Significance
Store-operated calcium entry (SOCE) is a widespread, essential signaling mechanism for cellular functions in both invertebrates and vertebrates and is controlled by two membrane proteins, STIM in the endoplasmic reticulum (ER) and Orai, in the plasma membrane (PM). How these proteins residing in two different compartments have evolved to interact with each other has not been elucidated. We show that Caenorhabditis elegans has a distinct mechanism of SOCE in which the 2–3 loop is regulated by STIM1 and the N and C termini of Orai1 by intramolecular interaction, differing from the previously reported mechanism of human SOCE. Therefore our studies suggest that, while the STIM–Orai interaction has been conserved from invertebrates to mammals, the gating mechanism for Orai has evolved considerably.
Keywords: Ca2+ signaling, STIM1, Orai1, SOCE, Caenorhabditis elegans
Abstract
Store-operated calcium entry (SOCE), an important mechanism of Ca2+ signaling in a wide range of cell types, is mediated by stromal interaction molecule (STIM), which senses the depletion of endoplasmic reticulum Ca2+ stores and binds and activates Orai channels in the plasma membrane. This inside-out mechanism of Ca2+ signaling raises an interesting question about the evolution of SOCE: How did these two proteins existing in different cellular compartments evolve to interact with each other? We investigated the gating mechanism of Caenorhabditis elegans Orai channels. Our analysis revealed a mechanism of Orai gating by STIM binding to the intracellular 2–3 loop of Orai in C. elegans that is radically different from Orai gating by STIM binding to the N and C termini of Orai in mammals. In addition, we found that the conserved hydrophobic amino acids in the 2–3 loop of Orai1 are important for the oligomerization and gating of channels and are regulated via an intramolecular interaction mechanism mediated by the N and C termini of Orai1. This study identifies a previously unknown SOCE mechanism in C. elegans and suggests that, while the STIM–Orai interaction is conserved between invertebrates and mammals, the gating mechanism for Orai channels differs considerably.
Ca2+ signaling is one of the most evolutionarily conserved and ubiquitous signaling mechanisms (1). A diverse array of calcium-signaling pathways regulates numerous cellular processes including secretion, muscle contraction, cellular motility, cell growth, and gene expression. The intracellular Ca2+ concentration is controlled by several calcium entry and extrusion proteins that are localized to the plasma membrane (PM) and intracellular organelles. Store-operated Ca2+ entry (SOCE) in the PM has recently been shown to serve as a crucial intracellular Ca2+ entry mechanism mediating cellular processes such as Ca2+ homeostasis and cell differentiation. SOCE is activated by the depletion of endoplasmic reticulum (ER) Ca2+ stores (2–4). The calcium release-activated calcium (CRAC) channel, which exhibits a high Ca2+ selectivity and low unitary conductance, is the most extensively studied store-operated calcium channel and the most evolutionarily conserved one between Caenorhabditis elegans and mammals (5–9).
It is thought that SOCE arose at an early stage during animal evolution, enabling diverse conserved physiological functions to be performed, but it remains unclear whether the molecular mechanisms that underlie SOCE are conserved among distant taxonomic clades.
In mammals, the identification of the two key mediators of SOCE, stromal interaction molecule (STIM) and Orai, has dramatically advanced our understanding of how SOCE is activated and inactivated. Stromal interaction molecule 1 (STIM1) is a single-transmembrane Ca2+ sensor dispersed in the ER (10–12). Orai1 is a PM Ca2+ channel with four transmembrane domains that comprises the pore subunit of SOCE (7, 13, 14). The molecular mechanisms by which ER Ca2+ depletion activates the CRAC channel have been well demonstrated in recent years. When ER Ca2+ stores are depleted by different cellular stimuli, STIM1 begins to form ordered oligomers, exposing its channel-activation domain (CAD, also known as “SOAR,” “OASF,” and “Ccb9”) (15–18) and moving to puncta closely localized to ER–PM junctions where it binds and activates Orai1. During this process, Orai1 also undergoes redistribution from a diffuse PM localization to colocalization in puncta with STIM1 upon STIM1 activation (5, 19–21). The Orai1 complex forms a hexameric structure comprising a trimer of dimers in which the C termini of the Orai1 dimers interact with each other in an antiparallel direction (22–25).
In C. elegans, mammalian STIM1 and Orai1 orthologs, namely C.STIM1 and C.Orai1, respectively, have been identified, and their expression and function have been explored in various tissues including neurons, gonadal sheath cells, intestines, and spermathecae (26, 27). The molecular mechanisms and biophysical properties of C.STIM1 and C.Orai1 are thought to be essential for intracellular Ca2+ signaling and the maintenance of ER Ca2+ stores, similar to the roles of STIM1 and Orai1 identified in mammalian SOCE. Nevertheless, some differences have been reported between C.STIM1 and mammalian STIM1 (28). For example, in contrast to human STIM1, which appears to be diffusely distributed in the ER before calcium store depletion, preassembled C.STIM1 shows a punctate distribution close to the PM before calcium store depletion (26, 27). In addition, C.STIM1 does not contain a polybasic sequence motif at its C terminus (CT); in mammals, this sequence contributes to the PM localization of STIM1 by promoting its interaction with phosphoinositides. These studies raise the question of whether the mechanisms regulating STIM1 function have been preserved during evolution, despite the high importance and conservation of SOCE in both C. elegans and humans.
In this study, we report a gating mechanism of C. elegans Orai1 channels that differs from the previously reported mechanism of mammalian SOCE. We found that the gating of Orai1 channels by STIM binding to the intracellular 2–3 loop (23L) in C. elegans is quite different from Orai gating by STIM binding to the CT of Orai in mammals. Moreover, the conserved hydrophobic amino acids in the intracellular 23L of C.Orai1 are critical for the oligomerization and gating of channels via an intramolecular interaction mechanism mediated by the N and C termini of C. elegans Orai1. This study identifies a previously unknown SOCE mechanism in C. elegans, indicating that the gating mechanisms underlying SOCE have diverged between invertebrates and mammals during evolution. Furthermore, the results provide a better understanding of the molecular mechanism of SOCE, which could be targeted for developing peptide or chemical regulators of SOCE in invertebrates.
Results
C. elegans STIM1 Activates C.Orai1 but Not Human Orai1.
We first examined the functional cross-reactivity between the vertebrate and invertebrate STIM and Orai proteins by investigating whether C.STIM1 could activate human Orai1 (hereafter, H.Orai1). HEK293 cells transiently expressing C.STIM1 and C.Orai1 generated an increase in the concentration of intracellular Ca2+ after the treatment of the cells with 1 μM thapsigargin (TG) to deplete internal ER Ca2+ stores (Fig. 1 A and B). However, HEK293 cells expressing C.STIM1 and H.Orai1 showed no store-operated Ca2+ influx, indicating that C.STIM1 cannot cross-interact with human Orai channels. The Ca2+ rise induced by C.STIM1 may reflect the influx of Ca2+ from sources other than PM C.Orai1 channels in our heterologous system. We thus went on to test directly for C.Orai1 channel activation by conducting whole-cell patch-clamp recordings from HEK293 cells transiently expressing C.STIM1 and either C.Orai1 or H.Orai1 and measuring CRAC currents (ICRAC). The treatment of transfected cells with EGTA to chelate calcium resulted in CRAC currents only in cells expressing C.STIM1 and C.Orai1 but not in those expressing C.STIM1 and H.Orai1. Additionally, in C.STIM1- and C.Orai1-expressing cells, the generated Ca2+ current was increased by the application of 10 mM Ca2+ and was blocked by 10 mM LaCl2, while no increased current was observed in cells expressing C.STIM1 and H.Orai1 (Fig. 1 C and D). Furthermore, human STIM1 (H.STIM1) could not induce an increase in the intracellular concentration of Ca2+ when coexpressed with C.Orai1 (Fig. S1 A–D).
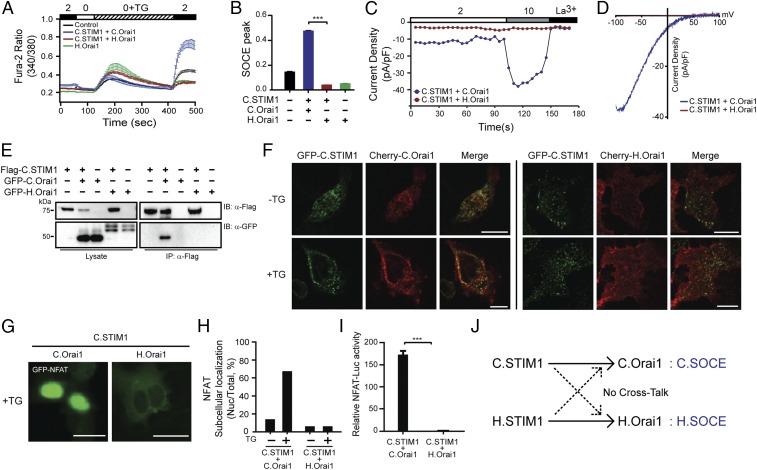
Fig. 1.
