase fractionation. By implementing a spiral plate motion during imaging, the integrity of spatial information was not lost with this method.27
Depth Profiling
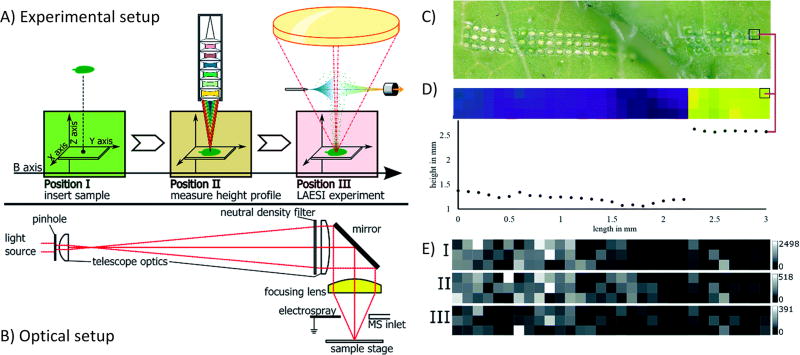
Another challenge specific to imaging is achieving uniform ionization over the surface of the sample section, something difficult to accomplish if the tissue is not perfectly flat. While extra care in sample preparation can help alleviate this to an extent in some sample types, often slight variations in the height of the tissue are unavoidable.91 To remedy this, modifications to instruments have been made that allow for height correction. For example, a novel LAESI source was recently developed that incorporated a confocal distance sensor that both moved the sample to a constant height and recorded the height information to generate a topography map.92 Figure 3 shows a schematic of the instrumental setup, both the acquisition workflow and optics, as well as example data indicating the information recorded about both sample height and spatial distributions of specific m/z values. Another method combined shear force microscopy with a nano-DESI source to measure and adjust the voltage magnitude to enable a stable feedback signal over surfaces with complex topographies.93 If a uniform sampling can be ensured over the surface of a tissue, it not only preserves spatial integrity throughout the plane of the sample but can also allow for three-dimensional (3D) imaging. With 3D imaging, it is imperative that the depth profile of the sample be preserved to ensure accurate record of the tissue profile. Several significant advances have been made in this respect in the area of elemental imaging, such as the development of a femtosecond laser ionization source for multielemental imaging with a 7 µm depth resolution.94 Submicrometer depth resolution, down to 20 nm, has been demonstrated using extreme ultraviolet laser light, allowing for 3D imaging of bacterial colonies.95 It is expected that these capabilities will continue to be developed and applied to 3D imaging of more complex systems.
Figure 3.
Overall setup and demonstration of capabilities of a LAESI source for samples with uneven surfaces. (A) Workflow representation of experimental setup related to the LAESI source, including sample insertion, measurement of height profile, and LAESI experiment for MS acquisition. (B) Schematic of the telescope optics implemented to focus the laser. (C) Optical image of R. sativus leaf after LAESI experiment, showing the laser ablation pattern. Note the even distance between ablation spots throughout sample despite uneven surface. (D) Topographical height profile of leaf surface along the ablation pattern, showing the change in height across the sample. The red line indicates identical positions on the sample surface. (E) MSI intensity maps of m/z values 418.051, 434.024, and 447.054, showing differences in spatial resolution. These m/z distributions were acquired in the same experiment as the topographical profile. Reproduced from Bartels, B.; Kulkarni, P.; Danz, N.; Bocker, S.; Saluz, H. P.; Svatos, A., RSC Advances 2017, 7 (15), 9045-9050 (ref 92), with permission (https://creativecommons.org/licenses/by/3.0/) of the Royal Society of Chemistry.
QUANTITATION
Comparison to LC-ESI-MS/MS: The Past
With the push for multimodal imaging (see below), it is clear that obtaining several pieces of information from a single tissue is imperative. While MSI is mainly qualitative, with the appropriate conditions, processing, and software, quantitative information can be extracted, although the degree of accuracy is under close scrutiny. Issues such as tissue heterogeneity, ion suppression, sample topography, etc. are all considered significant challenges in this field.96 Before the development of quantitative MSI, the analytes of interest were separately extracted from another tissue section and run on a LC-ESI-based instrument for quantitation. Once the absolute quantity of the analyte of calculated, these values can then be applied to the tissue of interest. This methodology is still in use widely, although it is more commonly utilized for confirmation or a starting point of a quantitative MSI study.42,97 This concept is similar to Western blot for other LC-MS quantitative results.33 Quantitative MSI is now necessary, as many application-based MSI publications focus on the comparison between two or more sample types. With proper sample preparation, comparisons can be made with the appropriate considerations.
Relative Quantitation
Direct Comparison (with or without Normalization)
As mentioned above, direct comparisons between different tissue sections is commonly done. While these “relative” comparison methods lean toward being “semi-quantitative,” several techniques and data processing strategies have perpetuated their use. For example, matrix effects and other interfering molecules tend to cause more deviation in the quantitative accuracy, although some researchers have shown that the correlation between MALDI-MSI and LC-MS/MS can be quantitative for fatty acids and protein.42,98 While these assessments of different molecules in a single tissue are interesting, ion suppression and ionization efficiencies between molecules should always be questioned. The addition of an internal standard can aid in the normalization of the signal.53 Normalization can also be done with the same molecules within different tissues, and this method still aids in more confident comparisons.53 The inclusion of a normalization procedure in pre- and postprocessing is now an expectation. This strategy is applicable for several other molecular species, including neurotransmitters, nucleotides, lipids, and tryptic peptides.1,28,30 Almost all software available for MSI provides the ability to normalize. For example, the use of SciLS software tools allow for normalization to the total ion current (TIC) before further statistical analysis.36 Using this method, several metabolites were found to be different between the cortex, outer medulla, and inner medulla of the rat kidney between control and furosemide-treated.36 It should be noted that care should be taken when comparing different regions of a tissue, as their biological matrixes can vary slightly.96 As expected, software is an important component in any imaging-based quantitative strategies, and Renslow et al. have further developed tools to nanoSIMS transition from qualitative to quantitative for element incorporation into biofilms.99
On-Tissue Labeling: Using Reporter Ions
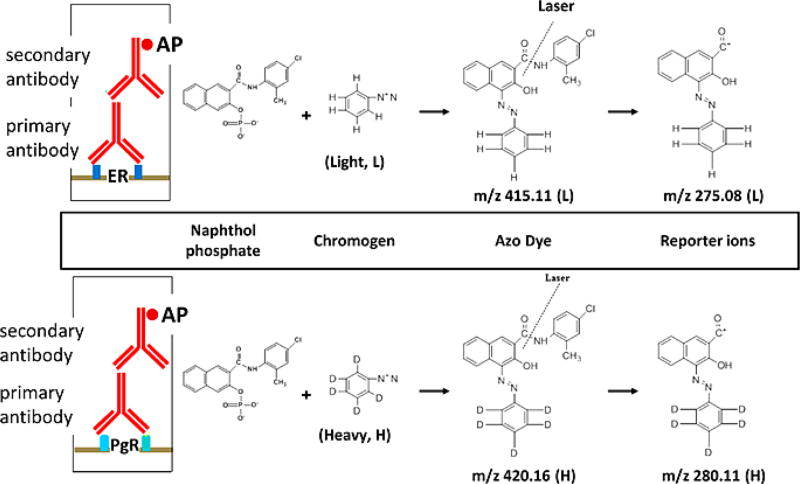
For LC-MS-based quantitation, two types of techniques are employed. Label-free methods directly compare samples in different runs, which is analogous to the “direct comparison” MSI described in the previous section. While label-free quantitation is commonly used in LC-MS and MSI applications, instrument variability, instrument limitations, and other factors lead to inconsistent and inaccurate comparisons. In contrast, the incorporation of stable isotopes (i.e., 2H, 13C, 15N, 18O) has allowed for same spectrum relative quantitation, although its application to MSI is extremely limited. One example in the literature entitled stable-isotope-label based mass spectrometric imaging (SILM-SI) utilizes light and heavy chromogens to differentiate between different cancer biomarkers of interest (Figure 4).100 After labeling with a primary and secondary antibody, the addition of the chromogen produces an azo dye that, when ionized by the laser, fragments into distinct, duplex reporter ions. The ratio of these reporter ions to another molecule can then be used to calculate their relative abundance, in this case the estrogen receptor compared to the progesterone receptor.100 While classically reporter ions can be seen in the MS/MS spectra via isobaric labeling, this same idea has not been implemented in MSI experiments, not only due to the poor fragmentation for singly charged ions but likely also due to the incompatibly of the methods for relative quantitation. In comparison, isotopic-based labeling methods can potentially be transitioned to on-tissue MSI applications, although the process of derivatizing molecules on-tissue has primarily been used for increasing ionization of different molecules.28,42,52
Figure 4.
Schematic of the general working principle of SILMSI. After incubating the section with a primary antibody for the biomarker of interest, in this case PgR and ER, a secondary antibody is applied that is conjugated with alkaline phosphatase (AP). AP cleaves naphthol from naphthol phosphate. The naphthol mixes with the heavy or light chromagen to form an azo dye precipitate on the tissue. The incubation of the two antibodies can be done on the same tissue with proper washing. In the instrument, the azo dye absorbs energy from the laser, creating fragments including characteristic reporter ions. The heavy and light reporter ions are separated by 5 Da in the MS spectrum. Reproduced from Wang, H.; DeGnore, J. P.; Kelly, B. D.; True, J.; Garsha, K.; Bieniarz, C., A technique for relative quantitation of cancer biomarkers in formalin-fixed, paraffin-embedded (FFPE) tissue using stable-isotope-label based mass spectrometry imaging (SILMSI), J. Mass Spectrom., Vol. 50, Issue 9 (ref 100). Copyright 2015 Wiley.
Absolute Quantitation
Internal Standard
While relative comparisons are commonplace, absolute quantitation is relatively underdeveloped. While obtaining the true concentration of a molecule is much more difficult, it is also more desirable since it allows for true comparisons between different molecular species without concerns about varying ionization efficiencies. As with LC-MS-based measurements, a straightforward method is to incorporate a deuterated internal standard into the sample. As explained previously, internal standards are now being used extensively to normalize MSI data sets, and the inclusion of a very specific standard (e.g., deuterated version of an analyte of interest) facilitates absolute quantitation of that analyte. This has been done primarily for DESI samples, with the standards incorporated into the solvent stream.2
Calibration Curve
In general, the creation of a calibration curve is the most confident way to obtain the absolute quantity of an analyte. This has been done with LC-MS in separate and the same runs.101 Initially, one may think producing an external, separately spotted calibration curve would work for MALDI-MSI, but the lack of sample matrix and matrix heterogeneity leads to inaccurate concentrations. Thus, researchers have adopted an on-tissue spotting technique that takes both of these considerations into account. The standards of interest (isotopic or nonisotopic) are spotted/applied on a separate, “control” section.28,32,33 This section is usually a serial section of the one being analyzed, as having the same matrix is important for accurate quantitation.96 For example, many researchers chose liver tissue for initial optimization or studies, as it is considered extremely homogeneous.33,96 Interestingly, in the case of elemental analysis, before spotting on the sample, the sections are washed to remove excess elements (e.g., sodium).32 To increase homogeneity of the areas where the standards are placed, researchers have developed methods where the standards are spiked into tissue homogenates themselves. These samples are then placed into a mold, frozen, sectioned, and placed near the imaged section, for which quantitation accuracy is similar, although it was noted that the dried droplet spotting method referenced above is much faster and easier.96 All of these methods require sophisticated computational tools, and several software packages exist for processing region of interest quantitation.102,103 msIQuant is an example software, which has been used to absolutely quantify drugs and neurotransmitters.103
DATA ANALYSIS
MSI data is difficult to process for a number of reasons, including the large size of the data files and the high degree of dimensionality, as acquisitions retain spatial information as well as other information. This is becoming more of a problem with the increase in spatial resolution causing an exponential growth in data file sizes. As such, key software developments have been made to address these challenges and ensure that effective analyses are being done without the loss of valuable information in the process. Figure 5 presents an overview of a typical workflow including several key data processing steps, all of which will be discussed below.
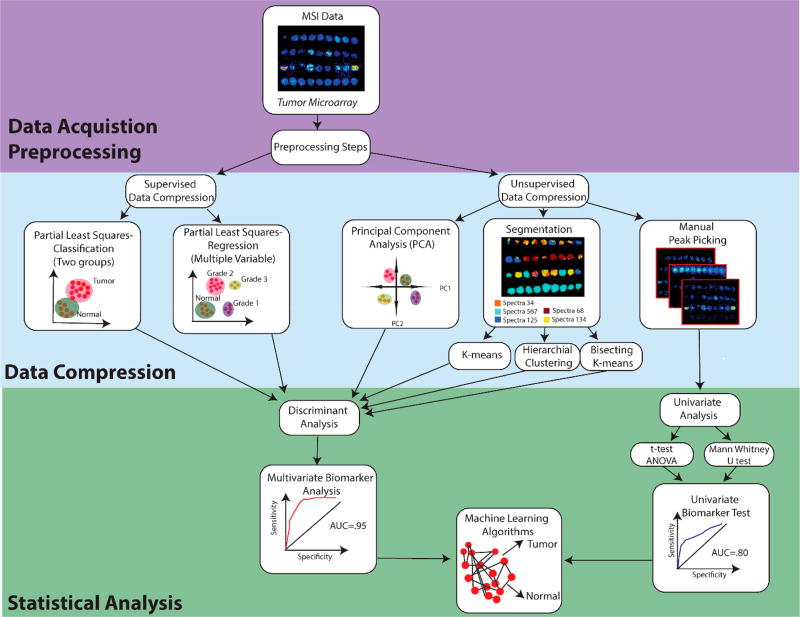
Figure 5.
Workflow for data processing and data analysis for MSI data. Following data acquisition, MSI data are subjected to preprocessing including normalization, baseline compression, smoothing, and spectral recalibration. Next, data are compressed to reduce computational load for statistical analysis. This includes supervised data compression, where the groups are defined. If two groups are used, it is known as classification, or if more than two groups are used, linear regression is used for analysis. Data can also be compressed without preclassifying the data through unsupervised data compression. Here, we describe three main methods: principal component analysis, segmentation, and manual peak picking. Unsupervised data compression includes k-means, hierarchical clustering, and bisecting k-means. Following compression of multiple variables, discriminant analysis is used to evaluate how well the chosen classification system separates groups of data. Manual peak picking helps pull out a few m/z peaks of interest. Univariate analysis can be done using either a t test or ANOVA (Gaussian distribution) or Mann-Whitney U test (non-Gaussian distribution) to test for significance between groups of data. If the user is interested in biomarker discovery, a specific m/z or group of m/z values can be used to conduct a biomarker analysis, where an AUC value closer to 1 indicates a perfect predictive biomarker. Following biomarker analysis, machine learning algorithms can then be used to predict the classification of new data sets into the existing data classifications.
Visualization
The most important information obtained from an imaging experiment is a visualization of the distribution of various molecules throughout the tissue. As each pixel of an imaging experiment contains an entire mass spectrum, special software is required to handle this specific need in the field. While there have been numerous advancements in this respect, the influx of progress caused there to be a lack of uniformity, making different software tools incompatible with each other. This means that typically the software could not be applied to large data sets, expensive commercial software would be required, or the software would require the end user to have some degree of programming knowledge to fit the data to the software input. However, recent efforts have been made to design open-source visualization tools that are user-friendly and applicable to multiple instrument platforms,102 particularly in the area of laser ablation-inductively coupled plasma mass spectrometry (LA-ICP MS), which is not as routinely implemented as MALDI-MSI or TOF-SIMS.104–106 MSiReader is a key player in open source visualization, providing both a graphic user interface and MATLAB open source code for users.102 Additionally, even open source microscopy imaging software like ImageJ have plug-in scripts capable of handling MSI data sets for visualization.107 These new tools show promise for making the processing of imaging data more widely accessible and customizable for the MSI community.
