Significance
This study describes the results of a large-scale case control analysis of copy number variants (CNVs) in a cohort of patients with congenital diaphragmatic hernia (CDH) and a large number of healthy population-matched controls. Using a customized array comparative genomic hybridization system, we have identified six CNVs that are associated with CDH with statistical significance (P < 0.05). These regions validate several hypothesized CDH candidate genes and identify additional genes and pathways that contribute to the pathogenesis of CDH. The estimated frequency of pathogenic CNVs in this cohort is 13%, which underscores the critical contribution of CNVs in CDH. This study also provides a model approach that is broadly applicable to other structural birth defects and identifies candidates for future functional studies.
Keywords: copy number variant, CNV, customized array, birth defects, congenital diaphragmatic hernia
Abstract
Congenital diaphragmatic hernia (CDH), characterized by malformation of the diaphragm and hypoplasia of the lungs, is one of the most common and severe birth defects, and is associated with high morbidity and mortality rates. There is growing evidence demonstrating that genetic factors contribute to CDH, although the pathogenesis remains largely elusive. Single-nucleotide polymorphisms have been studied in recent whole-exome sequencing efforts, but larger copy number variants (CNVs) have not yet been studied on a large scale in a case control study. To capture CNVs within CDH candidate regions, we developed and tested a targeted array comparative genomic hybridization platform to identify CNVs within 140 regions in 196 patients and 987 healthy controls, and identified six significant CNVs that were either unique to patients or enriched in patients compared with controls. These CDH-associated CNVs reveal high-priority candidate genes including HLX, LHX1, and HNF1B. We also discuss CNVs that are present in only one patient in the cohort but have additional evidence of pathogenicity, including extremely rare large and/or de novo CNVs. The candidate genes within these predicted disease-causing CNVs form functional networks with other known CDH genes and play putative roles in DNA binding/transcription regulation and embryonic development. These data substantiate the importance of CNVs in the etiology of CDH, identify CDH candidate genes and pathways, and highlight the importance of ongoing analysis of CNVs in the study of CDH and other structural birth defects.
Congenital diaphragmatic hernia (CDH) is one of the most common and lethal congenital anomalies. It has an incidence of ∼1 in 3,000 live births (1–4) and occurs when the diaphragm does not form properly, often resulting in displacement of the abdominal contents into the chest cavity. CDH arises during prenatal development and is almost always accompanied by lung hypoplasia and pulmonary hypertension, which are the major causes of morbidity and mortality. The lung hypoplasia is apparent before displacement of abdominal contents into the chest cavity in certain animal models, suggesting that a primary defect in lung development plays an important role in this disease. With advanced medical and surgical care, the survival rate for these infants can reach 80%, but worldwide mortality is 50%. Long-term morbidity among survivors is common (5).
The genetic contributions to congenital diaphragmatic defects are heterogeneous, with mutations in many different genes capable of generating the same phenotype (4), which makes interpretation of genomic data from patients with CDH more challenging. While some of the genetic mutations that result in CDH impair only formation of the diaphragm, several genes and pathways are also required for normal development of the lungs and its vasculature. More than 70 genes are now implicated in CDH based on analysis of the genomes of patients with CDH as well as the characterization of mutant mice (6–12). Many CDH genes discovered from mouse models were also later found to harbor rare and predicted pathogenic mutations in patients with CDH (8, 10, 13).
Numerous studies have implicated copy number variants (CNVs) in human disease with associated phenotypes ranging from cognitive disabilities to predispositions to obesity, cancer, and other diseases (14–17). Both inherited and de novo CNVs play a causative role in CDH (18–22), and at least 10% of CDH cases are estimated to be caused by genomic imbalances (21). Several recurrent genomic imbalances (i.e., deletions and duplications) have been found in specific chromosomal regions, including 1q41–42, 8p23.1, 8q23, and 15q26 (19, 22–25), and analysis of these regions has led to the identification of validated CDH genes (7, 8, 26). However, many other genomic imbalances have only been described in one or two patients with CDH, making conclusions about causation difficult, especially without the knowledge of the frequency of these CNVs among healthy individuals.
The aim of this study was to perform a systematic analysis of CNVs in a large number of patients with CDH alongside a large control cohort to provide further evidence for specific putative CDH genes in the etiology of this condition. We designed a customized high-resolution CNV array, targeting known and candidate genomic regions associated with diaphragmatic defects. This customized array was used to interrogate the genomes of 196 patients with CDH and 987 controls to identify CNVs that are significantly associated with CDH risk. An ethnically matched control cohort was used to provide additional statistical confirmation of significant association for each region. As a result, we have identified six CNVs that are significantly associated with CDH and encompass genes involved in DNA binding/transcription regulation and embryonic organ development. These findings support a critical role for CNVs in the pathogenesis of a significant subset of patients with CDH, indicating the importance of assessing CNVs as well as sequence variants in the genetic analyses of these patients.
Results
CDH Patient and Control Selection.