C. elegans has a distinct mode of SOCE. (A) Fura-2 Ca2+ measurements in HEK293 cells expressing C.STIM1 with either C.Orai1 (blue) or H.Orai1 (red). (B) Comparison between the SOCE peaks shown in A; ***P < 0.001, unpaired Student’s t test; n > 25. (C) Time course of ICRAC development in HEK293 cells after whole-cell break-in. (D) Current–voltage (I–V) relationships from the cells shown in C at peak store-operated calcium currents. (E) Western blots of whole-cell lysates (Left) or immunoprecipitates (Right) from cells expressing C.STIM1 and either C.Orai1 or H.Orai1. (F) Localization of GFP-C.STIM1 with Cherry-C.Orai1 (Left) or Cherry-H.Orai1 (Right). (G) Subcellular distribution of GFP-NFAT in HEK293 cells expressing C.STIM1 with C.Orai1 or H.Orai1 after store depletion. (H) Histograms of the mean abundance of nuclear-translocated NFAT under the indicated conditions. (I) NFAT-dependent luciferase activity in cells expressing C.STIM1 and either C.Orai1 or H.Orai1. Cells were treated with TG and with PMA. (J) A schematic drawing of the relationship between C.SOCE and H.SOCE. (All scale bars, 10 μm.) The results are typical of at least three independent experiments.
The observation that C.STIM1 activates only C.Orai1 and not H.Orai1 led us to test whether this difference was due to differences in the interactions between C.STIM1 and the two Orai1 channels. We thus labeled STIM1 and Orai1 with FLAG and GFP tags, respectively, and expressed the tagged proteins in HEK293T cells. The immunoprecipitation of FLAG-tagged C.STIM1 resulted in the coimmunoprecipitation of GFP-C.Orai1 in cells expressing both proteins but not in cells coexpressing C.STIM1 and H.Orai1 (Fig. 1E). This observation indicated that C.STIM1 preferentially binds to C.Orai1 and that this interaction may be mediated by a distinct domain in C.Orai1 with either a sequence or a structural conformation different from that in H.Orai1.
To further validate the differential binding of the C. elegans and human SOCE (H.SOCE) complex components in intact cells, we investigated the puncta formation of STIM1 and Orai1 following the application of TG. As previously reported, GFP-C.STIM1 showed prepuncta formation but did not colocalize to the PM with Cherry-C.Orai1 before store depletion (27–30). Upon TG treatment, we observed clear PM-localized puncta of GFP-C.STIM1 and Cherry-C.Orai1 (Fig. 1F, Left). In contrast, GFP-C.STIM1 did not form puncta with Cherry-H.Orai1 even after store depletion with TG (Fig. 1F, Right), and H.STIM1 did not form a complex with C.Orai1 (Fig. S1E), indicating that C.STIM1 preferentially binds to C.Orai1 in puncta, activating C. elegans SOCE (C.SOCE) after store depletion via a distinct mechanism.
To test whether C.SOCE induced by C.STIM1 and C.Orai1 expression in heterologous cells can induce downstream Ca2+ signaling, we introduced GFP-NFAT, which is a well-known SOCE-responsive transcription factor, into HEK293 cells. Before store depletion, 40% of C.STIM1- and C.Orai1-coexpressing cells showed nuclear translocation of GFP-NFAT, compared with 5% of cells expressing C.STIM1 and H.Orai1 (Fig. 1 G and H). This prenuclear-localized GFP-NFAT observed in HEK293 cells expressing C.STIM1 and C.Orai1 is likely caused by a leaking Ca2+ influx from overexpressed C.SOCE proteins. The treatment with TG increased the fraction of C.STIM1- and C.Orai1-expressing cells to 70%, indicating that C.STIM1 can induce downstream Ca2+ signaling in complex with C.Orai1 but not with H.Orai1. To further interrogate whether the nuclear translocation of NFAT induced by C.SOCE could induce the expression of NFAT target genes including IL2, we introduced an NFAT transcriptional reporter plasmid in which the IL2 promoter was fused to firefly luciferase into HEK293T cells. Treating cells with TG and a protein kinase C activator (phorbol 12-myristate 13-acetate, PMA) caused enhanced firefly luciferase activity driven by nuclear-translocated NFAT in cells expressing both C.STIM1 and C.Orai1 but not in cells expressing C.STIM1 and H.Orai1 (Fig. 1I), H.STIM1 and C.Orai1 (Fig. S1 F–H), C.STIM1 alone, or C.Orai1 alone (Fig. S1I).
Collectively, these results demonstrated that the C.SOCE complex can generate intracellular Ca2+ elevations and activate downstream Ca2+ signaling pathways. The inability of C.STIM1 and C.Orai1 to interact with members of the H.SOCE complex gave rise to the hypothesis that the binding or activation mechanisms of C.SOCE evolved differently from those of mammalian SOCE (Fig. 1J).
C.STIM1 Contains a Minimal Activation Domain That Resembles the H.STIM1 CAD.
We previously reported that during the activation of mammalian SOCE STIM1 binds and activates Orai1 directly through the CAD (15). To determine the mechanism by which C.STIM1 selectively activates C.Orai1, we first mapped and isolated the C.STIM1 CAD (C.CAD, amino acids 286–388) (Fig. S2A) corresponding to the H.STIM1 CAD region and examined whether C.CAD alone could activate C.Orai1. In the absence of store depletion by TG, C.CAD caused increased Ca2+ elevations when expressed with C.Orai1 in HEK293 cells but failed to result in calcium influx when expressed with H.Orai1 (Fig. 2A).
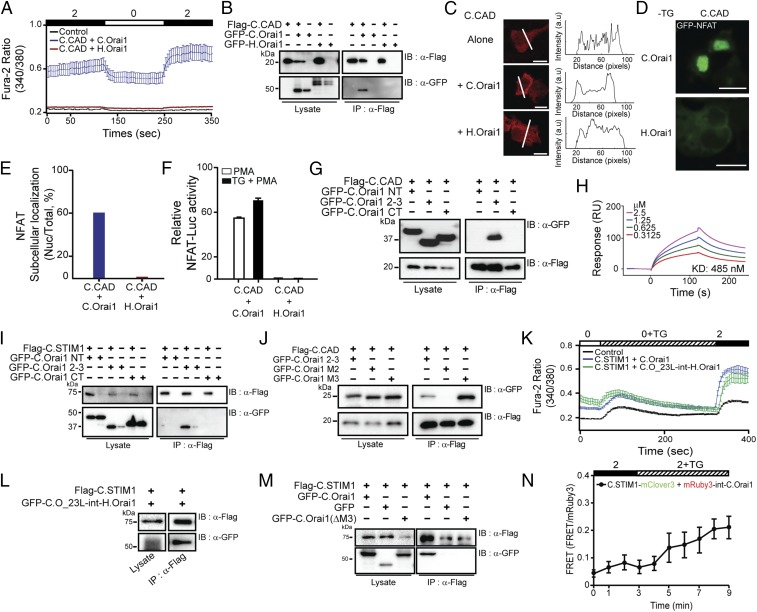
Fig. 2.