In addition to improving accessibility, new methods have also been explored for expanding the capabilities of visualization tools. For example, 3D MALDI imaging has been limited by inabilities to reconstruct 3D images, but Patterson and colleagues designed an open-source method for 3D reconstruction using multivariate segmentation.108 Others have expanded the way data is visualized in a different direction. Instead of using imaging to track a single molecule, they developed a tool to view the localization of biological indices (e.g., energy charge index), mapping the relationship between several specified molecules.109
An important note with visualization of data in MSI is that it is critical to ensuring that the image shown is an accurate representation of the molecular distribution. It has been found that cropping images to eliminate background can cause the emergence of distribution patterns not observed in the entire image. As a result, data can become skewed if the analyzed area is too small and does not contain sufficient background area for reference.110 With MSI making an increasing presence in biomedical applications as a diagnostic tool, appropriate representation of visual data is essential.
Preprocessing
Prior to data processing, several steps can be used to ensure accurate and efficient data analysis. These steps include normalization, baseline correction, spectra recalibration, smoothing, and data compression (unsupervised and supervised).111 Normalization is expected to be incorporated into data analysis, while other steps are frequently omitted. However, these additional steps may be necessary, depending upon chosen statistical analysis and the MS instrumentation used to collect the data as well as other experimental parameters and conditions. The inclusion of preprocessing steps in the data analysis workflow can also depend upon the specific goals of an individual project. Overall, preprocessing can help to reduce experimental variance within the data set, extract relevant information from large data sets, and draw meaningful conclusions from subsequent statistical analysis.
Normalization is used to remove systematic artifacts that can affect the mass spectra. Sample preparation, matrix application, ion suppression, and differential ionization efficiencies in complex samples can influence the intensity peaks of mass spectra. Some of these random effects in data acquisition can be minimized by proper normalization. Not applying normalization can lead to misleading artifacts and ultimately depict inaccurate ion distributions, statistical analyses, and conclusions about biological significance. There are a few different methods for normalization for MSI data sets based on the purpose of the analysis. Normalization to the TIC is the most commonly implemented method.112 Normalization to the TIC ensures that all spectra have the same integrated area and is based on the assumption that there is a comparable number of signals in each spectrum.111,113 However, in an imaging experiment, it cannot always be assumed that this condition is met since selection of the area is variable run-to-run. TIC normalization can improve the ability to compare expression levels across samples with similar sample types; however, is not applicable when comparing very different tissue types.112 In addition to normalization to the TIC for similar sample types, the TIC normalized data can be further normalized to matrix related peaks for MALDI imaging experiments to correct for uneven matrix coating. This may be necessary depending on how the matrix is applied to the sample. For example, airbrush sprayed matrix applications cannot produce as homogeneous of crystals across the whole tissue as matrix applied with an automated sprayer or automated microspotter.114 For samples with different tissue types, such as whole body imaging, an externally applied internal standard similar to the compound of interest should ideally be applied before or during matrix application (see above). For this normalization method, each spectrum is normalized to the intensity of the reference molecule for analysis. Normalization to an internal standard reduces the impact of ion suppression that arises from tissue inhomogeneity and improves pixel-to-pixel variability. TIC normalization is not recommended for whole body imaging or for different sample compositions, where internal standard normalization is considered the gold-standard normalization methodology.115 Other options include normalization to an endogenous molecule that is expected to be consistently expressed throughout the whole tissue, such as a phospholipid headgroup. Additionally, some researchers have calculated tissue extinction coefficients or relative response factors to determine the relative amount of a compound in whole body imaging or different tissue types. This tissue extinction coefficient takes into account ion suppression related to the compound of interest and the tissue of interest and is then compared to LC-MS/MS data.116 The tissue extinction coefficients were evaluated for the drugs proponolol and olanzapin on rat whole tissue sections, where kidney, lung, liver, brain, and stomach were chosen as tissues of interest. For both drugs, stomach has the highest extinction coefficient, while the stomach and brain experience the highest variation, likely because of tissue heterogeneity in these organs.116 The advantage of this method is that no expensive, labeled standards are needed of the compounds of interest, although accuracy of tissue extinction coefficients is still being investigated.
Following normalization, additional preprocessing steps are often taken to ensure accurate interpretation of the data. These include steps typically found in conventional MS workflows (e.g., baseline subtraction and spectral recalibration).111 Furthermore, to better visualize the data and increase the signal-to-noise ratio, smoothing algorithms are often applied, such as Savitsky Golay Smoothing49,117 or Boxcar Smoothing.118 Smoothing is especially important for imaging data to remove sudden fluctuations between pixels that do not necessarily represent the in vivo distributions. These preprocessing steps help to ensure that accurate interpretation of the MSI data.
Data Compression
Unsupervised Data Compression
As MSI acquisitions tend to create large data files (up to several terabytes per sample), data processing becomes more difficult and requires more strenuous computational methods. To alleviate this problem and make the data files easier to handle and distribute, several compression strategies have been implemented to reduce the size of data while still retaining the important information. Binning mass spectra for each pixel of an imaged tissue and compression based on region of interest (ROI) are the most successful methods, with ROI compression requiring the least amount of computational power.119 Autoencoders have also been useful for unsupervised nonlinear dimensionality reduction of imaging data by reducing each pixel one at a time to its core features.120 Once the size of data has been reduced, it can be more easily processed in subsequent steps of the processing pipeline.
Unsupervised clustering of the data is also used to compress data into features for statistical analysis. Unsupervised analysis can be divided into (1) manual, (2) component, or (3) segmentation analysis. (1) Manual analysis is carried out by selecting m/z values unique to the region of interest and generating an image for each m/z value. (2) Component analysis requires a statistical or machine learning algorithm to cluster the data. Principal component analysis (PCA) is used to reduce the dimensionality of the data set by converting possibly correlated variables into a set of linearly uncorrelated values, which are called principal components.121 PCA is an unsupervised statistical method to distinguish principal components that cause the greatest variance in the data. PCA plots the component that causes the greatest variation on the x-axis and the component that causes the second greatest amount of variation on the y-axis to induce groupings of related pixels in the data sets.122 While used as a data compression method, PCA can also be combined with discriminant analysis for statistical analysis of imaging data sets (see statistics section below). PCA can also be used to remove signals which are poorly connected with variability between groups, removing noise. (3) The last method, spatial segmentation, bins together similar spectra into regions of interests and identifies colocalized m/z values. Hierarchical clustering, a type spatial segmentation, partitions the image into its constituent regions at hierarchical levels. This only requires knowledge of the similarity between groups of data points and does not take into account spatial position during analysis. Hierarchical clustering is frequently used to rearrange multiple variables to visualize possible groups in the data.123 Another segmentation method is k-means clustering. k-Means clusters the number of partitions, n, into k number of clusters, where each cluster is based on the spatial distances between mass spectra. Following k-means clustering, each observation now belongs to the cluster with the nearest mean.124 Another method, bisecting k-means, is a combination of k-means and hierarchical clustering, although it is computationally more complex. Bisecting k-means is a hierarchical clustering method that uses k-means repeatedly on the parent cluster to determine the best possible split to obtain the next two daughter clusters.125 All of these methods can be used to compress the data into important features and are chosen based on the goal of the study. Some studies will even use more than one unsupervised data compression method. For example, Mourino-Alvarez et al. used both hierarchical clustering and PCA to differentiate proteins in calcified areas and collagen deposits in aortic value tissue. By applying a hierarchical clustering following PCA, they were able to observe several layers that surrounded the calcified deposits that differed in protein expression from other tissue regions.117
Supervised Data Compression
Supervised clustering is better suited when a specified set of classes is known and the ultimate goal is to classify new data set into one of those classes. Supervised data compression uses predefined classes or categories, while unsupervised data compression uses similarity between spectra to generate classes to reduce data size.126 Partial least-squares regression (PLS) is a supervised classification method, where classes of data are annotated with known labels.127 Partial least-squares regression is similar to PCA; however, instead of separating into components based on the maximum variance, it uses a linear regression or classification model to project predicted variables and observable variables to a new space, mathematically speaking. Classification refers to decisions among a typically small and discrete set of choices (tumor vs normal tissue), while regression refers to an estimation of possibly continuous-valued output variables (diagnosis of the severity of disease). This type of supervised clustering requires a training data set for the classification of groups. Like PCA, PLS can also later be coupled to discriminant analysis for statistical analysis but is described in this section as a supervised data compression method. PLS was used to reduce data into different tumor areas that were histology annotated including stroma, smooth muscle, submucosa, fibrous tissue, tumor, healthy mucosa (tumor adjacent), and healthy mucosa (tumor remote) in colorectal cancer. In this case, PLS was used in combination with discriminant analysis to accurately separate changes in lipids between tumor adjacent and tumor-remote healthy mucosa, supporting the idea that cancer influences the local tissue environment.128
Both supervised and unsupervised classification methods reduce data down to the most important m/z value distributions. Data compression projects the data to a lower dimension subspace, while maintaining the essence of the data for statistical analysis. With the large degree of dimensionality associated with MSI data, especially of biomedical samples, extracting important, relevant features becomes increasingly difficult. Machine learning algorithms for feature detection applied to LC-MS data can be limiting with imaging data, as they do not account for differences in spatial regions of the tissue of interest. A context aware feature mapping machine learning algorithm was recently developed that takes into account the spatial region of features when ranking.129
Statistical Analysis
Tests of Significance
Statistical analysis of large imaging data sets is incredibly important for the implementation and utility of MSI. Interpreting detected differences between samples involves statistical hypothesis testing to determine if there is a certain difference that exists between samples or between spatial regions within a sample. The choice of appropriate univariate analysis tests that one m/z, identifying to a compound of interest, will depend on the data set. If the data has a Gaussian distribution, a t test can be used to determine the difference between two samples with ANOVA being used to determine if there is any difference in a group of samples.46,130 For MSI data sets, t tests can be performed to compare m/z relative intensities between two different regions and ANOVA between three or more regions of interest. For example, peptides and metabolites were evaluated in the cortex region of the brain comparing wild type mice and transgenic mice carrying a missense mutation causing cortical spreading disease, which causes migraines. A t test was used to evaluate specific changes between the cortex of the wild type mice and the transgenic mice.131 Unfortunately, a Gaussian distribution of mean intensities cannot be assumed for clinical samples, but mean values may still be used if the central limit theorem is satisfied. If the data has a non-Gaussian distribution, nonparametric tests like the Mann-Whitney U-test can be used as a statistical test of the hypothesis. These tests are useful for finding peaks with an observable change between different regions or experimental conditions.
Discriminant Analysis
Data reduction methods such as PCA or PLS are preprocessing steps to discriminant analysis (DA). These analyses are commonly performed together and abbreviated as PCA-DA or PLS-DA, respectively. DA is a statistical tool to assess the adequacy of a classification system. For any kind of DA, the groups need to be assigned beforehand or in the case of PCA, preprocessed prior to discriminant analysis. DA is particularly useful in determining whether a set of variables is effective in predicting category membership. This is different from an ANOVA or multiple ANOVA, which is used to predict one or multiple continuous dependent variables by one or more independent categorical variables. DA is used in MSI to see how well components separate regions of interest in the data set. For example, PCA-DA was applied to colon spheroids to successfully differentiate the outer, middle, and inner regions of the sample.132 Additionally, PLS-DA was applied in histology driven data mining of lipid differences between colorectal cancer liver metastasis biopsies, where normal vs tumor were preselected as regions of interest prior to analysis.133
Biomarker Tests
Even if statistical differences exist between two conditions for a single m/z, this does not necessarily mean that this m/z value can act as a biomarker to distinguish the two classes. For univariate biomarker analysis to confirm if a m/z can be used as a diagnostic test to distinguish two regions of interests, a receiver operator characteristic (ROC) curve analysis is performed. In ROC analysis, the true positive rate (sensitivity) is plotted in function of the false positive rate (specificity).134–136 The area under the curve (AUC) in these plots can distinguish whether the m/z marker can be used for diagnostics. This is a test of accuracy, where an AUC value between 0.90 and 1 is excellent, 0.80–0.90 is good, 0.70–0.80 is fair, 0.60–0.70 is poor, and 0.50–0.60 is a failed test. This test is used to discriminate the ability of a specific marker (m/z) to correctly classify groups of interest. MALDI imaging was used to reveal thymosin beta-4 as an independent biomarker in flash frozen colorectal cancer compared with normal tissue using ClinPro Tools software to perform ROC analysis with an AUC of 0.80. 137
However, often in biomarker discovery, one biomarker is not able to correctly classify groups with a high enough AUC for clinical diagnostics. In this case, multiple biomarkers (multiple m/z values) are used for analysis. This is known as multivariate analysis. Here, machine learning algorithms are used to examine multiple biomarkers to search for correlated m/z values in the mass spectra that also correlate with the target outcome. This multivariate analysis provides a single ROC curve that is derived from multiple biomarkers. Additionally, an indicator of how much each m/z contributes to the score from the resulting algorithm is calculated for each m/z value.138,139 For regression-based methods such as PLS, the importance of an m/z value is a direct result of the model’s loading vector. Additionally, colocalization of two individual m/z in a tissue can be calculated in a correlation analysis to see how well m/z components of the multivariate analysis align based on spatial distributions.140
One problem for MSI analysis is that salt adducts of the m/z values of interest are identified separately. Therefore, in biomarker analysis, it would be ideal to combine m/z values identifying to the same molecular compounds into a single peak for analysis. For instance, m/z values can shift based on the presence of a sodium ion, potassium ion, the loss of ammonia, the loss of water, oxidation of methionine, and other common modifications. This can complicate identification and statistical analysis as well as univariate and multivariate biomarker analysis. For MALDI, Alexandrov introduced a method called masses alignment which is used to group masses corresponding to a single peak and then represent them as one m/z value.141 This also reduces the size of the data set, making computation and biological understanding of the data more attainable. It also links m/z values that belong to the same biomolecule together for statistical analysis.
Machine Learning Algorithms
Machine learning is starting to play a larger role in developing algorithms to quantify relationships in MSI and then using these identified data to make predictions for new data sets. First, the data set is converted from a population of profiles into a “n by m” data matrix, where “n” is individuals, and “m” is the biomolecules of interest.142 Following conversion, they can be analyzed using different algorithms that look for correlated structures in the measured data that also correlate with a target outcome. Neuronal networks, support vector machine algorithms, recursive maximum margin criterion, and genetic algorithms are used to build statistical models that use training data to predict the classification of new data sets. This is currently being implemented for automated decision making, modeling, computer aided diagnosis, and can be applied for tumor classification for pathology detection.143 Specifically, in one example, a PCA support vector machine was used for early detection of ovarian cancer with about 90% accuracy.144
Complete Data Analysis Pipelines
Because processing imaging data requires numerous different treatments compared to conventional LC-MS data, software with complete data analysis pipelines are useful for streamlining the entire data analysis process. While there are numerous open source and freely available software packages for processing data, functionality tends to be restricted, and there are typically no export options for the data. A widely used software package, MSiReader, has seen rapid developments toward incorporating various aspects of data analysis, including visualization, quantitation, and annotation in a streamlined, easy-to-use platform.102 A new MSI software package, SpectralAnalysis, aims to expand the reach of data processing by incorporating all processing steps from preprocessing to multivariate analysis, within a single package, allowing for the analysis of single experiments as well as large-scale experiments spanning multiple instruments and modalities.145 Improved data processing pipelines are also being developed in efforts to make full use of the spatial information unique to imaging experiments. One such pipeline, EXIMS, strives to reveal significant molecular distribution patterns by treating the data set as a collection of intensity images for various m/z values. The process incorporates preprocessing, sliding window normalization, denoising and contrast enhancement, spatial distribution-based peak-picking, and clustering of intensity images.146 massPix, an R statistical program, is able to perform multivariate analysis (PCA and clustering) and has integrated lipid feature annotation into the automated pipeline.147 Another software pipeline is SCiLS, which is available commercially from Bruker. SCiLS software can be used to analyze multiple imaging data sets, performing comparative analysis, colocalization analysis, spatial segmentation, and classification model calculations based on training data sets, and it contains numerous other highly useful features. Additionally, ImageQuest software from ThermoFisher Scientific is frequently used for visualization, normalization, and creation of two-and three-dimensional maps of analyzed tissue, and similar features are offered by High Definition Imaging software by Waters. Following m/z mapping to a biological compound, platforms that can handle high-dimensional biology data sets, such as Clustergrammer, a Web-based tool, help to visualize biology changes in heatmap formats that retain the high-dimensionality of the biological data.148 These pipelines emphasize the importance of special treatment for imaging data compared to LC-MS data.