We analyzed DNA samples from 196 patients with CDH (SI Appendix, Table S1). A total of 96 individuals (49%) had isolated CDH, and 80 individuals (40%) had complex phenotypes including additional congenital anomalies or other unusual features. The remaining cases did not have sufficient phenotype information to classify as isolated or complex. Among the complex cases, congenital heart disease and neurodevelopmental disorders were the most common co-occurring morbidities. Left-sided Bochdalek hernia was the most common specified type of diaphragm defect (n = 70). Because our intention was to detect all CNVs for an unbiased statistical analysis, patients with confirmed findings on clinical or research arrays were not excluded from the study.
To select proper normal control samples to minimize the ethnic background bias for this study, we performed a principal-component analysis (PCA) using whole-exome sequencing (WES) data on CDH cases (8) and whole-genome sequencing data from presumed healthy population samples from the 1000 Genomes Project (27). By this method, 987 population controls were selected for our study to match ancestries between cases and controls (SI Appendix, Fig. S1). These controls represented 26 populations, although since we collected the majority of our patients with CDH from North America, more ethnically matched controls from Europe (494) and admixed Americans (120) were selected for this study.
CNVs Detected from Customized Array.
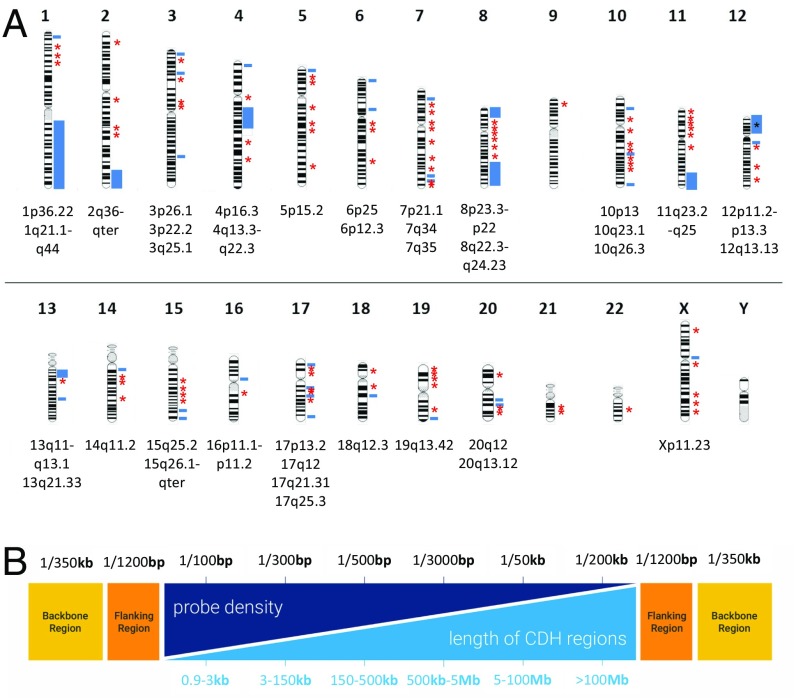
A customized array comparative genome hybridization (aCGH) platform was designed covering 140 known and candidate CDH regions (Fig. 1 and SI Appendix, Table S2) (see details in SI Appendix, Supplemental Materials and Methods). We completed aCGH experiments on 196 patients with CDH, 987 population-matched control samples, and 109 unaffected parents for a total of 1,292 individuals. In total, 234 gains and 437 losses were detected from all of the samples (SI Appendix, Table S3). The median number of CNVs detected per sample was 13, and they ranged in size from 0.4 kb to 95 Mb (median, 2.1 kb; average, 119 kb). The majority of CNVs identified were within the 1- to 10-kb range, and there was no significant difference in the size distribution among CDH patients, their parents, and population controls (SI Appendix, Fig. S2).
Fig. 1.
Customized array targeted for CDH candidate regions. (A) Map of genes and genomic regions covered by custom array. Blue bars indicate known and candidate genomic regions implicated in human CDH. Red asterisks indicate other CDH candidate genes. (B) The light blue section shows different lengths of the targeted CDH regions, and the dark blue section represents different probe densities designed for the targeted regions. The orange bars represent regions flanking CDH targets, and the yellow bars represent the backbone for the rest of the human genome.
Identification of Statistically Significant CNVs Enriched in Patients with CDH.
