Abstract
This work reports the second part of a review intending to give the state of the art of major metabolic phenotyping strategies. It particularly deals with inherent advantages and limits regarding data analysis issues and biological information retrieval tools along with translational challenges. This Part starts with introducing the main data preprocessing strategies of the different metabolomics data. Then, it describes the main data analysis techniques including univariate and multivariate aspects. It also addresses the challenges related to metabolite annotation and characterization. Finally, functional analysis including pathway and network strategies are discussed. The last section of this review is devoted to practical considerations and current challenges and pathways to bring metabolomics into clinical environments.
Keywords: Omics, Metabolomics, Metabolome, Mass spectrometry, Nuclear magnetic resonance, Chemometrics
Introduction
Addressing biology as an informational science is a key driver to translate biological data into actionable knowledge. This requires innovative tools that allow information extraction from high dimensional data. Bioinformatics is the field that was born to tackle this challenge (Hogeweg 2011). Bioinformatics applies informatics techniques such as applied mathematics, computer science, and statistics to retrieve the organized biological information. In short, bioinformatics is a management information system for a biological system (Luscombe et al 2001). The metabolomic data requires adapted statistical tools to retrieve as much chemical information as possible to translate it into biological knowledge. The major challenge is to reduce the dimensionality by selecting informative signals from the noise. To achieve this goal, chemometric tools are widely used. Chemometrics is the science of extracting useful information from chemical systems using data-driven means (Brereton 2014). It is inherently interdisciplinary, borrowing methods from data-analytic disciplines such as statistics, signal processing, and computer science. Descriptive and predictive problems could be addressed using chemical data. This second part of the review intends to give the state of the art of metabolomics data handling strategies along with their inherent advantages and limits regarding data analysis issues. Furthermore, biological information retrieval tools and their translational challenges into actionable results are described. Finally, practical considerations and current challenges to bring metabolomics into the clinical environment are discussed. The general metabolomics workflow is presented in Fig. 1.
Fig. 1.
General metabolomics workflow. Metabolomics is divided into two main strategies. A targeted metabolomics is a quantitative analysis or a semiquantitative analysis of a set of metabolites that might be linked to common chemical classes or a selected metabolic pathway. An untargeted metabolomics approach is primarily based on the qualitative or semiquantitative analysis of the largest possible number of metabolites from diverse chemical and biological classes contained in a biological sample. The generated data undergo the data analysis step (univariate and multivariate) and functional analysis to get actionable biological insight
Biological information recovery
The analytical performance improvements associated with metabolomics platforms have led to the generation of complex and high-dimensional data sets. Handling the huge amount of generated data in a smoothly high-throughput fashion is a very important issue for transforming the data into clinically actionable knowledge.
Preprocessing
Targeted metabolomics aims to process data sets retrieved from a subset of the metabolome. It contains predefined, chemically characterized and biochemically annotated metabolites. The main advantages of targeted metabolomics are that no analytical artifacts are carried throughout the downstream analysis; only a set of selected metabolites are analyzed. However, in untargeted metabolomics, data analysis is quite time-consuming. Different automated processes have been developed (Tsugawa et al 2013, 2014; Cai et al 2015) along with commercial solutions from instrument vendors. In contrast, the untargeted approach attempts a comprehensive analysis of all measurable metabolites in a given sample, including unknowns. It requires a holistic analysis of high-dimensional raw data sets, which in turn requires reducing the data into more computationally manageable formats without significantly compromising the contained chemical information. Because of noise, sample variation, or analytical/instrument factors, NMR and MS spectra often show differences in width, position, and peak shape. The goal of preprocessing is to correct these differences for better quantification of metabolites and enhanced intersample comparability. Data preprocessing includes some or all of the following steps: noise filtering, baseline correction, peak detection, peak alignment, and spectral deconvolution. Several preprocessing considerations and methods can be applied to both NMR and MS data (Vettukattil 2015; Szymanska et al 2016; Yi et al 2016). MS data preprocessing includes some or all of the following steps: noise filtering, baseline correction, peak detection, peak alignment, and spectral deconvolution. The order of the steps may differ between algorithms. Noise filtering is often applied to MS data to improve peak detection. Many different noise filters exist, including Gaussian, Savitzky–Golay, and wavelet-based filters (Yi et al 2016). The aim of the peak detection and deconvolution step is to identify and quantify the signals that correspond to the analytes (metabolites) in a given sample. Peak detection algorithms follow two strategies: derivative techniques or matched filter response (Szymanska et al 2016; Yi et al 2016). A deconvolution step is used to separate overlapping peaks in order to improve peak detection (Johnsen et al 2017). Furthermore, a de-isotoping step is used to cluster the isotopic peaks corresponding to the same chemical feature to clean the data matrix. Alignment of the detected features across different samples aims to remove intersample shifts, and several alignment algorithms have been developed (Smith et al 2013; Szymanska et al 2016). The data dimensionality has to be reduced to make them applicable to instruments paired with MS. Different strategies enable data compression such as binning and the “search of regions of interest (ROI)” methods that are the most adequate hyphenated MS data sets. A comparison of some peak-picking algorithms used in untargeted MS-based metabolomics have been reported (Rafiei and Sleno 2015). XCMS is an open access mass spectrometry data processing software. It is widely used in the metabolomics community. It was developed in response to the growing need for user-friendly software to process complex untargeted metabolomic data (Smith et al 2006; Gowda et al 2014). It has been designed as a solution for the entire untargeted metabolomic workflow ranging from the raw data processing to direct metabolite assignment through integrated and automated METLIN database queries. The platform has been recently upgraded with data streaming capabilities to support high-throughput, cloud-based data processing, and systems biology analyses (Huan et al 2017). NMR data preprocessing typically includes baseline correction, alignment, and binning. Baseline correction aims to correct systematic baseline distortion. Some spectral regions, such as that of water, are often removed. Peak shifts due to differences in instrumental factors such as salt concentrations, temperature, and pH changes can be corrected by alignment procedures (Smolinska et al 2012). Binning or bucketing is a dimension reduction method that splits the spectra into segments or bins and assigns a representative value to each bin. However, binning can hamper spectral resolution. The typical output of the preprocessing step is a data matrix that contains the detected features and the corresponding intensity (abundance) in each sample.
Normalization
As with other omics, metabolomics data have several intrinsic characteristics, such as their asymmetric distribution (De Livera et al 2012) and a substantial proportion of instrumental, analytical, and biological noise (Grun et al 2014; Mak et al 2015). Thus, the goal of data normalization is to eliminate experimental biases related to the abundance of detected features between samples without compromising biological variations. Most of the methods are inspired by previous omic strategies (genomics and transcriptomics) that suffer from similar experimental biases (Tebani et al 2016). Indeed, the chemical diversity of metabolites and interindividual variations lead to changes in extraction and MS ionization yields, making it difficult to distinguish changes of biological interest from analytical biases (instrumentation, operators, and reagents). Strategies for normalization of metabolomics data can be divided into statistical approaches and chemical approaches. Statistical approaches are based on statistical models that define correction factors specific to each sample from the complete data set (Li et al 2016), such as normalization by standard deviation (Scholz et al 2004), mean global intensity (Wang et al 2003), quantile normalization (Lee et al 2012), probabilistic quotient normalization (Dieterle et al 2006), cyclic loess (Dudoit et al 2002), QC-robust spline batch correction (Kirwan et al 2013) or support vector regression (Shen et al 2016). Chemical approaches are based on one or more reference compounds (Hermansson et al 2005; Bijlsma et al 2006; Sysi-Aho et al 2007), internal standards, or endogenous or exogenous compounds that are used to normalize the entire chromatogram (single compound) or certain regions of the chromatogram by normalizing each zone according to a standard that is eluted in that region. Other strategies based on the characteristics of the studied matrix, such as dry mass of the samples, volume (e.g., 24-h urine), and osmolality. Protein or creatinine levels can also be used (Wu and Li 2016). A comprehensive comparison of state-of-the-art normalization techniques was recently reported (Li et al 2016).