Repositories
Finally, data storage and sharing of the final results allow for the community to move forward and build upon the ever-growing wealth of knowledge. In order to further drive this, imaging repositories are necessary for allowing researchers access to imaging data for comparison of results and for discovering new answers to biological questions. Previously, such repositories were difficult to implement due to the requirements of large space and computational power, but technological advancements have allowed for the emergence of at least one such repository,149 with the promise of more becoming available in the near future. Another currently being developed is METASPACE for bioinformatics for spatial metabolomics, an online engine based on big-data technologies that automatically translates millions of ion images to molecular annotations.150 The estimated completion time for this project is June 2018.
MULTIMODAL IMAGING SYSTEMS
While MSI is useful for analyzing the spatial distributions of several molecular species, it lacks the molecular depth that other methods provide. The combination of MSI with other imaging modalities is sure to evolve into a comprehensive analysis tool to answer biological questions that could otherwise not be answered with a single imaging modality. Multimodal technologies are very commonly implemented in diagnostic imaging techniques,151 and the concept has been expanded into MSI analysis pipelines.152 Because MSI has high chemical specificity but lower spatial resolution compared with other imaging modalities, it is typically combined with modalities that complement these features. For example, MSI is combined with imaging modalities that have high spatial resolution or tissue structural information. This can be done with a single section, from which the complementary data can be powerful and enable greater, more significant discoveries.153
Multimodal imaging can be approached by either acquiring images at different times (asynchronous), where the images are fused in data processing step, or by simultaneously acquiring images (synchronous) and merging them during the data acquisition step.154 Asynchronous postprocessing can present some difficulties which arise from the positioning of the same samples between different imaging modality scans at different times, which could cause difficulties in coregistering images for analysis.155 Co-registration is especially difficult if data acquisitions are not acquired at the same spatial resolutions; however, advances in computational annotation help to improve image analysis.156 Often, voxels from the lower resolution modality are combined to form the voxel size of the higher spatial resolution.157 Image coregistration can be achieved by aligning known regions of interest, using calibration points to perform a rigid regression or by selecting a variety of points to perform moving least-squares registration between the images.158 Additionally, different imaging platforms have distinct sample preparation protocols, which can cause interference for different imaging modalities. For example, flash frozen tissue samples are ideal for MSI, although the embedding media that they are frequently stored in, which allow for optimal sectioning and staining, consist of the polymer polyethylene glycol can cause interfering peaks in the mass spectra. Therefore, it is important that the sample preparation used for other imaging modalities also are compatible with MSI. Because of this, not all ideal multimodal systems can be easily combined without changing the sample preparation workflow, depending on the desired analysis. Synchronous imaging is advantageous because consistency is achieved in both time and space, however combining instrumentation to accommodate synchronous acquisitions can require advanced skill and be very expensive, especially for MS instrumentation. Hybrid systems integrating MSI are currently being developed, although it should be noted that some method combinations are not possible due to sample incompatibility. One multimodal instrumentation example integrated a commercial optical microscope, laser micro-dissection instrument (capable of both bright-field and fluorescent imaging) with an electrospray ionization mass spectrometer capable of submicrometer MSI.56 In general, data analysis becomes even more difficult when integrating quantitative information from multiple existing functional modalities to create composites of three, four, or even five imaging modalities into a single data analysis pipeline. Multiplexing image modalities presents a “big data” computational challenge, making MSI data compression especially important for multimodal system integration. To handle the versatility of multimodal systems, workflow based analysis platforms that integrate existing methods are gaining popularity to handle big data problems for imaging systems.159 These programs allow separate steps that can be rearranged for customizable workflows and do not require computer programming knowledge. KNIME and Galaxy are commonly used workflows for multimodal imaging analysis.160,161 Bouslimani et al. created a method to merge microbial 16s rRNA amplicon sequences onto a 3D MSI map of the human skin. Computational tools used for this analysis are available as a workflow for KNIME, which can be modified and applied for future use for multimodal analysis.162 It is also clear though that advances in multimodal technology and instrumentation will allow for synchronous integration to be expanded for multiple imaging modalities, a few of which will be discussed below.
It is safe to predict that this overview of multimodal imaging is only the beginning of imaging combinations that will be possible in the future. Multimodal systems are rapidly expanding into multiple imaging modalities. Here, selected multimodal systems are highlighted, but other imaging techniques not described here, such as near-infrared microscopy, electron microscopy, coherent antistokes Raman spectroscopy, Fourier-transform infrared spectroscopy, etc. are also being integrated with MSI for multimodal analysis.163–167 With the largest hurdle in multimodal imaging systems being coregistration and data analysis, there will likely be an increase in the development of integrated systems. Additionally, before multimodal integrated systems will be used regularly, robust coregistration analysis algorithms need to be incorporated into software platforms to handle complex multimodal data compression and analysis.
Microscopy Multimodality
MSI is often combined with microscopy to provide high-resolution morphological and structural information in complement to MSI’s ability to visualize and identify distributions of specific molecules. For example, Plas et al. describes a method for fusing microscopy with MSI data to enable prediction of a molecular distribution both at high chemical specificity and at high spatial resolution. This is done post-data acquisition using the microscopy data to sharpen and perform out-of-sample predictions.168 Here, we focus on using optical light microscopy, including bright-field, fluorescence microscopy, and phase-contrast, to evaluate tissue structure and specific markers. Microscopy is the most common multimodal system currently paired with MSI and is very useful for identifying regions of interest for statistical probing or supervised classifications.
Histology
Although tissue sections used for MSI can be scanned by the computer during analysis to produce an optical overlay, important structural information on the cellular level is obtained from histological analysis of a sample using light microscopy. In general, light microscopy is used to enlarge details and portions of a tissue section. Samples are stained with a specific dye to highlight tissue structures of interest. Histological overlay is the most common multimodal imaging system combined with MSI currently applied in the literature.71,169,170 In the discussion below, the focus will primarily be on mammalian tissue stains, although plants and other organisms can also be studied in this fashion. For example, one study used a nonspecific dye stain of gallotannins and ellagitannins in the root of Paeonia lactiflora to overlay the structural histology with MSI identification of specific galltonnins and monoterpene glucosides.171
The most traditional histological stain is hematoxylin and eosin (H&E) stain, which distinguishes nucleic acids from proteins with blue and red colorings, respectively.172 This allows the user to visualize the differences between cells and the surrounding extracellular matrix.173 Other commonly used stains include Masson’s trichrome stain used for connective tissue, Alcian Blue for mucins, and periodic acid-Schiff reactions used for staining carbohydrate rich tissue regions.174 When stained, several key characteristics are analyzed, including, tissue morphology, cell structure, and staining distribution. In a clinical setting, trained pathologists use stained slides to identify different disease states of the tissues to stratify patient specimens and provide diagnostic indices. With the high magnification capabilities of modern microscopes, very fine resolution of spatial features can be achieved, which is complementary to MSI’s comparatively low spatial resolution. Combining histology with MS asynchronously can allow for the analysis of spatial molecular arrangements without the need for target-specific reagents, which allows, for example, the discovery of diagnostic and prognostic markers of different cancer types.175,176 It should be noted that typically a sample should first be analyzed using MSI (with a nondestructive ionization method) and then stained, as stained slides increase spectral complexity, and histological processes can cause degradation or sample loss if done prior to MSI analysis.177 MS compatible dyes do exist, such as cresyl violet and methylene blue, which can allow histology first followed by MSI analysis; however, tissue degradation and diffusion of molecules are still elements of concern.178 For destructive ionization mechanisms, a serial section is typically used for histological analysis and is coregistered to its adjacent section. However, with serial sections, artifacts such as physical destruction from cutting and fixations and staining artifacts can cause complex distortion effects, also complicating image registration.179 Although a serial section may not contain the exact same molecular structure as the section of interest, most structural features are well conserved section to section. Serial sections are commonly used for asynchronous multimodal imaging.
One interesting example applies the H&E stain serial tissue section to direct analysis of specific tissue regions on the section prepared for MSI. Regions of interests, including nontumor, undifferentiated tumor, moderately differentiated tumor, and well differentiated tumor, were selected from the H&E stained sections and only those individual regions of interest of the tumor were analyzed. The differentiation of the cells in the tumor region help the pathologist determine cancer staging, where differentiated cells are most similar to the normal tissue.180 By imaging only the regions of interest, the user can save significantly on instrument time. This application is particularly useful for high-resolution MSI, where data acquisition times can be very long and increased data throughput is needed.
Fluorescence Microscopy
Another microscopy technique uses fluorophore conjugated antibodies to label a molecule of interest, known as immunohistochemistry (IHC). First, a molecule of interest is labeled with a primary antibody. Then, a secondary, fluorescent antibody is added that recognizes the primary antibody. The secondary antibody is conjugated with a fluorophore, which, when excited, will provide a measurable light emission. Because multiple secondary antibodies can bind to a single primary antibody, the signal of the protein of interest can be amplified greatly to increase the sensitivity of the assay.181 Depending on the fluorophore of choice, special emission and excitation filters are needed to visualize the signal using a fluorescent microscope. While less common, the secondary antibody can also be coupled with a colorimetric assay. Using IHC, researchers can confirm spatial distributions seen in MSI experiments. For example, Heeren and co-workers looked at the hypoxia marker, Pimonidazole, in a breast tumor model using MSI, for which the distribution was confirmed using a secondary antibody conjugated to a horseradish peroxidase colorimetric assay.182 Some cells and certain molecules possess intrinsic fluorescence. Examples of proteins or small molecules that naturally fluoresce include NADH, tryptophan, chlorophyll, or green fluorescence proteins. Becker et al. uses the natural fluorescence of stillbenes to discriminate the stillbene region from the rest of the grapevine leaves. This feature was then combined with MSI to study metabolite changes in stillbene regions of grapevine leaves after P. viticola infection.183 Alternatively, molecular biology and genetic engineering techniques can label proteins, nucleic acids, lipids, or small molecules with an extrinsic fluorophore. These techniques can be applied in systems ranging from cell culture to human systems. Chughtai et al. genetically engineered mice to express tandem dimer tomato red fluorescent protein under control of a hypoxia response element in hypoxic regions of the tumor. They then used MSI to compare lipids and proteins in hypoxic and normoxic regions of the tumor.184 While the examples described above are asynchronous examples of multimodal imaging modalities, many are working on the integration of optical microscopy directly with MSI. Some of the limitations of combining fluorescent imaging with MSI is that often fluorescence signal requires in vivo imaging because of degradation of naturally fluorescent material. For instance, NADPH, a source of autofluorescence, has a half-life on the order of minutes and is sensitive to degradation at high pH conditions. Therefore, fluorescence of NADPH is ideally processed using in vivo systems, while commonly used MSI systems process samples ex vivo.185 Logistical sample preparations can also create issues with coregistration because differences in orientation during acquisition can be difficult to correct for during data analysis. However, if the fluorescent label is stable and can also be ionized in a mass range of interest for MSI, registration algorithms are not needed for multimodal analysis.184
Although there are many examples where fluorescence has been multiplexed with MSI, substantial opportunities for growth still exist in fluorescence and optical microscopy multi-imaging systems for MSI. For example, fluorescence technologies including fluorescence lifetime imaging microscopy (FLIM), which measures the amount of time a molecule fluoresces as its primary metabolic readout,186 or spectral lifetime imaging microscopy (SLIM), which is used to create optical molecular fingerprints based on spectra and lifetime of a fluorescent signal,187 have never been combined with MSI. Additionally, combining MSI with multiphoton microscopy, which relies on multiple photons to optically section through a tissue slice by acquiring clear images at multiple focal planes at different depths in a tissue,188 has not been used and could serve as a unique way to connect serial sections for 3D MSI.
Allen Brain Atlas Integration: Virtual Multimodality
Although not necessarily another imaging modality, MSI of the brain and histology images can be coregistered with the Allen Brain Atlas as a virtual multimodality. The Allen Brain Atlas is a publicly accessible collection of brain anatomy compiled high-resolution histology images and genome based on brain regions.189–191 Brain atlases exist for a variety of species, including mouse, nonhuman primate, and human, and have also been expanded to accommodate different neurological diseases and developmental states. These virtual maps were constructed as a 3D biochemical architecture of the brain, where the anatomy of the brain is paired with genome analysis of each region which averaged between many organisms. For the MSI field, MATLAB has been used to conduct automatic alignment of a MSI data set of a mouse or rat brain to the position in the 3D map atlas. Figure 6 demonstrates how the Allen Brain Atlas can be used to align m/z values of interest from MSI to a specific region of the brain; in this case, m/z 863 is localized in the striatum of the brain. This atlas can also be used for genomic insight for biological interpretation of the data, as the Allen Brain Atlas currently has projects mapping the genetic geography of the brain and cell type in each region and has compiled region-specific electrophysiology studies. The development of automatic alignment and annotation tools has increased the throughput for MSI data analysis. It has also enabled MSI distributions to be compared in the same coordinate space, although improvement upon these tools will be necessary to handle the increased file sizes.192,193 This atlas is a unique example of virtual multimodality that is extremely useful for neuroscience researchers working in the MSI field. Additionally, although integrating MSI is not currently an advertised focus of the Allen Brain Atlas, the development of MSI integrated atlases could be useful for multiomics integration.
Figure 6.
MSI acquired on a mouse brain is registered to corresponding histology and then to the Allen Brain Atlas (ABA) to understand where m/z values colocalize with anatomical brain regions. Shown here are three coronal sections from three different mouse brains (M1, M2, and M3) and their respective MSI images. (a, b, and c) Distribution of m/z 863 before preprocessing and registration. (d, e, and f) Samples after preprocessing and registration to histology, (g, h, and i) Distribution of m/z 863 after registration to histology, where histology image is removed. (j, k, and l) Registered images and registered MSI images are superimposed to display the visual distribution of ion m/z 863. (m, n, and o) Registered images with the MSI distribution are then registered again with the ABA. On the basis of the alignment, it appears that m/z 863 is expressed mainly in the striatum of the brain for these brain sections. Reproduced from Abdelmoula, W. M.; Carreira, R. J.; Shyti, R.; Balluff, B.; van Zeijl, R. J.; Tolner, E. A.; Lelieveldt, B. F.; van den Maagdenberg, A. M.; McDonnell, L. A.; Dijkstra, J. Analytical Chemistry 2014, 86 (8), 3947-3954 (ref 192). Copyright 2014 American Chemical Society.
Analytical Multimodalities Systems
Multimodal Uses for MSI
Different types of MSI ionization or biomolecule targets can be multiplexed to analyze compounds simultaneously. Because of the range of ionization mechanisms, MSI can actually be multiplexed with itself to provide a more comprehensive analysis of the sample by using different ionization mechanisms to analyze different groups of molecular compounds. An example of this is the combination of DESI and subsequent MALDI analysis to analyze lipid and protein distributions, respectively, on the same section.194 Another study combined MSI ionization techniques to complement the high spatial resolution of TOF-SIMS with the high mass resolution, high mass accuracy, and MS/MS of an AP-MALDI source coupled to an Orbitrap mass spectrometer to study the distribution of lipids in a colon cancer tissue section. First, TOF-SIMS analyzed the lipid distribution at 1 µm spatial resolution. Then, matrix was applied to the same tissue section and AP-MALDI was used to identify lipids at high mass accuracy and also to perform on-tissue MS/MS.195 Additionally, the same ionization mechanisms can be used for multimodal systems for analyzing different molecular species. For example, MALDI-MSI can be used to analyze both N-glycans and proteins using sequential analysis of the PNGaseF enzyme to release glycans followed by application of trypsin to analyze proteins from the same tissue section using FFPE tissues.47 This is considered multimodal imaging as two separate enzymatic and matrix applications are necessary for analysis.