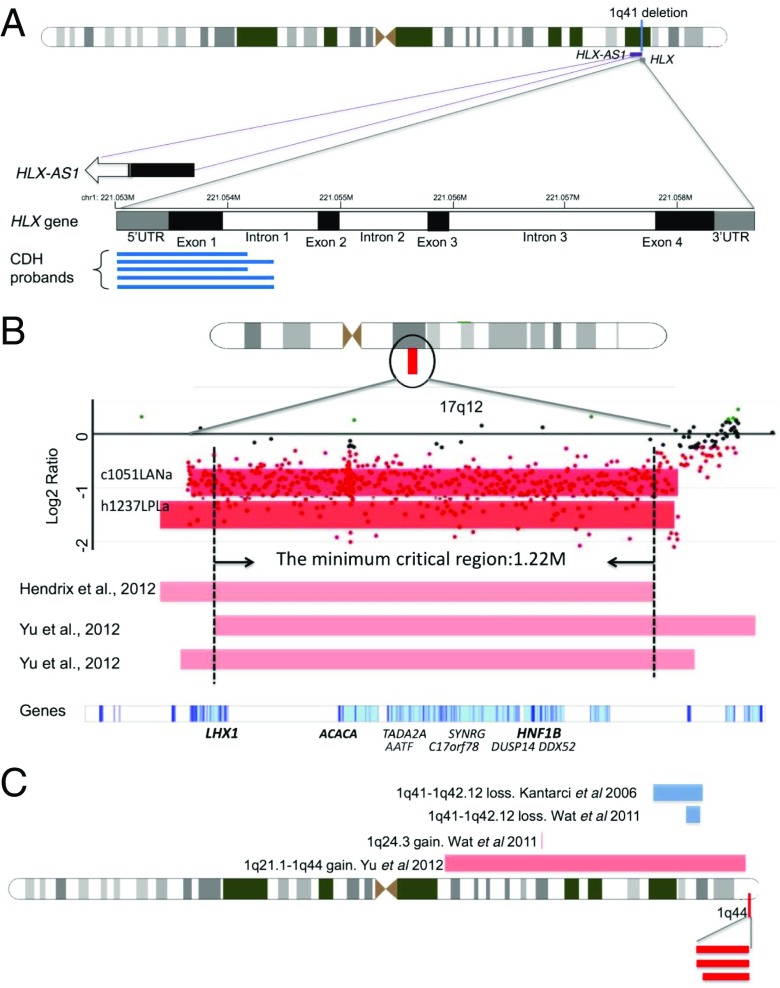
To identify significant CNVs associated with CDH, we compared the frequency of CNVs in patients with the frequency in ethnically matched controls. We identified six CNVs that were present in two or more patients and at a significantly higher frequency in cases than in controls (Table 1). Three of these CNVs were found in two or more patients with CDH but not in any population controls. First, a 1.6-kb gain involving the HLX gene was found in five patients with CDH (Table 1, P = 0.00058, Fisher’s exact test). This duplication encompasses most of exon 1 and part of intron 1 (Fig. 2A). HLX homozygous null mice have been previously reported to have diaphragmatic defects and the gene is expressed in the developing murine diaphragm (28, 29). Second, a 1.5-Mb loss within the chromosome 17q12 target region was found in two unrelated patients with CDH (Fig. 2B). This deletion is predicted to cause haploinsufficiency for 15 protein-coding genes, two noncoding transcripts, and two microRNAs (Fig. 2B and Table 1). The P value of this CNV slightly missed the significance threshold (P = 0.06) due to a relatively small sample size, although we still include it as a significant candidate because it is large in size, has been described previously in three other published cases of CDH (21, 30), and it is absent from control CNV databases. Third, a 113-kb loss from chromosome region 1q44 was found in three patients (P = 0.016), encompassing the ZNF672, ZNF692, and PGBD2 genes (Fig. 2C and Table 1). We also identified three CNVs in multiple patients with CDH and in one or more controls, but at frequencies significantly higher among patients compared with ethnically matched controls (Table 1). A 2.4-Mb gain at 16p11.2 was detected in 4 of 196 patients with CDH and 4 of 987 controls and was significantly associated with CDH (P = 0.047). This region encompasses the protein-coding genes TP53TG3E, TP53TG3B, TP53TG3F, and TP53TG3C, two noncoding transcripts, and several pseudogenes. Also, a 131-kb loss at 4p16.3 was identified in five patients with CDH and one ethnically matched control (P = 0.001), overlapping two zinc finger genes of unknown function, ZNF595 and ZNF718. Larger deletions, including this region, have been reported to be associated with Wolf–Hirschhorn syndrome with CDH (31, 32). Finally, a 79-kb loss at 5p15.2 was found in three patients with CDH and three controls (P = 0.04) and encompasses one noncoding RNA gene, LINC01194. The function of LINC01194 is unknown, but it is reported to be expressed in both skeletal muscle and lung, relevant tissue types for the pathogenesis of CDH (GTex) (33). This CNV was also detected in several controls from different ethnicities, suggesting that it may have a higher frequency in other populations without a significant association with CDH. We compared the phenotypes of patients with significant recurrent CNVs (SI Appendix, Table S4). Two of three patients with 1q44 deletions had colobomas in addition to CDH, although we did not identify any other major genotype–phenotype correlations.
Table 1.
Summary of the significant CNVs detected in multiple patients with CDH
| No. of samples affected | |||||||
| Region (hg19) | Size, bp | Cytoband | CNV type | Proband (n = 196) | Population (n = 987) | P value | Description/gene(s) |
| i) CNVs detected in two or more patients but not in population controls | |||||||
| chr1:221052740–221054346 | 1,606 | 1q41 | Gain | 5 | 0 | 0.00058** | Most of exon 1 and part of intron 1 of the HLX gene, as well as noncoding RNA gene HLX-AS1 |
| chr17:34813719–36278623 | 1,464,904 | 17q12 | Loss | 2 | 0 | 0.06 | AATF, ACACA, DDX52, DUSP14, GGNBP2, HNF1B, LHX1, MYO19, DHRS11, MRM1, c17orf78, PIGW, SYNRG, TADA2A, and ZNHIT3 |
| chr1:249126046–249238916 | 112,870 | 1q44 | Loss | 3 | 0 | 0.016* | ZNF672, ZNF692, and PGBD2 (overlaps terminal region of 1q21.1–q44 duplication) |
| ii) CNVs found in multiple patients with CDH but at frequency higher than ethnically matched control populations | |||||||
| chr16:32403182–34759850 | 2,356,668 | 16p11.2 | Gain | 4 | 4 | 0.047* | TP53TG3E, TP53TG3B, TP53TG3F, TP53TG3C |
| chr4:11942–143314 | 131,372 | 4p16.3 | Loss | 5 | 1 | 0.001** | ZNF595 and ZNF718. Overlaps with Wolf–Hirschhorn critical region |
| chr5:12674767–12754177 | 79,410 | 5p15.2 | Loss | 3 | 3 | 0.036* | LINC01194 (noncoding) |
The reference genes that overlap with the significant CNVs are in bold. Significance level: *P < 0.05 and **P < 0.01.
