Significance
HLA-I molecules play a central role in immune recognition of infected or cancer cells. They bind short intracellular peptides of 9 to 12 amino acids and present them to T cells for immune recognition. For many years, the confinement of HLA-I ligand has been a central dogma in immunology. Combing analysis of mass spectrometry data with novel algorithms, X-ray crystallography, and T cell recognition assays, we show that a substantial fraction of HLA-I molecules bind peptides extending beyond the C terminus of canonical ligands, and that these peptides can be recognized by CD8 T cells. Our ability to accurately predict such epitopes will help studying their role in infectious diseases or cancer immunotherapy.
Keywords: HLA-I–peptide interactions, HLA peptidomics, T cell epitope, HLA-I structures, computational immunology
Abstract
HLA-I molecules play a central role in antigen presentation. They typically bind 9- to 12-mer peptides, and their canonical binding mode involves anchor residues at the second and last positions of their ligands. To investigate potential noncanonical binding modes, we collected in-depth and accurate HLA peptidomics datasets covering 54 HLA-I alleles and developed algorithms to analyze these data. Our results reveal frequent (442 unique peptides) and statistically significant C-terminal extensions for at least eight alleles, including the common HLA-A03:01, HLA-A31:01, and HLA-A68:01. High resolution crystal structure of HLA-A68:01 with such a ligand uncovers structural changes taking place to accommodate C-terminal extensions and helps unraveling sequence and structural properties predictive of the presence of these extensions. Scanning viral proteomes with the C-terminal extension motifs identifies many putative epitopes and we demonstrate direct recognition by human CD8+ T cells of a 10-mer epitope from cytomegalovirus predicted to follow the C-terminal extension binding mode.
HLA class I (HLA-I) molecules play a major role in immune defense mechanisms by presenting to T cells peptides from the intracellular matrix. Peptides presented on HLA-I molecules originate mainly from proteasomal degradation of self or pathogen-derived proteins. These peptides are first translocated to the endoplasmic reticulum. There, they can load on HLA-I molecules provided their sequence is compatible with HLA-I binding motifs. Peptide–HLA-I complexes are then transported to the cell surface where they can elicit T cell recognition, primarily upon presentation of nonself peptides.
Most HLA-I alleles preferentially bind 9- to 12-mer peptides (1–5), and the majority of alleles accommodate peptides with anchor residues at the second and last positions. From a structural point of view, anchor residues point directly into the HLA-I peptide binding groove. Their importance for HLA-I–peptide interactions is also reflected at the sequence level, where alignments of HLA-I ligands display clear specificity at the second and last positions for most alleles. Nine-mer HLA-I ligands are characterized by a linear binding mode. For longer peptides, numerous crystal structures have shown the presence of a bulge at middle positions, protruding outside of the HLA-I binding site to accommodate the additional residues between the two anchor positions (e.g., ref. 6).
Over the years, anecdotal evidences of C-terminal extensions beyond the last anchor position have been observed among HLA-A02:01 ligands and crystal structures with such ligands were published first in 1994 (7) and later in 2009 (8). More recently, analysis of HLA peptidomics data obtained by mass spectrometry (MS) from cell lines transfected with soluble HLA-A02:01 and infected with Toxoplasma gondii revealed several C-terminal extensions and showed that, for this allele, C-terminal extensions were mainly found among peptides coming from pathogens (9, 10). X-ray crystallography revealed distinct structural mechanisms in HLA-A02:01 to accommodate C-terminal extensions (9, 10). N-terminal extensions have also been recently observed in HLA-B57:01 (11) and HLA-B58:01 (12). However, N- or C-terminal extensions have not been much investigated in other HLA-I alleles based on unbiased HLA-I ligand datasets [see some results in mouse (15)]. As such, it remains unclear whether they occur frequently and, if so, whether they can be recognized by CD8 T cells, although the latter may be expected.
Here, we introduce a statistical approach to rigorously investigate N- and C-terminal extensions in large datasets of naturally presented HLA-I ligands obtained by in-depth HLA peptidomics profiling of cell lines and tissue samples covering more than 50 HLA-I alleles. Our work reveals widespread C-terminal extensions for at least eight HLA-I molecules (HLA-A02:03, HLA-A02:07, HLA-A03:01, HLA-A31:01, HLA-A68:01, HLA-A68:02, HLA-B27:05, and HLA-B54:01), and we identify both sequence and structural features in HLA-I alleles predictive of the presence of C-terminal extensions. A crystal structure of HLA-A68:01 in complex with such a ligand uncovers structural changes to accommodate the C-terminal extensions. Scanning viral proteomes with our motifs describing C-terminal extensions further enabled us to demonstrate direct CD8 T cell recognition of an HLA-A03:01 restricted epitope from cytomegalovirus (CMV) predicted to follow the C-terminal extension binding mode.