Raman Spectroscopy
Raman spectroscopy signal is based on the vibrational structure of the molecules within each sample. Each biological sample consists of multiple molecules that form its complex structure and thus vibrational spectrum, which is known as the molecular fingerprint. Raman can be spatially resolved with microscopy for label-free chemical analysis, known as confocal Raman microscopy (CRM). CRM is a nondestructive technique that has high spatial resolution and allows for 3D analysis,196 where the samples can be optically sectioned taking different z-stacks of the tissue. CRM, however, can be insensitive without special enhancement and is not capable of molecular identification.197 MSI can thus be combined with CRM to assist with identification of specific components of a molecular fingerprint. Additionally, because Raman is a nondestructive technique, MSI can easily be performed post-Raman analysis. Correlated MSI and confocal Raman microscopy was used to study the structural and chemical diversity of three-dimensional cell cultures.157 The necrotic core of the spheroids experienced greater Raman variability and correlated with the principal component causing the greatest variance in the data.157 Additionally, CRM was also correlated with C60-SIMS to show consistent distribution of quinolone disruptions between the new analytical techniques, with specific molecular identities determined through tandem MS.198
Magnetic Resonance Imaging
Magnetic resonance imaging (MRI) is a medical imaging technique that uses strong magnetic fields and radio waves to generate detailed images of cross sections from tissues of the target sample.152 MRI enables researchers to obtain a 3D anatomical structure of a sample of interest with significant resolution and gives a precise sample shape. Additionally, MRI is noninvasive, although a contrast material is typically administered to enhance the signal-to-noise ratio for the tissue of interest. MRI complements MSI very well because it allows detailed structural information, while MSI provides chemical probing of the sample. In one example, MRI and MSI were used to study distributions of alkaloids in two structurally distinct regions in maturing areca nuts (seed of Areca catechu).199 Additionally, another study used MSI for the detection of the gadoteridol (MRI contrast agent) in human gliomas via DESI-MS imaging following MRI analysis. Detection of a compound by both modalities, as in the example above, is particularly useful for improvements in coregistration between imaging modalities.200 Overall, MRI, while underutilized with MSI in the literature, provides a powerful tool that could be more integrated with other systems, including with the Allen Brain Atlas described above. Scalable Brain Atlases based on MRI data are being developed for nonrigid spatial registration of MSI to MRI data.193 Additionally, new computation pipelines are also being developed to better integrate MRI, 3D MSI data, and histology.201
Positron Emission Tomography (PET)
Positron emission technology (PET) is commonly used clinically to visualize tumors and tumor metastases in 3D as a noninvasive imaging technique to provide metabolic assessment of a region, for example, a cancerous tumor. By administering a radiolabeled metabolite, commonly 18F-fluorodeoxyglucose (FDG), PET monitors the body’s uptake of FDG. Tumors tend to have altered, typically increased, metabolism compared to the regions surrounding it.202 PET can be combined with MSI to understand tumor heterogeneity and to show additional metabolic alterations in regions of interest. Biomap software allows for multimodality imaging processing between PET scans and MSI data sets. In one study, MALDI-MSI was actually used to assist in the development of a new PET radioligand by using MSI to image the PET molecule itself as well as the biological receptors for the PET molecule.203
Atomic Force Microscopy
Atomic force microscopy (AFM) is able to measure the topography with nanometer spatial resolution by raster scanning the tip of a scanning probe along the (x,y,z) position of a sample. Since tissue sections may not be completely flat, it can be beneficial to collect the surface topography of a tissue section using AFM for MSI. This can allow signal intensities to be correlated with surface topography, as uneven surfaces can affect uniform instrument ionization. Using AFM, the morphology of the section is translated into specific quantitative features, such as height, width, area, and volume. AFM and MSI can actually be obtained synchronously where the same probe is used without moving the sample from the system as demonstrated by Ovchinnikova et al. using proximal probe thermal desorption/ ionization MS.204 This eliminates the need for postacquisition coregistration, which can be particularly difficult for AFM and MSI multimodal systems.
Multimodal imaging systems couple additional structural, biological, morphological, and chemical information with MSI analysis. Optimized sample preparation for multimodalities, advances in coregistration, and improvements in computational workflows are required to advance the use of MSI integrated multimodal imaging systems.
BIOLOGICAL APPLICATIONS
It is clear that the increased molecular capabilities of MSI have caught the attention of the biological community. The expansion beyond peptides and proteins, MALDI-MSI, and even fresh tissue sections has broadened the general use of MSI in unforeseen directions. In the following sections, several interesting studies will be highlighted, along with specialty applications in the field, all of which have incorporated the advancements described in this review. It should be noted that MSI has applicability beyond the biological systems (e.g., imaging of dyes on banknotes), so individuals should not feel limited in the systems where MSI can be applied.167,205,206
Expansion of Molecular Species of Interest
Initially this technique was used to localize proteins and other peptides within a sample, with the first applications toward tissue samples.130 At first, MALDI was the core technique for imaging, and it is still by far the most popular method for analyzing peptide and protein rich samples.3,9,207 As an example, neuropeptides have been primarily imaged with MALDI,208 which has been applied to characterizing distribution changes in the pericardial organ of the green crab (red and green morph) after being exposed to salinity stress.3 In particular, the development of on-tissue digestion with trypsin has increased protein coverage by allowing not only higher molecular weight species to be analyzed but also by letting other more sensitive, higher resolution instrumental platforms become available for protein analysis.9,47 Animal tissues are not the only sample types to have benefitted from this technique, as plant peptides have been imaged to compare those found in seedling and mature Medicago truncatula.209 Alternative ionization techniques beyond MALDI have been applied to better image proteins and peptides. For example, matrix-enhanced SIMS has produced high spatial resolution images of neuropeptides.210 Furthermore, matrix-free, ambient ionization methods have also found popularity, as both DESI and nanoDESI have just recently been shown that they are capable of imaging global protein distributions.48,88 While proteins and peptides maintain their popularity due to their obvious biological roles, it is apparent that many more recent MSI studies have focused on mapping the distributions of other molecular species.
The advent of on-tissue digestion with trypsin along with the use of PNGaseF for release of glycans has prompted the analysis of N-glycans with MALDI-MSI. As discussed previously, on-tissue digestion sequentially with PNGaseF and trypsin enabled MSI analyses of both released N-glycans and tryptic protein fragments on a single tissue.47 It should be noted though that native glycan imaging has been performed.211 N-glycans have been analyzed in variety of samples, including kidney, bone, and cancerous tissues.38,212–214 To highlight one example, different bone marrow samples that had either no bone lesions or with various stages of bone lesions were compared, and the glycan (NeuAc)2(Hex)2(HexNAc)2 + (Man)3(GlcNAc)2 was shown to be upregulated in the Stage 1 bone lesion affected marrow.214 Notably, much of the recent literature has focused on FFPE tissue, which is expected with their increased availability compared to other clinically relevant tissue. In order to assess different N-glycans, derivatization methods are beginning to emerge, including for sialic acid-containing N-glycans, for which images are shown in Figure 2.52 Beyond derivatization, tissue engineering-based approaches can be used to develop specific probes against different glycan groups to improve ionization and thus detection of these important molecules, though this technique is still in the “proof-of-principle” stage.215 The methodology for N-glycan imaging is still underdeveloped and the availability of more derivatization agents, expansion to other ionization sources, and improved sample preparation steps are expected to be developed.
Because of their diagnostic power, lipids are becoming more targeted compounds for MSI analysis.1,216,217 For example, folic acid distribution appears to increase in prostate tumor tissue after intravenous administration.216 Lipids also have the potential to show tumor boundaries, specifically in breast cancer.217 Because of their diverse structure, several common ionization sources, including MALDI,1,20,42,97 DESI,22,217,218 SIMS,219–221 and IR-MALDESI,18,19 have found success in lipid imaging. Notably, newly developed laser spray ionization (LSI), with its simple sample preparation, is becoming more commonly used for lipids as well.20,222 As an example, division of normal and cancerous tissue was easily achieved for a few dozen lipid species, including diglyceride DG (18:1/20:0), which was indicated as a potential biomarker for renal cancer.222 While tissue sections have obvious popularity in the literature, lipids can be measured in a variety of sources, including fingerprints223 and plants.224 Finally, because of their comprehensive distribution, whole body imaging of lipids has been done on the Anopheles stephensi mosquito.15 One caveat of lipid imaging is the difficulty of identification due to lipid diversity, and care should be taken in ensuring proper procedures to confirm their identity, especially if the information is intended to be utilized in the clinic.
Surprisingly, elemental analysis has been done more frequently to distinguish biological samples. By looking at metal species, researchers have the unique advantages of low background noise along with good quantitative dynamic range, unlike most protein or peptide studies.225 Unfortunately, only select ionization sources are capable of imaging metals, and each method is limited in which metals they can image.226 Currently, laser ablation-inductively coupled plasma (LA-ICP) MS32,225,227–230 and SIMS24,231 have been the only methods available to analyze elemental metals. With the use of nanoSIMS, mapping at a subcellular resolution (300 nm) has been achieved for copper, phosphorus, iron, and calcium, allowing the discrimination between the cell wall and different vacuoles in a C. reinhardtii cell. Notably, this technique can also be useful for drug visualization if metal-based drugs (e.g., cisplatin) are involved by directly analyzing their metal core and not the drug itself.232 With the recognition that all molecular species can have an effect on human health, metal imaging will likely become more popular in the upcoming years.
By far the most imaged molecular species in the current literature are small molecules. This may come with their broad diversity, as they include TCA cycle components,1,218,233 neurotransmitters,2,28 drugs,29 and even fungicides.234 Importantly, these diverse small molecule metabolites could be indicative of disease state.152,218 Some of the more notable applications include imaging tetrahydrocannabinol (THC) in hair strands (Figure 7),29 determination of antimicrobial metabolites for biocontrol against F. oxysporum,235 and metabolomic distribution analysis of kidney metabolites after rats being treated with furosemide.36 Because of their structure, metabolites can have poor ionization efficiency, which has prompted the development of many derivatization schemes to increase access to metabolites in samples.28,31,53 Just like lipids, a wide variety of ionization techniques lends well to metabolites, including MALDI,12,14,20,28,29,31,36,137,233–236 DESI,2,218,237 and LAESI,69 although, based upon citations alone, MALDI is by far the most popular. This is actually surprising, as most matrixes used for MALDI analysis are small organic compounds, meaning they can have their own interfering ions in the metabolite mass range at high concentrations, masking some possible signals of interest. Care should be taken in understanding which peaks come from the matrix, and alternatives, such as deuterated matrixes should be sought out if necessary.238
Figure 7.
MALDI-MSI MS/MS was utilized to confidently identify THC drugs in single hair samples. (A) Parent ion images (m/z 406.2). (B) Characteristic fragment ion (m/z 110.0). The distributions match each other for all hair samples, indicating they belong to the same ion. Reproduced from Beasley, E.; Francese, S.; Bassindale, T. Analytical Chemistry 2016, 88 (20), 10328-10334 (ref 29). Copyright 2016 American Chemical Society.
Expansion of Sample Sources
It is clear that the expansion of molecular species that can be analyzed by MSI has also sparked its application to new and unusual sample sources. Biological samples (e.g., tissue) will likely continue to be utilized for method development and proof-of-principle experiments due to their natural sample complexity and variety.31,33,222,229,239 This trend is especially true for clinically inventoried samples, since strategies for analyzing FFPE tissue samples are in high demand.49,51,100,213 For example, tissue microarrays, which are usually FFPE samples, provide unique opportunities for researchers to perform high-throughput screening of different diseases with little variation in preparation.51,211 Beyond FFPE, one unique application of MSI on a mammalian tissue sample has been where cannabinoids, such as THC, and other drugs have been imaged with MS and MS/MS on single hair samples, which has promise for integration into toxicology-based screenings (Figure 7).29,240 Fingerprints have also been targeted for MSI analysis.45,241
In addition to mammalian systems, other model organisms, such as crustaceans, grasshoppers, and ants, have also been imaged using MSI techniques.3,91,236 Furthermore, the use of plants as an alternative research system is commonplace so, naturally, researchers have also transitioned to imaging plant-based samples.234,242 In the last 2 years, food products, such as onions,243 cucumbers,18 and citrus peel extracts,237 have been analyzed, but it is clear that theoretically any item can be imaged with the appropriate equipment.244,245 For example, DESI-MSI was utilized to understand the thin-layer chromatography (TLC) separation of the citrus peel extracts directly on the TLC plates (Figure 8). Notably, methods have even been developed to image microbial systems, many of which can provide insight into antimicrobial agents or characterization of microbial interactions.235,246,247
Figure 8.
Extracted ion images from a positive ion mode DESI-MS imaged high-performance TLC plate, showing the separation of a variety of metabolic compounds. (A) m/z 138, (B) m/z 152, (C) m/z 166, (D) m/z 168, (E) m/z 261, (F) m/z 303, (G) m/z 373, (H) m/z 403, (I) m/z 433, (J) m/z 463, (K) m/z 579, (L) m/z 581, (M) m/z 609, (N) m/z 611, (O) m/z 625, (P) m/z 667, (Q) m/z 725, (R) m/z 741, (S) m/z 755. Reproduced from High-performance thin-layer chromatography/desorption electrospray ionization mass spectrometry imaging of the crude extract from the peels of Citrus aurantium L. (Rutaceae), Bagatela, B. S.; Lopes, A. P.; Cabral, E. C.; Perazzo, F. F.; Ifa, D. R., Rapid Commun. Mass Spectrom., Vol. 29, Issue 16 (ref 237). Copyright 2015 Wiley.
Unique Applications
Clinical Incorporation
With the streamlining of sample preparation, collection, and processing, integration of MSI into the clinic is becoming more accepted, which has been highlighted throughout this review. Briefly, MSI has shown applicability to determining metal accumulation,226 discriminating tumor regions,41,49,51,212,213 disease diagnosis,218,248 and even intraoperative usage.5,6 MS has already found its way into real-time surgeries with the invention of the iKnife,70 and it is only a matter of time before MSI is utilized in a similar fashion.
3D MSI
The construction of 3D MS-based images has expanded in the past decade with advancements in the throughput of MS instrumentation. 3D analysis incorporates volumetric molecular distribution into the equation. Currently, 3D imaging is commonly done through serial sectioning of a sample, where each section is analyzed in 2D and computational reconstruction of a 3D model based on the distance between section distances is performed.249–252 For example, Seeley et al. utilized a multimodality approach with MRI and MSI to rebuild 3D image of a mouse leg with a bone tumor.249 However, emerging techniques, such as 2.5D (i.e., single section localization within the tissue volume using other imaging modalities), surface, or ablation-based imaging, are also being proposed.253 Sectioning is not always necessary with ablation-based methods, such as LA-ICP MS, as “layer-by-layer” of the sample can be analyzed.254 This idea of “depth profiling” can also be used for samples that cannot be sectioned (e.g., minerals, 3D cell cultures).94,95,254,255 Limitations exist for 3D imaging though, including throughput,253 alignment between serial tissue sections, and the computational limits for data size, for which some 3D MSI are 200–300 gigabytes.256 Data compression and/or clustering, supervised or unsupervised, is necessary for each 2D tissue section prior to alignment and reconstruction.132,250,253,256
It should be noted that even when sectioned, samples may not lay flat on the slide or they may have density differences along the tissue that could expand or contract during further sample processing. With the slight variations in tissue height, inaccurate quantitative information can be obtained, bringing the significance of any results into question. The advent of topography-integrated MSI instrumentation allows for the system to correct based on tissue height.92,93 This new modality will surely be incorporated into future instrumentation in the years to come with the goal to obtain more quantitative measures and accurate 3D images from MSI data sets.
Single Cell Analysis
While superior in determining vast chemical information, MSI is oftentimes considered as lacking in spatial resolution compared to other imaging techniques. Over the past few years, the achievable spatial resolution of MSI, especially for MALDI imaging, has improved greatly.54,55 With the development of new instrumentation (see above), a lateral resolution of 5–10 µm is readily achievable with commercial equipment, specifically MALDI, which is within the reaches of assessing individual mammalian cells.257 While this resolution is technologically impressive, biological heterogeneity is a major concern, and being able to distinguish variation at the single cell level will only make MSI more powerful.258 For example, with the use of high-resolution ionization (LA-ICP) coupled to cytometry by time-of-flight (e.g., CyTOF, mass cytometry), the cellular subpopulation heterogeneity can be easily highlighted.259,260 When reaching these very small pixel sizes, care should be taken that enough material is actually being ionized and thus detected at a significant level. While the development of higher spatial resolution instrumentation, matching optics and mass analyzers must be implemented as well to move single cell MSI forward.