Fig. 2.
Depiction of significant CNV regions. For CNVs, red bars represent loss and blue bars represent gain. (A) The 1q41 CNVs involving the HLX gene. Black boxes represent exons, white boxes are introns, and gray boxes are the untranslated regions. The numbers on the top of the genes indicate chromosomal coordinates (hg19/GRCH37). Blue lines indicate the relative positions of the duplications in our five patients with CDH. The 3′ end of the noncoding RNA gene, HLX-AS1, overlaps with the 1q41 duplication as shown. The open arrow indicates the 5′ of this gene is beyond the range of this figure. (B) The 17q12 deletions from two patients (dark red bars) in our study as well as three patients (light red bars) from previous studies (21, 30). The x axis indicates the genomic location, and the y axis indicates the log2 ratio from the array. The dots indicate probe intensities in this region of the array. LHX1, ACACA, and HNF1B (in bold) are candidate CDH genes in this CNV. (C) The 1q44 deletions from three patients (dark red bars) in our study. Four published cases of deletions (blue bars) and duplications (light red bars) in the neighboring region are also indicated.
The custom array also detected eight chromosomal aberrations greater than 1 Mb in size in patients that were absent from controls. Each of these CNVs was present only in a single patient and thus did not reach statistical significance. However, we feel that they are likely to be pathogenic based on size alone (Table 2). Not surprisingly, these patients tended to have other malformations or medical problems in addition to CDH. All of these chromosomal abnormalities were confirmed by clinical karyotype, clinical microarray, digital droplet PCR (ddPCR), and/or read depth data from WES.
Table 2.
Specific singleton CNVs (>1 Mb)
| Genomic coordinates (hg19) | Loss/gain | Size, Mb | Patient ID | Phenotype | Confirmatory testing |
| chr4:43284–49244506 | Gain | 49.2 | C119 | Hypoplastic left heart syndrome, stillbirth | Karyotype: 47, XX, +der(4)t(4,5)(q21; q35) |
| chr4:52742992–70765489 | Gain | 18.2 | |||
| chr5:177047523–180699183 | Gain | 3.6 | |||
| chr11:65440233–114273551 | Gain | 48.8 | C146 | Multiple congenital anomalies, additional details unknown | Confirmed by ddPCR and read depth data from WES |
| chr18:52948800–77967778 | Loss | 25.0 | C5 | Morgagni hernia, cleft palate, brachydactyly, dysmorphisms, short stature, microcephaly, moderate developmental delay | Confirmed by ddPCR and karyotype: 46, XX, del(18)(q21.2) |
| chr3:66478–12304371 | Loss | 12.2 | C86 | Seizures, partial agenesis of corpus callosum, dysmorphic facial features | Confirmed by ddPCR and clinical aCGH: del 3p26.3-p25.2 |
| chr15:93825922–102465355 | Loss | 8.6 | C148 | Multiple congenital anomalies, additional details unknown | Karyotype: 46, XX, der(15)t(1;15)(q44; q26.3)mat |
| chr16:14714766–18844674 | Gain | 4.1 | C158 | No phenotypic data available | Confirmed by ddPCR and read depth data from WES |
To assess the performance of the customized array, we tested all 6 significantly associated CNVs from Table 1, 4 large singleton CNVs from Table 2, and 14 other randomly chosen CNVs for validation using ddPCR; all 6 significant CNVs and 4 large CNVs were validated (SI Appendix, Fig. S3). In total, ∼87.5% (21/24) of the chosen CNVs were successfully validated. The CNVs that failed ddPCR were relatively small deletions or duplications with a limited number (7, 5) of probes, which is low coverage compared with most other candidate regions on the array. Overall, our results indicate that CNVs determined by ddPCR were highly correlated with the CNV data obtained by the custom array.
Including the six statistically significant CNVs (Table 1), the estimated frequency of likely causative CNVs in this cohort is 19/196 (9.7%). If we also include large (>1 Mb) CNVs that are present in only one patient but are suspected to be pathogenic based on their size alone (Table 2), then this frequency is 25/196 (13%). This frequency is slightly higher than reported in previous studies (21), although this may be at least partially explained by the higher resolution over CDH candidate regions compared with conventional aCGH.
Analysis of de Novo CNVs in Patients with CDH.