Transformation, centering, and scaling
Statistical methods assume that the data under analysis have a specific type of probability distribution. Thus, the inferences made from the data depend on the chosen distribution. If the data under examination do not exhibit that distribution, then the inferences could be false or misleading. Most parametric methods in metabolomics assume that the data have a Gaussian distribution. However, in metabolomics, MS and NMR data are hampered by noise from different sources. Furthermore, the feature distributions can be skewed. So, transformations aim to correct for heteroscedasticity and skewness before statistical analysis. This allows building of statistically meaningful and interpretable models in metabolomics. Different mathematical transformations can be used, such as log transformation and power transformation (van den Berg et al 2006). Multivariate analytical methods are based on latent variable projections that extract information from the data by projecting observations onto the direction of the maximum variance. Hence, NMR and MS data analysis by these methods mainly focuses on the average spectrum. This approach may mask underlying biological variation because more abundant metabolites will exhibit high values in the data matrix and subsequently show large differences among samples compared to less abundant metabolites. Data scaling methods divide each data point for a given feature by a scaling factor that is a measure of data dispersion for that feature. Therefore, scaling the data aims to remove the offset from the data and focus on the biological variation regarding similarities and dissimilarities of samples. There are several scaling methods such as auto-scaling (unit variance scaling), in which the mean and the standard deviation of the feature are calculated. The aim of auto-scaling is to give equal weights to all features, but this method is very sensitive to large deviations from the sample mean. Thus, pareto scaling is the most popular alternative in metabolomics. In pareto-scaling, each observation in the mean-centered feature is divided by the square root of the standard deviation. Pareto scaling is a compromise between mean-centering and auto-scaling (van den Berg et al 2006).
Data analysis
Univariate data analysis
Univariate statistical methods can be used in metabolomics. Their main limitation is that they consider only one variable at a time, which may not be convenient for high-dimensional data. Parametric tests such as Student’s t-test and ANOVA are commonly applied to assess the differences between two or more groups, respectively, provided that the normality assumption is verified (Broadhurst and Kell 2006). Otherwise, if normality is not assumed, a nonparametric test such as Mann–Whitney U test or Kruskal–Wallis one-way ANOVA can be used. Another important issue is that applying multiple univariate tests in parallel with a high-dimensional data set raises the multiple testing problem. Since a large number of features are simultaneously analyzed in metabolomics, the probability of accidentally finding a statistically significant difference (i.e., true positive) is high. Different correction methods can be used to handle this multiple testing issue. In the Bonferroni correction, the significance level for a hypothesis is divided by the number of hypotheses simultaneously being tested (Broadhurst and Kell 2006). Hence, the Bonferroni correction is considered a conservative correction method. Less conservative methods are available and are based on lowering the false-discovery rate (FDR). Less restrictive approaches FDR-based methods minimize the expected proportion of false positives among the total number of positives (Benjamini and Hochberg 1995). It should be noted that potential confounding factors such as sex, age, or diet may lead to spurious results if not properly addressed. Furthermore, the main disadvantage of univariate methods is their lack of feature correlations and insights about interactions. Hence, advanced multivariate approaches are more suitable for in-depth inferences.
Multivariate data analysis
Bioinformatics a field that permits data collection, analysis, parsing, and contextual interpretation, and it supports decision-making on those bases. Bioinformatics can be defined as conceptualizing biology in terms of molecular components and by applying “informatics techniques” borrowed from disciplines such as applied mathematics, computer science, and statistics to understand and organize information on a large scale (Luscombe et al 2001). The major challenge is to reduce the dimensionality by selecting informative metabolic signals from the highly noisy raw data. Chemometric tools are widely used to achieve this goal. Chemometrics is defined as the science of extracting useful information from chemical systems by data-driven means (Brereton 2014). It may be applied to solve both descriptive and predictive problems, using biochemical data. In multivariate methods, representative samples are presented as points in the space of the initial variables. The samples can then be projected into a lower dimensionality space based on components or latent variables, such as a line, a plane, or a hyperplane, which can be seen as the shadow of the initial data set viewed from its best perspective. The sample coordinates of the newly defined latent variables are the scores, while the directions of variance to which they are projected are the loadings. The loadings vector for each latent variable contains the weights of each of the initial variables (metabolites) for that latent variable. Unsupervised methods attempt to reveal patterns or clustering trends in the data that underpin relationships between the samples. These methods also highlight the variables that are responsible for these relationships, using visualization means. Chemometrics methods are mainly divided into unsupervised and supervised methods. In unsupervised methods, no assumptions are made about the samples and the aim is mainly exploratory. In metabolomics data, metabolic similarity shapes the observed clustering. Principal component analysis (Hotelling 1933) is a widely used pattern recognition method; it is a projection-based method that reduces the dimensionality of the data by creating components. Principal component analysis allows a two- or three-dimensional visualization of the data. Because it contains no assumptions on the data, it is used as an initial visualization and exploratory tool to detect trends, groups, and outliers. It allows simpler global visualization by representing the variance in a small number of uncorrelated latent variables. Independent component analysis (ICA) is another unsupervised method that is a blind source separation method that separates multivariate signals into additive subcomponents (Bouveresse and Rutledge 2016). Its interpretation is similar to PCA, but instead of orthogonal components, it calculates non-Gaussian and mutually independent components (Wang et al 2008; Al-Saegh 2015). Compared to PCA, ICA as a linear method could provide potential benefits for untargeted metabolomics. ICA has been successfully used in metabolomics (Li et al 2012; Monakhova et al 2015; Liu et al 2016). Other unsupervised methods, such as clustering, aim to identify naturally occurring clusters in the data set by using similarity measures defined by distance and linkage metrics (Wiwie et al 2015). A dendrogram or a heat map can be created to visualize the similarities between samples. Commonly used clustering methods include correlation matrix, k-means clustering (Hartigan and Wong 1979), hierarchical cluster analysis (Johnson 1967), and self-organizing maps (Kohonen 1990; Goodwin et al 2014). In supervised methods, samples are assigned to classes or each sample is associated with a specific outcome value, and the aim is mainly explanatory and predictive. When the variables are discrete (e.g., control group versus diseased group), the task is called classification. When the variables are continuous (e.g., metabolite concentration) the task is called regression. The main purposes of supervised techniques are (i) to determine the association between the response variable and the predictors (metabolites) and (ii) to make accurate predictions based on the predictors. In metabolomics biomarker discovery, within the modeling process, it is important to find the simplest combination of metabolites that can produce a suitably effective predictive outcome. The biomarker discovery process involves two parameters, the biomarker utility and the number of metabolites used in the predictive model. The main challenges are therefore predictor selection and the evaluation of the fitness and predictive power of the built model. Predictor selection aims to identify important metabolites from among the detected ones that best explain and predict the biological or clinical outcome. Different predictor selection techniques have been described. Some of these suggested strategies are based on univariate or multivariate statistical proprieties of variables used as filters (loading weights, variable importance on projection scores, or regression coefficients), while others are based on optimization algorithms (Saeys et al 2007; Yi et al 2016). Recently, another elegant method has been reported that essentially combines estimation of Mahalanobis distances with principal component analysis and variable selection using a penalty metric instead of dimension reduction (Engel et al 2017). This method was successfully applied for inherited metabolic diseases (IMD) screening purposes. Finally, we need goodness-of-fit metrics to assess the model predictive power. Commonly used statistics may include root mean square error (RMSE) for regression problems and sensitivity, specificity, and the area under the receiver-operating characteristic (ROC) curve for classification models. To have independent test data sets, sometimes, data collection may be expensive or hampered by limited samples such as in rare diseases which is the case in IMD. In this case, various resampling methods are used to efficiently use the available data set, such as cross-validation, bootstrapping, and jackknifing (Westad and Marini 2015). Regarding the supervised methods, various techniques can be used in metabolomics. Some of the most used techniques include linear discriminant analysis (LDA) (Balog et al 2013; Ouyang et al 2014) and partial least squares (PLS) methods such as PLS-discriminant analysis (PLS-DA) (Wold et al 2001) and orthogonal-PLS-DA (OPLS-DA) (Trygg and Wold 2002; Manwaring et al 2013), as well as support vector machines (Cortes and Vapnik 1995; Lin et al 2011) and random forest (Breiman 2001; Huang et al 2015). Recently, Habchi et al proposed an innovative supervised method based on ICA called IC-DA. This method has been successfully applied to analyze DIMS metabolomics data that could be useful for high throughput screening (Habchi et al 2017). Furthermore, new methods based on topology data analysis are drawing interest and seem promising for data analysis because of their intrinsic flexibility and exploratory and predictive abilities (Liu et al 2015; Offroy and Duponchel 2016). Recently, a new method, called statistical health monitoring (SHM), has been adapted from industrial statistical process control; an individual metabolic profile is compared to a healthy one in a multivariate fashion. Abnormal metabolite patterns are thus detected, and more intelligible interpretation is enabled (Engel et al 2014). This approach has been successfully applied in IMD investigations (Engel et al 2017). The aim of metabolomics studies and the data analysis strategy are highly interdependent. Moreover, multivariate and univariate data analysis pipelines are not mutually exclusive, and they are often used together to enhance the quality of the information recovery. For further details on data analysis techniques and tools used in metabolomics, the reader may refer to recent reviews on this issue (Gromski et al 2015; Ren et al 2015; Misra and van der Hooft 2016).