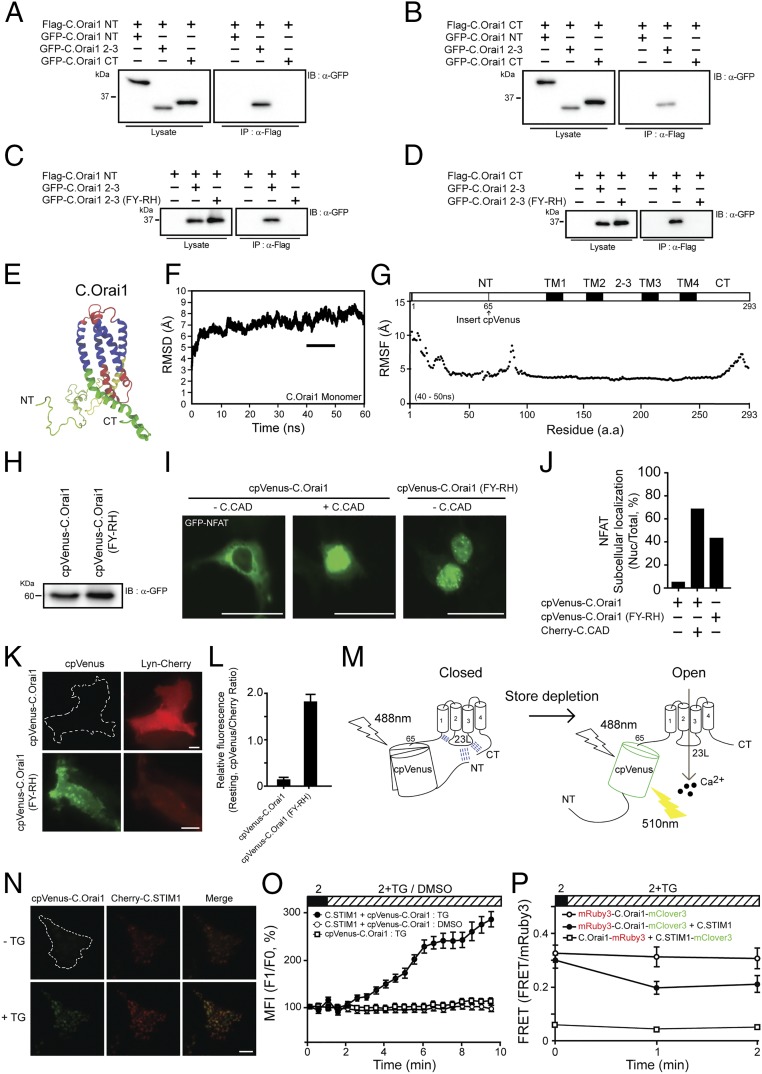
The CRAC CAD of STIM1 binds to the C.Orai1 23L. (A) Fura-2 Ca2+ measurements in HEK293 cells expressing YFP (black), C.CAD and C.Orai1 (blue), or C.CAD and H.Orai1 (red). The C.CAD-induced constitutive Ca2+ influx was measured with 2 mM Ca2+ in the presence of C.Orai1 without store depletion. (B) FLAG-tagged C.CAD coimmunoprecipitated with GFP-C.Orai1 but not with H.Orai1. (C) Localization of Cherry-C.CAD alone (Top), with C.Orai1 (Middle), and with H.Orai1 (Bottom). Cherry-C.CAD accumulates at the PM when coexpressed with C.Orai1. (D) C.CAD induced the nuclear translocation of NFAT when it was expressed with C.Orai1 but not when it was expressed with H.Orai1. (E) Histograms showing the mean abundance of nuclear-translocated NFAT under the indicated conditions. (F) NFAT-dependent luciferase activity in cells expressing C.CAD and either C.Orai1 or H.Orai1. Cells were treated with PMA or with TG + PMA as indicated. (G and I) FLAG-tagged C.CAD and C.STIM1 coimmunoprecipitated with the GFP-23L of C.Orai1 but not with the NT or CT of C.Orai1. (H) Quantitative SPR measurements of the immobilized GST-23L interacting with various concentrations of C.CAD. The Kd value was determined to be 485 nM. (J) Immunoprecipitation of C.CAD with the M3 domains of C.Orai1. (K) Fura-2 Ca2+ measurements in HEK293 cells expressing C.STIM1 with either C.Orai1 (blue) or C.O_23L-int-H.Orai1 (green). (L) FLAG-tagged C.STIM1 coimmunoprecipitated with GFP-C.O_23L-int-H.Orai1. (M) FLAG-tagged C.STIM1 coimmunoprecipitated with GFP-C.Orai1 but not with GFP-C.Orai1 (ΔM3). (N) FRET experiments of C.STIM1-mClover3 with mRuby3-int-C.Orai1. (All scale bars, 10 μm.) The results are typical of at least three independent experiments.
The observed selective activation of C.Orai1 by C.CAD may be due to altered binding between C.CAD and the two Orai1 channels. We thus examined whether C.CAD binds only to C.Orai1 and not to H.Orai1. To test this idea, we labeled the C.CAD and Orai1 channels with FLAG and GFP tags, respectively, and subsequently expressed the labeled proteins in HEK293T cells. The immunoprecipitation of FLAG-tagged C.CAD resulted in the coimmunoprecipitation of GFP-C.Orai1 only in cells expressing both proteins but not in cells expressing H.Orai1 (Fig. 2B), indicating that C.STIM1-CAD is a potent activator of C.Orai1 channels through direct binding (Fig. 2H).
To further confirm the suggested preferential binding between the C.CAD and C.Orai1 channels, we performed colocalization experiments. CAD is known to localize to the PM upon binding to Orai1 when they are coexpressed in HEK293 cells (Fig. S2C). We first expressed Cherry-C.CAD alone or with either C.Orai1 or H.Orai1 in HEK293 cells. Like human CAD (H.CAD), Cherry-C.CAD was localized diffusely in the cytoplasm in the absence of C.Orai1 either without (Fig. 2C, Top) or with (Fig. 2C, Bottom) the presence of H.Orai1, but it moved to the PM when C.Orai1 was expressed (Fig. 2C, Middle). This result indicated that C.Orai1 is a binding target of C.CAD and that they form a complex upon binding that localizes to the PM.
We next performed various experiments to validate the activity of C.CAD. An elevated Ca2+ influx driven by C.CAD and C.Orai1 in HEK293 cells resulted in the nuclear localization of GFP-labeled NFAT (Fig. 2 D and E) and an enhanced transcriptional activity of NFAT (Fig. 2F) before store depletion. In contrast, we failed to see either nuclear translocation or enhanced transcriptional activity of NFAT in cells expressing either C.CAD or H.Orai1 (Fig. 2 D–F), further supporting the idea that C.CAD preferentially binds and activates C.Orai1, comparable to full-length C.STIM1.
C.CAD Binds to the 23L of C.Orai1.
Our results thus far set forth the idea that the selective activation of C.Orai1 channels by C.CAD may be due to differences in the sequence or structural conformation of C.Orai1. We therefore mapped C.CAD target domains within C.Orai1 using immunoprecipitation. We first isolated three cytosolic domains, the N terminus (NT, amino acids 1–121), the 23L (amino acids 170–199), and the CT (amino acids 249–293) of C.Orai1. Then we transiently coexpressed GFP-tagged versions of these intracellular domains of C.Orai1 in HEK293T cells together with FLAG-tagged C.CAD (Fig. 2G) or C.STIM1 (Fig. 2I). To our surprise, both FLAG-tagged C.STIM1 and C.CAD were coimmunoprecipitated with the 23L of C.Orai1 and not with the NT or CT (Fig. 2 G and I).
To determine whether the binding of C.STIM1 and C.CAD to 23L is direct or indirect, we performed a surface plasmon resonance (SPR) experiment using purified GST-tagged 23L and histidine-tagged C.CAD. We found that C.CAD binds directly to the 23L of C.Orai1 (Fig. 2H). To further map the interaction domain in more detail, we subdivided the 23L of C.Orai1 into two subdomains (Fig. S2G), the M2 peptide (the N-terminal half, amino acids 170–183) and the M3 peptide (the C-terminal half, amino acids 184–199), and tested which domain coimmunoprecipitated with FLAG-tagged C.CAD. Our findings demonstrated that C.CAD interacted with the M3 peptide of the 23L of C.Orai1 (Fig. 2J).
We next performed various experiments to confirm that the 23L of C.Orai1 is the binding and activation domain of C.STIM1 in full-length proteins. First, we generated a chimeric H.Orai1 channel (C.O_23L-int-H.Orai1) replacing the 23L of H.Orai1 with the 23L of C.Orai1 and checked whether this grafting could recruit C.STIM1 into the 23L of the chimeric H.Orai1. When expressed with C.STIM1 in HEK293 cells, chimeric H.Orai1 harboring the 23L of C.Orai1 caused increased Ca2+ elevations after store depletion, as in C.Orai1 (Fig. 2K). The immunoprecipitation of FLAG-tagged C.STIM1 resulted in the coimmunoprecipitation of GFP-chimeric Orai1 (Fig. 2L). Second, we generated the C.STIM1-binding domain, M3, and deleted Orai1 (C.Orai1-ΔM3). The immunoprecipitation of FLAG-tagged C.STIM1 failed to result in the coimmunoprecipitation of GFP-C.Orai1-ΔM3 (Fig. 2M), did not increase Ca2+ elevation, and failed to form the puncta with C.STIM1 in the cells expressing C.Orai1-ΔM3 (Fig. S4 B and C). Last, we generated the mRuby3-int-C.Orai1 by inserting mRuby3 between amino acids 178 and 179 in the 23L of C.Orai1 as a FRET probe for the interaction of the 23L and STIM1. HEK293 cells expressing C.STIM1 and mRuby3-int-C.Orai1 generated increased Ca2+ elevations after store depletion, as in C.Orai1 (Fig. S2I). As expected, we could detect a change in FRET signal when C.STIM1-mClover3 came close to mRuby3 in the 23L of C.Orai1 after store depletion (Fig. 2N). These results in the 23L grafting channel, the deleted STIM1-binding domain channel, and a FRET probe channel confirmed that the 23L of C.Orai1 is the bona fide binding domain of C.STIM1 in intact cells.