To detect de novo CNVs, we also used the custom array to study parental samples from a subset of patients, providing data on a total of 37 proband-parent trios. Among the de novo CNVs detected were several of statistically significant CNVs, including a deletion at 1q44 and a duplication at 16p11.2 (SI Appendix, Table S4). In addition, a 48.8-Mb duplication at 11q13.2–q23.2 and a 12.3-Mb deletion at 3p26.3–p25.2 (Table 2) were validated as de novo CNVs by ddPCR in this study. The observation that some of these CDH-associated CNVs are de novo further supports their pathogenicity. We detected several other de novo CNVs, each present in one patient and absent in controls, although these were small and in gene-poor regions so their clinical significance is unclear. Several other CNVs that appeared de novo in probands with CDH were also found in many control samples, suggesting that these may be at regions of high mutation rate (i.e., hot spots for CNV formation).
Pathway Analysis and Functional Interaction Network Study.
To understand better the biological functions of the genes within significantly CDH-associated CNVs, we performed Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on 41 unique RefSeq genes within the six CNVs significantly associated with CDH (Table 1). Key functions of these genes included DNA binding/gene transcription regulation (e.g., AATF, HLX, HNF1B, LHX1, TADA2A, and ZNF718), embryonic organ development (e.g., HLX, TADA2A, HNF1B, and LHX1), and metabolic pathways (e.g., ACACA and PIGW) (SI Appendix, Table S5).
In addition, we examined the direct protein–protein interactions and indirect functional associations involving these 41 genes by STRING analysis (34), as well as protein–protein interactions using InWeb (35). We identified two networks using STRING analysis (SI Appendix, Fig. S4), one consisting of 13 genes also identified by the pathway analysis and the second including the LHX1 and HNF1B genes. Eleven genes have transcription regulatory function but do not have known interactions with any other genes within the cluster (data not shown). Protein–protein interaction network analysis using InWeb (35) identified two candidate genes from the 17q12 region that interact with other known CDH genes or pathways: LHX1 (which interacts with the known CDH genes GLI2/3) and ACACA (which interacts with the cohesin complex, which includes the causative genes for the CDH-associated Cornelia de Lange syndrome).
Integration with Gene Expression Datasets and WES Data.
We integrated these results with gene expression datasets from the developing mouse diaphragm and lung (11, 36). This approach has been highly successful in the identification of CDH-causing genes (10, 11). Of the protein-coding genes within the significant CNV regions, several are highly expressed in the developing murine diaphragm as well as in murine lungs during critical stages of development (Table 3).
Table 3.
Selected prioritized candidate genes from CDH-associated CNV regions
| Region | Gene | Embryonic diaphragm expression | Lung expression (peak stage) | Constraint (ExAC pLI) | No. sequence variants (in 275 probands)* |
| dup 1q41 | HLX | High | Alveolar | 0.56 | 1 |
| del 17q12 | ACACA | Low | Embryonic/alveolar | 1.00 | 0 |
| DUSP14 | High | Alveolar | 0.77 | 0 | |
| GGNBP2 | High | — | 1.00 | 1 | |
| HNF1B | Low | Saccular/alveolar | 1.00 | 0 | |
| LHX1 | Low | — | 0.29 | 0 | |
| SYNRG | Low | Alveolar | 0.97 | 2 |
Diaphragm expression indicates whether a gene is expressed above (“high”) or below (“low”) the median normalized hybridization intensities for intronic probes based on expression microarrays from the embryonic mouse diaphragm (11). Lung expression indicates the peak developmental stage of expression levels in mouse lungs, and “—” indicates this gene is not highly expressed in lung (36).
Number of sequence variants in each gene that are rare (<0.1% in control populations) and predicted pathogenic (by SIFT, PolyPhen) in a cohort of 275 patients with CDH studied with WES.
We also examined WES data on a cohort of 275 patients with CDH (8) to identify sequence variants in genes within recurrent CNVs (Table 3). Eleven of the protein-coding genes, and one noncoding gene (ZNF595) harbor rare (<0.1% allele frequency in control populations), predicted pathogenic variants (including frameshift, stop gain, essential splice site variants, as well as missense variants predicted to be pathogenic by multiple in silico algorithms) (Table 3 and SI Appendix, Table S6), providing additional support that these genes may function independently as CDH risk genes. The 17q12 deletion region contains several highly constrained genes predicted to be intolerant of haploinsufficiency (ExAc pLi > 0.9). Among these constrained genes, sequence variants were also identified in GGNBP2 and SYNRG (Table 3).
Discussion
In this study, we have developed a high-resolution customized CNV array targeting genomic regions that contain known or candidate CDH genes, and have identified six recurrent CNVs that are significantly enriched in patients with CDH compared with ethnically matched controls. This customized array was chosen to provide improved coverage of CDH candidate regions to overcome the relatively low resolution of commercially available genome-wide aCGH platforms, allowing for identification of small CNVs that would have been missed on a standard aCGH (such as the recurrent 1.6-kb duplication at the HLX locus). This customized array also provided a cost-effective approach that allowed direct case control comparisons using the same platform to validate with statistical significance of the contribution of these genomic regions to CDH. By studying 196 patients with CDH and 987 normal controls, this is the largest patient cohort study evaluating genomic structural abnormalities associated with CDH undertaken to date. The significant CNVs identified in this study validate several CDH genes and genomic regions and identify additional candidate genes and pathways that contribute to the pathogenicity of CDH.