Metabolite annotation and characterization
The identification of the discriminant metabolites is an important step in metabolomics. The introduction of high-resolution mass spectrometers and accurate mass measurements that facilitate access to the chemical formula of the detected peaks has considerably accelerated this step. The combined use of quadrupole ion traps for sequential fragmentation experiments provides additional structural information needed to identify metabolites of interest. However, MS using soft ionization techniques such as electrospray methods, exhibits high variability in the fragmentation profiles generated on different devices due to the lack of standardized ionization conditions, thus limiting the construction of universal spectral data banks such as those obtained by electron ionization or NMR (Cui et al 2008). This issue could be addressed using standardized ionization conditions such as electron based ionization techniques that are highly reproducible across MS systems worldwide and across different vendors. Indeed, in MS, one or more chemical formulas can be generated if high-resolution instruments are used, which provides a first element for carrying out an interrogation of the existing databases. The acquisition of fragmentation spectra at this stage enables us to discriminate the responses obtained previously on the basis of the produced ions or neutral losses, characteristic of chemical groups. Given the importance of the identification step, standardization elements have been proposed to harmonize metabolite identification data. Thus, identification standards have been defined within the framework of the Metabolomics Standards Initiative according to the available information on the metabolite to be characterized (Sumner et al 2007). Computational tools such as CAMERA (Kuhl et al 2012), ProbMetab (Silva et al 2014), AStream (Alonso et al 2011), and MetAssign (Daly et al 2014) have been developed for metabolite annotation. These methods mainly use m/z, retention time, adduct patterns, isotope patterns, and correlation methods for metabolite annotation. However, in MS the detected m/z ion and MS database matching is insufficient for unambiguous charcterization. Although retention time prediction are still used to improve identification confidence, complementary orthogonal information is required for reliable assignment of chemical identity, such as retention time matching and molecular dissociation patterns compared to authentic standards (Sumner et al 2007). For reliable characterization, a solution may be in a multidimensionnal framework based on orthogonal information integration, which may include accurate mass m/z, chromatographic retention time, MS/MS spectra patterns, CCS, chiral form, and peak intensity. Furthermore, hybrid strategies, including pathway network and analysis methods, could enhance metabolite characterization through different metrics integration, including data-driven network topology, chemical features correlation, omics data, and biological databases. Such a multidimensional approach may permit the chemical characterization by merging both extended chemical information and biological context. The Human Metabolome Database (HMDB) was first introduced in 2007 and is currently the most comprehensive, organism-specific metabolomic database. It contains NMR and MS spectra, quantitative, analytical, and physiological information about human metabolites. It also contains associated enzymes or transporters and disease-related properties. The HMDB is a fully searchable database with many built-in tools for viewing, sorting and extracting metabolites information features. In addition, the HMDB also supports the direct identification of potential diagnostic biomarkers based on their accurate mass, mass spectra or NMR spectra. Hence, the HMDB is a valuable support for translational metabolomics to support biomarker discovery. Perhaps, the HMDB (Wishart et al 2013) is one of the most valuable databases for IMD investigations. Other databases are presented in Table 1.
Table 1.
Biological databases and functional analysis tools
| Tools | Websites | References |
|---|---|---|
| Biological databases | ||
| KEGG (Kyoto Encyclopedia of Genes and Genomes) | http://www.genome.jp/kegg | (Kanehisa et al 2016) |
| HumanCyc (Encylopedia of Human Metabolic Pathways) | http://humancyc.org | (Romero et al 2005) |
| MetaCyc (Encyclopedia of Metabolic Pathways) | http://metacyc.org | (Caspi et al 2008) |
| Reactome (A Curated Knowledgebase of Pathways) | http://www.reactome.org | (Vastrik et al 2007) |
| SMPDB (Small Molecule Pathway Database) | http://www.smpdb.ca | (Jewison et al 2014) |
| Virtual Metabolic Human Database | https://vmh.uni.lu | (Thiele et al 2013) |
| Wikipathways | http://www.wikipathways.org | (Kelder et al 2012) |
| Pathway and networks analysis and visualization | ||
| BioCyc—Omics Viewer | http://biocyc.org | (Caspi et al 2016) |
| iPath | http://pathways.embl.de | (Yamada et al 2011) |
| MetScape | http://metscape.ncibi.org | (Karnovsky et al 2012) |
| Paintomics | http://www.paintomics.org | (Garcia-Alcalde et al 2011) |
| Pathos | http://motif.gla.ac.uk/Pathos | (Leader et al 2011) |
| Pathvisio | http://www.pathvisio.org | (Kutmon et al 2015) |
| VANTED | http://vanted.ipk-gatersleben.de | (Rohn et al 2012) |
| IMPaLA | http://impala.molgen.mpg.de | (Kamburov et al 2011) |
| MBROLE 2.0 | http://csbg.cnb.csic.es/mbrole2 | (Lopez-Ibanez et al 2016) |
| MPEA | http://ekhidna.biocenter.helsinki.fi/poxo/mpea | (Kankainen et al 2011) |
| Mummichog | http://clinicalmetabolomics.org/init/default/software | (Li et al 2013) |
| PIUMet | http://fraenkel-nsf.csbi.mit.edu/PIUMet/ | (Pirhaji et al 2016) |
| 3Omics | http://3omics.cmdm.tw/ | (Kuo et al 2013) |
| InCroMAP | http://www.ra.cs.uni-tuebingen.de/software/InCroMAP/ | (Wrzodek et al 2013) |
| Multifunctional tools | ||
| MetaboAnlayst | http://www.metaboanalyst.com | (Xia et al 2015) |
| XCMS online | https://xcmsonline.scripps.edu | (Tautenhahn et al 2012) |
| MASSyPup | http://www.bioprocess.org/massypup | (Winkler 2015) |
| Workflow4Metabolomics | http://workflow4metabolomics.org | (Giacomoni et al 2015) |
| Metabox | https://github.com/kwanjeeraw/metabox | (Wanichthanarak et al 2017) |
Functional analysis: translating information into knowledge