To date, there has been no report of STIM1 binding to the 23L of Orai1 to activate SOCE. As it is known that H.STIM1 binds Orai1 channels at the NT and CT, this finding presents a previously unrecognized mechanism of SOCE activation in C. elegans.
Mutating Conserved Amino Acids Phe and Tyr in the 23L Produces a Constitutively Active Orai1 Channel.
Several lines of evidence from our experiments indicated that the 23L of C.Orai1 is the critical domain for C.STIM1 binding and function. To validate which amino acids are important for SOCE activation, we aligned and compared the amino acid sequences of the 23L domains of various Orai1 isoforms from representative species ranging from invertebrates (C. elegans) to vertebrates (human) (Fig. 3A). We found that two hydrophobic amino acids (F192 and Y193 [FY] or WY) within the 23L of C.Orai1 were conserved among invertebrates, while the corresponding amino acids in vertebrate Orai1 channels had changed to positively charged residues (R170 and H171) in H.Orai1 (Fig. 3A). This observation suggests that the hydrophobic amino acids Phe and Tyr may play a conserved role in the functions of Orai1 in C. elegans and other invertebrates and may explain the evolutionarily divergent regulatory mechanisms of SOCE in C. elegans and humans.
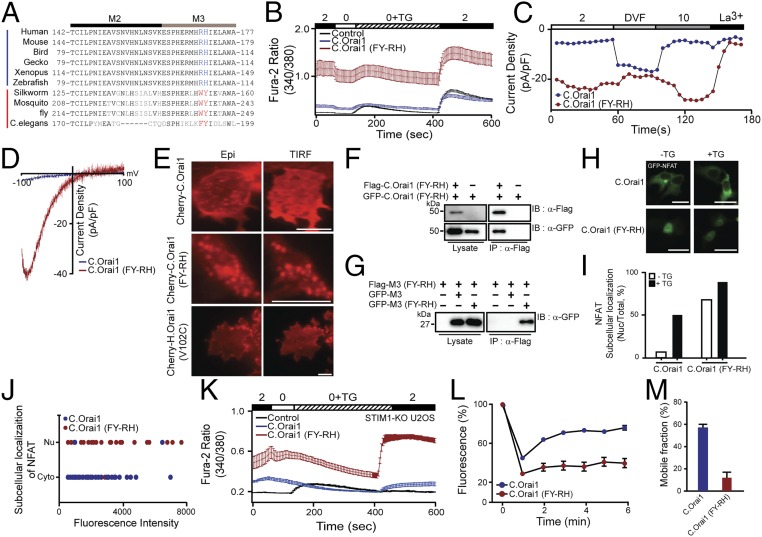
Fig. 3.
The FY-RH mutation in the 23L produces a constitutively active C.Orai1 channel. (A) Sequence alignment of the 23L of Orai1 channels from C. elegans to human, showing identical or conserved amino acids (black) and nonconserved amino acids (gray). The conserved amino acids essential for this study in invertebrates and vertebrates are shown in bold red and blue fonts, respectively. (B) Fura-2 Ca2+ measurements in HEK293 cells expressing YFP (black), C.Orai1 (blue), or C.Orai1 (FY-RH) (red). (C) Whole-cell ICRAC measurements on HEK293 cells expressing C.Orai1 (blue) or C.Orai1 (FY-RH) (red). The time course of ICRAC development was measured after whole-cell break-in. (D) I–V relationships from the cells expressing C.Orai1 (blue) or C.Orai1 (FY-RH) (red) shown in C at peak store-operated currents in 10 mM Ca2+ Ringer’s solution. (E) Cellular localization of Cherry-C.Orai1, C.Orai1 (FY-RH), or H.Orai1 (V102C). C.Orai1 (FY-RH) alone made prepuncta without C.STIM1. (F) FLAG-tagged C.Orai1 (FY-RH) coimmunoprecipitated with GFP-tagged C.Orai1 (FY-RH) but not with WT C.Orai1. (G) The FLAG-tagged M3 peptide (FY-RH) coimmunoprecipitated with the GFP-tagged M3 peptide (FY-RH) but not the WT M3 peptide of C.Orai1. (H) Cellular distribution of GFP-NFAT in HEK293 cells expressing C.Orai1 or C.Orai1 (FY-RH) alone. GFP-NFAT translocated into the nucleus in the cells expressing C.Orai1 (FY-RH) even before store depletion by TG. (I) Histograms showing the mean abundance of nuclear-translocated NFAT under the indicated conditions. (J) A graph showing the relationship between the proportion of nuclear-translocated NFAT and the expression level of C.Orai1 in the absence of C.STIM1. The x axis of the scatter plot represents the expression level of C.Orai1 (mean fluorescence intensity), and the y axis represents the subcellular localization of NFAT in either the cytosol (Cyto) or the nucleus (Nu). (K) Fura-2 Ca2+ measurements in STIM1-KO U2OS cells expressing YFP (control, black), C.Orai1 (blue), or C.Orai1 (FY-RH) (red). (L) Time course of FRAP from single cells expressing C.Orai1 or C.Orai1 (FY-RH). (M) Mobile fractions of GFP-C.Orai1 or C.Orai1 (FY-RH) alone; data are shown as means ± SE; n > 13. (All scale bars, 10 μm.) The results are typical of at least three independent experiments.
To examine whether this change from hydrophobic to positively charged residues (from FY to RH) affects the function of C.Orai1, we first measured intracellular Ca2+ rises in cells with mutant C.Orai1 (FY-RH). We expressed either WT or mutant C.Orai1 (FY-RH) in HEK293 cells without expressing C.STIM1. To our surprise, the cells expressing mutant C.Orai1 (FY-RH) showed constitutive Ca2+ influx even before store depletion, whereas WT C.Orai1 alone did not affect C.SOCE compared with the endogenous HEK293 SOCE (Fig. 3B). We checked whether FY-deleted C.Orai1 (C.Orai1-ΔFY) could be activated by C.STIM1. The cells expressing C.Orai1-ΔFY with C.STIM1 did not show any elevated SOCE compared with the control (Fig. S4A), indicating that the conserved FY amino acids are essential for the function of C.Orai1. We next tested whether this Ca2+ rise was dependent on C.Orai1 (FY-RH) using the whole-cell patch-clamp technique and confirmed that a constitutive Ca2+ current was evoked by C.Orai1 (FY-RH) without C.STIM1 expression or store depletion (Fig. 3 C and D).
We thus explored how C.Orai1 (FY-RH) induces intracellular Ca2+ rises even in the absence of C.STIM1. First, we expressed either Cherry-C.Orai1 or Cherry-C.Orai1 (FY-RH) alone in HEK293 cells and examined the expression pattern of C.Orai1 channels. While WT C.Orai1 was distributed evenly in the PM, C.Orai1 (FY-RH) appeared to form puncta before store depletion and in the absence of C.STIM1 (Fig. 3E). This observation was unexpected, because all published data showed the diffusely distributed pattern of Orai1 channels in PM with full store or in the absence of STIM1. We therefore compared the puncta formation of C.Orai1 (FY-RH) with that in another constitutively active Orai1 mutant, V102C. We expressed Cherry-tagged constitutively active V102C Orai1 channels alone in HEK293 cells and confirmed the diffusely distributed expression pattern of V102C Orai1 in the absence of STIM1 (Fig. 3E, Bottom), as previously reported (31).