This study establishes 17q12 as a significant CNV associated with CDH, providing additional evidence that this is a true chromosomal hot spot for CDH. The 17q12 deletions from our study have more than 80% overlap with deletions reported by previous studies (21, 30, 37). By comparing these cases, we were able to narrow the minimum critical region to 1.22 Mb (Fig. 2B), containing nine protein-coding genes (LHX1, AATF, ACACA, C17orf78, TADA2A, DUSP14, SYNRG, DDX52, and HNF1B). Lim Homeobox 1 (LHX1) was initially suggested to be associated with CDH based on gene enrichment analysis (21). Our protein network analyses show multiple interactions between genes within this deletion hot spot as well as between these and other known CDH genes and pathways. For example, LHX1 may interact with HNF1 homeobox B (HNF1B) (SI Appendix, Fig. S4). HFN1B is involved in the WNT signaling pathway (38), which is important in mesodermal differentiation, a key component of proper diaphragm formation (39). Both LHX1 and HNF1B are homeobox genes that play multiple critical roles in organ development (40–42). The network analyses support a model in which the genes within the 17q12 hot spot may function together and/or in cooperation with other CDH genes and pathways in the orchestration of diaphragm and/or lung development.
The most common CDH-associated CNV in this study was a recurrent duplication in the HLX gene found in five patients but no controls, or ∼2.5% of this cohort. HLX is a member of the homeobox family of transcription factors that is located within the chromosome 1q41–1q42 region, a deletion hot spot for CDH. There are multiple lines of evidence suggesting that HLX is an important CDH candidate gene, including developmental expression patterns (11, 36) and bioinformatics functional association analyses (43). In addition, HLX homozygous null mice have diaphragmatic defects (28, 29, 44). HLX is a putative target for NR2F2 (COUP-TFII) (ENCODE), a transcriptional repressor of the retinoic acid signaling pathway (SI Appendix, Fig. S5), and an established CDH risk gene in both humans and mouse models (13, 45). We, and others, have shown previously that sequence variants in the HLX gene are present at low frequency in patients with CDH (8, 44), but this study demonstrates that duplications of this gene may be a common mutation mechanism important to the etiology of CDH. The mechanism by which recurrent HLX duplications may result in CDH is still unclear, although we hypothesize that the partial gene duplication may disrupt the coding region and/or splicing. There is also an antisense transcript in this region (HLX-AS1) that may have regulatory functions. Further experiments will be necessary to delineate the functional consequence of these duplications on HLX and HLX-AS1 expression and to assess for perturbations of relevant developmental pathways including retinoic acid signaling (SI Appendix, Fig. S5).
The remaining four statistically significant CDH-associated CNVs from this study identify several other novel candidate genes including ZNF672, ZNF692, PGBD2, TP53TG3, and ZNF595. There is very little information known about the function or developmental expression patterns of these genes in the embryonic diaphragm or lung, although ZNF672, ZNF692, PGBD2, and ZNF595 are expressed in adult skeletal muscle and lung (GTex) (33). Further studies will be necessary to understand if and how they play a role in the pathogenesis of CDH.
The results of this study support the hypothesis that CNVs make a significant contribution to the pathogenesis of diaphragmatic defects and provide a model that may be applied to other structural birth defects. Future analyses of CNVs in CDH, either through application of this customized targeted array or through other platforms such as whole-genome sequencing, will be applied to additional CDH cohorts to validate the CNVs identified in this manuscript and to identify other novel CNVs. Also, we propose that functional studies on the CDH candidate genes prioritized through this CNV analysis will provide additional insight into the pathogenesis of CDH and identify pathways for the future development of targeted therapeutics for this severe and often lethal birth defect.
Materials and Methods
Patient and Control Sample Selection.
In total, 196 patient samples and 109 parental samples were collected as part of the study “Gene Mutation and Rescue in Congenital Diaphragmatic Hernia” and were recruited at Massachusetts General Hospital and Boston Children’s Hospital, and via outside referrals from other clinicians or family support groups. Informed consent, blood, and tissue samples were obtained according to Partners Human Research Committee and Boston Children’s Hospital clinical investigation standards (Protocol 2000P000372 and 05-07-105R, respectively). Each individual underwent a detailed phenotypic analysis including review of the medical record, family history (46), and, whenever possible, physical examination by the study geneticists.
The 987 population control samples were chosen from the phase 3 DNA samples of the 1000 Genomes Project (27). Samples were selected as ethnically matched to the patient samples based on PCA of variants in already available whole-exome and/or whole-genome data. Concordant SNPs present in both groups were selected using the selectvariant function from GATK (47) and converted into binary file by Plink (48). PCA weights were computed by Plink and GCTA software (49).The first 20 principal components from all samples were generated and plotted in an R program (50) (see Results for more details). Population control samples were purchased from the Coriell Institute for Medical Research.
Customized aCGH Design, Hybridization, and Data Analysis.