One of the fundamental difficulties in pathophysiological studies is that diseases might be caused by various genetic and environmental factors and their combinations. In addition, if a disease is caused by a combinatorial effect of many factors, the individual effects of each component might be low and thus hard to unveil. So, considering systems approaches to get deeper and informative biological insights is appealing. Any biological network can be pictured as a collection of linked nodes. The nodes may be genes, proteins, metabolites, diseases, or even individuals. The links or edges represent the interactions between the nodes: metabolic reactions, protein–protein interactions, gene–protein interactions, or interactions between individuals. The distribution of nodes ranges from random to highly clustered. However, biological networks are not random. They are collections of nodes and links that evolve as clusters; therefore, biological networks are referred to as scale-free, which means that they contain few highly-connected nodes called hubs. The core idea of the biological network theory is the modularity structure. Three distinct modules can be defined: topological, functional, and disease modules (Barabasi et al 2011). A topological module represents a local subset of nodes and links in the network; in this module, nodes have a higher tendency to link to nodes within the same local neighborhood. A functional module is a collection of nodes with similar or correlated function in the same network zone. Finally, a disease module represents a group of network components that together contribute to a cellular function whose disruption results in a disease phenotype. Of note, these three modules are correlated and overlap. Computational biology is gaining increasingly more space in modern biology to embrace this new network perspective. It can be divided into two main fields: knowledge discovery (or data-mining) and simulation-based analysis. The former generates hypotheses by extracting hidden patterns from high-dimensional experimental data. However, the latter tests hypotheses with in silico experiments, yielding predictions to be confirmed by in vitro and in vivo studies (Kitano 2002). Thus, pathway and network analysis strategies rely on the information generated by metabolomics studies for biological inference (Thiele et al 2013; Cazzaniga et al 2014). Both approaches exploit the interrelationships contained in the metabolomic data. Network modeling and pathway-mapping tools help to decipher the roles of metabolite interactions in a biological disturbance (Cazzaniga et al 2014). Biological databases are important for mapping different metabolic pathways (Table 1). Conceptual framework of pathway analysis is illustrated in Fig. 2. Indeed, pathway analysis or metabolite set enrichment analysis (MSEA) are methodologically based on the gene set enrichment analysis approach, previously developed for pathway analysis of gene-expression data (Khatri et al 2012; Garcia-Campos et al 2015). There are three distinct methods for performing MSEA: overrepresentation analysis (ORA), quantitative enrichment analysis (QEA), and single-sample profiling (SSP) (Xia and Wishart 2010; Garcia-Campos et al 2015; Xia et al 2015). An important advantage of computational metabolomics lies in the use of correlations among feature signals to map chemical identity. Since metabolites are interconnected by a series of biochemical reactions to build the network of metabolites, they can be interrogated using network-based analytical tools. In metabolomics, network analysis uses the high degree of correlation in metabolomics data to build metabolic networks based on the complex relationships of the measured metabolites. Based on the observed relationship patterns in the experimental data, correlation-based methods allow building metabolic networks in which each metabolite represents a node. However, unlike the pathway analysis, the links between nodes denote the level of mathematical correlation between each metabolite pair and are called edge (Krumsiek et al 2011; Valcarcel et al 2011; Do et al 2015). These data-driven strategies have been successfully applied for the reconstruction of metabolic networks from metabolomics data (Krumsiek et al 2011; Shin et al 2014; Bartel et al 2015). Biological inference often needs prior identification of metabolites. Since this step is challenging, a novel approach, named Mummichog, has been proposed by Li et al to reboot the conventional metabolomic workflow (Li et al 2013). This method predicts biological activity directly from MS-based untargeted metabolomics data without a priori identification of metabolites. The idea behind this strategy is combining network analysis and metabolite prediction under the same computational framework, which significantly reduces the metabolomics workflow time. Based on spectral peaks, the computational prediction of metabolites yields several hits; thus, a “null” distribution can be estimated by how these predicted metabolites, retrieved from a metabolomics experiment, map to all known metabolite reactions through interrogating databases. Despite most annotations being false, the biological meaning underpinning the data drives enrichment of the metabolites. The metabolite enrichment pattern of real metabolites compared to the null distribution is then statistically assessed. This method has been elegantly illustrated in an exploration of innate immune cell activation, which revealed that glutathione metabolism is modified by viral infection driven by constitutive nitric oxide synthases (Li et al 2013). Recently, Mummichog has been used for metabolic pathway analysis in a population by untargeted metabolomics. Hoffman et al identified metabolic pathways linked to age, sex, and genotype, including glycerophospholipid, neurotransmitters, metabolism carnitine shuttle, and amino acid metabolism (Hoffman et al 2016). Tyrosine metabolism was found to be associated with nonalcoholic fatty liver (Jin et al 2016). Pirhaji et al described a new network-based approach using a prize-winning Steiner forest algorithm for integrative analysis of untargeted metabolomics (PIUMet). This method infers molecular pathways via integrative analysis of metabolites without prior identification. Furthermore, PIUMet enabled elucidating putative identities of altered metabolites and inferring experimentally undetected, disease-associated metabolites and dysregulated proteins (Pirhaji et al 2016). Compared to Mummichog, PIUMet also allows system-level inference by integrating other omics data. Contextualization of metabolomics information is also important in pathophysiological investigations. From a metabolic network stand point, flux is defined as the rate (i.e., quantity per unit time) at which metabolites are converted or transported between different compartments (Aon and Cortassa 2015). Thus, metabolic fluxes, or the fluxome, represent a unique and functional readout of the phenotype (Cascante and Marin 2008; Aon and Cortassa 2015). Thus, from a network view of metabolism, one or more metabolic fluxes could be altered in a given metabolic disorder depending on the complexity of the disease (Lanpher et al 2006). To interrogate these fluxes, fluxome network modeling can be achieved using constraints of mass and charge conservation along with stoichiometric and thermodynamic limitations (Cortassa and Aon 2012; Winter and Kromer 2013; Kell and Goodacre 2014; Aurich and Thiele 2016). Based on the stoichiometry of the reactants and products of biochemical reactions, flux balance analysis can estimate metabolic fluxes without knowledge about the kinetics of the participating enzymes (Cascante and Marin 2008; Aon and Cortassa 2015). Recently, Cortassa et al suggested a new approach, distinct from flux balance analysis or metabolic flux analysis, that takes into account kinetic mechanisms and regulatory interactions (Cortassa et al 2015).
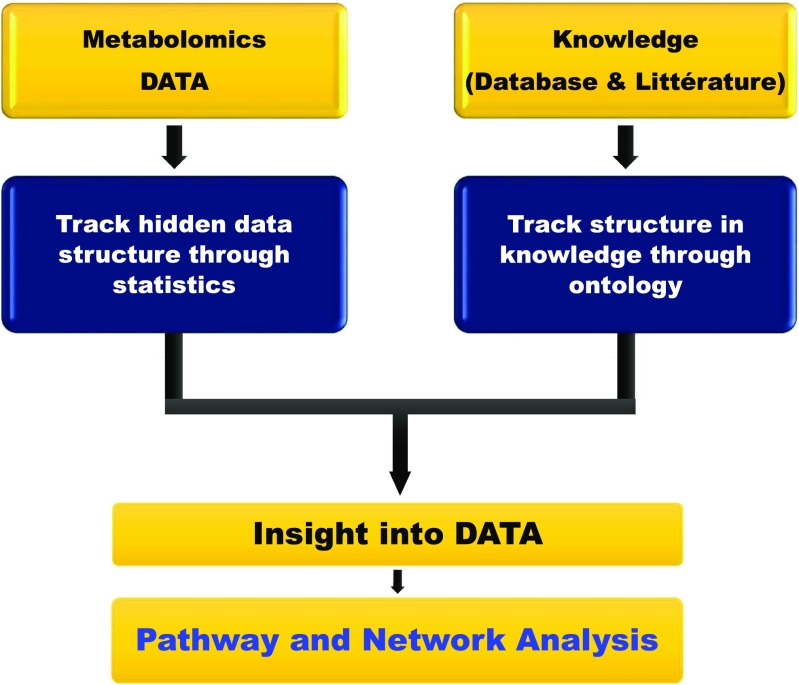
Fig. 2.