Naturally high spatial resolution instrumentation, such as SIMS, has easily been able to image cells with laser beam focuses of 500 to 30 nm.255,261–263 A facet of SIMS, multi-isotope mass spectrometry (MIMS), has been able to reach 50 nm spatial resolution to visualize human adipose tissue and its age-related plasticity loss.264 Unfortunately, SIMS is unable to ionize most peptides or proteins, although ME-SIMS and the combination of SILAC and TOF-SIMS has allowed for imaging of peptides, lipids, and newly synthesized proteins.210,265 In general, though, developments for other ionization sources are a must for these major biomolecules. Comparatively, MALDI is far away from being able to reach comparable spatial resolution levels to SIMS, but in the past few years, researchers have gotten closer to routinely utilizing 5- to 10-µm laser spot sizes (example shown in Figure 9).177,207,266 Other ionization techniques that can provide subcellular resolution include LA-ICPMS,267,268 single-probe MS,269 and laser desorption/ ionization droplet delivery (LDI-DD).270 Notably, LDI-DD is able to reach 2.4-µm spatial resolution for an ink printed pattern and 3-µm for mouse brain,270 boasting the importance of developing new ionization techniques for bioanalysis.
Figure 9.
Demonstration of 5-µm, subcellular resolution MALDI-MS images for several lipid species (rows) overlaid on their optical image across four different genotypes of maize leaves (columns) at the midpoint and distal regions. The scale bar for the images is 50 µm for all images, and it is noted that the Mo17 and Mo17 × B73 have slightly larger scale bars. Reproduced from High spatial resolution mass spectrometry imaging reveals the genetically programmed, developmental modification of the distribution of thylakoid membrane lipids among individual cells of maize leaf, Duenas, M. E.; Klein, A. T.; Alexander, L. E.; Yandeau-Nelson, M. D.; Nikolau, B. J.; Lee, Y. J., Plant J., Vol. 89, Issue 4 (ref 224) Copyright 2017 Wiley.
CONCLUSION
The field of MSI has expanded drastically in recent years as its utility has been recognized for a wide variety of applications. Because of its ability to analyze thousands of biomolecules without any form of labeling, MSI is being increasingly used as an analytical technique both as a complement and replacement to other imaging methods. However, with its rapid expansion, there is an urgent need for improvements in reproducibility at the sample preparation level and for extra care to be taken in ensuring correct interpretation of results. Recent literature has begun to address this, but we are still far from a uniform standard for reproducibility between users, laboratories, and biological samples. The literature also reflects a push toward improved spatial resolution, and this will likely continue to be an area of focus for the foreseeable future. With spatial resolution already approaching submicrometer levels and 3D MSI becoming more commonplace, the number of spectra acquired per experiment has increased exponentially and will only get larger as spatial resolution continues to be improved. As a result, there is a growing need for more sophisticated data analysis tools that can handle the large amounts of data being produced, both in extracting meaningful information and storing data for community access. The incorporation of open-source software has done a remarkable job addressing this challenge, but future efforts need to be made in ensuring ease-of-use for the end user. Ideally, software will be designed in ways that make it possible for various tools from different sources to be incorporated into a single data analysis pipeline, allowing the researcher to customize software for individual imaging experiments. By improving upon sample preparation protocols, instrumental throughput and resolution capabilities, and streamlined data analysis and quantitation, it is anticipated that MSI will become routinely utilized in clinical settings.
Acknowledgments
The authors gratefully acknowledge support from the National Institutes of Health (NIH) through Grants R56MH110215 and R01 DK071801. A.R.B. acknowledges the National Institute of General Medical Sciences F31 Fellowship (Grant 1F31GM119365) for funding. K.D. acknowledges a predoctoral fellowship supported by the NIH, under Ruth L. Kirschstein National Research Service Award T32 HL 007936 from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center. L.L. and J.J. would like to thank the University of Wisconsin Carbone Cancer Center (UWCCC) Pancreas Cancer Task Force for funding support. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Janis Apinis Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Biographies
Amanda Rae Buchberger is a chemistry Ph.D. candidate and NRSA Graduate Research Fellow in the research group of Professor Lingjun Li at the University of Wisconsin-Madison. She received her B.S. in Chemistry from the University of Wisconsin-Eau Claire in 2013. She is currently obtaining her Ph.D. in Analytical Chemistry at University of Wisconsin-Madison. Her research focuses on the development of quantitative approaches for the analysis of crustacean neuropeptides and their changes due to environmental stress using mass spectrometry.
Kellen DeLaney received her B.S. in Chemistry from The Pennsylvania State University in 2015 and is currently working toward a Ph.D. in Analytical Chemistry in the research group of Professor Lingjun Li at the University of Wisconsin-Madison. Her research involves the development and application of a multifaceted mass spectrometry platform to study neuropeptide modulation in the crustacean nervous system.
Jillian Johnson received her B.S. in Biology from Rensselaer Polytechnic Institute in 2013. She is currently working toward her Ph.D. in Pharmaceutical Sciences in the research group of Professor Lingjun Li at the University of Wisconsin-Madison. Her research focuses on using mass spectrometry to understand cancer cell-tumor microenvironment interactions in pancreatic cancer.
Lingjun Li is a Vilas Distinguished Achievement Professor and the Janis Apinis Professor of Pharmaceutical Sciences and Chemistry at the University of Wisconsin-Madison. Dr. Li received her B.E. degree in Environmental Analytical Chemistry from Beijing University of Technology, China, and her Ph.D. degree in Analytical Chemistry/ Biomolecular Chemistry from the University of Illinois at Urbana-Champaign under Dr. Jonathan Sweedler in 2000. She then did joint postdoctoral research at the Pacific Northwest National Laboratory (with Dr. Richard Smith) and Brandeis University (with Dr. Eve Marder) before joining the faculty at UW-Madison in December 2002. Dr. Li’s research interests include the development of novel mass spectrometry (MS)-based tools to advance biological discovery with special focus on neuropeptides in model organisms and protein biomarkers in neurodegenerative diseases. Dr. Li received numerous awards, including the ASMS Research Award, NSF CAREER Award, Sloan Fellowship, PittCon Achievement Award, and ASMS Biemann Medal and was named one of the Top 50 most influential women in the analytical sciences and was included in the 2016 Analytical Scientist Power List. Dr. Li currently serves as an Associate Editor for the Journal of The American Society for Mass Spectrometry
Footnotes
The authors declare no competing financial interest.
References
- 1.Miyamoto S, Hsu C-C, Hamm G, Darshi M, Diamond-Stanic M, Decleves A-E, Slater L, Pennathur S, Stauber J, Dorrestein PC, Sharma K. EBioMedicine. 2016;7:121–134. doi: 10.1016/j.ebiom.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman HM, Lundin E, Andersson M, Lanekoff I. Analyst. 2016;141(12):3686–95. doi: 10.1039/c5an02620b. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Buchberger A, Muthuvel G, Li L. Proteomics. 2015;15:3969. doi: 10.1002/pmic.201500256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y, Li B, Aharoni A. Trends Plant Sci. 2016;21(8):686–698. doi: 10.1016/j.tplants.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 5.St John ER, Rossi M, Pruski P, Darzi A, Takats Z. TrAC, Trends Anal. Chem. 2016;85:2–9. [Google Scholar]
- 6.Huang KT, Ludy S, Calligaris D, Dunn IF, Laws E, Santagata S, Agar NY. Adv. Cancer Res. 2017;134:257–282. doi: 10.1016/bs.acr.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Kooijman PC, Kok SJ, Weusten JJ, Honing M. Anal. Chim. Acta. 2016;919:1–10. doi: 10.1016/j.aca.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 8.Carter CL, Jones JW, Farese AM, MacVittie TJ, Kane MA. Anal. Chem. 2016;88(9):4788–4794. doi: 10.1021/acs.analchem.6b00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gessel M, Spraggins JM, Voziyan P, Hudson BG, Caprioli RM. J. Mass Spectrom. 2015;50(11):1288–1293. doi: 10.1002/jms.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill EL, Yost RA, Vedam-Mai V, Garrett TJ. Anal. Chem. 2017;89(1):576–580. doi: 10.1021/acs.analchem.6b04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke MB, Raymond BBA, Djordjevic SP, Padula MP. Rapid Commun. Mass Spectrom. 2015;29(7):637–644. doi: 10.1002/rcm.7138. [DOI] [PubMed] [Google Scholar]
- 12.Klein AT, Yagnik GB, Hohenstein JD, Ji Z, Zi J, Reichert MD, MacIntosh GC, Yang B, Peters RJ, Vela J, Lee YJ. Anal. Chem. 2015;87(10):5294–301. doi: 10.1021/acs.analchem.5b00459. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Bai H, Cai Z, Gao D, Jiang Y, Liu J, Liu H. Electrophoresis. 2016;37(13):1956–66. doi: 10.1002/elps.201600027. [DOI] [PubMed] [Google Scholar]
- 14.Holzlechner M, Reitschmidt S, Gruber S, Zeilinger S, Marchetti-Deschmann M. Proteomics. 2016;16(11–12):1742–6. doi: 10.1002/pmic.201500510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil SM, Rompp A, Pretzel J, Becker K, Spengler B. Anal. Chem. 2015;87(22):11309–16. doi: 10.1021/acs.analchem.5b02781. [DOI] [PubMed] [Google Scholar]
- 16.Bai H, Wang S, Liu J, Gao D, Jiang Y, Liu H, Cai Z. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016;1026:263–271. doi: 10.1016/j.jchromb.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Lietz CB, Li L. J. Am. Soc. Mass Spectrom. 2014;25(12):2177–80. doi: 10.1007/s13361-014-0986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekelof M, McMurtrie EK, Nazari M, Johanningsmeier SD, Muddiman DC. J. Am. Soc. Mass Spectrom. 2017;28(2):370–375. doi: 10.1007/s13361-016-1541-7. [DOI] [PubMed] [Google Scholar]
- 19.Nazari M, Muddiman DC. Analyst. 2016;141(2):595–605. doi: 10.1039/c5an01513h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan NTN, Mohammadi AS, Pour MD, Ewing AG. Anal. Chem. 2016;88(3):1734–1741. doi: 10.1021/acs.analchem.5b03942. [DOI] [PubMed] [Google Scholar]
- 21.Alberici RM, Vendramini PH, Eberlin MN. Anal. Methods. 2017;9(34):5029–5036. [Google Scholar]
- 22.Vendramini PH, Gattaz WF, Schmitt A, Falkai P, Eberlin MN, Martins-de-Souza D. Schizophr Res. 2016;177(1–3):67–69. doi: 10.1016/j.schres.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Angerer TB, Dowlatshahi Pour M, Malmberg P, Fletcher JS. Anal. Chem. 2015;87(8):4305–13. doi: 10.1021/ac504774y. [DOI] [PubMed] [Google Scholar]
- 24.Vanbellingen QP, Fu T, Bich C, Amusant N, Stien D, Della-Negra S, Touboul D, Brunelle A. J. Mass Spectrom. 2016;51(6):412–423. doi: 10.1002/jms.3762. [DOI] [PubMed] [Google Scholar]
- 25.Korsgen M, Pelster A, Dreisewerd K, Arlinghaus HF. J. Am. Soc. Mass Spectrom. 2016;27(2):277–284. doi: 10.1007/s13361-015-1275-y. [DOI] [PubMed] [Google Scholar]
- 26.Shi F, Archer JJ, Levis RJ. Methods. 2016;104:79–85. doi: 10.1016/j.ymeth.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 27.OuYang C, Chen B, Li L. J. Am. Soc. Mass Spectrom. 2015;26(12):1992–2001. doi: 10.1007/s13361-015-1265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shariatgorji M, Nilsson A, Goodwin RJA, Kallback P, Schintu N, Zhang XQ, Crossman AR, Bezard E, Svenningsson P, Andren PE. Neuron. 2014;84(4):697–707. doi: 10.1016/j.neuron.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Beasley E, Francese S, Bassindale T. Anal. Chem. 2016;88(20):10328–10334. doi: 10.1021/acs.analchem.6b03551. [DOI] [PubMed] [Google Scholar]
- 30.Heijs B, Tolner EA, Bovee JV, van den Maagdenberg AM, McDonnell LA. J. Proteome Res. 2015;14(12):5348–54. doi: 10.1021/acs.jproteome.5b00849. [DOI] [PubMed] [Google Scholar]
- 31.Barre FPY, Flinders B, Garcia JP, Jansen I, Huizing LRS, Porta T, Creemers LB, Heeren RMA, CIllero-Pastor B. Anal. Chem. 2016;88(24):12051–12059. doi: 10.1021/acs.analchem.6b02491. [DOI] [PubMed] [Google Scholar]
- 32.Shariatgorji M, Nilsson A, Bonta M, Gan J, Marklund N, Clausen F, Kallback P, Loden H, Limbeck A, Andren PE. Methods. 2016;104:86–92. doi: 10.1016/j.ymeth.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 33.Chumbley CW, Reyzer ML, Allen JL, Marriner GA, Via LE, Barry CE, III, Caprioli RM. Anal. Chem. 2016;88(4):2392–2398. doi: 10.1021/acs.analchem.5b04409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quiason CM, Shahidi-Latham SK. J. Am. Soc. Mass Spectrom. 2015;26(6):967–73. doi: 10.1007/s13361-015-1132-z. [DOI] [PubMed] [Google Scholar]
- 35.Moening TN, Brown VL, He L. Anal. Methods. 2016;8(46):8234–8240. [Google Scholar]
- 36.Jung JW, Lee MS, Choi HJ, Jung S, Lee YJ, Hwang GS, Kwon TH. Am. J. Physiol Renal Physiol. 2016;310(11):F1317–27. doi: 10.1152/ajprenal.00524.2015. [DOI] [PubMed] [Google Scholar]