In addition, the immunoprecipitation of FLAG-C.Orai1 (FY-RH) showed that it could induce oligomerization with GFP-C.Orai1 (FY-RH) (Fig. 3F). Furthermore, M3 peptide (FY-RH) resulted in the coimmunoprecipitation of the M3 peptide (FY-RH) but not the WT M3 peptide (Fig. 3G). These results suggested that C.Orai1 (FY-RH) induced cytosolic Ca2+ rises directly by forming C.Orai1 (FY-RH) puncta through 23L oligomerization. To confirm this idea, we next examined whether C.Orai1 (FY-RH) alone could activate the SOCE-dependent downstream signaling cascade leading to the nuclear localization and transcriptional activation of cytosolic NFAT. When we introduced GFP-NFAT into cells expressing Cherry-C.Orai1 (FY-RH), 70% of cells showed the nuclear translocation of NFAT before store depletion, and this proportion was further increased to 90% with store depletion by TG (Fig. 3 H and I). To validate the effect of C.Orai1 (FY-RH), we compared the expression level of Cherry-C.Orai1 (WT) or C.Orai1 (FY-RH) to the proportion of nuclear-localized NFAT. While the cells with a higher expression of WT C.Orai1 alone did not show the nuclear translocation of NFAT, most of the cells expressing C.Orai1 (FY-RH) showed the nuclear translocation of NFAT regardless of their level of C.Orai1 expression (Fig. 3J). However, to eliminate the effect of endogenous STIM proteins in HEK293 cells on the function of C.Orai1 proteins, we validated the constitutive function of C.Orai1 (FY-RH) in STIM1-KO U2OS cell lines (32). We measured intracellular Ca2+ rises in STIM1-KO U2OS cells with mutant C.Orai1 (FY-RH). The cells expressing mutant C.Orai1 (FY-RH) showed a constitutive Ca2+ influx even before store depletion and significantly increased SOCE after store depletion in the absence of endogenous STIM1, while the control cells without STIM1 expression did not show the developing SOCE after store depletion (Fig. 3K). These results indicated that C.Orai1 (FY-RH) is a potent and constitutively active channel subunit that can form the SOCE complex even in the absence of C.STIM1 and is independent of endogenous STIM1.
Fluorescence recovery after photobleaching (FRAP) experiments were used to measure the diffusion rates of C.Orai1 and C.Orai1 (FY-RH) in the cell membranes of HEK293 cells. We expressed either WT C.Orai1 or C.Orai1 (FY-RH) and measured the diffusion coefficient (D) by conducting FRAP in HEK293 cells. Our FRAP measurements of GFP-C.Orai1 alone in HEK293 cells suggested that a fraction (56.5 ± 4.9%) of the GFP-labeled proteins were mobile in the membrane, demonstrating that C.Orai1 channels are freely diffusible and quite mobile (Fig. 3 L and M). However, in cells expressing GFP-C.Orai1 (FY-RH), the mobile fraction was dramatically reduced (11.8 ± 5.4%), and C.Orai1 (FY-RH) diffusion was slowed by a factor of five (Fig. 3 L and M). The significant slowing of C.Orai1 (FY-RH) diffusion is consistent with the formation of puncta and constitutively active channels.
C.STIM1 Binding Regulates Constitutively Permeant C.Orai1 (FY-RH).
Orai1 channels are highly Ca2+-selective and inward-rectifying channels. The pore-lining hydrophobic gate (F99 or V102) mutant Orai1 channels are constitutive ion-permeant channels without STIM1 and showed less Ca2+ selectivity than in the presence of STIM1 (31).
To investigate whether C.STIM1 regulated the inward currents and gating properties of C.Orai1 (FY-RH) channels, we expressed C.STIM1 and C.Orai1 (FY-RH) in HEK293 cells and measured Ca2+ currents by whole-cell patch clamping. To our surprise, a constitutive Ca2+ current evoked by C.Orai1 (FY-RH) in the absence of C.STIM1 (Fig. 3C, red trace) was reduced by C.STIM1 expression (Fig. 4A, red trace). When we compared the expression level of C.Orai1 (FY-RH) proteins in the presence and absence of C.STIM1, there was no difference in the expression level of C.Orai1 (Fig. 4A, Right). Additionally, there was no difference in the Ca2+ gating and permeability of C.Orai1 (FY-RH) when it was coexpressed with C.STIM1 (Fig. 4 A and B), indicating that a constitutive Ca2+ current evoked by C.Orai1 (FY-RH) can be regulated by C.STIM1 or C.CAD. Collectively, these observations led us to interrogate the binding and localization of both proteins. To examine whether C.STIM1 binds to C.Orai1 (FY-RH), we first explored the interaction between the two proteins by total internal reflection fluorescence (TIRF) microscopy. HEK293 cells expressing Cherry-C.STIM1 and GFP-C.Orai1 (FY-RH) showed prepuncta formation before store depletion (Fig. 4C), indicating that constitutively active C.Orai1 (FY-RH) could be directly regulated by C.STIM1 binding. Next, we expressed FLAG-tagged C.CAD with GFP-Orai1 or Orai1 (FY-RH), and the immunoprecipitation of the FLAG-tagged C.CAD resulted in the coimmunoprecipitation of both GFP-C.Orai1 and C.Orai1 (FY-RH) (Fig. 4D). Together, these results imply that the binding of C.STIM1-CAD to the 23L of C.Orai1 (FY-RH) changes the conformational alignment of ion-selective filters in the C.SOCE complex, leading to a closed or intermediate state. Furthermore, these findings confirmed that the binding and activation of the C.SOCE complex is a dissociable and separate step in C. elegans SOCE, much as in vertebrate SOCE.
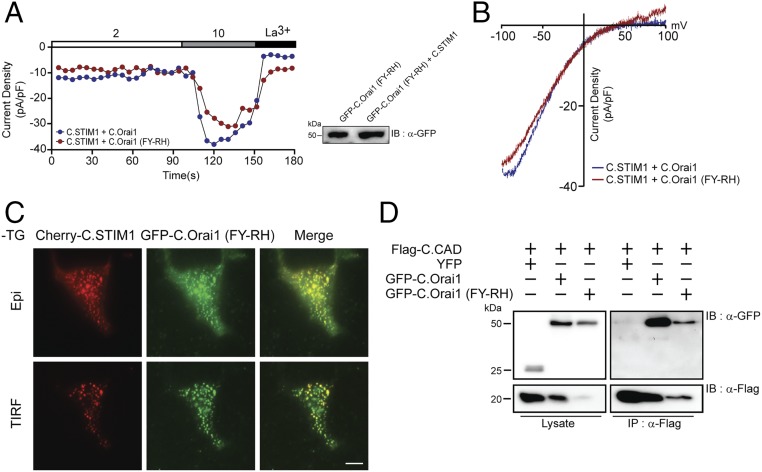
Fig. 4.
Gating of the C.Orai1 (FY-RH) channel is regulated by C.STIM1 binding. (A) The time course of ICRAC development in cells expressing C.STIM1 with either C.Orai1 (blue) or C.Orai1 (FY-RH) (red) after whole-cell break-in. The Ca2+ permeability of the C.Orai1 (FY-RH) channel is regulated by C.STIM1 binding. (B) I–V relationships in the cells expressing C.STIM1 and either C.Orai1 (blue) or C.Orai1 (FY-RH) (red) shown in A at peak store-operated currents in 10 mM Ca2+ Ringer’s solution. (C) Cellular localization of Cherry-C.STIM1 and GFP-C.Orai1 (FY-RH) before store depletion. C.Orai1 (FY-RH) formed prepuncta with C.STIM1. All images of the cell footprint were taken with epifluorescence and TIRF microscopes. (Scale bar, 10 μm.) (D) FLAG-tagged C.CAD coimmunoprecipitated with both GFP-tagged C.Orai1 and C.Orai1 (FY-RH) showing that C.Orai1 (FY-RH) can be regulated by C.STIM1 or C.CAD binding.
The Gating Mode of Orai1 Is Orchestrated by the Interaction of Intracellular Domains in C. elegans.
Unlike the proposed mechanism of mammalian SOCE, gated by STIM1 binding to the CT of Orai1 (33), C.SOCE appears to be regulated by a different and unexplored mechanism involving the 23L of C.Orai1. However, the molecular mechanism by which the 23L of C.Orai1 orchestrates C.SOCE remains elusive. To explore the differences between these channels, we first asked whether the 23L of C.Orai1 interacts with other cytosolic domains of C.Orai1. Immunoprecipitation results showed that the NT and CT of C.Orai1 were able to bind to WT 23L but not to the 23L of C.Orai1 (FY-RH) (Fig. 5 A–D), indicating that the closed channel was bound by other cytosolic domains but the mutated, constitutively open channel was unbound. This binding preference led us to propose an intramolecular interaction model in which the accessibility of the 23L is regulated by the other cytosolic domains of C.Orai1, and this may contribute to the gating and activation mechanism of C.SOCE.