An illustration of pathway analysis strategies. Metabolome pathway analysis is designed to uncover significant pathway–phenotype relationships within a large data set. On one hand, it unveils hidden data structure in experimental data through differential expression using statistical metrics. On the other hand, it uses prior knowledge retrieved through biological databases and literature. Pathway analysis combines these two pillars to interpret the experimental findings
Since metabolites are often involved in multiple pathways, biologically-mediated labeling is particularly informative in such cases. Given the dynamics and compartmentation that characterize the metabolism, isotopic labeling is poised as an appealing approach to unambiguously track metabolic events. Advances in atom-tracking technologies and related informatics are disruptive for metabolomics-based investigations thanks to their contextual high throughput information retrieval. Among these technologies, stable isotope resolved metabolomics (SIRM) is a method that allows tracking individual atoms through compartmentalized metabolic networks which allowed highly resolved investigations of disease-related metabolomes (Higashi et al 2014; Fan et al 2016; Kim et al 2016). A wide variety of software tools are available for analyzing metabolomic data at the pathway and network levels. Table 1 presents different functional analysis tools for both pathway analysis and visualization.
Metabolomics and other omics cross-talk
Since IMD are associated with a genetic defect, their current characterization addresses both the mutated gene and its products. Currently, understanding of genetic variation effects on phenotypes is limited in most IMD which leads to consider the influence of genetic or environmental modifying factors and the impact of an altered pathway on metabolic flux as a whole. These diseases are related to the disruption of specific interactions in a highly organized metabolic network (Sahoo et al 2012; Argmann et al 2016). Thus, the impact of a given disruption is not easily predictable (Lanpher et al 2006; Cho et al 2012). Therefore, functional overview, integrating both space and time dimensions, is needed to assess the actors of the altered pathway and the potential interactions of each actor (Aon 2014). Thus, metabolomics combined with genome-wide association studies (mGWAS) track genetic influences on metabotypes which underpin the human’s metabolic individuality (Suhre et al 2016). Unveiling the genetically influenced metabolic variations could raise huge potential pathophysiological studies (Shin et al 2014). This includes functional understanding of clinical outcomes and genetic variation associations, designing targeted therapies for metabolic disorders and also identification of genetic modifiers underlying metabolic disease biomarkers. Different studies have reported genetic influences of metabotypes, disease-risk biomarkers or drug response variations (Suhre et al 2016). In a recent study, Rhee et al analyzed the association between exome variants and 217 plasma metabolites in 2076 participants in the Framingham Heart Study, with replication in 1528 individuals of the Atherosclerosis Risk in Communities Study. They identified an association between guanosine monophosphate synthase and xanthosine using single variant analysis and associations between histidine amonia lyase (HAL) and histidine, phenylalanine hydoxylase (PAH) and phenylalanine, and ureidopropionase (UPB1) and ureidopropionate using gene-based tests, which highlights novel coding variants that may unveil inborn errors of metabolism (Rhee et al 2016). Shin et al reported a comprehensive study exploring genetic loci influences on human metabotypes in 7824 individuals from two European cohorts, KORA (Germany) and Twins (UK), using MS-based metabolomics. They mapped significant associations at 145 loci and their metabotype connectivity through more than 400 blood metabolites. The built model unveiled information on heritability, gene expression and overlap with known complex disorders and inborn errors of metabolism loci. The data were used to build an online database for data mining and visualization (Shin et al 2014). The effectiveness of multi-omic approaches has been recently illustrated by van Karnebeek et al. The authors reported a disruption of the N-acetylneuraminic acid pathway in patients with severe developmental delay and skeletal dysplasia using both genomics and metabolomics approaches. Variations in the NANS gene encoding the synthase for N-acetylneuraminic acid were identified (van Karnebeek et al 2016). This elegantly highlights how systemic approaches may address IMD complexity and allow their diagnosis (Argmann et al 2016). For more details on mGWAS studies, the reader may refer to recent reviews (Kastenmuller et al 2015; Suhre et al 2016). Figure 3 shows how laboratory workflow using high-throughput analytical technologies, integrative bioinformatics, and computational frameworks will reshape IMD investigations. This integrative approach will allow intelligible molecular and clinical information recovery for a more effective medical decision-making in IMD.
Fig. 3.
Paradigm shift in inherited metabolic diseases investigation. High-throughput analytical technologies, integrative bioinformatics, and medical computational frameworks will allow intelligible molecular and clinical information recovery and effective medical decision-making
Perspectives in clinical metabolomics translation
Despite spectral information becoming available in the literature or in spectral databases, metabolite identification is still a challenging task (Goodacre et al 2007). However, metabolite identification remains a central issue in metabolomics prior to embracing complete clinical translation. No software is currently available to automate the identification step. Furthermore, metabolite identification is mandatory for absolute quantitation especially in MS-based methods requiring the use of stable isotope-labeled internal standards. Some data-driven alternatives have been developed to elucidate metabolite structure associations such as correlation-based network and modularity analysis. The association structure can be used to identify MS ions derived from the same metabolite (Broeckling et al 2014) or to identify biotransformations (Kind and Fiehn 2010). However, these knowledge-based approaches may be hampered by their limits for addressing the entire chemical space and limited coverage of metabolome databases. Another limitation lies in the cost for targeted analyses, which cannot reasonably be expected to support measurement of tens of thousands of chemicals in large populations. Thus, more efforts are needed to overcome this issue. However, in IMD a few hundred key metabolites may be defined for large-scale screening. Standardized and validated protocols are a prerequisite for metabolic phenotyping technologies. Harmonization of the sample preparation, processing, analysis, and reporting, using validated and standardized protocols, is mandatory (Chitayat and Rudan 2016; Kohler et al 2016). Standardized protocols are particularly helpful for untargeted metabolomics. In targeted methods, since each analyte is known and quantified, technology versatility is less important. Despite substantial efforts to standardize untargeted metabolomics methods, there are still no universally adopted protocols, particularly for MS-based strategies. This situation is due to the diverse and ever-changing analytical platform. The community and journals may take a lead in standardization by aligning it to community-published standards, such as the Metabolomics Standards Initiative (Sumner et al 2007), and data repisotories to encourage open metabolomic data, such as MetaboLights database at the EBI. All these endeavors aim to develop infrastructures and frameworks standardize terminology, data structure, and analytical workflows (Levin et al 2016). Finally, addressing these standardization issues is essential for regulatory compliance, which is a prerequisite for any clinical implementation. Automation at different stages, at instrument and pre- and post-analytic levels, is an important issue for broader use of metabolomics technologies. Automation enhances throughput, reproducibility, and reliability. Direct infusion MS-based methods are currently used for newborn screening in routine clinical practice (Therrell et al 2015; Ombrone et al 2016). Moreover, they are also taking the lead from a translational perspective, such as the iKnife, which would allow real-time cancer diagnosis (Balog et al 2013), and breathomics strategies for lung and respiratory diseases based on breath signatures (Hauschild et al 2015). Furthermore, metabolomics generates a huge amount of data that require comprehensive analysis and integration with other omics and metadata to infer the topology and dynamics of the underlying biological networks. Advanced statistical and computational tools along with effective data visualization are required to smoothly handle the diversity and quantity of the data and metabolite mapping (Alyass et al 2015; Ritchie et al 2015). In this regard, combining genomic and metabolic information may enhance biological inference and even clinical diagnostics (Tarailo-Graovac et al 2016; van Karnebeek et al 2016). Despite these promising steps, further advances in computational tools are needed for more efficient storage and integration (Perez-Riverol et al 2017).
