Abstract
Background:
Klebsiella pneumoniae (K. pneumoniae) is an opportunistic pathogen that could be resistant to many antimicrobial agents. Resistance genes can be carried among gram-negative bacteria by integrons. Enzymatic inactivation is the most important mechanism of resistance to aminoglycosides. In this study, the frequencies of two important resistance gene aac(6′)-IIa and ant(2″)-I, and genes coding integrase I and II, in K. pneumoniae isolates resistant to aminoglycosides were evaluated.
Methods:
In this cross-sectional study, an attempt was made to assess the antibiotic susceptibility of 130 K. pneumoniae isolates obtained from different samples of patients hospitalized in training hospitals of Yazd evaluated by disk diffusion method. The frequencies of aac(6′)-IIa, ant(2″)-I, intl1, and intl2 genes were determined by PCR method. Data were analyzed by chi-square method using SPSS software (Ver. 16).
Results:
our results showed that resistance to gentamicin, tobramycin, kanamycin, and amikacin were 34.6, 33.8, 43.8, and 14.6%, respectively. The frequencies of aac (6′)-IIa, ant(2″)-I, intl1, and intl2 genes were 44.6, 27.7, 90, and 0%, respectively.
Conclusion:
This study showed there are high frequencies of genes coding aminoglycosides resistance in K. pneumoniae isolates. Hence, it is very important to monitor and inhibit the spread of antibiotic resistance genes.
Keywords: Aminoglycosides, Drug resistance, Integrons, Klebsiella pneumoniae, Microbial
Introduction
Klebsiella pneumoniae (K. pneumoniae) is an opportunistic pathogen that mainly causes nosocomial infections1. This bacterium can cause serious illnesses, such as septicemia, pneumoniae, bacteremia, urinary tract and soft tissue infections. Emergence of antibiotic resistant strains challenges the efficacy of antibiotic drugs; and the number of antibiotic resistant strains is increasing2.
Aminoglycosides are a group of antibiotics that are very effective against a broad spectrum of gram-positive and gram-negative aerobic bacteria. These antibiotics are the most widely used drugs for treatment of infections caused by facultative anerobic gram-negative rods3. Despite widespread usage and clinical success of aminoglycosides, administration of these anti-biotics for controlling bacterial infections has been challenged by emergence of resistant strains. Inactivation by a wide range of enzymes known as Aminoglycosides Modifying Enzymes (AMEs), is the most common mechanism of inactivation of aminoglycosides.
AMEs inactivate aminoglycosides through several molecular mechanisms including phosphorylation, acetylation and adenylation, which are catalyzed by ATP-dependent O-phosphotransferase, actyl-CoA dependent N-acetyltransferase, and ATP-dependent O-nucleotidyl-transferase, respectively4. ANT(2″)-Ia and AAC(6′)-II are two important aminoglycosides modifying enzyme in K. pneumoniae5,6. ANT(2″)-Ia was isolated from K. pneumoniae for the first time7, and nowadays, has become one of the most important enzymes causing antibiotic resistances in gram-negative pathogens8–10. On the other hand, AAC(6′) is the most important and common enzyme in the AAC super-family. More than 40 kinds of this enzyme have been identified. ACC(6′) exists in both gram-negative and gram-positive bacteria11.
Integrons play an important role in increasing antibiotic resistance, particularly in gram-negative pathogens. There are five classes of integrons in which class 1 and class 2 are the most common among other classes, and mainly associated with hospital acquired resistance, which often includes resistance to aminoglycosides and beta-lactams, trimethoprim, ampicillin and chloramphenicol12. Class 1 integron, which is the main cause of antibiotic resistance spreading, has been studied in many microorganisms. Studies suggest that this integron can be seen in 22–59% of gram-negative rods, especially K. pneumoniae13.
The number of aminoglycosides resistant K. pneumoniae strains is increasing. In addition, genes coding AMEs can be carried by integrons which can be involved in aminoglycosides resistant bacteria. Thus, in this study, the frequency of aac(6′)-II and ant(2″)-Ia genes and integrons class 1 and 2 were investigated in K. pneumoniae isolated from clinical samples of hospitalized patients in Yazd.