The customized aCGH includes 140 target regions containing known or candidate genes for CDH (Fig. 1A and SI Appendix, Table S6). The regions targeted included the following: (i) regions of previously published recurrent chromosomal deletions or duplications in two or more patients with CDH, usually detected by lower resolution platforms (19, 21, 22, 24, 51–55); (ii) genes identified through human genomic studies on CDH and monogenic syndromic disorders associated with CDH (4, 6–9); (iii) genomic regions from preliminary studies from our laboratory; (iv) genes identified as causative of diaphragm defects and/or lung hypoplasia in mouse models; and (v) candidate genes prioritized from analyses of gene expression in normal mouse embryonic diaphragms (11) and protein–protein interaction analyses with known CDH genes and pathways (8).
The customized 60K Agilent aCGH platform was designed to obtain optimal coverage and yield high-quality data. The probe density and distribution varied in accordance with the target region size (Fig. 1B and SI Appendix, Supplemental Materials and Methods). The array design was tested for reproducibility and quality before use. Protocols for aCGH hybridization and data analysis are available in SI Appendix, Supplemental Materials and Methods.
CNV Validation.
To assess the performance of the customized array, we applied the ddPCR technology (SI Appendix, Supplemental Materials and Methods). Several CNVs were also confirmed by analyzing read depth data from WES using the exome hidden Markov model (56). All validated CNV calls using this method had a quality control score >90.
Supplementary Material
Acknowledgments
We thank the patients and their families for participating in this study, as well as the physicians at MassGeneral Hospital for Children and Boston Children’s Hospital for their continued support: T. Buchmiller, J. Zalieckas, C. C. Chen, D. Doody, S. J. Fishman, A. Goldstein, L. Holmes, T. Jaksic, R. Jennings, C. Kelleher, D. Lawlor, C. W. Lillehei, P. Masiakos, D. P. Mooney, K. Papadakis, R. Pieretti, M. Puder, D. P. Ryan, R. C. Shamberger, C. Smithers, J. Vacanti, and C. Weldon. We also thank Ms. Jane Cha for improvements on the graphs and for discussions of the manuscript. Funding was provided by National Institute of Child Health and Human Development/NIH (https://www.nichd.nih.gov/) Grant P01HD068250. C.L. is a distinguished Ewha Womans University Professor supported in part by Ewha Womans University Research Grant 2016/7.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap (accession nos. phs000783.v1.p1 and phs000783.v2.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714885115/-/DCSupplemental.
References
- 1.Torfs CP, Curry CJR, Bateson TF, Honoré LH. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- 2.Skari H, Bjornland K, Haugen G, Egeland T, Emblem R. Congenital diaphragmatic hernia: A meta-analysis of mortality factors. J Pediatr Surg. 2000;35:1187–1197. doi: 10.1053/jpsu.2000.8725. [DOI] [PubMed] [Google Scholar]
- 3.Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C:158–171. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pober BR. Genetic aspects of human congenital diaphragmatic hernia. Clin Genet. 2008;74:1–15. doi: 10.1111/j.1399-0004.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohseni-Bod H, Bohn D. Pulmonary hypertension in congenital diaphragmatic hernia. Semin Pediatr Surg. 2007;16:126–133. doi: 10.1053/j.sempedsurg.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Kantarci S, Donahoe PK. Congenital diaphragmatic hernia (CDH) etiology as revealed by pathway genetics. Am J Med Genet C Semin Med Genet. 2007;145C:217–226. doi: 10.1002/ajmg.c.30132. [DOI] [PubMed] [Google Scholar]
- 7.Yu L, et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2013;132:285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longoni M, et al. Molecular pathogenesis of congenital diaphragmatic hernia revealed by exome sequencing, developmental data, and bioinformatics. Proc Natl Acad Sci USA. 2014;111:12450–12455. doi: 10.1073/pnas.1412509111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, et al. Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J Med Genet. 2014;51:197–202. doi: 10.1136/jmedgenet-2013-101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longoni M, et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet. 2017;136:679–691. doi: 10.1007/s00439-017-1774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell MK, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci USA. 2012;109:2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coles GL, Ackerman KG. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2013;110:E1898–E1905. doi: 10.1073/pnas.1222797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.High FA, et al. De novo frameshift mutation in COUP-TFII (NR2F2) in human congenital diaphragmatic hernia. Am J Med Genet A. 2016;170:2457–2461. doi: 10.1002/ajmg.a.37830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39(7 Suppl):S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 15.Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craddock N, et al. Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 18.Klaassens M, et al. Congenital diaphragmatic hernia associated with duplication of 11q23-qter. Am J Med Genet A. 2006;140:1580–1586. doi: 10.1002/ajmg.a.31321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wat MJ, et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2010;149A:1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veenma DCM, de Klein A, Tibboel D. Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr Pulmonol. 2012;47:534–545. doi: 10.1002/ppul.22553. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, et al. De novo copy number variants are associated with congenital diaphragmatic hernia. J Med Genet. 2012;49:650–659. doi: 10.1136/jmedgenet-2012-101135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longoni M, et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A. 2012;158A:3148–3158. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borys D, Taxy JB. Congenital diaphragmatic hernia and chromosomal anomalies: Autopsy study. Pediatr Dev Pathol. 2004;7:35–38. doi: 10.1007/s10024-003-2133-7. [DOI] [PubMed] [Google Scholar]