Conclusion
Translating metabolomic data into actionable knowledge is the ultimate goal. Particular attention should be paid to computational tools for multidimensional data processing. There is an urgent need for more databases with validated and curated MRM transitions for targeted metabolites. Furthermore, for untargeted metabolomics, larger libraries and curated MS/MS spectra for metabolite identification are needed. Hybrid strategies including pathway and network analysis methods could enhance metabolite characterization through integration of different metrics, including data-driven network topology, chemical features correlation, omics data, and biological databases. Such multidimensional approaches may improve the chemical characterization by combining both extended chemical information and biological context. With all the high-dimensional data management issues, like other omics, metabolomics clinical implementation should be tackled using big data handling strategies for efficient storage, integration, visualization, and sharing of metabolomics data. To achieve the promise of the Precision Medicine era, it is crucial to combine expertise from multiple disciplines, including clinicians, medical laboratory professionals, data scientists, computational biologists, and biostatisticians. This raises the urgent need to think about new teams with new skill sets and overlapping expertise for more effective medical interactions across all healthcare partners for the management of IMD. Training the next generation medical workforce to manage and interpret omics data is a way to go and inception of such thinking has already started (Henricks et al 2016).
Acknowledgments
This work was supported by the Normandy University, the Institut National de la Santé et de la Recherche Médicale (INSERM), the Conseil Régional de Normandie, Labex SynOrg (ANR-11-LABX-0029), and the European Regional Development Fund (ERDF 31708).
Compliance with ethical standards
Conflict of interest
A. Tebani, C. Afonso, and S. Bekri declare that they have no conflict of interest.
Animal rights
This article does not contain any studies with human or animal subjects performed by the any of the authors.
References
- Alonso A, Julia A, Beltran A, et al. AStream: an R package for annotating LC/MS metabolomic data. Bioinformatics. 2011;27:1339. doi: 10.1093/bioinformatics/btr138. [DOI] [PubMed] [Google Scholar]
- Al-Saegh A (2015) Independent component analysis for separation of speech mixtures: a comparison among thirty algorithms. Iraqi J Electr Electron Eng 11(1):1–9
- Alyass A, Turcotte M, Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genet. 2015;8:1–12. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aon MA (2014) Complex systems biology of networks: the riddle and the challenge. In: Systems biology of metabolic and signaling networks. Springer, Berlin, p 19–35
- Aon MA, Cortassa S. Systems biology of the Fluxome. PRO. 2015;3:607–618. [Google Scholar]
- Argmann CA, Houten SM, Zhu J, Schadt EE. A next generation multiscale view of inborn errors of metabolism. Cell Metab. 2016;23:13–26. doi: 10.1016/j.cmet.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurich MK, Thiele I. Computational Modeling of human metabolism and its application to systems biomedicine. Methods Mol Biol. 2016;1386:253–281. doi: 10.1007/978-1-4939-3283-2_12. [DOI] [PubMed] [Google Scholar]
- Balog J, Sasi-Szabo L, Kinross J, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med. 2013;5:11. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- Barabasi A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel J, Krumsiek J, Schramm K, et al. The human blood Metabolome-Transcriptome Interface. PLoS Genet. 2015;11:e1005274. doi: 10.1371/journal.pgen.1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- Bijlsma S, Bobeldijk I, Verheij ER, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- Bouveresse DJ-R, Rutledge D (2016) Independent components analysis: theory and applications. Resolving spectral mixtures: with applications from ultrafast time-resolved spectroscopy to super-resolution imaging, vol 30. Elsevier, Amsterdamn, p 7225
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Brereton RG. A short history of chemometrics: a personal view. J Chemom. 2014;28:749–760. [Google Scholar]
- Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2006;2:171–196. [Google Scholar]
- Broeckling CD, Afsar FA, Neumann S, Ben-Hur A, Prenni JE. RAMClust: a novel feature clustering method enables spectral-matching-based annotation for Metabolomics data. Anal Chem. 2014;86:6812–6817. doi: 10.1021/ac501530d. [DOI] [PubMed] [Google Scholar]
- Cai Y, Weng K, Guo Y, Peng J, Zhu Z-J. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics. 2015;11:1575–1586. [Google Scholar]
- Cascante M, Marin S. Metabolomics and fluxomics approaches. Essays Biochem. 2008;45:67–82. doi: 10.1042/BSE0450067. [DOI] [PubMed] [Google Scholar]
- Caspi R, Foerster H, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga P, Damiani C, Besozzi D, et al. Computational strategies for a system-level understanding of metabolism. Meta. 2014;4:1034–1087. doi: 10.3390/metabo4041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitayat S, Rudan JF (2016) Phenome centers and global harmonization, chap. 10. In: Metabolic phenotyping in personalized and public healthcare. Academic, Boston, p 291–315
- Cho D-Y, Kim Y-A, Przytycka TM. Chapter 5: network biology approach to complex diseases. PLoS Comput Biol. 2012;8:e1002820. doi: 10.1371/journal.pcbi.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortassa S, Aon MA. Computational modeling of mitochondrial function. Methods Mol Biol. 2012;810:311–326. doi: 10.1007/978-1-61779-382-0_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortassa S, Caceres V, Bell LN, O’Rourke B, Paolocci N, Aon MA. From metabolomics to fluxomics: a computational procedure to translate metabolite profiles into metabolic fluxes. Biophys J. 2015;108:163–172. doi: 10.1016/j.bpj.2014.11.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- Cui Q, Lewis IA, Hegeman AD, et al. Metabolite identification via the Madison Metabolomics consortium database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- Daly R, Rogers S, Wandy J, Jankevics A, Burgess KE, Breitling R. MetAssign: probabilistic annotation of metabolites from LC-MS data using a Bayesian clustering approach. Bioinformatics. 2014;30:2764. doi: 10.1093/bioinformatics/btu370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Livera AM, Dias DA, De Souza D, et al. Normalizing and integrating Metabolomics data. Anal Chem. 2012;84:10768–10776. doi: 10.1021/ac302748b. [DOI] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Do KT, Kastenmüller G, Mook-Kanamori DO, et al. Network-based approach for analyzing intra- and Interfluid metabolite associations in human blood, urine, and saliva. J Proteome Res. 2015;14:1183–1194. doi: 10.1021/pr501130a. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–139. [Google Scholar]
- Engel J, Blanchet L, Engelke UF, Wevers RA, Buydens LM. Towards the disease biomarker in an individual patient using statistical health monitoring. PLoS One. 2014;9:e92452. doi: 10.1371/journal.pone.0092452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J, Blanchet L, Engelke UFH, Wevers RA, & Buydens LMC (2017) Sparse statistical health monitoring: A novel variable selection approach to diagnosis and follow-up of individual patients. Chemom Intell Lab Syst 164:83–93
- Fan TW, Lane AN, Higashi RM (2016) Stable isotope resolved metabolomics studies in ex vivo tissue slices. Bio Protoc 6(3). pii:e1730 [DOI] [PMC free article] [PubMed]
- Garcia-Alcalde F, Garcia-Lopez F, Dopazo J, Conesa A. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–139. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Campos MA, Espinal-Enriquez J, Hernandez-Lemus E. Pathway analysis: state of the art. Front Physiol. 2015;6:383. doi: 10.3389/fphys.2015.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomoni F, Le Corguille G, Monsoor M, et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31:1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodacre R, Broadhurst D, Smilde AK, et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics. 2007;3:231–241. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CR, Sherrod SD, Marasco CC, et al. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal Chem. 2014;86:6563–6571. doi: 10.1021/ac5010794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda H, Ivanisevic J, Johnson CH, et al. Interactive XCMS online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem. 2014;86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromski PS, Muhamadali H, Ellis DI, et al. A tutorial review: Metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Meth. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Habchi B, Alves S, Jouan-Rimbaud Bouveresse D, et al. An innovative chemometric method for processing direct introduction high resolution mass spectrometry metabolomic data: independent component–discriminant analysis (IC–DA) Metabolomics. 2017;13:45. [Google Scholar]