- 37.Lai YH, Cai YH, Lee H, Ou YM, Hsiao CH, Tsao CW, Chang HT, Wang YS. J. Am. Soc. Mass Spectrom. 2016;27(8):1314–21. doi: 10.1007/s13361-016-1406-0. [DOI] [PubMed] [Google Scholar]
- 38.Skraskova K, Claude E, Jones EA, Towers M, Ellis SR, Heeren RM. Methods. 2016;104:69–78. doi: 10.1016/j.ymeth.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Malys BJ, Owens KG. Rapid Commun. Mass Spectrom. 2017;31(9):804–812. doi: 10.1002/rcm.7848. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Zhang Y, Liu Ja, Han J, Guan M, Yang H, Lin Y, Xiong S, Zhao Z. Sci. Rep. 2016;6:37903. doi: 10.1038/srep37903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Wang Y, Zhou D, Li Z. Anal. Chem. 2015;87(12):5860–5. doi: 10.1021/ac504761t. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q, Comi TJ, Li B, Rubakhin SS, Sweedler JV. Anal. Chem. 2016;88(11):5988–95. doi: 10.1021/acs.analchem.6b01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanek A, Holzer S, Knop K, Schubert US, von Eggeling F. Anal. Bioanal. Chem. 2016;408(14):3769–81. doi: 10.1007/s00216-016-9465-4. [DOI] [PubMed] [Google Scholar]
- 44.Stoeckli M, Staab D. J. Am. Soc. Mass Spectrom. 2015;26(6):911–914. doi: 10.1007/s13361-015-1099-9. [DOI] [PubMed] [Google Scholar]
- 45.Kamanna S, Henry J, Voelcker N, Linacre A, Kirkbride KP. Rapid Commun. Mass Spectrom. 2017;31(22):1927–1937. doi: 10.1002/rcm.7986. [DOI] [PubMed] [Google Scholar]
- 46.Oetjen J, Lachmund D, Palmer A, Alexandrov T, Becker M, Boskamp T, Maass P. Anal. Bioanal. Chem. 2016;408(24):6729–40. doi: 10.1007/s00216-016-9793-4. [DOI] [PubMed] [Google Scholar]
- 47.Heijs B, Holst S, Briaire-de Bruijn IH, van Pelt GW, de Ru AH, van Veelen PA, Drake RR, Mehta AS, Mesker WE, Tollenaar RA, Bovee JVMG, Wuhrer M, McDonnell LA. Anal. Chem. 2016;88(15):7745–7753. doi: 10.1021/acs.analchem.6b01739. [DOI] [PubMed] [Google Scholar]
- 48.Hsu CC, Chou PT, Zare RN. Anal. Chem. 2015;87(22):11171–11175. doi: 10.1021/acs.analchem.5b03389. [DOI] [PubMed] [Google Scholar]
- 49.Panderi I, Yakirevich E, Papagerakis S, Noble L, Lombardo K, Pantazatos D. Rapid Commun. Mass Spectrom. 2017;31(2):160–170. doi: 10.1002/rcm.7776. [DOI] [PubMed] [Google Scholar]
- 50.Pietrowska M, Gawin M, Polanska J, Widlak P. Proteomics. 2016;16(11–12):1670–7. doi: 10.1002/pmic.201500424. [DOI] [PubMed] [Google Scholar]
- 51.Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, Janssen KP, Kuppen PJ, van de Velde CJ, Weirich G, Erlmeier F, Langer R, Aubele M, Zitzelsberger H, McDonnell L, Aichler M, Walch A. Nat. Protoc. 2016;11(8):1428–43. doi: 10.1038/nprot.2016.081. [DOI] [PubMed] [Google Scholar]
- 52.Holst S, Heijs B, Haan N, van Zeijl RJM, Briaire-de Bruijn IH, van Pelt GW, Mehta AS, Angel PM, Mesker WE, Tollenaar TA, Drake RR, Bovee JVMG, McDonnell LA, Wuhrer M. Anal. Chem. 2016;88(11):5904–5913. doi: 10.1021/acs.analchem.6b00819. [DOI] [PubMed] [Google Scholar]
- 53.Cobice DF, Livingstone DEW, Mackay CL, Goodwin RJA, Smith LB, Walker BR, Andrew R. Anal. Chem. 2016;88(21):10362–10367. doi: 10.1021/acs.analchem.6b02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kompauer M, Heiles S, Spengler B. Nat. Methods. 2017;14(1):90–96. doi: 10.1038/nmeth.4071. [DOI] [PubMed] [Google Scholar]
- 55.Feenstra AD, Duenas ME, Lee YJ. J. Am. Soc. Mass Spectrom. 2017;28(3):434–442. doi: 10.1007/s13361-016-1577-8. [DOI] [PubMed] [Google Scholar]
- 56.Cahill JF, Kertesz V, Van Berkel GJ. Anal. Chem. 2015;87(21):11113–11121. doi: 10.1021/acs.analchem.5b03293. [DOI] [PubMed] [Google Scholar]
- 57.Hieta JP, Vaikkinen A, Auno S, Raikkonen H, Haapala M, Scotti G, Kopra J, Piepponen P, Kauppila TJ. J. Am. Soc. Mass Spectrom. 2017;28(6):1060–1065. doi: 10.1007/s13361-016-1578-7. [DOI] [PubMed] [Google Scholar]
- 58.Li XX, Chen LF, Ouyang YZ, Feng F, Chen HW. Chinese J. Anal Chem. 2016;44(1):25–31. [Google Scholar]
- 59.Bajo K, Itose S, Matsuya M, Ishihara M, Uchino K, Kudo M, Sakaguchi I, Yurimoto H. Surf. Interface Anal. 2016;48(11):1190–1193. [Google Scholar]
- 60.O’Rourke MB, Raymond BBA, Padula MP. J. Am. Soc. Mass Spectrom. 2017;28(5):895–900. doi: 10.1007/s13361-017-1632-0. [DOI] [PubMed] [Google Scholar]
- 61.Zubair F, Prentice BM, Norris JL, Laibinis PE, Caprioli RM. Anal. Chem. 2016;88(14):7302–7311. doi: 10.1021/acs.analchem.6b01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fagerer SR, Rompp A, Jefimovs K, Bronnimann R, Hayenga G, Steinhoff RF, Krismer J, Pabst M, Ibanez AJ, Zenobi R. Rapid Commun. Mass Spectrom. 2015;29(11):1019–1024. doi: 10.1002/rcm.7191. [DOI] [PubMed] [Google Scholar]
- 63.Fincher JA, Korte AR, Reschke B, Morris NJ, Powell MJ, Vertes A. Analyst. 2017;142(17):3157–3164. doi: 10.1039/c7an00805h. [DOI] [PubMed] [Google Scholar]
- 64.Chen JX, Hu YJ, Lu QA, Wang PC, Zhan HQ. Analyst. 2017;142(7):1119–1124. doi: 10.1039/c6an02721k. [DOI] [PubMed] [Google Scholar]
- 65.Stopka SA, Rong C, Korte AR, Yadavilli S, Nazarian J, Razunguzwa TT, Morris NJ, Vertes A. Angew. Chem., Int. Ed. 2016;55(14):4482–4486. doi: 10.1002/anie.201511691. [DOI] [PubMed] [Google Scholar]
- 66.Prentice BM, Chumbley CW, Caprioli RM. J. Mass Spectrom. 2015;50(4):703–10. doi: 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellis SR, Cappell J, Potocnik NO, Balluff B, Hamaide J, Van der Linden A, Heeren RMA. Analyst. 2016;141(12):3832–3841. doi: 10.1039/c6an00169f. [DOI] [PubMed] [Google Scholar]
- 68.Fowble KL, Teramoto K, Cody RB, Edwards D, Guarrera D, Musah RA. Anal. Chem. 2017;89(6):3421–3429. doi: 10.1021/acs.analchem.6b04137. [DOI] [PubMed] [Google Scholar]
- 69.Zou J, Talbot F, Tata A, Ermini L, Franjic K, Ventura M, Zheng JZ, Ginsberg H, Post M, Ifa DR, Jaffray D, Miller RJD, Zarrine-Afsar A. Anal. Chem. 2015;87(24):12071–12079. doi: 10.1021/acs.analchem.5b02756. [DOI] [PubMed] [Google Scholar]
- 70.Alexander J, Gildea L, Balog J, Speller A, McKenzie J, Muirhead L, Scott A, Kontovounisios C, Rasheed S, Teare J, Hoare J, Veselkov K, Goldin R, Tekkis P, Darzi A, Nicholson J, Kinross J, Takats Z. Surg Endosc. 2017;31(3):1361–1370. doi: 10.1007/s00464-016-5121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calligaris D, Feldman DR, Norton I, Olubiyi O, Changelian AN, Machaidze R, Vestal ML, Laws ER, Dunn IF, Santagata S, Agar NY. Proc. Natl. Acad. Sci. U. S. A. 2015;112(32):9978–83. doi: 10.1073/pnas.1423101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calligaris D, Feldman DR, Norton I, Brastianos PK, Dunn IF, Santagata S, Agar NYR. Int. J. Mass Spectrom. 2015;377:690–8. doi: 10.1016/j.ijms.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH, Orringer DA, Gill KK, Dunn IF, Dias-Santagata D, Ligon KL, Jolesz FA, Golby AJ, Cooks RG, Agar NY. Proc. Natl. Acad. Sci. U. S. A. 2014;111(30):11121–6. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luxembourg SL, Vaezaddeh AR, Amstalden ER, Zimmermann-Ivol CG, Hochstrasser DF, Heeren RM. Rapid Commun. Mass Spectrom. 2006;20(22):3435–42. doi: 10.1002/rcm.2747. [DOI] [PubMed] [Google Scholar]
- 75.Kiss A, Jungmann JH, Smith DF, Heeren RM. Rev. Sci. Instrum. 2013;84(1):013704. doi: 10.1063/1.4772396. [DOI] [PubMed] [Google Scholar]
- 76.Jungmann JH, MacAleese L, Buijs R, Giskes F, de Snaijer A, Visser J, Visschers J, Vrakking MJ, Heeren RM. J. Am. Soc. Mass Spectrom. 2010;21(12):2023–30. doi: 10.1016/j.jasms.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 77.Vanbellingen QP, Elie N, Eller MJ, Della-Negra S, Touboul D, Brunelle A. Rapid Commun. Mass Spectrom. 2015;29(13):1187–1195. doi: 10.1002/rcm.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shon HK, Yoon S, Moon JH, Lee TG. Biointerphases. 2016;11(2):02A321. doi: 10.1116/1.4941447. [DOI] [PubMed] [Google Scholar]
- 79.Tian H, Maciazek D, Postawa Z, Garrison BJ, Winograd N. J. Am. Soc. Mass Spectrom. 2016;27(9):1476–1482. doi: 10.1007/s13361-016-1423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fisher GL, Bruinen AL, Potocnik NO, Hammond JS, Bryan SR, Larson PE, Heeren RMA. Anal. Chem. 2016;88(12):6433–6440. doi: 10.1021/acs.analchem.6b01022. [DOI] [PubMed] [Google Scholar]
- 81.Fisher GL, Hammond JS, Larson PE, Bryan SR, Heeren RMA. J. Vac. Sci. Technol., B: Nanotechnol. Microelectron.: Mater., Process., Meas., Phenom. 2016;34(3):03H126. [Google Scholar]
- 82.Seki T, Kusakari M, Fujii M, Aoki T, Matsuo J. Nucl. Instrum. Methods Phys. Res., Sect. B. 2016;371:189–193. [Google Scholar]
- 83.Suzuki K, Kusakari M, Fujii M, Seki T, Aoki T, Matsuo J. Surf. Interface Anal. 2016;48(11):1119–1121. [Google Scholar]
- 84.Donnarumma F, Cao F, Murray KK. J. Am. Soc. Mass Spectrom. 2016;27(1):108–116. doi: 10.1007/s13361-015-1249-0. [DOI] [PubMed] [Google Scholar]
- 85.Fischer JL, Lutomski CA, El-Baba TJ, Siriwardena-Mahanama BN, Weidner JF, Allen MJ, Trimpin S. J. Am. Soc. Mass Spectrom. 2015;26(12):2086–2095. doi: 10.1007/s13361-015-1233-8. [DOI] [PubMed] [Google Scholar]
- 86.Xu L, Kliman M, Forsythe JG, Korade Z, Hmelo AB, Porter NA, McLean JA. J. Am. Soc. Mass Spectrom. 2015;26(6):924–33. doi: 10.1007/s13361-015-1131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Griffiths RL, Creese AJ, Race AM, Bunch J, Cooper HJ. Anal. Chem. 2016;88(13):6758–6766. doi: 10.1021/acs.analchem.6b01060. [DOI] [PubMed] [Google Scholar]
- 88.Feider CL, Elizondo N, Eberlin LS. Anal. Chem. 2016;88(23):11533–11541. doi: 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li H, Smith BK, Márk L, Nemes P, Nazarian J, Vertes A. Int. J. Mass Spectrom. 2015;377:681–689. [Google Scholar]
- 90.Lamont L, Baumert M, Ogrinc Potocnik N, Allen M, Vreeken R, Heeren RMA, Porta T. Anal. Chem. 2017;89(20):11143–11150. doi: 10.1021/acs.analchem.7b03512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gemperline E, Horn HA, DeLaney K, Currie CR, Li L. ACS Chem. Biol. 2017;12(8):1980–1985. doi: 10.1021/acschembio.7b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartels B, Kulkarni P, Danz N, Bocker S, Saluz HP, Svatos A. RSC Adv. 2017;7(15):9045–9050. [Google Scholar]
- 93.Nguyen SN, Liyu AV, Chu RK, Anderton CR, Laskin J. Anal. Chem. 2017;89(2):1131–1137. doi: 10.1021/acs.analchem.6b03293. [DOI] [PubMed] [Google Scholar]
- 94.He M, Meng Y, Yan S, Hang W, Zhou W, Huang B. Anal. Chem. 2017;89(1):565–570. doi: 10.1021/acs.analchem.6b03540. [DOI] [PubMed] [Google Scholar]
- 95.Kuznetsov I, Filevich J, Dong F, Woolston M, Chao W, Anderson EH, Bernstein ER, Crick DC, Rocca JJ, Menoni CS. Nat. Commun. 2015;6:6944. doi: 10.1038/ncomms7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hansen HT, Janfelt C. Anal. Chem. 2016;88(23):11513–11520. doi: 10.1021/acs.analchem.6b02711. [DOI] [PubMed] [Google Scholar]
- 97.Kuchar L, Faltyskova H, Krasny L, Dobrovolny R, Hulkova H, Ledvinova J, Volny M, Strohalm M, Lemr K, Kryspinova L, Asfaw B, Rybova J, Desnick RJ, Havlicek V. Anal. Bioanal. Chem. 2015;407(8):2283–91. doi: 10.1007/s00216-014-8402-7. [DOI] [PubMed] [Google Scholar]
- 98.Théron L, Centeno D, Coudy-Gandilhon C, Pujos-Guillot E, Astruc T, Rémond D, Barthelemy JC, Roche F, Feasson L, Hébraud M, Béchet D, Chambon C. Proteomes. 2016;4(4):32. doi: 10.3390/proteomes4040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Renslow RS, Lindemann SR, Cole JK, Zhu Z, Anderton CR. Biointerphases. 2016;11(2):02A322. doi: 10.1116/1.4941764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang H, DeGnore JP, Kelly BD, True J, Garsha K, Bieniarz CJ. Mass Spectrom. 2015;50(9):1088–1095. doi: 10.1002/jms.3623. [DOI] [PubMed] [Google Scholar]
- 101.Greer T, Lietz CB, Xiang F, Li LJ. J. Am. Soc. Mass Spectrom. 2015;26(1):107–119. doi: 10.1007/s13361-014-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robichaud G, Garrard KP, Barry JA, Muddiman DC. J. Am. Soc. Mass Spectrom. 2013;24(5):718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kallback P, Nilsson A, Shariatgorji M, Andren PE. Anal. Chem. 2016;88(8):4346–4353. doi: 10.1021/acs.analchem.5b04603. [DOI] [PubMed] [Google Scholar]
- 104.Sforna MC, Lugli F. J. Anal. At. Spectrom. 2017;32(5):1035–1043. [Google Scholar]
- 105.Lopez-Fernandez H, Pessoa GD, Arruda MAZ, Capelo-Martinez JL, Fdez-Riverola F, Glez-Pena D, Reboiro-Jato M. J. Cheminf. 2016;8:10. doi: 10.1186/s13321-016-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uerlings R, Matusch A, Weiskirchen R. Int. J. Mass Spectrom. 2016;395:27–35. [Google Scholar]
- 107.Gormanns P, Reckow S, Poczatek JC, Turck CW, Lechene C. PLoS One. 2012;7(2):e30576. doi: 10.1371/journal.pone.0030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patterson NH, Doonan RJ, Daskalopoulou SS, Dufresne M, Lenglet S, Montecucco F, Thomas A, Chaurand P. Proteomics. 2016;16(11–12):1642–1651. doi: 10.1002/pmic.201500490. [DOI] [PubMed] [Google Scholar]
- 109.Hayakawa E, Fujimura Y, Miura D. Bioinformatics. 2016;32(24):3852–3854. doi: 10.1093/bioinformatics/btw548. [DOI] [PubMed] [Google Scholar]
- 110.O’Rourke MB, Padula MP. Proteomics: Clin. Appl. 2017;11(3–4):1600098. doi: 10.1002/prca.201600098. [DOI] [PubMed] [Google Scholar]
- 111.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM. Int. J. Mass Spectrom. 2007;260(2–3):212–21. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deininger SO, Cornett DS, Paape R, Becker M, Pineau C, Rauser S, Walch A, Wolski E. Anal. Bioanal. Chem. 2011;401(1):167–81. doi: 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, Holmes E. Anal. Chem. 2012;84(3):1310–9. doi: 10.1021/ac201767g. [DOI] [PubMed] [Google Scholar]