Fig. 5.
C.SOCE is orchestrated through the interaction between the NT and the 23L of C.Orai1. (A and B) FLAG-tagged C.Orai1 NT and CT coimmunoprecipitated with the GFP-23L. (C and D) FLAG-tagged NT and CT coimmunoprecipitated with the GFP-23L but not with the 23L (FY-RH). (E–G) The predicted 3D structure (E), rmsd (F), and RMSF probability analysis (G) of the C.Orai1 monomer between 40–50 ns of MD simulation. (H) Western blots of whole-cell lysates from the cells expressing cpVenus-C.Orai1 and cpVenus-C.Orai1 (FY-RH). (I) Nuclear translocation of GFP-NFAT in cells expressing cpVenus-C.Orai1 alone (Left) or together with C.CAD (Middle) or cpVenus-C.Orai1 (FY-RH) alone (Right). (J) Histograms show the mean abundance of nuclear-translocated NFAT under the indicated conditions. (K) Representative images of cpVenus fluorescence in HEK293 cells expressing cpVenus-C.Orai1 (Upper) and cpVenus-C.Orai1 (FY-RH) (Lower). PM-localized Lyn-Cherry was visualized on the same focus plane. (L) Mean fluorescence intensity of cpVenus-C.Orai1 and cpVenus-C.Orai1 (FY-RH). (M) A schematic representation depicting the mechanism by which cpVenus fluorescence is altered by the conformational change of C.Orai1. (N) Representative images of cpVenus-C.Orai1 before and after store depletion. (O) The time course of the fluorescence intensity of cpVenus-C.Orai1 in cells expressing Cherry-C.STIM1. Store depletion by TG increases the fluorescent intensity of cpVenus-C.Orai1 only in the presence of C.STIM1. (P) FRET experiments of mRuby3-C.Orai1-mClover3 with or without C.STIM1 and C.Orai1-mRuby3 with C.STIM1-mClover3. (All scale bars, 10 μm.)
To understand the molecular gating mechanism of C.SOCE in detail, we assumed that C.Orai1 and C.Orai1 (FY-RH) had different conformational structures that could represent the closed and open states of C.Orai1. Therefore, we applied molecular dynamics (MD) simulations using models of the predicted C.Orai1 monomer block modified from the crystal structure of the Drosophila Orai1 [Protein Data Bank (PDB) ID code 4HKR]. The simulation system reached a steady state after 40 ns (Fig. 5 E and F). We measured and compared the structural fluctuation patterns for 10 ns (from 40 to 50 ns). The root-mean-squared fluctuation (RMSF) results showed that there were high fluctuations in the NT and CT but not in other domains of C.Orai1 (Fig. 5G). Furthermore, the RMSF plot of the NT showed that two regions were undergoing high fluctuations, namely the distal NT (amino acids 1–64) and the proximal NT (amino acids 65–121) (Fig. 5G). We designed a circularly permuted Venus (cpVenus) fluorescent protein that is sensitive to changes in pH or in protein conformation and generated cpVenus-C.Orai1 by inserting cpVenus at amino acid 65 between the two fluctuating portions in the NT of C.Orai1. We expected to detect a change in cpVenus fluorescence upon the conformational change between the open and closed states of Orai1 channels (34–36).
We first measured the expression levels of cpVenus-C.Orai1 and cpVenus-C.Orai1 (FY-RH) proteins by Western blot in HEK293 cells expressing one of these proteins. We observed similar expression levels of both proteins (Fig. 5H). We next checked the effect of the cpVenus insertion on the activity of these channels. Both cpVenus-C.Orai1 channels seemed to be fully functional. Expressing either cpVenus-C.Orai1 with C.CAD or cpVenus-C.Orai1 (FY-RH) alone resulted in an elevated abundance of nuclear-localized NFAT (Fig. 5 I and J), indicating that adding cpVenus in the NT of C.Orai1 did not change its conformational state.
We expressed either cpVenus-C.Orai1 or cpVenus-C.Orai1 (FY-RH) with Lyn-Cherry as a PM marker in HEK293 cells and measured the fluorescence intensity after TG treatment. Interestingly, cells expressing cpVenus-C.Orai1 showed weak fluorescence intensity regardless of store depletion. However, most cells expressing cpVenus-C.Orai1 (FY-RH) showed strong fluorescence intensity before store depletion (Fig. 5 K and L). This result led us to hypothesize that the difference between the fluorescence intensities of cpVenus-C.Orai1 and cpVenus-C.Orai1 (FY-RH) could arise from the various states of C.Orai1 (i.e., closed and open states) (Fig. 5M) and not from the expression level (Fig. 5H).
We next expressed cpVenus-C.Orai1 with Cherry-C.STIM1 in HEK293 cells and measured the fluorescence intensity after TG treatment using TIRF microscopy. To our surprise, cpVenus-C.Orai1 showed an increasing fluorescence intensity and formed puncta with Cherry-C.STIM1 soon after store depletion (Fig. 5 N and O). These results suggest that the NT of C.Orai1 changed its orientation from inward facing, close to the 23L, to outward facing when C.STIM1-CAD came into proximity as a result of store depletion and that this motion likely affected the cpVenus fluorescence. In contrast, the NT of constitutively active C.Orai1 (FY-RH) seems to be present in an outward-facing orientation, and therefore the fluorescence intensity was unchanged.
We measured the interaction of C.STIM1 and C.Orai1 and the intracellular domains of C.Orai1 using FRET. We first expressed mRuby3-C.Orai1-mClover3 with or without C.STIM1 in HEK293 cells and measured the fluorescence intensity after TG treatment using confocal microscopy. The cells expressing mRuby3-C.Orai1-mClover3 and C.STIM1 showed decreased FRET signal after TG treatment (Fig. 5P, closed circles). However, the cells without STIM1 expression did not show any change in FRET signal (Fig. 5P, open circles). These results indicated that both the NT and CT of C.Orai1 were close enough to show FRET signal at the resting state but then displaced each other soon after C.STIM1 binding to the 23L (Fig. 6H). However, the internal FRET signal of C.Orai1 was not changed in the absence of C.STIM1 (Fig. 5P, open circle). This conformational change of C.Orai1 was confirmed by the FRET measurement expressing C.Orai1-mRuby3 with C.STIM1-mClover3 in HEK293 cells. As we expected, the FRET signal was very low at the resting state and was not changed by TG treatment (Fig. 5P, open rectangle). This might be because, unlike the interaction between mammalian STIM1 and Orai1 proteins, the interaction between C.STIM1 and the 23L of C.Orai1 did not affect the relative FRET efficiency of the CT of either of the two proteins.
Fig. 6.
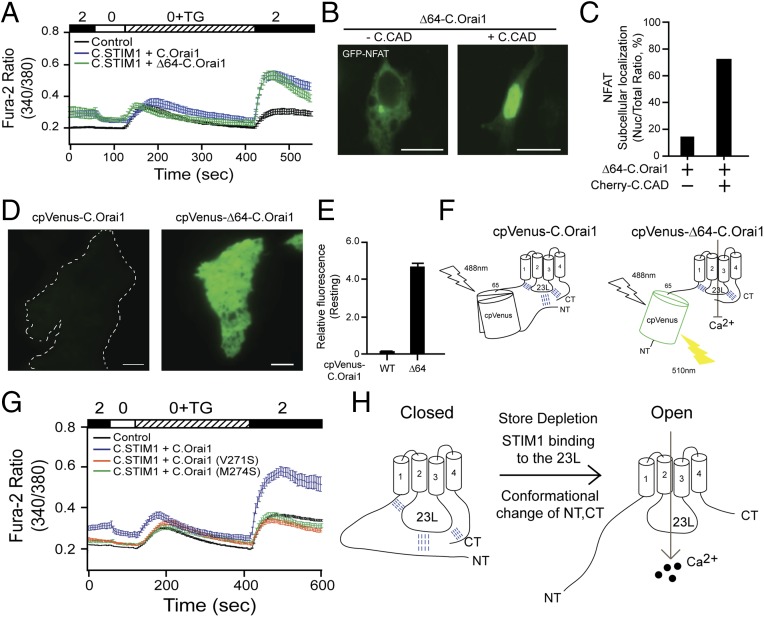
C.SOCE is regulated by the hydrophobic interaction between the NT and the 23L of C.Orai1. (A) Fura-2 Ca2+ measurements in HEK293 cells expressing YFP (black), C.STIM1 and C.Orai1 (blue), or C.STIM1 and Δ64-C.Orai1 (green). (B) Nuclear translocation of GFP-NFAT in cells expressing Δ64-C.Orai1 either alone (Left) or together with C.CAD (Right). (C) Histograms showing the mean abundance of nuclear-translocated NFAT under the indicated conditions. (D) Representative images of cpVenus fluorescence in HEK293 cells expressing cpVenus-C.Orai1 or cpVenus-Δ64-C.Orai1. (E) Fluorescence intensity measurements in cells expressing cpVenus-C.Orai1 or cpVenus-Δ64-C.Orai1. (F) A schematic depicting the mechanism by which the deletion of the distal NT of C.Orai1 recovers the fluorescence of cpVenus-Δ64-C.Orai1 but the proximal NT and CT still maintain the closed form of C.Orai1 (G) Fura-2 Ca2+ measurements in HEK293 cells expressing C.STIM1 with C.Orai1 (blue), C.Orai1 (V271S) (orange), or C.Orai1 (M274S) (green). (H) A schematic of the molecular gating mechanism of the C. elegans SOCE. (All scale bars, 10 μm.)