- Hartigan JA, Wong MA. Algorithm AS 136: a K-means clustering algorithm. J R Stat Soc: Ser C: Appl Stat. 1979;28:100–108. [Google Scholar]
- Hauschild AC, Frisch T, Baumbach JI, Baumbach J. Carotta: revealing hidden confounder markers in metabolic breath profiles. Meta. 2015;5:344–363. doi: 10.3390/metabo5020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricks WH, Karcher DS, Harrison JH, et al. Pathology informatics essentials for residents: a flexible informatics curriculum linked to accreditation Council for Graduate Medical Education milestones. J Pathol Inform. 2016;7:27. doi: 10.4103/2153-3539.185673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, Uphoff A, Kakela R, Somerharju P. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- Higashi RM, Fan TW, Lorkiewicz PK, Moseley HN, Lane AN. Stable isotope Labeled tracers for metabolic pathway elucidation by GC-MS and FT-MS. Methods Mol Biol. 2014;1198:147–167. doi: 10.1007/978-1-4939-1258-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JM, Tran V, Wachtman LM, Green CL, Jones DP, Promislow DE. A longitudinal analysis of the effects of age on the blood plasma metabolome in the common marmoset, Callithrix Jacchus. Exp Gerontol. 2016;76:17–24. doi: 10.1016/j.exger.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeweg P. The roots of bioinformatics in theoretical biology. PLoS Comput Biol. 2011;7:e1002021. doi: 10.1371/journal.pcbi.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H (1933) Analysis of a complex of statistical variables into principal components. Warwick & York, Baltimore
- Huan T, Forsberg EM, Rinehart D, et al. Systems biology guided by XCMS online metabolomics. Nat Methods. 2017;14:461–462. doi: 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-H, Fu L, Li B, et al. Distinguishing the serum metabolite profiles differences in breast cancer by gas chromatography mass spectrometry and random forest method. RSC Adv. 2015;5:58952–58958. [Google Scholar]
- Jewison T, Su Y, Disfany FM, et al. SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res. 2014;42:D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Banton S, Tran VT, et al. Amino acid metabolism is altered in adolescents with nonalcoholic fatty liver disease-an untargeted, high resolution Metabolomics study. J Pediatr. 2016;172:14–19.e15. doi: 10.1016/j.jpeds.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen LG, Skou PB, Khakimov B, Bro R. Gas chromatography mass spectrometry data processing made easy. J Chromatogr A. 2017;1503:57–64. doi: 10.1016/j.chroma.2017.04.052. [DOI] [PubMed] [Google Scholar]
- Johnson SC. Hierarchical clustering schemes. Psychometrika. 1967;32:241–254. doi: 10.1007/BF02289588. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Gopalacharyulu P, Holm L, Oresic M. MPEA--metabolite pathway enrichment analysis. Bioinformatics. 2011;27:1878–1879. doi: 10.1093/bioinformatics/btr278. [DOI] [PubMed] [Google Scholar]
- Karnovsky A, Weymouth T, Hull T, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmuller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: an update. Hum Mol Genet. 2015;24:R93–r101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T, van Iersel MP, Hanspers K, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell DB, Goodacre R. Metabolomics and systems pharmacology: why and how to model the human metabolic network for drug discovery. Drug Discov Today. 2014;19:171–182. doi: 10.1016/j.drudis.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IY, Suh SH, Lee IK, Wolfe RR. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp Mol Med. 2016;48:e203. doi: 10.1038/emm.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal Rev. 2010;2:23–60. doi: 10.1007/s12566-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan J, Broadhurst D, Davidson R, Viant M. Characterising and correcting batch variation in an automated direct infusion mass spectrometry (DIMS) metabolomics workflow. Anal Bioanal Chem. 2013;405:5147–5157. doi: 10.1007/s00216-013-6856-7. [DOI] [PubMed] [Google Scholar]
- Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- Kohler I, Verhoeven A, Derks RJ, Giera M. Analytical pitfalls and challenges in clinical metabolomics. Bioanalysis. 2016;8:1509–1532. doi: 10.4155/bio-2016-0090. [DOI] [PubMed] [Google Scholar]
- Kohonen T. The self-organizing map. Proc IEEE. 1990;78:1464–1480. [Google Scholar]
- Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol. 2011;5:21. doi: 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C, Tautenhahn R, Bottcher C, Larson TR, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem. 2012;84:283. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T-C, Tian T-F, Tseng YJ. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol. 2013;7:64. doi: 10.1186/1752-0509-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, van Iersel MP, Bohler A, et al. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015;11:e1004085. doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanpher B, Brunetti-Pierri N, Lee B. Inborn errors of metabolism: the flux from Mendelian to complex diseases. Nat Rev Genet. 2006;7:449–460. doi: 10.1038/nrg1880. [DOI] [PubMed] [Google Scholar]
- Leader DP, Burgess K, Creek D, Barrett MP. Pathos: a web facility that uses metabolic maps to display experimental changes in metabolites identified by mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:3422–3426. doi: 10.1002/rcm.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Park J, Lim MS, et al. Quantile normalization approach for liquid chromatography-mass spectrometry-based metabolomic data from healthy human volunteers. Anal Sci. 2012;28:801–805. doi: 10.2116/analsci.28.801. [DOI] [PubMed] [Google Scholar]
- Levin N, Salek RM, Steinbeck C (2016) From databases to big data, chap. 11. In: Metabolic phenotyping in personalized and public healthcare. Academic, Boston, p 317–331
- Li X, Hansen J, Zhao X, et al. Independent component analysis in non-hypothesis driven metabolomics: improvement of pattern discovery and simplification of biological data interpretation demonstrated with plasma samples of exercising humans. J Chromatogr B. 2012;910:156–162. doi: 10.1016/j.jchromb.2012.06.030. [DOI] [PubMed] [Google Scholar]
- Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput Metabolomics. PLoS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tang J, Yang Q, et al. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted Metabolomics analysis. Sci Rep. 2016;6:38881. doi: 10.1038/srep38881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wang Q, Yin P, et al. A method for handling metabonomics data from liquid chromatography/mass spectrometry: combinational use of support vector machine recursive feature elimination, genetic algorithm and random forest for feature selection. Metabolomics. 2011;7:549–558. [Google Scholar]
- Liu W, Bai X, Liu Y, et al. Topologically inferring pathway activity toward precise cancer classification via integrating genomic and metabolomic data: prostate cancer as a case. Sci Rep. 2015;5:13192. doi: 10.1038/srep13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Smirnov K, Lucio M, Gougeon RD, Alexandre H, Schmitt-Kopplin P. MetICA: independent component analysis for high-resolution mass-spectrometry based non-targeted metabolomics. BMC Bioinf. 2016;17:1–14. doi: 10.1186/s12859-016-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ibanez J, Pazos F, Chagoyen M. MBROLE 2.0-functional enrichment of chemical compounds. Nucleic Acids Res. 2016;44:W201–W204. doi: 10.1093/nar/gkw253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Greenbaum D, Gerstein M. What is bioinformatics? A proposed definition and overview of the field. Methods Inf Med. 2001;40:346–358. [PubMed] [Google Scholar]