- 114.Gemperline E, Rawson S, Li LJ. Anal. Chem. 2014;86(20):10030–10035. doi: 10.1021/ac5028534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba AD, Muddiman DC. Anal. Bioanal. Chem. 2015;407(8):2073–84. doi: 10.1007/s00216-014-8220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J. J. Proteomics. 2012;75(16):4952–61. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 117.Mourino-Alvarez L, Iloro I, de la Cuesta F, Azkargorta M, Sastre-Oliva T, Escobes I, Lopez-Almodovar LF, Sanchez PL, Urreta H, Fernandez-Aviles F, Pinto A, Padial LR, Akerstrom F, Elortza F, Barderas MG. Sci. Rep. 2016;6:27106. doi: 10.1038/srep27106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. J. Proteomics. 2012;75(16):4962–89. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 119.Bedia C, Tauler R, Jaumot J. J. Chemom. 2016;30(10):575–588. [Google Scholar]
- 120.Thomas SA, Race AM, Steven RT, Gilmore IS, Bunch J. 2016 IEEE Symposium Series on Computational Intelligence (SSCI) Athens: Greece; 2016. Dimensionality reduction of mass spectrometry imaging data using autoencoders; p. 7. [DOI] [Google Scholar]
- 121.Bro R, Smilde A. Anal. Methods. 2014;6:2812–2831. [Google Scholar]
- 122.Jones EA, van Remoortere A, van Zeijl RJ, Hogendoorn PC, Bovee JV, Deelder AM, McDonnell LA. PLoS One. 2011;6(9):e24913. doi: 10.1371/journal.pone.0024913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken CJ. Proteome Res. 2008;7(12):5230–6. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 124.Sarkari S, Kaddi CD, Bennett RV, Fernandez FM, Wang MD. Conf Proc. IEEE Eng. Med. Biol. Soc. 2014;2014:4771–4. doi: 10.1109/EMBC.2014.6944691. [DOI] [PubMed] [Google Scholar]
- 125.Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, Strehlow J, Aichler M, Kobarg JH, Oetjen J, Dyatlov A, Heldmann S, Walch A, Thiele H, Maass P, Alexandrov T. Anal. Chem. 2012;84(14):6079–87. doi: 10.1021/ac300673y. [DOI] [PubMed] [Google Scholar]
- 126.Jirasko R, Holcapek M, Khalikova M, Vrana D, Student V, Prouzova Z, Melichar B. J. Am. Soc. Mass Spectrom. 2017;28(8):1562–1574. doi: 10.1007/s13361-017-1644-9. [DOI] [PubMed] [Google Scholar]
- 127.Ilin Y, Kraft ML. Curr. Opin. Biotechnol. 2015;31:108–16. doi: 10.1016/j.copbio.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mirnezami R, Spagou K, Vorkas PA, Lewis MR, Kinross J, Want E, Shion H, Goldin RD, Darzi A, Takats Z, Holmes E, Cloarec O, Nicholson JK. Mol. Oncol. 2014;8(1):39–49. doi: 10.1016/j.molonc.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Minhas FUA, Asif A, Arif M. Comput. Biol. Med. 2016;79:68–79. doi: 10.1016/j.compbiomed.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 130.Marczyk M, Drazek G, Pietrowska M, Widlak P, Polanska J, Polanski A. Procedia Comput. Sci. 2015;51(2015):693–702. [Google Scholar]
- 131.Carreira RJ, Shyti R, Balluff B, Abdelmoula WM, van Heiningen SH, van Zeijl RJ, Dijkstra J, Ferrari MD, Tolner EA, McDonnell LA, van den Maagdenberg AM. J. Am. Soc. Mass Spectrom. 2015;26(6):853–61. doi: 10.1007/s13361-015-1136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weaver EM, Hummon AB, Keithley RB. Anal. Methods. 2015;7(17):7208–19. doi: 10.1039/C5AY00293A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thomas A, Patterson NH, Marcinkiewicz MM, Lazaris A, Metrakos P, Chaurand P. Anal. Chem. 2013;85(5):2860–6. doi: 10.1021/ac3034294. [DOI] [PubMed] [Google Scholar]
- 134.Grund B, Sabin C. Curr. Opin. HIV AIDS. 2010;5(6):473–9. doi: 10.1097/COH.0b013e32833ed742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Metz CE. Radiol Phys. Technol. 2008;1(1):2–12. doi: 10.1007/s12194-007-0002-1. [DOI] [PubMed] [Google Scholar]
- 136.Scheipers U, Perrey C, Siebers S, Hansen C, Ermert H. Ultrason Imaging. 2005;27(3):181–98. doi: 10.1177/016173460502700304. [DOI] [PubMed] [Google Scholar]
- 137.Gemoll T, Strohkamp S, Schillo K, Thorns C, Habermann JK. Oncotarget. 2015;6(41):43869–80. doi: 10.18632/oncotarget.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shultz EK. Clin. Chem. 1995;41(8Pt 2):1248–1255. [PubMed] [Google Scholar]
- 139.Wang MC, Li S. Lifetime Data Anal. 2013;19(2):257–77. doi: 10.1007/s10985-012-9233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McDonnell LA, van Remoortere A, van Zeijl RJ, Deelder AM. J. Proteome Res. 2008;7(8):3619–27. doi: 10.1021/pr800214d. [DOI] [PubMed] [Google Scholar]
- 141.Alexandrov T. BMC Bioinf. 2012;13(Suppl16):S11. doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Galli M, Zoppis I, Smith A, Magni F, Mauri G. Expert Rev. Proteomics. 2016;13(7):685–96. doi: 10.1080/14789450.2016.1200470. [DOI] [PubMed] [Google Scholar]
- 143.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Comput. Struct. Biotechnol. J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu J, Ji Y, Zhao L, Ji M, Ye Z, Li S. Comput. Math Methods Med. 2016;2016:6169249. doi: 10.1155/2016/6169249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Race AM, Palmer AD, Dexter A, Steven RT, Styles IB, Bunch J. Anal. Chem. 2016;88(19):9451–9458. doi: 10.1021/acs.analchem.6b01643. [DOI] [PubMed] [Google Scholar]
- 146.Wijetunge CD, Saeed I, Boughton BA, Spraggins JM, Caprioli RM, Bacic A, Roessner U, Halgamuge SK. Bioinformatics. 2015;31(19):3198–3206. doi: 10.1093/bioinformatics/btv356. [DOI] [PubMed] [Google Scholar]
- 147.Bond NJ, Koulman A, Griffin JL, Hall Z. Metabolomics. 2017;13(11):128. doi: 10.1007/s11306-017-1252-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma’ayan A. Sci. Data. 2017;4:170151. doi: 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rompp A, Wang R, Albar JP, Urbani A, Hermjakob H, Spengler B, Vizcaino JA. Anal. Bioanal. Chem. 2015;407(8):2027–33. doi: 10.1007/s00216-014-8357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alexandrov T. [accessed September 17, 2017];Metaspace. http://metaspace2020.eu.
- 151.Cherry SR. Semin. Nucl. Med. 2009;39(5):348–53. doi: 10.1053/j.semnuclmed.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ho YN, Shu LJ, Yang YL. Wiley Interdiscip Rev. Syst. Biol. Med. 2017;9(5):e1387. doi: 10.1002/wsbm.1387. [DOI] [PubMed] [Google Scholar]
- 153.Vogler N, Heuke S, Bocklitz TW, Schmitt M, Popp J. Annu. Rev. Anal. Chem. 2015;8:359–87. doi: 10.1146/annurev-anchem-071114-040352. [DOI] [PubMed] [Google Scholar]
- 154.Marti-Bonmati L, Sopena R, Bartumeus P, Sopena P. Contrast Media Mol. Imaging. 2010;5(4):180–9. doi: 10.1002/cmmi.393. [DOI] [PubMed] [Google Scholar]
- 155.Meyer C, Ma B, Kunju LP, Davenport M, Piert M. Eur. J. Nucl. Med. Mol. Imaging. 2013;40(S1):72–78. doi: 10.1007/s00259-013-2382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Eliceiri KW, Berthold MR, Goldberg IG, Ibanez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, Stuurman N, Swedlow JR, Tomancak P, Carpenter AE. Nat. Methods. 2012;9(7):697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ahlf DR, Masyuko RN, Hummon AB, Bohn PW. Analyst. 2014;139(18):4578–4585. doi: 10.1039/c4an00826j. [DOI] [PubMed] [Google Scholar]
- 158.Huhdanpaa H, Hwang DH, Gasparian GG, Booker MT, Cen Y, Lerner A, Boyko OB, Go JL, Kim PE, Rajamohan A, Law M, Shiroishi MS. J. Digit Imaging. 2014;27(3):369–79. doi: 10.1007/s10278-013-9655-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wollmann T, Erfle H, Eils R, Rohr K, Gunkel M. J. Biotechnol. 2017;261:70. doi: 10.1016/j.jbiotec.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 160.Afgan E, Baker D, Coraor N, Chapman B, Nekrutenko A, Taylor J. BMC Bioinf. 2010;11(Suppl 12):S4. doi: 10.1186/1471-2105-11-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Berthold MR, Cebron N, Dill F, Gabriel TR, Kotter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B. KNIME: The Konstanz Information Miner. In: Preisach C, Burkhardt H, Schmidt-Thieme L, Decker R, editors. Data Analysis, Machine Learning, and Applications. Springer; Berlin, Heidelberg, Germany: 2008. [Google Scholar]
- 162.Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, Berg-Lyon D, Ackermann G, Moeller Christensen GJ, Nakatsuji T, Zhang L, Borkowski AW, Meehan MJ, Dorrestein K, Gallo RL, Bandeira N, Knight R, Alexandrov T, Dorrestein PC. Proc. Natl. Acad. Sci. U. S. A. 2015;112(17):E2120–9. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vollnhals F, Audinot JN, Wirtz T, Mercier-Bonin M, Fourquaux I, Schroeppel B, Kraushaar U, Lev-Ram V, Ellisman MH, Eswara S. Anal. Chem. 2017;89(20):10702–10710. doi: 10.1021/acs.analchem.7b01256. [DOI] [PubMed] [Google Scholar]
- 164.Aoyagi S, Abe K, Yamagishi T, Iwai H, Yamaguchi S, Sunohara T. Anal. Bioanal. Chem. 2017;409(27):6387–6396. doi: 10.1007/s00216-017-0578-1. [DOI] [PubMed] [Google Scholar]
- 165.Balbekova A, Bonta M, Török S, J O, Döme; Limbeck BA, Lendl B. Anal. Methods. 2017;9(37):5464–5471. [Google Scholar]
- 166.Marty F, Rago G, Smith DF, Gao X, Eijkel GB, MacAleese L, Bonn M, Brunner E, Basler K, Heeren RMA. Anal. Chem. 2017;89(18):9664–9670. doi: 10.1021/acs.analchem.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Silva LMA, Alves Filho EG, Simpson AJ, Monteiro MR, Cabral E, Ifa D, Venancio T. Talanta. 2017;173:22–27. doi: 10.1016/j.talanta.2017.05.060. [DOI] [PubMed] [Google Scholar]
- 168.Van de Plas R, Yang J, Spraggins J, Caprioli RM. Nat. Methods. 2015;12(4):366–72. doi: 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taverna D, Pollins AC, Sindona G, Caprioli RM, Nanney LB. J. Proteome Res. 2015;14(2):986–96. doi: 10.1021/pr5010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rauser S, Marquardt C, Balluff B, Deininger SO, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, Schmitt M, Aubele M, Hofler H, Walch A. J. Proteome Res. 2010;9(4):1854–63. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 171.Li B, Bhandari DR, Rompp A, Spengler B. Sci. Rep. 2016;6:36074. doi: 10.1038/srep36074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eberlin LS, Tibshirani RJ, Zhang J, Longacre TA, Berry GJ, Bingham DB, Norton JA, Zare RN, Poultsides GA. Proc. Natl. Acad. Sci. U. S. A. 2014;111(7):2436–41. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fischer AH, Jacobson KA, Rose J, Zeller R. Cold Spring Harbor Protocols. 2008 2008, pdb.prot4991. [Google Scholar]
- 174.Lewin K. J. Anat. 1969;105(Pt 1):171–176. [PMC free article] [PubMed] [Google Scholar]
- 175.Sans M, Gharpure K, Tibshirani R, Zhang J, Liang L, Liu J, Young JH, Dood RL, Sood AK, Eberlin LS. Cancer Res. 2017;77(11):2903–2913. doi: 10.1158/0008-5472.CAN-16-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hinsch A, Buchholz M, Odinga S, Borkowski C, Koop C, Izbicki JR, Wurlitzer M, Krech T, Wilczak W, Steurer S, Jacobsen F, Burandt EC, Stahl P, Simon R, Sauter G, Schluter H. J. Mass Spectrom. 2017;52(3):165–173. doi: 10.1002/jms.3916. [DOI] [PubMed] [Google Scholar]
- 177.Deutskens F, Yang J, Caprioli RM. J. Mass Spectrom. 2011;46(6):568–71. doi: 10.1002/jms.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Anal. Chem. 2004;76(4):1145–55. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 179.Wang CW, Ka SM, Chen A. Sci. Rep. 2015;4:6050. doi: 10.1038/srep06050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Heijs B, Abdelmoula WM, Lou S, Briaire-de Bruijn IH, Dijkstra J, Bovee JV, McDonnell LA. Anal. Chem. 2015;87(24):11978–83. doi: 10.1021/acs.analchem.5b03610. [DOI] [PubMed] [Google Scholar]
- 181.Levenson RM, Borowsky AD, Angelo M. Lab. Invest. 2015;95(4):397–405. doi: 10.1038/labinvest.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mascini NE, Cheng M, Jiang L, Rizwan A, Podmore H, Bhandari DR, Rompp A, Glunde K, Heeren RMA. Anal. Chem. 2016;88(6):3107–3114. doi: 10.1021/acs.analchem.5b04032. [DOI] [PubMed] [Google Scholar]
- 183.Becker L, Bellow S, Carre V, Latouche G, Poutaraud A, Merdinoglu D, Brown SC, Cerovic ZG, Chaimbault P. Anal. Chem. 2017;89(13):7099–7106. doi: 10.1021/acs.analchem.7b01002. [DOI] [PubMed] [Google Scholar]
- 184.Chughtai K, Jiang L, Post H, Winnard PT, Jr, Greenwood TR, Raman V, Bhujwalla ZM, Heeren RM, Glunde K. J. Am. Soc. Mass Spectrom. 2013;24(5):711–7. doi: 10.1007/s13361-012-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu JT, Wu LH, Knight JA. Clin. Chem. 1986;32(2):314–319. [PubMed] [Google Scholar]
- 186.Walsh AJ, Skala MC. Biomed. Opt. Express. 2015;6(2):559–73. doi: 10.1364/BOE.6.000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Provenzano PP, Rueden CT, Trier SM, Yan L, Ponik SM, Inman DR, Keely PJ, Eliceiri KW. J. Biomed. Opt. 2008;13(3):031220. doi: 10.1117/1.2940365. [DOI] [PubMed] [Google Scholar]
- 188.Provenzano PP, Eliceiri KW, Keely PJ. Clin. Exp. Metastasis. 2009;26(4):357–70. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- 189.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 190.Jones AR, Overly CC, Sunkin SM. Nat. Rev. Neurosci. 2009;10(11):821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- 191.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, David Daly B, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Abdelmoula WM, Carreira RJ, Shyti R, Balluff B, van Zeijl RJ, Tolner EA, Lelieveldt BF, van den Maagdenberg AM, McDonnell LA, Dijkstra J. Anal. Chem. 2014;86(8):3947–54. doi: 10.1021/ac500148a. [DOI] [PubMed] [Google Scholar]