There have been many previous reports analyzing the bimolecular interaction of Orai1 and STIM1 using bimolecular fluorescence complementation (BiFC) or FRET. However, our study used circularly permuted fluorescent proteins to visualize the different conformational states or motions of the intracellular domains of the Orai1 channel during the gating process of Orai1 channels in vivo.
Both the NT and the CT of C.Orai1 Are Necessary for the Gating of C.Orai1.
The CT of C.Orai1 also seemed to be involved in the interaction with the 23L of C.Orai1 (Fig. 5B). However, we have not isolated the putative interacting domain in the CT, since, so far, none of mutations we have tried has produced functional channels.
We have shown that both the NT and the CT bind to the 23L at rest (Fig. 5 A–D) and induce the conformational change after the store depletion (Fig. 5M), and we also constructed NT- and/or CT-truncated C.Orai1 channels (ΔNT, ΔCT). However, these truncated C.Orai1 channels were not activated by C.STIM1 (Fig. S4A) and did not form the puncta with C.STIM1 (Fig. S4D), indicating that other cytosolic domains are also essential for the function of C.Orai1.
We next asked which portions of the C.Orai1 NT and CT are responsible for the conformational change related to the binding of the 23L. In parallel with the MD simulation showing high fluctuations in the NT and CT (Fig. 5 E–G), we made cpVenus-C.Orai1 channels in which 64 N-terminal amino acids (amino acids 1–64) were deleted (cpVenus-Δ64-C.Orai1) and measured intracellular Ca2+ increases in cells with Δ64-C.Orai1. We expressed either WT or Δ64-C.Orai1 in HEK293 cells with C.STIM1. As we expected, cells expressing either WT or Δ64-C.Orai1 showed a basal Ca2+ level before store depletion and an elevated Ca2+ influx after store depletion (Fig. 6A). We next checked the function of the cpVenus-Δ64-C.Orai1 channels by measuring the nuclear translocation of GFP-NFAT. We expressed cpVenus-Δ64-C.Orai1 with or without C.CAD in HEK293 cells. Like the cells expressing the full-length cpVenus-Orai1, those expressing cpVenus-Δ64-C.Orai1 alone did not show any NFAT nuclear translocation, but the cells expressing cpVenus-Δ64-C.Orai1 with C.CAD showed an elevated abundance of nuclear-localized NFAT (Fig. 6 B and C). We also measured the fluorescence intensity of cpVenus by expressing either cpVenus-C.Orai1 or cpVenus-Δ64-C.Orai1 alone in HEK293 cells by TIRF microscopy. To our surprise, cpVenus-Δ64-C.Orai1 showed a strong fluorescence intensity and was localized diffusely in the PM even before store depletion in the absence of C.STIM1 (Fig. 6 D and E), while full-length cpVenus-C.Orai1 did not show fluorescence at rest (Fig. 5K). These results indicate that the deletion of 64 distal N-terminal amino acids did not affect the function, that the closed conformation of Δ64-C.Orai1 was maintained by other intramolecular interactions between the 23L and the proximal NT and CT (Fig. 6F), and that its gating still required C.STIM1 binding to develop C.SOCE and was regulated by the other portions of the NT and CT. (Fig. 6H).
To check whether the CT is responsible for the conformational change related to the binding of the 23L and the gating of C.Orai1, we made several mutant C.Orai1 channels (V271 and M274, equivalent to residues L273 and L276 in H.Orai1). The immunoprecipitation of FLAG-tagged C.STIM1 resulted in the coimmunoprecipitation of all CT-mutant C.Orai1 channels (Fig. S4E) but failed to result in calcium influx when expressed with C.Orai1 (Fig. 6G), indicating that the gating of C.Orai1 still required the functional conformation of the CT and the displacement of NT and CT from the 23L by C.STIM1 binding or store depletion.
Taken together, these results implied that the NT and the CT of C.Orai1 bind to the 23L of the channel, forming a closed channel in the resting state, but it undergoes a conformational change into an open channel upon the binding of STIM1 to activate SOCE in C. elegans (Fig. 6H).
Discussion
The unexpected finding of distinct mechanisms of SOCE activation in mammals and the nematode C. elegans revealed that C.Orai1 has features that are entirely different from those of the well-studied mammalian Orai1 proteins. Several recent studies have established that the C-terminal domain of Orai1 is important for the STIM1 binding and channel gating (33, 37). Our results, however, showed a previously unreported view of Orai1 channel gating, demonstrating that C.Orai1 has a central domain within the 23L for STIM1 binding, channel gating, and oligomerization. Moreover, this work might, in theory, provide evidence that Orai channels have evolved with a shift of both the STIM-binding site and the channel dimerization domain as a component of SOCE from the internal 23L to the external NT and CT. This shift might allow Orai channels to be regulated more finely by multiple binding proteins with easier accessibility.
To distinguish the proposed functions of the 23L of C.Orai1, we have tried to abrogate the STIM1 binding by specific mutations and deletions without affecting C.Orai1 channel gating or oligomerization. We found that the STIM1-binding domain overlapped closely with the amino acids critical for channel gating and oligomerization (Fig. S2 G and H), because several mutations and deletions did not make Orai1 channels functional (Fig. S4A). However, it would be helpful in further studies to clarify which C.Orai1 channels are gated following C.STIM1 binding by determining which amino acids are essential for STIM1 binding and channel gating.
2-Aminoethocyldiphenyl borate (2-APB) has been shown to facilitate all mammalian Orai1, Orai2, and Orai3 channels at low micromolar concentrations and to inhibit Orai1 and Orai2, but not Orai3, at higher concentrations (38, 39). Therefore, we checked whether 2-APB affects the activity of C.Orai1 (Fig. S3A) and the cross-interactions between the C.STIM1 and H.Orai1 or between H.STIM1 and C.Orai1 (Fig. S3B) by Fura-2 Ca2+ imaging in HEK293 cells. The cells expressing C.STIM1 and C.Orai1 showed no significant effect and inhibited SOCE by 2-APB treatment (50 uM) compared with H.SOCE. This is consistent with or similar to the previously reported data (27). The cells expressing H.STIM1 and C.Orai1 showed similar elevated Ca2+ levels after store depletion compared with endogenous SOCE (control), while the cells expressing C.STIM1 and H.Orai1 did not show any SOCE due to the dominant inhibitory effect of H.Orai1 expression. In this condition, however, we could not see any increased cross-interaction between H.STIM1 and C.Orai1 induced by 2-APB (Fig. S3B).
Many groups have proposed several types of gates in ion channels, including the girdle model and electrostatic switch, which involve hydrophobic residues and polar residue salt bridges, respectively (40, 41). We hypothesize that the conserved hydrophobic amino acids within the 23L of C.Orai1 have a critical role in the gating mechanism of the C.Orai1 channel. A neutralizing mutation produced a constitutively active Orai1 channel in the absence of STIM1, implying that the process of opening and closing the pore might be regulated by the hydrophobic interaction providing a tapering path for ions at the resting state. However, the molecular mechanism by which the conformational change of the 23L propagates through the associated pore-lining helices to gate ions needs further study. Our results provide another regulation mechanism: that both the NT and CT of C.Orai1 are required and essential for channel gating through the intracellular interaction with the 23L of C.Orai1.