- Mak TD, Laiakis EC, Goudarzi M, Fornace AJ. Selective paired ion contrast analysis: a novel algorithm for analyzing Postprocessed LC-MS Metabolomics data possessing high experimental noise. Anal Chem. 2015;87:3177–3186. doi: 10.1021/ac504012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwaring V, Boutin M, Auray-Blais C. A metabolomic study to identify new globotriaosylceramide-related biomarkers in the plasma of Fabry disease patients. Anal Chem. 2013;85:9039–9048. doi: 10.1021/ac401542k. [DOI] [PubMed] [Google Scholar]
- Misra BB, van der Hooft JJ. Updates in metabolomics tools and resources: 2014-2015. Electrophoresis. 2016;37:86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- Monakhova YB, Godelmann R, Kuballa T, Mushtakova SP, Rutledge DN. Independent components analysis to increase efficiency of discriminant analysis methods (FDA and LDA): application to NMR fingerprinting of wine. Talanta. 2015;141:60–65. doi: 10.1016/j.talanta.2015.03.037. [DOI] [PubMed] [Google Scholar]
- Offroy M, Duponchel L. Topological data analysis: a promising big data exploration tool in biology, analytical chemistry and physical chemistry. Anal Chim Acta. 2016;910:1–11. doi: 10.1016/j.aca.2015.12.037. [DOI] [PubMed] [Google Scholar]
- Ombrone D, Giocaliere E, Forni G, Malvagia S, la Marca G. Expanded newborn screening by mass spectrometry: new tests, future perspectives. Mass Spectrom Rev. 2016;35:71–84. doi: 10.1002/mas.21463. [DOI] [PubMed] [Google Scholar]
- Ouyang M, Zhang Z, Chen C, Liu X, Liang Y. Application of sparse linear discriminant analysis for metabolomics data. Anal Methods. 2014;6:9037–9044. [Google Scholar]
- Perez-Riverol Y, Bai M, da Veiga Leprevost F, et al. Discovering and linking public omics data sets using the Omics discovery index. Nat Biotechnol. 2017;35:406–409. doi: 10.1038/nbt.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhaji L, Milani P, Leidl M, et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nat Methods. 2016;13:770–776. doi: 10.1038/nmeth.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiei A, Sleno L. Comparison of peak-picking workflows for untargeted liquid chromatography/high-resolution mass spectrometry metabolomics data analysis. Rapid Commun Mass Spectrom. 2015;29:119–127. doi: 10.1002/rcm.7094. [DOI] [PubMed] [Google Scholar]
- Ren S, Hinzman A, Kang E, Szczesniak R, Lu L. Computational and statistical analysis of metabolomics data. Metabolomics. 2015;11:1492–1513. [Google Scholar]
- Rhee EP, Yang Q, Yu B, et al. An exome array study of the plasma metabolome. Nat Commun. 2016;7:12360. doi: 10.1038/ncomms12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- Rohn H, Junker A, Hartmann A, et al. VANTED v2: a framework for systems biology applications. BMC Syst Biol. 2012;6:1–13. doi: 10.1186/1752-0509-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2–R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Sahoo S, Franzson L, Jonsson JJ, Thiele I. A compendium of inborn errors of metabolism mapped onto the human metabolic network. Mol BioSyst. 2012;8:2545–2558. doi: 10.1039/c2mb25075f. [DOI] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J. Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics. 2004;20:2447–2454. doi: 10.1093/bioinformatics/bth270. [DOI] [PubMed] [Google Scholar]
- Shen X, Gong X, Cai Y, et al. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics. 2016;12:89. [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RR, Jourdan F, Salvanha DM, et al. ProbMetab: an R package for Bayesian probabilistic annotation of LC-MS-based metabolomics. Bioinformatics. 2014;30:1336. doi: 10.1093/bioinformatics/btu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Smith R, Ventura D, Prince JT. LC-MS alignment in theory and practice: a comprehensive algorithmic review. Brief Bioinform. 2013;16:104–117. doi: 10.1093/bib/bbt080. [DOI] [PubMed] [Google Scholar]
- Smolinska A, Blanchet L, Buydens LM, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- Suhre K, Raffler J, Kastenmüller G. Biochemical insights from population studies with genetics and metabolomics. Arch Biochem Biophys. 2016;589:168–176. doi: 10.1016/j.abb.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinf. 2007;8:93. doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanska E, Davies A, Buydens L. Chemometrics for ion mobility spectrometry data: recent advances and future prospects. Analyst. 2016;141(20):5689–5708. doi: 10.1039/c6an01008c. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Shyr C, Ross CJ, et al. Exome sequencing and the Management of Neurometabolic Disorders. N Engl J Med. 2016;374:2246–2255. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS online: a web-based platform to process untargeted metabolomic data. Anal Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebani A, Afonso C, Marret S, Bekri S. Omics-based strategies in precision medicine: toward a paradigm shift in inborn errors of metabolism investigations. Int J Mol Sci. 2016;17(9):1555. doi: 10.3390/ijms17091555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrell BL, Padilla CD, Loeber JG, et al. Current status of newborn screening worldwide: 2015. Semin Perinatol. 2015;39:171–187. doi: 10.1053/j.semperi.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RM, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16:119–128. [Google Scholar]
- Tsugawa H, Arita M, Kanazawa M, Ogiwara A, Bamba T, Fukusaki E. MRMPROBS: a data assessment and metabolite identification tool for large-scale multiple reaction monitoring based widely targeted metabolomics. Anal Chem. 2013;85:5191–5199. doi: 10.1021/ac400515s. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Ohta E, Izumi Y, et al. MRM-DIFF: data processing strategy for differential analysis in large scale MRM-based lipidomics studies. Front Genet. 2014;5:471. doi: 10.3389/fgene.2014.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel B, Wurtz P, Seichalbasatena NK, et al. A differential network approach to exploring differences between biological states: an application to prediabetes. PLoS One. 2011;6:e24702. doi: 10.1371/journal.pone.0024702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Karnebeek CD, Bonafé L, Wen X-Y, et al. NANS-mediated synthesis of sialic acid is required for brain and skeletal development. Nat Genet. 2016;48(7):777–784. doi: 10.1038/ng.3578. [DOI] [PubMed] [Google Scholar]
- Vastrik I, D’Eustachio P, Schmidt E, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettukattil R. Preprocessing of raw Metabonomic data. In: Bjerrum JT, editor. Metabonomics: methods and protocols. New York: Springer; 2015. pp. 123–136. [DOI] [PubMed] [Google Scholar]
- Wang WX, Zhou HH, Lin H, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- Wang G, Ding Q, Hou Z. Independent component analysis and its applications in signal processing for analytical chemistry. TrAC Trends Anal Chem. 2008;27:368–376. [Google Scholar]
- Wanichthanarak K, Fan S, Grapov D, Barupal DK, Fiehn O. Metabox: a toolbox for Metabolomic data analysis, interpretation and integrative exploration. PLoS One. 2017;12:e0171046. doi: 10.1371/journal.pone.0171046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westad F, Marini F. Validation of chemometric models – a tutorial. Anal Chim Acta. 2015;893:14–24. doi: 10.1016/j.aca.2015.06.056. [DOI] [PubMed] [Google Scholar]
- Winkler R. An evolving computational platform for biological mass spectrometry: workflows, statistics and data mining with MASSyPup64. PeerJ. 2015;3:e1401. doi: 10.7717/peerj.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Kromer JO. Fluxomics — connecting ‘omics analysis and phenotypes. Environ Microbiol. 2013;15:1901–1916. doi: 10.1111/1462-2920.12064. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0--the human Metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwie C, Baumbach J, Rottger R. Comparing the performance of biomedical clustering methods. Nat Methods. 2015;12:1033–1038. doi: 10.1038/nmeth.3583. [DOI] [PubMed] [Google Scholar]
- Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109–130. [Google Scholar]
- Wrzodek C, Eichner J, Büchel F, Zell A. InCroMAP: integrated analysis of cross-platform microarray and pathway data. Bioinformatics. 2013;29:506–508. doi: 10.1093/bioinformatics/bts709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li L. Sample normalization methods in quantitative metabolomics. J Chromatogr A. 2016;1430:80–95. doi: 10.1016/j.chroma.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–W415. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Dong N, Yun Y, et al. Chemometric methods in data processing of mass spectrometry-based metabolomics: a review. Anal Chim Acta. 2016;914:17–34. doi: 10.1016/j.aca.2016.02.001. [DOI] [PubMed] [Google Scholar]