Materials and Methods
Sample obtaining and culture
In this cross-sectional study, from October 2015 to March 2016, a total of 130 K. pneumoniae isolates were obtained from different samples of hospitalized patients in training hospitals of Yazd. The samples were transferred to Microbiology Laboratory of Yazd Shahid Sadoughi University of Medical Sciences, and were cultured in EMB medium (Merck Darmstadt, Germany) and incubated at 37°C for 18–24 hr.
Bacteria identification
Lactose-positive colonies were identified by differential biochemical tests including glucose and lactose fermentation in TSI culture medium, indole production and motility on SIM medium, reaction in MR-VP medium, growth on Simmon-citrate medium, and urease production on urea agar (all media from Merck Darmstadt, Germany).
Antibiotic susceptibility testing
Susceptibility of K. pneumoniae isolates to aminoglycoside such as tobramycin (10 mg), gentamicin (10 mg), amikacin (30 mg) and kanamycin (5 mg) was performed using disk diffusion method (Kirby-Bauer) (Liofilchem, Italy) recommended by Clinical and Laboratory Standard Institute (CLSI) guideline14. In this test, the bacterial suspension with a turbidity which was equal to turbidity of McFarland tube 0.5 (1.5×108 CFU/ml) was used. E. coli ATCC 25922 was used as the control.
Measuring the Minimum Inhibitory Concentration (MIC) of gentamicin
E-test method (Liofilcam, Italy) was used to determine the Minimum Inhibitory Concentration (MIC) of gentamicin15. Based on CLSI guideline, MIC>16 was considered as resistant isolates to gentamicin14. Escherichia coli (E. coli) ATCC 25922 was used as control.
Genomic DNA extraction
Genomic DNA was extracted by salting out method16. The quality and quantity of obtained DNA was measured by 0.7% agarose gel electrophoresis and nano-drop, respectively. DNA samples were stored at −20°C until the examining day.
Polymerase Chain Reaction (PCR)
PCR was carried out using thermocycler (Eppendorf, Germany) to evaluate the presence of ant(2″)-Ia, aac(6′)-II, intl1, and intl2 genes in the isolates. Specific primers were as follows: 5′-GGTGTGGCGGGCTTCGTG-3′ and 5′-GCATCCTCGGTTTTCTGG-3′ as forward and reverse primer for intl1 (amplicon size: 480 bp)17; 5′-CTAGAATAGGCTGTATAGGCAGA-3′ and 5′-GAGTGACGAAATGTATGACAAG-3′ as forward and reverse primer for intl2 (amplicon size: 850 bp)18; 5′-CACAACGCAGGTCATT-3′ and 5′-CGCTAAGAATCCATAGTCCAA-3′ as forward and reverse primer for ant(2″)-Ia (amplicon size: 220 bp; designed using Primer3 software); and 5′-GCCGATGCTCCATGATTG-3′ and 5′-TCGAAGGCTTGTCGTGTT-3′ as forward and reverse primer for aac(6′)-II (amplicon size: 480 bp; designed using Primer3 software).
The final volume for PCR reaction was 20 ml (water 5 μl), PCR 2X master mix (Amplicon-Denmark) (10 μl), working primers with final concentration of 5 pmol (2 μl), and template DNA (3 μl).
The thermal profile was as follows: 94°C for 5 min as initial denaturation for one cycle; 94°C for 60 s, 52°C for 60 s, and 72°C for 60 s, for 30 cycles; with a final extension at 72°C for 5 min. The PCR reaction condition was the same for all tested genes, except intl1 that its annealing temperature was considered 50°C. 1% agarose gel electrophoresis was performed on PCR products, and sequenced for further confirmation.
Statistical analysis
Correlation between the genes and resistance to aminoglycosides were analyzed by chi-square method using SPSS software v. 16.0 (SPSS Inc, Chicago, IL, USA).