- 24.Klaassens M, et al. Congenital diaphragmatic hernia and chromosome 15q26: Determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet. 2005;76:877–882. doi: 10.1086/429842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srisupundit K, et al. Targeted array comparative genomic hybridisation (array CGH) identifies genomic imbalances associated with isolated congenital diaphragmatic hernia (CDH) Prenat Diagn. 2010;30:1198–1206. doi: 10.1002/pd.2651. [DOI] [PubMed] [Google Scholar]
- 26.Slavotinek AM, et al. Array comparative genomic hybridization in patients with congenital diaphragmatic hernia: Mapping of four CDH-critical regions and sequencing of candidate genes at 15q26.1–15q26.2. Eur J Hum Genet. 2006;14:999–1008. doi: 10.1038/sj.ejhg.5201652. [DOI] [PubMed] [Google Scholar]
- 27.Consortium 1000 Genomes Project Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hentsch B, et al. Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev. 1996;10:70–79. doi: 10.1101/gad.10.1.70. [DOI] [PubMed] [Google Scholar]
- 29.Lints TJ, Hartley L, Parsons LM, Harvey RP. Mesoderm-specific expression of the divergent homeobox gene Hlx during murine embryogenesis. Dev Dyn. 1996;205:457–470. doi: 10.1002/(SICI)1097-0177(199604)205:4<457::AID-AJA9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Hendrix NW, Clemens M, Canavan TP, Surti U, Rajkovic A. Prenatally diagnosed 17q12 microdeletion syndrome with a novel association with congenital diaphragmatic hernia. Fetal Diagn Ther. 2012;31:129–133. doi: 10.1159/000332968. [DOI] [PubMed] [Google Scholar]
- 31.Pober BR, et al. Infants with Bochdalek diaphragmatic hernia: Sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am J Med Genet A. 2005;138A:81–88. doi: 10.1002/ajmg.a.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casaccia G, Mobili L, Braguglia A, Santoro F, Bagolan P. Distal 4p microdeletion in a case of Wolf-Hirschhorn syndrome with congenital diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol. 2006;76:210–213. doi: 10.1002/bdra.20235. [DOI] [PubMed] [Google Scholar]
- 33.GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14:61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beauchemin KJ, et al. Temporal dynamics of the developing lung transcriptome in three common inbred strains of laboratory mice reveals multiple stages of postnatal alveolar development. PeerJ. 2016;4:e2318. doi: 10.7717/peerj.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goumy C, et al. Congenital diaphragmatic hernia may be associated with 17q12 microdeletion syndrome. Am J Med Genet A. 2015;167A:250–253. doi: 10.1002/ajmg.a.36840. [DOI] [PubMed] [Google Scholar]
- 38.Welters HJ, Oknianska A, Erdmann KS, Ryffel GU, Morgan NG. The protein tyrosine phosphatase-BL, modulates pancreatic β-cell proliferation by interaction with the Wnt signalling pathway. J Endocrinol. 2008;197:543–552. doi: 10.1677/JOE-07-0262. [DOI] [PubMed] [Google Scholar]
- 39.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C-C, Orvis GD, Kwan KM, Behringer RR. Lhx1 is required in Müllerian duct epithelium for uterine development. Dev Biol. 2014;389:124–136. doi: 10.1016/j.ydbio.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 42.Desgrange A, et al. HNF1B controls epithelial organization and cell polarity during ureteric bud branching and collecting duct morphogenesis. Development. 2017;144:4704–4719. doi: 10.1242/dev.154336. [DOI] [PubMed] [Google Scholar]
- 43.Rouillard AD, et al. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slavotinek AM, et al. Sequence variants in the HLX gene at chromosome 1q41–1q42 in patients with diaphragmatic hernia. Clin Genet. 2009;75:429–439. doi: 10.1111/j.1399-0004.2009.01182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You L-R, et al. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2005;102:16351–16356. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackerman KG, et al. Congenital diaphragmatic defects: Proposal for a new classification based on observations in 234 patients. Pediatr Dev Pathol. 2012;15:265–274. doi: 10.2350/11-05-1041-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team 2017 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at https://www.r-project.org/. Accessed August 1, 2017.
- 51.Keitges EA, et al. Prenatal diagnosis of two fetuses with deletions of 8p23.1, critical region for congenital diaphragmatic hernia and heart defects. Am J Med Genet A. 2013;161A:1755–1758. doi: 10.1002/ajmg.a.35965. [DOI] [PubMed] [Google Scholar]
- 52.Kantarci S, et al. Findings from aCGH in patients with congenital diaphragmatic hernia (CDH): A possible locus for Fryns syndrome. Am J Med Genet A. 2006;140:17–23. doi: 10.1002/ajmg.a.31025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott DA, et al. Genome-wide oligonucleotide-based array comparative genome hybridization analysis of non-isolated congenital diaphragmatic hernia. Hum Mol Genet. 2007;16:424–430. doi: 10.1093/hmg/ddl475. [DOI] [PubMed] [Google Scholar]
- 54.Brady PD, et al. Identification of dosage-sensitive genes in fetuses referred with severe isolated congenital diaphragmatic hernia. Prenat Diagn. 2013;33:1283–1292. doi: 10.1002/pd.4244. [DOI] [PubMed] [Google Scholar]
- 55.Wat MJ, et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J Med Genet. 2011;48:299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.