Detecting a conformational change of interest in a protein without perturbing the physiological environment in intact cells is an important process in research aimed at understanding the precise mechanisms underlying protein functions, such as interactions with other proteins and altered activation states. Several approaches, including FRET and BiFC, have been developed for monitoring changes in the conformation of proteins. However, detecting the changes in membrane-anchored ion channels in intact cells is a significant challenge due to their fast and small alternations between the open and closed states. We attempted to use FRET with double-labeled fluorescent proteins and BiFC with the split NT-half and CT-half of the Venus fluorescent protein at both ends of C.Orai1, but neither approach worked as we expected. However, adding cpVenus in the flexible region of the Orai1 NT (Fig. 5 H–O) or mRuby3 in the flexible region of the 23L (Fig. 2N) provided very informative insights regarding the different structural configurations of the Orai1 channel in the open and closed states. Other techniques, such as X-ray crystallography or NMR spectroscopy, will provide further evidence for the gating mechanism of C.Orai1 channels.
Based on our current findings, we propose a distinct gating mechanism of C. elegans store-operated calcium channels that has not previously been proposed. Our model hypothesizes that the C.Orai1 channel has two flexible configurations (open and closed) and that the binding of STIM1 to the 23L of C.Orai1 displaces both the NT and CT from the 23L and induces the Orai1 channel to switch conformation from the closed state to the open state.
Materials and Methods
HEK293 and HEK293T cells were cultured in DMEM supplemented with 10% FBS at 37 °C in 5% CO2. For patch clamping, Flp-In T-Rex stable HEK293 cells expressing GFP-myc-H.Orai1 and cherry-h.STIM1 (Invitrogen) were maintained with 50 μg/mL of hygromycin. For transient transfection, the cells were transfected at 70% confluency with 0.3–3 μg DNA using Lipofectamine 2000 (Life Technology) according to the manufacturer’s instructions. STIM1-KO U-2 OS cells were very kindly provided by Francisco Javier Martin-Romero (University of Extremadura, Badajoz, Spain) (32).
Additional information is available in SI Text.
Supplementary Material
Acknowledgments
We thank Drs. Anshul Rana (Stanford University), Amir Masoud Sadaghiani (Stanford University, now at Novartis Institutes), Georgia Panagiotakos (University of California, San Francisco), Won Do Heo (Korea Advanced Institute of Science and Technology), and Sergiu Pasca (Stanford University) for their careful and critical reading of the manuscript; Dr. Ricardo E. Dolmetsch (Stanford University, now at Novartis Institutes) for the initiation and development of our scientific projects; Dr. Francisco Javier Martin-Romero (University of Extremadura) for providing STIM1- and Orai1-KO cells (U2OS and PC3 cells); and Hyokeun Park (Hong Kong University of Science and Technology) for kindly providing mRuby3 and mClover3. This work was supported by National Research Foundation of Korea (NRF) Grant NRF-2015R1A1A1A05027490 (funded by the Korean Ministry of Education, Science, and Technology) (to C.Y.P.), by Korea Research Institute of Bioscience and Biotechnology Research Initiative Program KGM4251723, and by Basic Science Research Program Grant 2015R1D1A1A01059590 (funded by the Ministry of Education through the NRF) (to K.P.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714986115/-/DCSupplemental.
References
- 1.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Estevez AY, Roberts RK, Strange K. Identification of store-independent and store-operated Ca2+ conductances in Caenorhabditis elegans intestinal epithelial cells. J Gen Physiol. 2003;122:207–223. doi: 10.1085/jgp.200308804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parekh AB, Putney JW., Jr Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 4.Putney JW., Jr Muscarinic, alpha-adrenergic and peptide receptors regulate the same calcium influx sites in the parotid gland. J Physiol. 1977;268:139–149. doi: 10.1113/jphysiol.1977.sp011851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakriya M, Lewis RS. Store-operated calcium channels: Properties, functions and the search for a molecular mechanism. Advances in Molecular and Cell Biology. 2004;32:121–140. [Google Scholar]
- 7.Prakriya M, et al. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 8.Hoth M, Penner R. Calcium release-activated calcium current in rat mast cells. J Physiol. 1993;465:359–386. doi: 10.1113/jphysiol.1993.sp019681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoth M. Calcium and barium permeation through calcium release-activated calcium (CRAC) channels. Pflugers Arch. 1995;430:315–322. doi: 10.1007/BF00373905. [DOI] [PubMed] [Google Scholar]
- 10.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vig M, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeromin AV, et al. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muik M, et al. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: Local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, et al. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- 22.Stathopulos PB, et al. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4:2963. doi: 10.1038/ncomms3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothberg BS, Wang Y, Gill DL. Orai channel pore properties and gating by STIM: Implications from the Orai crystal structure. Sci Signal. 2013;6:pe9. doi: 10.1126/scisignal.2003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirado-Lee L, Yamashita M, Prakriya M. Conformational changes in the Orai1 C-terminus evoked by STIM1 binding. PLoS One. 2015;10:e0128622. doi: 10.1371/journal.pone.0128622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan X, et al. Function of a STIM1 homologue in C. elegans: Evidence that store-operated Ca2+ entry is not essential for oscillatory Ca2+ signaling and ER Ca2+ homeostasis. J Gen Physiol. 2006;128:443–459. doi: 10.1085/jgp.200609611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorin-Nebel C, Xing J, Yan X, Strange K. CRAC channel activity in C. elegans is mediated by Orai1 and STIM1 homologues and is essential for ovulation and fertility. J Physiol. 2007;580:67–85. doi: 10.1113/jphysiol.2006.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S, et al. Mechanism of different spatial distributions of Caenorhabditis elegans and human STIM1 at resting state. Cell Calcium. 2009;45:77–88. doi: 10.1016/j.ceca.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Jin H, Cai X, Li S, Shen Y. Structural and mechanistic insights into the activation of stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci USA. 2012;109:5657–5662. doi: 10.1073/pnas.1118947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strange K, Yan X, Lorin-Nebel C, Xing J. Physiological roles of STIM1 and Orai1 homologs and CRAC channels in the genetic model organism Caenorhabditis elegans. Cell Calcium. 2007;42:193–203. doi: 10.1016/j.ceca.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNally BA, Somasundaram A, Yamashita M, Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Guerrero AM, et al. Regulation of membrane ruffling by polarized STIM1 and ORAI1 in cortactin-rich domains. Sci Rep. 2017;7:383. doi: 10.1038/s41598-017-00331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, et al. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat Commun. 2016;7:13725. doi: 10.1038/ncomms13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Germond A, Fujita H, Ichimura T, Watanabe TM. Design and development of genetically encoded fluorescent sensors to monitor intracellular chemical and physical parameters. Biophys Rev. 2016;8:121–138. doi: 10.1007/s12551-016-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao BS, et al. A highly selective fluorescent probe for visualization of organic hydroperoxides in living cells. J Am Chem Soc. 2010;132:17065–17067. doi: 10.1021/ja1071114. [DOI] [PubMed] [Google Scholar]
- 36.Hsu S-TD, et al. Folding study of Venus reveals a strong ion dependence of its yellow fluorescence under mildly acidic conditions. J Biol Chem. 2010;285:4859–4869. doi: 10.1074/jbc.M109.000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palty R, Stanley C, Isacoff EY. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 2015;25:963–980. doi: 10.1038/cr.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peinelt C, Lis A, Beck A, Fleig A, Penner R. 2-Aminoethoxydiphenyl borate directly facilitates and indirectly inhibits STIM1-dependent gating of CRAC channels. J Physiol. 2008;586:3061–3073. doi: 10.1113/jphysiol.2008.151365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prakriya M, Lewis RS. Potentiation and inhibition of Ca(2+) release-activated Ca(2+) channels by 2-aminoethyldiphenyl borate (2-APB) occurs independently of IP(3) receptors. J Physiol. 2001;536:3–19. doi: 10.1111/j.1469-7793.2001.t01-1-00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat Chem Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 41.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 42.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribeiro JV, et al. QwikMD–Integrative molecular dynamics toolkit for novices and experts. Sci Rep. 2016;6:26536. doi: 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bajar BT, et al. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep. 2016;6:20889. doi: 10.1038/srep20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poteser M, et al. Live-cell imaging of ER-PM contact architecture by a novel TIRFM approach reveals extension of junctions in response to store-operated Ca2+-entry. Sci Rep. 2016;6:35656. doi: 10.1038/srep35656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.