Results
Among all 130 K. pneumoniae isolates, aminoglycosides resistance was as follows: 43.8% for kanamycin, 34.6% for gentamicin, 33.8% for tobramycin and 14.6% for amikacin. Generally, 54.6% of isolates were resistant at least to one aminoglycoside. Phenotypic results showed the highest simultaneous resistance of 29.2% for gentamicin-kanamycin, followed by 25.3% for gentamicin-tobramycin, 24.4% for kanamycin-tobramycin, and 9.23% for gentamicin-tobramycin (Table 1).
Table 1.
Results of antibiotic resistance and studied genes in 130 K. pneumoniae isolates
| Antibiotic resistance | Resistant n (%) | aac(6′)-IIa gene n (%) | ant(2″)-Ia gene n (%) | int1 gene n (%) |
|---|---|---|---|---|
| GM-K-TOB-AK | 12 (9.23) | 12 (9.23) | 4 (3) | 12 (9.23) |
| GM-TOB- K | 32 (24.6) | 32 (24.6) | 13 (10) | 32 (24.6) |
| GM-AK-K | 14 (10.7) | 14 (10.7) | 4 (3) | 14 (10.7) |
| GM-K | 38 (29.2) | 38 (29.2) | 16 (12.3) | 37 (28.4) |
| K-TOB | 37 (28.4) | 32 (24.6) | 16 (12.3) | 35 (26.9) |
| GM-TOB | 33 (25.3) | 33 (25.3) | 14 (10.7) | 32 (24.6) |
| K | 57 (43.8) | 45 (34.6) | 26 (20) | 54 (41.5) |
| GM | 45 (34.6) | 40 (30) | 17 (13) | 41 (31.5) |
| TOB | 44 (33.8) | 34 (26.1) | 20 (15.3) | 40 (30.7) |
| AK | 19 (14.6) | 17 (13) | 6 (4.6) | 17 (13) |
K: Kanamycin; TOB: Tobramycin; GM: Gentamicin; AK: Amikacin.
MIC of 45 isolates which were resistant to gentamicin by disc diffusion method was as follows: 37 resistant isolates (83%) and 8 sensitive isolates (18%). MIC was 32 μg/ml for 23.9% of isolates (Table 2).
Table 2.
Results of MIC for gentamicin by E test method
| MIC range (μg/ml) | 2> | 4 | 8 | 16 | 24 | 32 | 48 | 64 | 256 |
|---|---|---|---|---|---|---|---|---|---|
| No. | 8 | 0 | 0 | 3 | 7 | 11 | 8 | 3 | 5 |
| Percent | 17.7 | 0 | 0 | 6.6 | 15.5 | 24.4 | 17.7 | 6.6 | 11.1 |
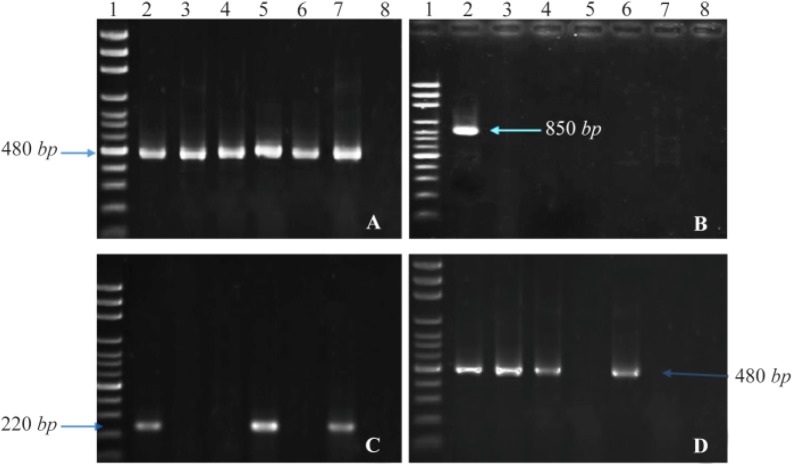
Our results showed that frequencies of aac(6′)-II, ant(2″)-Ia, intl1 in studied isolates were 44.6% (n=58), 27.7% (n=36), and 90% (n=117), respectively. But intl2 was not found in the isolates (Figure 1). Furthermore, the correlation between presence of these genes and resistance to one particular antibiotic or different mixtures of antibiotics have also been assessed. The results showed that in isolates which were resistant to gentamicin-kanamycin-tobramycin, the presence of intl1, ant(2″)-Ia, and aac(6′)-II was 25.3, 10 and 25.3%, respectively (Table 1). Other correlations are stated in table 1.
Figure 1.
PCR products of intl1 (A), intl2 (B), ant(2″)-Ia (C), and aac(6′)-IIa (D) on agarose gel electrophoresis visualized under UV light. Lane 1 shows DNA Marker; lane 2 shows positive control; lane 3–7 show PCR products of the amplicon amplified from K. pneumonia isolates; and lane 8 shows negative control of attributed genes.
Discussion
K. pneumoniae is an opportunistic pathogen of Enterobacteriaceae family. K. pneumoniae colonization is very common among hospitalized patients. This bacterium causes septicemia, bacteremia, meningitis, urinary and pulmonary tracts infection, and enteritis in children, elderly and patients with immunodeficiency disorders19. Aminoglycosides are one the most important treatments used against K. pneumoniae infections. Acetylation and adenylation are two important mechanisms of aminoglycosides resistance, catalyzed by actyl-CoA dependent N-acetyltransferase, and ATP-dependent O-nucleotidyltransferase, respectively4. In this study, resistance to aminoglycosides and the frequencies of aac (6′)-IIa and ant(2″)-Ia genes which encode the most important acetyl- and adenyl-transferases in nosocomial pathogens4, have been evaluated in K. pneumoniae isolates. Moreover, since integrons specially class 1 integron are one of the main causes of antibiotic resistance among bacterial populations13,20, also the frequencies of intl1 and intl2 genes were evaluated in K. pneumoniae isolates.
Our results showed that resistance of K. pneumoniae isolates to gentamicin, kanamycin, tobramycin, and amikacin was 34.6, 43.8, 33.8, and 14.6%, respectively. Moreover, according to E test results, most isolates (24.4%) had MIC at range of 32 for gentamicin. Najar-Peerayeh et al who studied 200 K. pneumoniae isolates revealed that the frequency of resistance to gentamicin, tobramycin, and amikacin were 36, 32, and 27%, respectively21. Our results about resistance to gentamicin and tobramycin were consistent with theirs, but percentage of resistance to amikacin was nearly doubled in their study (27 vs. 14.6%). Study of Mobarak-Qamsari et al on 104 K. pneumoniae isolates revealed that the frequency of resistance for kanamycin, tobramycin, gentamicin, and amikacin was 48, 28.6, 25.9, and 25.9%, respectively22. These results are nearly consistent with ours, except resistance percentage to amikacin, which was doubled in their study. In fact, these two studies that carried out in Tehran (Iran) showed same results about amikacin. The observed differences might be due to the geographical differences, and wide prescription of antibiotics in that area. However, Li et al showed that frequency of resistance to gentamicin, tobramycin, and amikacin in China, was 62.2, 71.1, and 31.4%, respectively. These results are very different from the results obtained from Iran. These results showed that frequency of resistance to aminoglycosides could vary in different countries, and even hospitals.
The relationship between aac(6′)-II and ant(2″)-I, two important genes coding aminoglycosides modifying enzymes, with resistance to the aminoglycosides demonstrates that aac(6′)-II gene is significantly associated with resistance to these four antibiotics (p= 0.000). 93.3% of resistant isolates were positive to aac(6′)-II gene, while only 18.82% of susceptible isolates contained this gene. Among kanamycin resistant isolates, 77.19% of them were aac(6′)-II gene positive, but 17.14% of susceptible isolates possess this gene. Moreover, this gene was present in amikacin and tobramycin resistant isolates at a frequency of 94.73 and 81.81%, respectively, while in susceptible isolates by 33.01, and 22.22% respectively. In the present study, aac(6′)-II gene was found in 44.6% of isolates. Interestingly, in Najar-Peerayeh et al21 study, this gene was found in 42.5% of isolates. Moreover, Liang et al23 reported that 30.25% of the isolates were positive for this gene. Thus, our results are consistent with these studies.
ant(2″)-I gene was found in gentamicin resistant and gentamicin susceptible isolates at the rate of 34.77, and 22.35%, respectively. The relationship between this gene and gentamicin resistance was not statically significant (p=0.068). But, there was a significant association between this gene with resistance to kanamycin (p=0.001), and 43.85% of the kanamycin resistant isolates had this gene, while just 15.71% of the susceptible ones had it. Our results also showed that there is a significant correlation between ant(2″)-I and resistance to tobramycin (p=0.003)(data in details are not shown here). The results showed that 47.72% of resistant isolates and 16.04% of susceptible isolates had this gene. Based on our study, there was no significant correlation between ant(2″)-I gene and resistance to amikacin (p=0.31) (data in details are not shown here). Generally, ant(2″)-I gene was found in 27.7% of the studied isolates. The presence of this gene was reported 13.59% in Liang et al’s23 study.
In the next section of the present study, the frequencies of intl1 and intl2 genes were evaluated. PCR results showed that 90% (n=117) of isolates had intl1 gene; but intl2 was not found in the studied isolates. Integron class 1 is considered as an important genetic element carrying resistance genes to antibiotics. Keeping this in mind, the correlation of intl1 gene with the mentioned resistance, and with ant(2″)-Ia and aac(6′)-II genes was evaluated. High frequency of intl1 gene was found in resistant isolates. The frequency of this gene in resistant isolates to gentamicin, kanamycin, amikacin, and tobramycin was 93.3, 94.73, 89.47, and 93.18%, respectively; on the other hand in susceptible isolates, the frequencies were 88.23, 85.71, 90.56, and 87.65%, respectively. There was no significant correlation between this gene and the mentioned resistance (p>0.05). Moreover, there was no significant association between intl1 gene and aac(6′)-II and ant(2″)-I genes (p=0.29) (data in details are not shown here).
The frequency of integron class 1 was reported 51.1% in the study of Li et al24. They showed that there is a significant correlation between intl1 and resistance to antibiotics. Despite high frequency of class 1 integron in our study, no significant correlation between intl1 and resistance to antibiotics was observed. Comparing our results with the study of Li et al, high frequency of intl1 gene in the studied isolates was found. These reports are really alarming, because class 1 integron is one of the most important genetic elements that cause expansion of antibiotic resistance in hospitals, and acquiring resistance to drugs. Given the high frequency of class 1 integron in this study and its effect on increasing resistance to antibiotics, especially aminoglycosides, it can be concluded that more attention is needed in this area because they can easily create resistance to the antibiotics, and reduce the sensitivities of isolates to aminoglycosides. Since class I interon was found in many susceptible isolates of this study, and given the fact that specific antibiotic resistance genes are carried by gene cassettes in integrons20, the correlation of cassettes and genes encoding resistance to antibiotics should also be examined, to figure out the association between intl1 gene and resistance to the aminoglycosides. Considering that intl2 gene was not found in these K. pneumoniae isolates, further studies are required.
Conclusion
Our results showed high frequencies of aac(6′)-IIa, ant(2″)-Ia, and intl1 genes in K. pneumoniae isolates which could be the results of unnecessary prescription of antibiotics. These results revealed the importance of antibiotic resistance gene. Moreover, selecting an antibiotic by antibiotic susceptibility testing has an important role in treatment and inhibiting the resistance to antibiotics. Since aminoglycosides are very important in treatment of K. pneumoniae infections, thus, the prevalence rate of this resistance should be evaluated annually in countries.
Acknowledgement
This paper is extracted from a Master’s thesis of International Campus, Shahid Sadoughi University of Medical Sciences, Yazd, Iran, and thereby we thank the personnel of Medical Microbiology Department of International Campus and we also thank the personnel of Central Laboratory of Yazd.
Footnotes
Conflict of Interest
We declare that there is no conflict of interest.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998;11 (4):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bbosa GS, Mwebaza N, Odda J, Kyegombe DB, Ntale M. Antibiotics/antibacterial drug use, their marketing and promotion during the post-antibiotic golden age and their role in emergence of bacterial resistance. Health 2014;6 (5):410–425. [Google Scholar]
- 3.Maurin M, Raoult D. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob Agents Chemother 2001;45(11):2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updates 2010;13(6):151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox G, Stogios PJ, Savchenko A, Wright GD. Structural and molecular basis for resistance to aminoglycoside antibiotics by the adenylyltransferase ANT (2″)-Ia. MBio 2015;6(1):e02180–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frasson I, Cavallaro A, Bergo C, Richter SN, Palù G. Prevalence of aac(6′)-Ib-cr plasmid-mediated and chromosome-encoded fluoroquinolone resistance in Entero-bacteriaceae in Italy. Gut Pathog 2011;3(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benveniste R, Davies J. R-factor mediated gentamicin resistance: a new enzyme which modifies aminoglycoside antibiotics. FEBS Lett 1971;14(5):293–296. [DOI] [PubMed] [Google Scholar]
- 8.Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 1993;57(1):138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu K, Kumada T, Hsieh WC, Chung HY, Chong YU, Hare RS, et al. Comparison of aminoglycoside resistance patterns in Japan, Formosa, and Korea, Chile, and the United States. Antimicrob Agents Chemother 1985;28(2):282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller GH, Sabatelli FJ, Hare RS, Glupczynski Y, Mackey P, Shlaes D, et al. The most frequent aminoglycoside resistance mechanisms--changes with time and geographic area: a reflection of aminoglycoside usage patterns? aminoglycoside resistance study groups. Clin Infect Dis 1997;24(Suppl 1):S46–62. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez MS, Nikolaidis N, Tolmasky M. Rise and dissemination of aminoglycoside resistance: the aac(6′)-Ib paradigm. Front Microbiol 2013;4:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge SR. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 2011;35 (5):820–855. [DOI] [PubMed] [Google Scholar]
- 13.Zarei-Yazdeli M, Eslami G, Zandi H, Mousavi SM, Kosha H, Akhavan F, et al. Relationship between antimicrobial resistance and class I integron in Pseudomonas aeruginosa isolated from clinical specimens in Yazd during 2012–2013. FEYZ 2014;18(1):60–67. [Google Scholar]
- 14.Patel JB, Tenover FC, Turnidge JD, Jorgensen JH. Susceptibility test methods: dilution and disk diffusion methods. In: Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, et al., editors. Manual of clinical microbiology. USA: American Society for Microbiology; 2015. [Google Scholar]
- 15.Citron DM, Ostovari MI, Karlsson A, Goldstein EJ. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J Clin Microbiol 1991;29(10):2197–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16(3):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousefi S, Nahaei M, Farajnia S, Ghojazadeh M, Akhi M, Sharifi Y, et al. Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: prevalence and antibiotic susceptibility. Iran J Microbiol 2010:2(3):115–121. [PMC free article] [PubMed] [Google Scholar]
- 18.Ohara M, Kouda S, Onodera M, Fujiue Y, Sasaki M, Kohara T, et al. Molecular Characterization of imipenem-resistant Pseudomonas aeruginosa in Hiroshima, Japan. Microbiol Immunol 2007;51(3):271–277. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol 2014;9(9):1071–1081. [DOI] [PubMed] [Google Scholar]
- 20.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev 2014;78(2):257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peerayeh SN, Rostami E, Eslami M, Rezaee MA. High frequency of extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates from male patients’ Urine. Arch Clin Infect Dis 2016;11(2): e60127. [Google Scholar]
- 22.Mobarak-Qamsari M, Ashayeri-Panah M, Eftekhar F, Feizabadi MM. Integron mediated multidrug resistance in extended spectrum beta-lactamase producing clinical isolates of Klebsiella pneumoniae. Braz J Microbiol 2013;44(3):849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C, Xing B, Yang X, Fu Y, Feng Y, Zhang Y. Molecular epidemiology of aminoglycosides resistance on Klebsiella pneumonia in a hospital in China. Int J Clin Exp Med 2015;8(1):1381–1385. [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Hu Y, Wang Q, Yi Y, Woo PC, Jing H, et al. Structural diversity of class 1 integrons and their associated gene cassettes in Klebsiella pneumoniae isolates from a hospital in China. PloS One 2013;8(9):e75805. [DOI] [PMC free article] [PubMed] [Google Scholar]