- 193.Verbeeck N, Spraggins JM, Murphy MJM, Wang HD, Deutch AY, Caprioli RM, Van de Plas R. Biochim. Biophys. Acta, Proteins Proteomics. 2017;1865(7):967–977. doi: 10.1016/j.bbapap.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 194.Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NY, Cooks RG. Anal. Chem. 2011;83(22):8366–71. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Desbenoit N, Walch A, Spengler B, Brunelle A, Römpp A. Rapid Commun. Mass Spectrom. 2017 doi: 10.1002/rcm.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li Z, Chu LQ, Sweedler JV, Bohn PW. Anal. Chem. 2010;82(7):2608–11. doi: 10.1021/ac100026r. [DOI] [PubMed] [Google Scholar]
- 197.Butler HJ, Ashton L, Bird B, Cinque G, Curtis K, Dorney J, Esmonde-White K, Fullwood NJ, Gardner B, Martin-Hirsch PL, Walsh MJ, McAinsh MR, Stone N, Martin FL. Nat. Protoc. 2016;11(4):664–87. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 198.Lanni EJ, Masyuko RN, Driscoll CM, Dunham SJ, Shrout JD, Bohn PW, Sweedler J. V. Anal. Chem. 2014;86(21):10885–91. doi: 10.1021/ac5030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Srimany A, George C, Naik HR, Pinto DG, Chandrakumar N, Pradeep T. Phytochemistry. 2016;125:35–42. doi: 10.1016/j.phytochem.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 200.Tata A, Zheng J, Ginsberg HJ, Jaffray DA, Ifa DR, Zarrine-Afsar A. Anal. Chem. 2015;87(15):7683–9. doi: 10.1021/acs.analchem.5b01992. [DOI] [PubMed] [Google Scholar]
- 201.Oetjen J, Aichler M, Trede D, Strehlow J, Berger J, Heldmann S, Becker M, Gottschalk M, Kobarg JH, Wirtz S, Schiffler S, Thiele H, Walch A, Maass P, Alexandrov T. J. Proteomics. 2013;90:52–60. doi: 10.1016/j.jprot.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 202.Zhu A, Lee D, Shim H. Semin. Oncol. 2011;38(1):55–69. doi: 10.1053/j.seminoncol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Goodwin RJ, Mackay CL, Nilsson A, Harrison DJ, Farde L, Andren PE, Iverson SL. Anal. Chem. 2011;83(24):9694–701. doi: 10.1021/ac202630t. [DOI] [PubMed] [Google Scholar]
- 204.Ovchinnikova OS, Kjoller K, Hurst GB, Pelletier DA, Van Berkel GJ. Anal. Chem. 2014;86(2):1083–90. doi: 10.1021/ac4026576. [DOI] [PubMed] [Google Scholar]
- 205.Correa DN, Zacca JJ, Rocha WFC, Borges R, de Souza W, Augusti R, Eberlin MN, Vendramini PH. Forensic Sci. Int. 2016;260:22–26. doi: 10.1016/j.forsciint.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 206.Karki V, Singh M. Int. J. Mass Spectrom. 2017;421:51–60. [Google Scholar]
- 207.Brentan Silva D, Aschenbrenner AK, Lopes NP, Spring O. Molecules. 2017;22(5):774. doi: 10.3390/molecules22050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Buchberger A, Yu Q, Li L. Annu. Rev. Anal. Chem. 2015;8(1):485–509. doi: 10.1146/annurev-anchem-071114-040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gemperline E, Keller C, Jayaraman D, Maeda J, Sussman M, Ane J, Li L. Anal. Chem. 2016;15(12):4403–4411. doi: 10.1021/acs.jproteome.6b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ogrinc Potocnik N, Fisher GL, Prop A, Heeren RMA. Anal. Chem. 2017;89(16):8223–8227. doi: 10.1021/acs.analchem.7b02573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kunzke T, Balluff B, Feuchtinger A, Buck A, Langer R, Luber B, Lordick F, Zitzelsberger H, Aichler M, Walch A. Oncotarget. 2017;8(40):68012–25. doi: 10.18632/oncotarget.19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Powers TW, Holst S, Wuhrer M, Mehta AS, Drake RR. Biomolecules. 2015;5(4):2554–72. doi: 10.3390/biom5042554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Everest-Dass AV, Briggs MT, Kaur G, Oehler MK, Hoffmann P, Packer NH. Mol. Cell. Proteomics. 2016;15:3003. doi: 10.1074/mcp.M116.059816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Briggs MT, Kuliwaba JS, Muratovic D, Everest-Dass AV, Packer NH, Findlay DM, Hoffmann P. Proteomics. 2016;16(11–12):1736–41. doi: 10.1002/pmic.201500461. [DOI] [PubMed] [Google Scholar]
- 215.He Z, Chen Q, Chen F, Zhang J, Li H, Lin J. Chem. Sci. 2016;7:5448–5452. doi: 10.1039/c6sc00215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lu Q, Hu Y, Chen J, Jin S. Anal. Chem. 2017;89(16):8238–8243. doi: 10.1021/acs.analchem.7b00140. [DOI] [PubMed] [Google Scholar]
- 217.Tata A, Gribble A, Ventura M, Ganguly M, Bluemke E, Ginsberg H, Jaffray DA, Ifa DR, Vitkin A, Zarrine-Afsar A. Chem. Sci. 2016;7(3):2162–2169. doi: 10.1039/c5sc03782d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Banerjee S, Zare RN, Tibshirani RJ, Kunder CA, Nolley R, Fan R, Brooks JD, Sonn GA. Proc. Natl. Acad. Sci. U. S. A. 2017;114(13):3334–3339. doi: 10.1073/pnas.1700677114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Phan NTN, Munem M, Ewing AG, Fletcher JS. Anal. Bioanal. Chem. 2017;409(16):3923–3932. doi: 10.1007/s00216-017-0336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cai L, Sheng L, Xia M, Li Z, Zhang S, Zhang X, Chen H. J. Am. Soc. Mass Spectrom. 2017;28(3):399–408. doi: 10.1007/s13361-016-1557-z. [DOI] [PubMed] [Google Scholar]
- 221.Tian H, Sparvero LJ, Amoscato AA, Bloom A, Bayir H, Kagan VE, Winograd N. Anal. Chem. 2017;89(8):4611–4619. doi: 10.1021/acs.analchem.7b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Niziol J, Ossolinksi K, Ossolinski T, Ossolinska A, Bonifay V, Sekula J, Dobrowolski Z, Sunner J, Beech I, Ruman T. Anal. Chem. 2016;88(14):7365–7371. doi: 10.1021/acs.analchem.6b01859. [DOI] [PubMed] [Google Scholar]
- 223.Gustafsson OJR, Guinan TM, Rudd D, Kobus H, Benkendorff K, Voelcker NH. Rapid Commun. Mass Spectrom. 2017;31(12):991–1000. doi: 10.1002/rcm.7869. [DOI] [PubMed] [Google Scholar]
- 224.Duenas ME, Klein AT, Alexander LE, Yandeau-Nelson MD, Nikolau BJ, Lee YJ. Plant J. 2017;89(4):825–838. doi: 10.1111/tpj.13422. [DOI] [PubMed] [Google Scholar]
- 225.Pluhacek T, Petrik M, Luptakova D, Benada O, Palyzova A, Lemr K, Havlicek V. Proteomics. 2016;16(11–12):1785–92. doi: 10.1002/pmic.201500487. [DOI] [PubMed] [Google Scholar]
- 226.Susnea I, Weiskirchen R. Mass Spectrom. Rev. 2016;35(6):666–686. doi: 10.1002/mas.21454. [DOI] [PubMed] [Google Scholar]
- 227.Niedzwiecki MM, Austin C, Remark R, Merad M, Gnjatic S, Estrada-Gutierrez G, Espejel-Nunez A, Borboa-Olivares H, Guzman-Huerta M, Wright RJ, Wright RO, Arora M. Metallomics. 2016;8(4):444–52. doi: 10.1039/c6mt00010j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bishop DP, Clases D, Fryer F, Williams E, Wilkins S, Hare DJ, Cole N, Karst U, Doble PA. J. Anal. At. Spectrom. 2016;31:197–202. [Google Scholar]
- 229.Bussweiler Y, Borovinskaya O, Tanner M. Spectroscopy. 2017;32(5):14–20. [Google Scholar]
- 230.Bonta M, Hegedus B, Limbeck A. Anal. Chim. Acta. 2016;908:54–62. doi: 10.1016/j.aca.2015.12.048. [DOI] [PubMed] [Google Scholar]
- 231.Penen F, Malherbe J, Isaure MP, Dobritzsch D, Bertalan I, Gontier E, Le Coustumer P, Schaumloffel D. J. Trace Elem. Med. Biol. 2016;37:62–68. doi: 10.1016/j.jtemb.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 232.Lee RFS, Theiner S, Meibom A, Koellensperger G, Keppler BK, Dyson PJ. Metallomics. 2017;9(4):365–381. doi: 10.1039/c6mt00231e. [DOI] [PubMed] [Google Scholar]
- 233.Feenstra AD, Hansen RL, Lee YJ. Analyst. 2015;140(21):7293–304. doi: 10.1039/c5an01079a. [DOI] [PubMed] [Google Scholar]
- 234.Annangudi SP, Myung K, Avila Adame C, Gilbert JR. Environ. Sci. Technol. 2015;49(9):5579–83. doi: 10.1021/es506334y. [DOI] [PubMed] [Google Scholar]
- 235.Araujo FD, Araujo WL, Eberlin MN. J. Am. Soc. Mass Spectrom. 2017;28(5):901–907. doi: 10.1007/s13361-017-1610-6. [DOI] [PubMed] [Google Scholar]
- 236.Crecelius AC, Michalzik B, Potthast K, Meyer S, Schubert US. Anal. Bioanal. Chem. 2017;409(15):3807–3820. doi: 10.1007/s00216-017-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bagatela BS, Lopes AP, Cabral EC, Perazzo FF, Ifa DR. Rapid Commun. Mass Spectrom. 2015;29(16):1530–4. doi: 10.1002/rcm.7246. [DOI] [PubMed] [Google Scholar]
- 238.Shariatgorji M, Nilsson A, Goodwin RJ, Svenningsson P, Schintu N, Banka Z, Kladni L, Hasko T, Szabo A, Andren PE. Anal. Chem. 2012;84(16):7152–7. doi: 10.1021/ac301498m. [DOI] [PubMed] [Google Scholar]
- 239.de Macedo CS, Anderson DM, Schey KL. Talanta. 2017;174:325–335. doi: 10.1016/j.talanta.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 240.Flinders B, Beasley E, Verlaan RM, Cuypers E, Francese S, Bassindale T, Clench MR. J. Am. Soc. Mass Spectrom. 2017;28(11):2462–2468. doi: 10.1007/s13361-017-1766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bécue A. Anal. Methods. 2016;8(45):7983–8003. [Google Scholar]
- 242.Takahashi K, Kozuka T, Anegawa A, Nagatani A, Mimura T. Plant Cell Physiol. 2015;56(7):1329–38. doi: 10.1093/pcp/pcv083. [DOI] [PubMed] [Google Scholar]
- 243.Sekula J, Niziol J, Misiorek M, Dec P, Wrona A, Arendowski A, Ruman T. Anal. Chim. Acta. 2015;895:45–53. doi: 10.1016/j.aca.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 244.Yoshimura Y, Goto-Inoue N, Moriyama T, Zaima N. Food Chem. 2016;210:200–211. doi: 10.1016/j.foodchem.2016.04.096. [DOI] [PubMed] [Google Scholar]
- 245.Li B, Dunham SJB, Dong Y, Yoon S, Zeng M, Sweedler J. Trends Food Sci. Technol. 2016;47:50–63. [Google Scholar]
- 246.Stasulli NM, Shank EA. FEMS Microbiol Rev. 2016;40(6):807–813. doi: 10.1093/femsre/fuw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hoffmann T, Dorrestein PC. J. Am. Soc. Mass Spectrom. 2015;26(11):1959–62. doi: 10.1007/s13361-015-1241-8. [DOI] [PubMed] [Google Scholar]
- 248.Taverna D, Norris JL, Caprioli RM. Anal. Chem. 2015;87(1):670–676. doi: 10.1021/ac503479a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Seeley EH, Wilson KJ, Yankeelov TE, Johnson RW, Gore JC, Caprioli RM, Matrisian LM, Sterling JA. Bone. 2014;61:208–16. doi: 10.1016/j.bone.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Inglese P, McKenzie JS, Mroz A, Kinross J, Veselkov K, Holmes E, Takats Z, Nicholson JK, Glen RC. Chem. Sci. 2017;8(5):3500–3511. doi: 10.1039/c6sc03738k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Liu X, Hummon AB. Anal. Chem. 2015;87(19):9508–19. doi: 10.1021/acs.analchem.5b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Morosi L, Giordano S, Falcetta F, Frapolli R, Licandro SA, Matteo C, Zucchetti M, Ubezio P, Erba E, Visentin S, D’Incalci M, Davoli E. Clin. Pharmacol. Ther. 2017;102(5):748–751. doi: 10.1002/cpt.786. [DOI] [PubMed] [Google Scholar]
- 253.Palmer AD, Alexandrov T. Anal. Chem. 2015;87(8):4055–62. doi: 10.1021/ac504604g. [DOI] [PubMed] [Google Scholar]
- 254.Chirinos JR, Oropeza DD, Gonzalez JJ, Hou H, Morey M, Zorba V, Russo RE. J. Anal. At. Spectrom. 2014;29:1292–1298. [Google Scholar]
- 255.Vanbellingen QP, Castellanos A, Rodriguez-Silva M, Paudel I, Chambers JW, Fernandez-Lima FA. J. Am. Soc. Mass Spectrom. 2016;27(12):2033–2040. doi: 10.1007/s13361-016-1485-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Thiele H, Heldmann S, Trede D, Strehlow J, Wirtz S, Dreher W, Berger J, Oetjen J, Kobarg JH, Fischer B, Maass P. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844(1):117–137. doi: 10.1016/j.bbapap.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 257.Ogrinc Potocnik N, Porta T, Becker M, Heeren RM, Ellis SR. Rapid Commun. Mass Spectrom. 2015;29(23):2195–203. doi: 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- 258.Gough A, Stern AM, Maier J, Lezon T, Shun TY, Chennubhotla C, Schurdak ME, Haney SA, Taylor DL. SLAS Discov. 2017;22(3):213–237. doi: 10.1177/2472555216682725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Immunity. 2012;36(1):142–52. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B. Nat. Methods. 2014;11(4):417–22. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 261.Bloom AN, Tian H, Schoen C, Winograd N. Anal. Bioanal. Chem. 2017;409(12):3067–3076. doi: 10.1007/s00216-017-0262-5. [DOI] [PubMed] [Google Scholar]
- 262.Wu K, Jia F, Zheng W, Luo Q, Zhao Y, Wang F. JBIC, J. Biol. Inorg. Chem. 2017;22(5):653–661. doi: 10.1007/s00775-017-1462-3. [DOI] [PubMed] [Google Scholar]
- 263.Tian H, Six DA, Krucker T, Leeds JA, Winograd N. Anal. Chem. 2017;89(9):5050–5057. doi: 10.1021/acs.analchem.7b00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Guillermier C, Fazeli PK, Kim S, Lun M, Zuflacht JP, Milian J, Lee H, Francois-Saint-Cyr H, Horreard F, Larson D, Rosen ED, Lee RT, Lechene CP, Steinhauser ML. JCI Insight. 2017;2(5):e90349. doi: 10.1172/jci.insight.90349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sheng L, Cai L, Wang J, Li Z, Mo Y, Zhang S, Xu J-J, Zhang X, Chen H-Y. Int. J. Mass Spectrom. 2017;421:238–244. [Google Scholar]
- 266.Korte AR, Yandeau-Nelson MD, Nikolau BJ, Lee YJ. Anal. Bioanal. Chem. 2015;407(8):2301–9. doi: 10.1007/s00216-015-8460-5. [DOI] [PubMed] [Google Scholar]
- 267.Mueller L, Herrmann AJ, Techritz S, Panne U, Jakubowski N. Anal. Bioanal. Chem. 2017;409(14):3667–3676. doi: 10.1007/s00216-017-0310-1. [DOI] [PubMed] [Google Scholar]
- 268.Herrmann AJ, Techritz S, Jakubowski N, Haase A, Luch A, Panne U, Mueller L. Analyst. 2017;142(10):1703–1710. doi: 10.1039/c6an02638a. [DOI] [PubMed] [Google Scholar]
- 269.Rao W, Pan N, Yang ZBJ. Visualized Exp. 2016;(112):119. doi: 10.3791/53911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee JK, Jansson ET, Nam HG, Zare RN. Anal. Chem. 2016;88(10):5453–5461. doi: 10.1021/acs.analchem.6b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]



 Lingjun Li:
Lingjun Li: