Significance
The cuticle, mainly composed of chitin and cuticular proteins (CPs), is a multifunctional structure of arthropods. CPs usually account for >1% of the total insect proteins encoded in the genome. Why does an insect need so many different CPs? In this study, we use comprehensive large-scale technologies to study the full complement of CPs and their functions in the brown planthopper (BPH). A total of 32 of the 140 BPH CP genes are found to be essential for nymph/adult development, egg production, or embryo development; in addition, redundant and complementary functions of CPs are revealed.
Keywords: insect cuticular proteins, transcriptomic analysis, proteomic analysis, large-scale RNAi, brown planthopper
Abstract
Cuticle, mainly composed of chitin and cuticular proteins (CPs), is a multifunctional structure of arthropods. CPs usually account for >1% of the total insect proteins. Why does an insect encode so many different CP genes in the genome? In this study, we use comprehensive large-scale technologies to study the full complement of CPs (i.e., the CP-ome) of the brown planthopper (BPH), Nilaparvata lugens, a major rice plant pest. Eight CP families (CPR, CPF, TWDL, CPLCP, CPG, CPAP1, CPAP3, and CPAPn) including 140 proteins in BPH, in which CPAPn is a CP family that we discovered. The CPG family that was considered to be restricted to the Lepidoptera has also been identified in BPH. As reported here, CPLCP family members are characterized by three conserved sequence motifs. In addition, we identified a testis protein family with a peritrophin A domain that we named TPAP. We authenticated the real existence of 106 proteins among the 140 CPs. RNA interference (RNAi) experiments were conducted against 135 CP genes in early- and late-instar nymphs and newly emerged female adults, demonstrating that 32 CPs were essential for BPH normal development or egg production. Combined RNAi experiments suggested redundant and complementary functions of the large number of CPs. Transcriptomic data revealed that the CP genes were expressed in a tissue-specific manner, and there were four clusters of developmental expression patterns. This study gives a comprehensive understanding of the roles of CPs in an insect cuticle.
The cuticle is the outermost layer of an insect’s body. Toward the inside, it maintains the internal environment and supports the body shape. Toward the outside, it protects the organism against dehydration and invasion of pathogens and pesticides. The insect cuticle is made from three horizontal layers: the envelope, the epicuticle, and the procuticle (1). The procuticle, composed of the exocuticle and endocuticle, is the region of a chitin–protein matrix, the composition of which varies between body regions and species (2). Indeed, much of the cuticular diversity is due to the variation in protein composition entailing differences in molecular architecture of the procuticle (3).
Cuticular protein (CP) genes usually account for >1% of the total genes in an insect genome. Although hundreds of CPs belonging to 13 CP families have been reported in different insects (4–6), why an insect needs so many different CPs has not been comprehensively understood yet. The brown planthopper (BPH), Nilaparvata lugens (Hemiptera: Delphacidae), is one of the most destructive rice plant pests in the world. In this study, we thoroughly searched and identified all CPs of BPH at both genomic and proteomic levels, and most of the CPs were authenticated at the protein level. To better understand the CP function in hemimetabolous insects, we performed large-scale RNAi experiments and did temporal and spatial expression analyses on all of the CPs from BPH. This study provides comprehensive understanding into the roles of CPs in insect cuticle.
Results
BPH Has 140 CPs.
In total, 140 CPs were identified in BPH and classed into 8 CP families: CPR (CPs with the R&R consensus), 96 CPs (NlugCpr1-36: RR-1 subgroup; NlugCpr37-93: RR-2 subgroup; NlugCpr94-96: RR-3 subgroup); CPF (CPs with a 42–44 amino acid motif), 3 CPs; TWDL (Tweedle, CPs with four conserved regions), 3 CPs; CPLCP (CPs of low complexity with proline-rich), 6 CPs; CPG (CPs with glycine-rich), 4 CPs; CPAP1 (CPs analogous to peritrophins with one conserved peritrophin A domain), 17 CPs; CPAP3 (CPs analogous to peritrophins with three conserved peritrophin A domains), 8 CPs; and CPAPn (CPs analogous to peritrophins with no fixed number of conserved peritrophin A domains), 3 CPs; in which CPR is the biggest CP family. The findings of several CP families from BPH should be highlighted here. As reported, CPLCP family members are characterized by three conserved sequence motifs. The CPG family was considered to be restricted to the Lepidoptera and has also been identified in BPH. CPAPn is a CP family that we discovered. In addition, we identified a testis protein family that shares the same chitin-binding domain as CPAP1, CPAP3, and CPAPn that we named TPAP (testis proteins analogous to peritrophins). For detailed information about BPH CP families, see SI Appendix, section 1.
Proteomic Analysis by Ultra-Performance Liquid Chromatography-Tandem MS Authenticated the CPs.
To authenticate the N. lugens CPs that were conceptually translated from cDNA and genomic sequences, first, 135 of the above 140 N. lugens CP gene sequences were successfully verified and revised through PCR, RACE, and gene clone sequencing (Dataset S1). To further validate and authenticate the CPs of N. lugens at the protein level, proteins from four tissue samples were collected and digested with trypsin. The resulting peptides were analyzed by shotgun ultra-performance liquid chromatography (UPLC)-tandem MS (MS/MS). The four protein samples were cast first-instar nymph cuticles (CC1), cast third- to fifth-instar nymph cuticles (CC3–5), adult long wings (LW), and fifth-instar nymph abdominal cuticles (AC).
Only proteins with at least two peptide-spectrum matches and a score of >4.0 were considered reliable. Under these conditions, we identified a total of 519; 686; 1,341; and 710 proteins from CC1, CC3-5, LW, and AC protein samples, respectively (Dataset S2 A–D). It is possible that not all peptides could be detected because they were too long for detection or because cuticle components were cross-linked to each other (7). Different CPs might have different solubility (8), and sclerotization could also make some CPs unextractable (9).
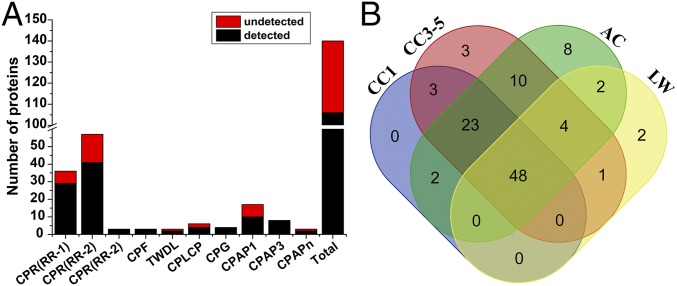
To confirm the existence of the 140 BPH putative CPs, manual annotation of the peptide sequences was carried out to verify or correct the predictions from the National Center for Biotechnology Information (NCBI). Together, in four MS experiments, 106 CPs (75.71%) were detected in at least one MS result, 93 CPs (66.43%) were detected in at least two MS results, and 48 CPs (34.29%) were detected in all four MS results (Fig. 1 and SI Appendix, Table S1). For each MS result, 76; 92; 57; and 97 CPs were identified in CC1, CC3-5, LW, and AC samples, respectively. For CPF, CPG, and CPAP3 family proteins, all proteins were detected in at least one of the four MS results. The five CP genes that failed to be authenticated through PCR or RACE (NlugCpr41, NlugCpr43, NlugCpr53, NlugCPF4, and NlugCPG4) were all detected in at least one of the MS results. See SI Appendix, section 2 for functional classification of four MS/MS results according to Gene Ontology.
Fig. 1.
Number of BPH putative CPs detected and verified by UPLC-MS/MS. (A) MS results in eight BPH CP families. The black bar indicates the number of detected proteins (detected in at least one of the four MS experiments) in each family, while the red bar indicates the number of undetected proteins (not detected in any of the MS experiments) in each family. All proteins of three families (CPF, CPG, and CPAP3) were detected. (B) A Venn diagram indicates the number of detected CPs in the CC1, CC3-5, LW, and AC samples.
A Total of 17 and 20 CPs Are Indispensable for Normal Nymphs/Adults Development and Egg Production/Embryo Development, Respectively.
To determine the functions of BPH CPs, we conducted RNAi experiments. Double-stranded RNA (dsRNA) for 135 of the 140 CP genes had been successfully synthesized (Dataset S1). Three groups of BPHs were treated with dsRNAs targeting BPH CP genes: 0- to 12-h second-instar nymphs (representing early instar nymphs), 0- to 12-h fifth-instar nymphs (representing late-instar nymphs), and 0- to 1-h female adults (for reproduction and embryos). To avoid off-target effects, RNAi experiments were replicated by choosing two nonoverlapping regions as targets to the fifth-instar nymphs (SI Appendix, section 3). The results showed that no off-target effects had happened in the dsRNA injection experiments. Quantitative RT-PCR (RT-qPCR) assays confirmed that all target genes were efficiently suppressed by the dsRNA molecules (Dataset S3).
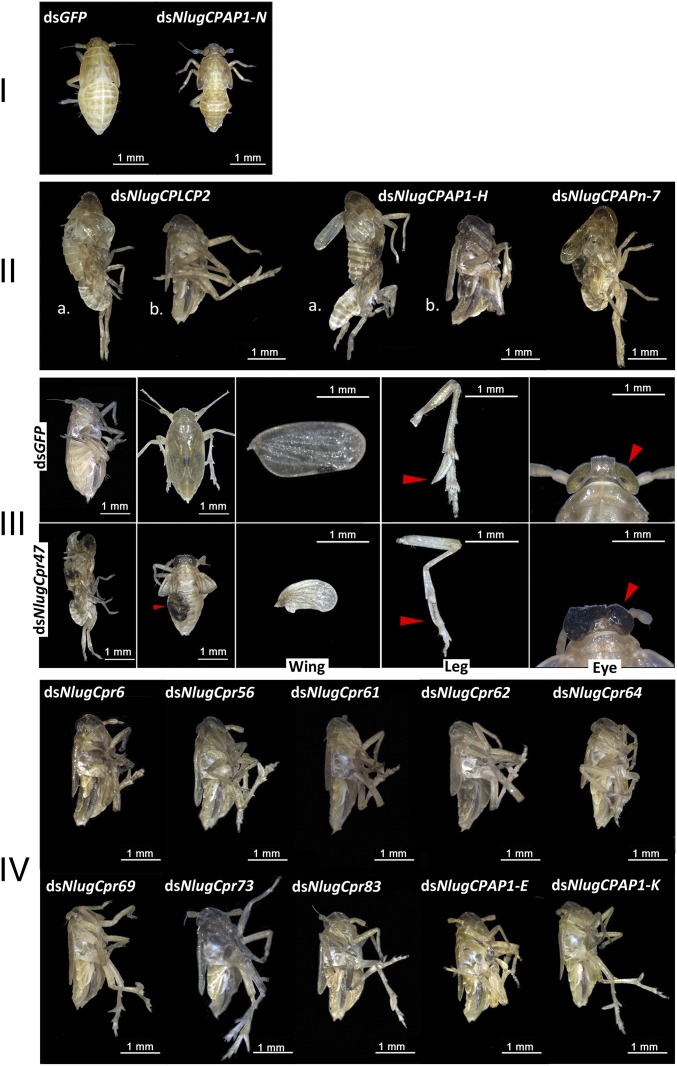
All 135 CP dsRNAs were injected into the thorax of fifth-instar nymphs. In general, the injection of two different dsRNAs targeting one gene caused the same phenotype. RNAi against 15 CP genes (NlugCpr6, NlugCpr47, NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, NlugCpr69, NlugCpr73, and NlugCpr83 from the CPR family; NlugCPLCP2 from the CPLCP family; NlugCPAP1-E, -H, -K, and -N from the CPAP1 family; and NlugCPAPn-7 from the CPAPn family) led to lethal phenotypes with high mortality, indicating their essential roles in BPH integument structure and normal development. The survival rate of the treated BPHs was <20% 9 d after dsRNA treatments, compared with ∼90% survival rate of BPHs treated with dsRNA targeting the mock gene encoding green fluorescent protein (dsGFP) (Fig. 2). Intriguingly, the lethal phenotypes after injection of these 15 dsRNAs could be divided into four distinct classes, phenotypes I–IV: phenotype I, treated BPHs died before ecdysis; phenotype II, molting difficulties and adults with thin body shape; phenotype III, treated BPHs died during ecdysis or had abnormal organs after adult emergence; and phenotype IV, adults died with thin body shape (Fig. 3 and SI Appendix, section 3). The body weight as well as the triglyceride content of the dsRNA-treated female adults showing lethal phenotype IV was significantly decreased compared with the dsGFP-treated group; and honeydew excretion measurement suggested that the decreased feeding activity could have been an important reason for lethal phenotype IV (SI Appendix, section 3). In addition, RNAi against one gene (NlugCPAP3-D1) led to a much thinner body shape similar to phenotype IV, but only ∼27.3% lethality 9 d after injection. The knockdown of other CP genes did not cause any distinguishable physiological malfunction or high death rate in BPHs, compared with insects that were treated with dsGFP.
Fig. 2.
Survival rate of BPHs in 9 d after knockdown of 15 CP genes in late-instar nymphs. dsRNAs against the CP genes were injected at early stage fifth-instar BPH nymphs (0–12 h). dsGFP was injected as a negative control for the nonspecific effects of dsRNA; n = 100 insects. The values were calculated from three biological replicates (means ± SEM). Red line, dsGFP; green line, dsNlugCPAP1-N from phenotype I; orange line, dsNlugCPLCP2 and dsNlugCPAP1-H from phenotype II; purple line, dsNlugCPAPn-7 from phenotype II; blue line, dsNlugCpr47 from phenotype III; black line, dsNlugCpr6, dsNlugCpr56, dsNlugCpr61, dsNlugCpr62, dsNlugCpr64, dsNlugCpr69, dsNlugCpr73, dsNlugCpr83, dsNlugCPAP1-E, and -K from phenotype IV.
Fig. 3.
Lethal phenotypes of BPHs injected with dsRNA for 15 essential CP genes at late-instar nymphs. dsRNAs against the CP genes were injected at early stage fifth-instar BPH nymphs (0–12 h). dsGFP was injected as negative control for the nonspecific effects of dsRNA. The control group (female adult BPH injected with dsGFP) used for phenotypes II and IV was the same as used for phenotype III in the figure.
Injection of all 135 CP dsRNAs against second-instar nymphs showed that all genes that had important functions in late nymphs or adults also played important roles during early nymph developmental stages. However, the respective phenotypes were not necessarily the same after injection with the same dsRNA into second-instar nymphs (Table 1). For example, knockdown of NlugCPAP1-E in fifth-instar nymphs resulted in lethal phenotype IV, while knockdown of NlugCPAP1-E in second-instar nymphs resulted in second-third nymph molting difficulties and high lethality. Only one gene, NlugCPAP3-C (encoding two alternative splice transcripts, NlugCPAP3-C5a and -C5b), which did not lead to distinguishable physiological malfunction or high death rate after RNAi treatment at fifth-instar nymph, showed lethal phenotype after injection of dsRNA in second-instar nymphs. Knockdown of the common region of NlugCPAP3-C5a and -C5b or of the specific region of NlugCPAP3-C5b resulted in very thin body shape and ∼90% death rate after adult emergence, but knockdown directed against the specific region of NlugCPAP3-C5a showed no obvious phenotype (see more information in SI Appendix, section 3).
Table 1.
Summary of phenotypes after RNAi for 32 essential CP genes at three developmental stages in N. lugens
| Gene | dsRNA injection time and phenotypes | ||
| Fifth instar | Second instar | Female adults* | |
| NlugCpr3 | LEH† | ||
| NlugCpr6 | IV‡ (T-en) | DR | |
| NlugCpr8 | LEH | ||
| NlugCpr10 | LEH | ||
| NlugCpr15 | LEP | ||
| NlugCpr24 | LEH | ||
| NlugCpr36 | LEH | ||
| NlugCpr47 | III‡ (L-en) | ME | LEP, LEH |
| NlugCpr51 | LEH† | ||
| NlugCpr52 | LEH† | ||
| NlugCpr54 | LEH | ||
| NlugCpr56 | IV‡ (T-en, T-ex) | DR‡ | |
| NlugCpr58 | LEH† | ||
| NlugCpr61 | IV‡ (T-en) | DR‡ | |
| NlugCpr62 | IV‡ (T-en, T-ex) | DR‡ | |
| NlugCpr64 | IV‡ (T-en, T-ex) | DR | |
| NlugCpr69 | IV‡ (T-en, T-ex) | DR‡ | |
| NlugCpr73 | IV‡ (T-en, T-ex) | DR‡ | LEH |
| NlugCpr83 | IV‡ (T-en, T-ex) | DR‡ | |
| NlugCpr90 | LEH | ||
| NlugCpr94 | LEH | ||
| NlugTwdlE3 | LEH | ||
| NlugCPLCP2 | II‡ (T-en, T-ex) | DR‡ | |
| NlugCPAP1-E | IV‡ (T-en) | NM‡ | LEH |
| NlugCPAP1-H | II‡ (T-en) | NM‡ | LEP, LEH |
| NlugCPAP1-I | LEH | ||
| NlugCPAP1-K | IV‡ (T-en, T-ex) | DR | |
| NlugCPAP1-N | I‡ (T-en) | NM‡ | |
| NlugCPAP3-B | LEH† | ||
| NlugCPAP3-C5b | IV‡ (T-en) | ||
| NlugCPAP3-D1 | IV (T-en) | IV | LEH† |
| NlugCPAPn-7 | II‡ (C-en, T-ex) | NM‡ | |
The insects were treated as described in Materials and Methods. DR, developmental retardation; LEH, low egg hatchability; LEP, low egg production ability; ME, malformed eyes; NM, second-third nymph molting. Phenotypes in the brackets indicate TEM observation results: C-en, curved and disordered endocuticle; L-en, loose and thick endocuticle; T-en, thin and disordered endocuticle; T-ex, thin exocuticle.
The phenotypes listed were significantly decreased according to statistical test (Dataset S4). Phenotypes I–IV were classified as shown in Fig. 3.
Egg hatchability was <10%.
The phenotypes were lethal.
Since parental RNAi has been demonstrated to occur in BPHs (10), we conducted the RNAi experiments on the newly emerged female adults before their ovaries became mature to observe the phenotypes of egg production and embryos from the next generation. A total of 68 CP genes (either relatively highly expressed in the ovary or expressed obviously during embryogenesis) were selected for RNAi experiments (Dataset S4). The average number of total eggs produced by one female adult and the average egg hatchability after RNAi were used to assess the function of each corresponding CP. The results showed that 20 CP genes were indispensable. The reproductive ability (total number of eggs) significantly decreased after knockdown of three genes (NlugCpr15, NlugCpr47, and NlugCPAP1-H), and the egg hatchability significantly decreased after knockdown of 19 different CP genes (Table 1). Notably, after RNAi against the 19 essential CP genes, no first-instar nymph hatched after knockdown of NlugCPAP3-B, and the egg hatchability was <10% after knockdown of NlugCpr3, NlugCpr51, NlugCpr52, NlugCpr58, and NlugCPAP3-D1. Knockdown of two CP genes, NlugCpr47 and NlugCPAP1-H, led to both low reproductive ability and low egg hatchability. Interestingly, among the total number of 20 indispensable CP genes for embryogenesis, only five (NlugCpr47, NlugCpr73, NlugCPAP1-E, -H, and NlugCPAP3-D1) also played important roles in nymphs and adults.
These results indicate that the functions of CPs during nymphal stages through the adult stage were relatively constant and consistent. However, the functions of CPs at egg/embryo developmental stage were dramatically different from that of nymphal or adult stages. All above RNAi phenotypes for the total 32 indispensable CP genes in BPH are summarized in Table 1.
CP Functions Revealed by Transmission Electron Microscopy.
Because of the diversity of BPH lethal phenotypes upon knockdown of the 17 indispensable CP genes (NlugCpr6, NlugCpr47, NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, NlugCpr69, NlugCpr73, NlugCpr83, NlugCPLCP2, NlugCPAP1-E, -H, -K, -N, NlugCPAP3-C5b, -D1, and NlugCPAPn-7) for normal nymph/adult development, we were interested in their integument ultrastructure defects. By transmission electron microscopy (TEM) observation, we found that BPHs treated with dsGFP displayed a normal structure of integument. However, BPHs treated with dsRNA for each of the above 17 CP genes had distinctly different integument structure compared with the control group (Fig. 4).
Fig. 4.
TEM observation of the integument of dsRNA-treated BPHs. BPHs injected with dsNlugCPAP3-C5b were treated at second-instar nymphs (0–12 h). BPHs injected with dsRNAs targeting the other 16 genes (NlugCpr6, NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, NlugCpr69, NlugCpr73, NlugCpr83, NlugCPLCP2, NlugCPAP1-E, -H, -K, NlugCPAP3-D1, NlugCpr47, NlugCPAP1-N, and NlugCPAPn-7) were treated at fifth-instar nymphs (0–12 h). All of the treated BPHs were raised for the following experiments. BPHs treated with dsNlugCpr47, dsNlugCPAP1-N, and dsNlugCPAPn-7 were collected for TEM at the end of fifth-instar nymphal stage (72–78 h) when the insects were still alive; BPHs treated with dsRNA for the other genes were collected for TEM at early stage of adult females (24–30 h) while the insects were still alive. BPHs treated with dsGFP were collected at the same time as negative control animals. Images show parts of an abdominal tergum taken from the fourth to the eighth segment.
In BPHs that died with a thin body shape after adult emergence after knockdown of 14 different CP genes (NlugCpr6, NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, NlugCpr69, NlugCpr73, NlugCpr83, NlugCPLCP2, NlugCPAP1-E, -H, -K, NlugCPAP3-C5b, and -D1), the endocuticles were significantly thinner and disordered compared with that of dsGFP. The well-ordered lamellar structure of the endocuticle almost did not exist in the treated BPHs, suggesting that these 14 CPs might be involved in the formation of the endocuticle. Note that in the BPHs treated with dsNlugCpr56, dsNlugCpr62, dsNlugCpr64, dsNlugCpr69, dsNlugCpr73, dsNlugCpr83, dsNlugCPLCP2, and dsNlugCPAP1-K, the exocuticle was also thinner than in dsGFP-treated BPHs, suggesting that these eight proteins might also be involved in the formation of BPH exocuticle.
In the BPHs that died before or during ecdysis after RNAi treatment (phenotypes I, II, and III), the malformation of the integument structure was different. After dsNlugCPAP1-N treatment, the well-ordered endocuticle structure almost disappeared. After dsNlugCPAPn-7 treatment, the exocuticle became thinner, and the lamellae of the endocuticle became curved and disordered. Following dsNlugCpr47 treatment, the procuticle structure seemed to be loose and became thicker than the control group (Fig. 4). TEM observation clearly indicated that these CPs are indispensable for the formation and maintenance of structure stability in the BPH procuticle.
Complementary Functions of BPH CPs Revealed by Combined RNAi Experiments.
The large number of proteins in the BPH CPR family was intriguing. Apart from the nine CPR genes that provoke lethal phenotypes after RNAi, we also carried out combined RNAi experiments on the remaining of the CPR genes. Fifteen groups were divided according to their homology in the phylogenetic tree (SI Appendix, Fig. S1). The grouped dsRNAs were injected into fifth-instar nymphs (0–12 h). Surprisingly, no high lethal rate was observed 9 d after injection. Only BPHs treated with dsRNAs for Group13 had obvious wing malformations (SI Appendix, Fig. S2). This result suggests that CPR family proteins might have compensatory effects on each other.
BPH CP Genes Are Highly Tissue-Specific and Have Four Distinct Developmental Expression Patterns.
We estimated the expression level of each transcript using FPKM (fragments per kilobase of exons per million fragments mapped) values (Dataset S5) and found that BPH CP genes had highly tissue-specific expression patterns (FPKM). A total of 126 genes (90.0%) were highly expressed in the integument (Fig. 5A, branch d). The remaining 14 genes (10.0%) had their highest expression level in other tissues: 7 in the ovaries, 4 in the testis, and 3 in the gut (Fig. 5A, branches a, b, and c). Intriguingly, knockdown of four genes (NlugCpr3, NlugCpr8, NlugCpr10, and NlugCpr90) highly expressed in the ovaries significantly decreased egg hatchability.
Fig. 5.
Tissue specificity and developmental expression patterns of all genes from eight N. lugens CP families. FPKM data of each row were normalized by Z-score, and groups were clustered by hierarchical clustering. (A) Transcript abundance of all CP-coding genes in five tissues (ovary, testis, fat body, gut, and integument). Branch a, genes relatively highly expressed in the ovary; branch b, genes relatively highly expressed in the testis; branch c, genes relatively highly expressed in the gut; branch d, genes relatively highly expressed in the integument. (B) Transcript abundance of all CP-coding genes across four stages (egg, fourth and fifth instars, and adult) with 24 time points. Green triangles indicate CP genes essential for egg production/embryo development; blue asterisks indicate CP genes essential for endocuticle formation; orange hashtags indicate CP genes essential for exocuticle formation.
To study the developmental expression patterns of N. lugens CP genes, we selected four time periods (egg, fourth-instar, fifth-instar, and adult stages) covering three developmental stages (egg, nymph, and adult). Analysis of the FPKM transcript data among the four time periods revealed four distinct expression patterns from four clusters of CP genes (clusters I–IV) (Fig. 5B and SI Appendix, Fig. S3). The results showed that transcripts of most BPH CP genes were present in all three developmental stages.
Cluster I genes were highly expressed in early to midstage embryos. Only eight genes (5.7%) belong to cluster I. Among these eight genes, six of them (NlugCpr1, NlugCpr2, NlugCpr3, NlugCpr8, NlugCpr35, and NlugCpr90) came from the same branch in Fig. 5A (branch a). Cluster III contains 98 genes and is thereby the largest cluster (70.0%). Among the genes that caused lethal phenotypes after RNAi experiments, most of them belong to this cluster, including NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, NlugCPAP1-E, -H, etc. The expression pattern of cluster III genes had two features. First, they had their expression levels peaking near the end of each nymph stage and reached the minimum near the middle of each nymph stage. Secondly, genes from cluster III were highly expressed at the end of embryogenesis, indicating that these genes were also important in late embryonic integument formation. Their gene expression levels during adult stage remained low.
The expression peak of cluster II genes occurred earlier than the peak of cluster III genes. Ten genes in cluster II (7.1%) reached their highest expression level at early to mid stage of each nymphal stage. Expression of cluster IV genes peaked later than cluster III just at ecdysis. A total of 24 genes (17.1%) belong to this cluster. Both clusters II and IV had relatively low gene-expression levels during embryonic development. The tissue and developmental expression patterns were further confirmed by RT-qPCR (SI Appendix, Fig. S3).
Discussion
Insect cuticle proteins are coded by fast-evolving genes. Their copy number may change dramatically within taxonomic families. For instance, the genome of the Culcidae Anopheles gamibae encodes 156 CPR proteins compared with the 240 copies found in another Culcidae, Aedes aegypti (11). To learn about insect cuticle biology, it is therefore mandatory to systematically identify and characterize these proteins in many different species, including agricultural pests. Here, we assessed the number of CPs in BPH and gave valuable clues to answering the question of why there are so many CPs encoded in an insect genome. In particular, we showed the importance of 32 CP genes in BPH viability and demonstrated the complementarity among the large quantity of CPs. However, we have yet to understand how the CPs interact with one another in complementary ways and how the CPs interact with other components of the cuticle.
Apparently, CPs have important functions, and mutants show severe defects in one insect species, while knockdown of its orthologous genes does not cause similar phenotypes in other insects. For example, in the fly Drosophila melanogaster Obstructor-E (Obst-E) was reported to affect the whole body shape and directly control the mechanical property of the exoskeleton (12). Conversely, as an orthologous gene to Obst-E, knockdown of NlugCPAP3-E did not affect the normal development of BPH. Another Drosophila CPAP3, Obst-A, is needed for chitin fiber maturation and protection in the assembly zone of the cuticle adjacent to the apical plasma membrane of respective cells (13). However, silencing of its orthologous gene NlugCPAP3-A1 did not affect BPH’s normal development either.
In Tribolium, the dsTcCPR4-treated adults exhibited no distinguishable phenotypes from the control insects. NlugCpr22 was an orthologous gene to TcCPR4, with 43% protein sequence identity. Knockdown of NlugCpr22 did not lead to distinguishable phenotypes either. Transcript levels of TcCPR4 dramatically increased in 3-d-old pupae when adult cuticle synthesis begins, while transcript levels of NlugCpr22 also dramatically increased in 2.5-d fifth-instar nymphs when adult cuticle synthesis begins. However, ultrastructural analysis showed that RNAi against TcCPR4 resulted in an amorphous material in the lumen of pore canals instead of fibers in the elytral cuticle, which is massive (14). Further studies are needed to elucidate whether the knockdown of NlugCpr22 and other CP genes which did not lead to distinguishable phenotypes or high death rate in BPHs would lead to minor changes in ultrastructure and weaken their adaptability to environmental hazards.
The expression patterns of 140 genes from eight N. lugens CP families underscored the complicated relationship between timing and the type of cuticle being formed. In insects, if a transcript does not appear until after ecdysis, the respective protein will be used in forming the endocuticle. Conversely, if a transcript appears before ecdysis, the corresponding protein probably will be used in forming the exocuticle (4). Based on this definition, cluster III proteins might be involved in forming the exocuticle, while cluster II and IV proteins might be involved in forming the endocuticle (Fig. 5B and SI Appendix, Fig. S3). This simple view contradicts our findings that many genes from cluster III (such as NlugCpr56, NlugCpr61, NlugCpr62, NlugCpr64, etc.) resulted in malformed endocuticle or endocuticle and exocuticle structures (Fig. 4). Thus, we recommend that the developmental expression pattern should only be one of the references to investigate the localization and function of CPs. For example, the knockdown of NlugCpr56, NlugCpr62, NlugCpr64, NlugCpr69, NlugCpr73, and NlugCpr83 (all from the RR-2 subgroup) led to thin endocuticles and thin exocuticles. Combined with their respective mRNA expression patterns, we could deduce that they may contribute to the structure of both exocuticle and endocuticle. However, EM immunodetection experiments are needed to study the precise localization and function of the individual proteins (14, 15).
This comprehensive CP-ome study unifying genomic, transcriptomic, and proteomic aspects in N. lugens, together with analyses of gene function and expression profiles, contributes substantially to the insect cuticle research field. These findings may furthermore stimulate the design and development of insecticides specifically targeting cuticle proteins.
Materials and Methods
Insects.
BPHs used in this study were originally obtained from Hangzhou (30°16′N, 120°11′E), China, in 2008. The insects were reared on fresh rice seedlings (strain: Xiushui 134) in a walk-in chamber at 26 ± 0.5 °C and 50 ± 5% relative humidity under a photoperiod of 16:8 h (light:dark).
Gene Identification.
Insect CP amino acids and gene sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/), EnsemblMetazoa (metazoa.ensembl.org/), CutProtFam (aias.biol.uoa.gr/CutProtFam-Pred/home.php) (16), and CuticleDB (bioinformatics2.biol.uoa.gr/cuticleDB/) (17). The common motifs of CPs from D. melanogaster, Bombyx mori, A. gambiae, Tribolium castaneum, Apis mellifera, and Locusta migratoria were used as queries against the BPH genome (https://www.ncbi.nlm.nih.gov/, BioProject PRJNA177647; 27,571 coding protein sequences) and transcriptomic databases (https://www.ncbi.nlm.nih.gov/sra, accession no. SRX023419; 21,908 coding protein sequences). The full-length cDNA sequences were obtained from transcriptomic databases, and most of them were confirmed by RT-PCR. The full cDNA sequences of some CP genes were cloned by using RACE core sets (catalog nos. 6107 and 6106; TaKaRa) according to the manufacturer’s instructions. The primers used are shown in Dataset S1.
Sequence Analysis.
The ORF prediction was performed on the Softberry website. The SMART program (smart.embl-heidelberg.de/) was used for the identification of modular domains. The signal peptide prediction program SignalP 4.1 server (www.cbs.dtu.dk/services/SignalP/) was used to predict N-terminal signal peptides. The multiple sequence alignment of the CPLCP family proteins was carried out by using ClustalX software (18). The phylogenetic trees were constructed via the neighbor-joining method by using the MEGA6.06 program (19). Homologous relationships were determined by bootstrap analysis based on 1,000 or 5,000 replications, as each legend describes.
Cuticle Protein Sample Preparation for UPLC-MS/MS.
Four different BPH cuticle tissue samples were prepared for UPLC-MS/MS analysis. Cast first-instar nymph cuticles (CC1) were collected from ∼12,000 first-instar nymphs. Cast third- to fifth-instar nymph cuticles (CC3–5) were collected from ∼700 fifth-instar BPHs, 200 fourth-instar BPHs, and 200 third-instar BPHs. The BPHs were maintained in a polycarbonate box of 20 cm in length, 14 cm in width, and 9 cm in height with fresh rice seedlings. The cast cuticles were collected with forceps, and the CC1 samples were examined under the stereomicroscope (Leica S8AP0) to avoid contamination. Adult long wings (LWs) were carefully dissected under the stereomicroscope from ∼1,200 randomly selected living adult long-winged BPHs. Fifth-instar nymph abdominal cuticles (ACs) were dissected from ∼800 randomly selected living fifth-instar BPHs. The adherent tissues of the cuticle were carefully removed with forceps as thoroughly as possible under the stereomicroscope. All four cuticle tissue samples were rinsed with water (Aladdin) at least three times and then processed under the same sample preparation procedures described in SI Appendix, section 5.
RNAi Experiments.
There were two regions of each gene selected for dsRNA synthesis to overcome possible off-target effects. The regions were designed to have no other similar sequences in the genome (SI Appendix, section 3). The purified DNA template was used to produce dsRNAs, and the primers were designed to contain the T7 RNA polymerase promoter at both ends (Dataset S1). The dsRNA was then synthesized through RT-PCR amplification by using the MEGA script T7 High Yield Transcription Kit (catalog no. AM1334; Ambion), according to the manufacturer’s instructions. The concentration and quality of dsRNAs were measured with NanoDrop 2000 (Thermo Fisher Scientific), and the size of the dsRNAs was verified via electrophoresis in a 1% agarose gel. The quantities of dsRNA injected into second- and fifth-instar nymphs and adults were ∼25, 200, and 250 ng, respectively.
Female adult BPHs treated with dsRNAs were crossed to healthy male BPHs for 3 d and then transferred onto fresh rice seedlings in glass tubes. One female together with two males were in one tube to produce offspring for 3 d. Subsequently, the females were removed, and the eggs/seedlings were maintained for 10 d for counting the number of hatched offspring. The leaves and stems of the rice seedlings were dissected under the microscope to count the number of eggs failing to hatch. Each target gene was carried out for 10 biological replicates. For the method of microinjection with dsRNA, see SI Appendix, section 5.
TEM Observation.
The dorsal abdominal integument of BPH for examining the ultrastructure was separated carefully under the microscope, and the integument between the fourth and eighth segments was dissected for sample preparation and final observation. The time for insect collecting was described in the previous sections. The sample preparation was performed as reported (20), and specimen sections were observed on a Hitachi Model H-7650 TEM.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 31630057 and 31471765.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. MF942728–MF942869).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716951115/-/DCSupplemental.
References
- 1.Locke M. The Wigglesworth lecture: Insects for studying fundamental problems in biology. J Insect Physiol. 2001;47:495–507. doi: 10.1016/s0022-1910(00)00123-2. [DOI] [PubMed] [Google Scholar]
- 2.Neville AC. Biology of the Arthropod Cuticle. Springer; New York: 1975. [Google Scholar]
- 3.Andersen SO. Insect cuticular sclerotization: A review. Insect Biochem Mol Biol. 2010;40:166–178. doi: 10.1016/j.ibmb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Willis JH. Structural cuticular proteins from arthropods: Annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vannini L, Bowen JH, Reed TW, Willis JH. The CPCFC cuticular protein family: Anatomical and cuticular locations in Anopheles gambiae and distribution throughout Pancrustacea. Insect Biochem Mol Biol. 2015;65:57–67. doi: 10.1016/j.ibmb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ. Cuticular proteins. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic; London: 2012. pp. 134–166. [Google Scholar]
- 7.Andersen SO, Højrup P, Roepstorff P. Insect cuticular proteins. Insect Biochem Mol Biol. 1995;25:153–176. doi: 10.1016/0965-1748(94)00052-j. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. Properties of the cuticular proteins of Anopheles gambiae as revealed by serial extraction of adults. PLoS One. 2017;12:e0175423. doi: 10.1371/journal.pone.0175423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins TL, Krchma LJ, Ahmad SA, Kramer KJ. Pupal cuticle proteins of Manduca sexta: Characterization and profiles during sclerotization. Insect Biochem Mol Biol. 2000;30:19–27. doi: 10.1016/s0965-1748(99)00091-0. [DOI] [PubMed] [Google Scholar]
- 10.Xu HJ, et al. Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) Insect Mol Biol. 2013;22:635–647. doi: 10.1111/imb.12051. [DOI] [PubMed] [Google Scholar]
- 11.Cornman RS. Molecular evolution of Drosophila cuticular protein genes. PLoS One. 2009;4:e8345. doi: 10.1371/journal.pone.0008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajiri R, Ogawa N, Fujiwara H, Kojima T. Mechanical control of whole body shape by a single cuticular protein Obstructor-E in Drosophila melanogaster. PLoS Genet. 2017;13:e1006548. doi: 10.1371/journal.pgen.1006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pesch YY, Riedel D, Behr M. Obstructor A organizes matrix assembly at the apical cell surface to promote enzymatic cuticle maturation in Drosophila. J Biol Chem. 2015;290:10071–10082. doi: 10.1074/jbc.M114.614933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y. Tribolium castaneum RR-1 cuticular protein TcCPR4 is required for formation of pore canals in rigid cuticle. PLoS Genet. 2015;11:e1004963. doi: 10.1371/journal.pgen.1004963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vannini L, Willis JH. Localization of RR-1 and RR-2 cuticular proteins within the cuticle of Anopheles gambiae. Arthropod Struct Dev. 2017;46:13–29. doi: 10.1016/j.asd.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ. CutProtFam-Pred: Detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem Mol Biol. 2014;52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magkrioti CK, Spyropoulos IC, Iconomidou VA, Willis JH, Hamodrakas SJ. cuticleDB: A relational database of Arthropod cuticular proteins. BMC Bioinformatics. 2004;5:138. doi: 10.1186/1471-2105-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HJ, et al. Rice ragged stunt virus-induced apoptosis affects virus transmission from its insect vector, the brown planthopper to the rice plant. Sci Rep. 2015;5:11413. doi: 10.1038/srep11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.