Results
Unbiased Investigation of C- and N-Terminal Extensions.
To investigate the presence of C- and N-terminal extensions in HLA-I ligands, we collected recent pooled and monoallelic HLA peptidomics datasets from seven studies covering 43 samples, 54 different HLA-I alleles, and 109,953 unique peptides (2, 13, 14, 16–19) (SI Appendix and Dataset S1). These datasets were all generated with <1% false discovery rate and were not filtered with any predictor. We hypothesized that C- or N-terminal extensions among naturally presented endogenous HLA-I ligands may be determined by identifying peptides of 10 or more amino acids that do not match motifs expected for ligands following the bulge model (Fig. 1A). For pooled HLA peptidomics data, 9-mer binding motifs were identified and annotated with our recent motif deconvolution algorithm (4, 19) (Fig. 1B and SI Appendix). Importantly, these motifs were identified without relying on HLA-I ligand predictors and, therefore, represent a fully unbiased view of the binding specificity of HLA-I alleles. Focusing on the first three and the last two positions in the 9-mer motifs, we then built position weight matrices (PWM) modeling bulges, N-terminal extensions, or C-terminal extensions for each of the 9-mer motifs (SI Appendix). For 10-mers, bulges were modeled by incorporating five nonspecific positions between the first three and the last two residues. C- and N-terminal extensions were modeled by adding four nonspecific positions in the middle and one nonspecific position at the C and N terminus, respectively (Fig. 1C and SI Appendix).
Fig. 1.
(A) General description of the pipeline developed in this work to identify and validate N- or C-terminal extensions. (B) Example of 9-mer motifs identified in HLA peptidomics data from Mel_15 (16). The number of peptides assigned to each motif is shown in parentheses. (C) Illustration of the different models built from the 9-mer motifs (bulge, C- and N-terminal extension) to investigate noncanonical binding modes among 10-mers.
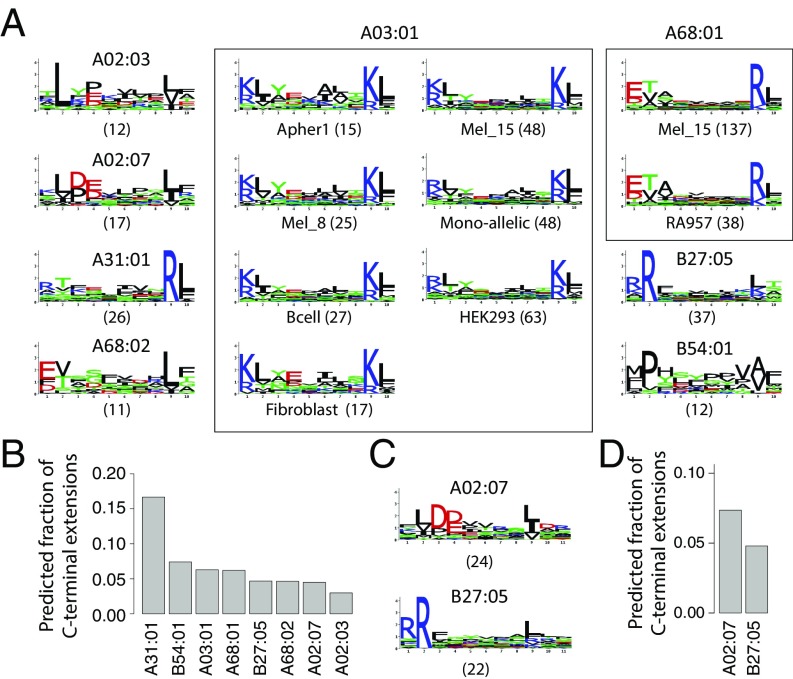
Using the three models derived from each 9-mer motif, we then scored all 10-mer peptides (Fig. 1C and SI Appendix). Peptides that displayed a significantly higher score for exactly one allele and one model were assigned to this allele and the corresponding model (SI Appendix). For monoallelic cell lines, comparison was only performed among bulge, N- and C-terminal extension models of the same allele. To determine statistical significance of N- or C-terminal extensions, we developed a null model representing the expected 10-mer HLA-I ligands assuming only bulges (SI Appendix). We finally retrieved all sets of peptides predicted to follow N- or C-terminal extensions associated to a given allele in a given sample that passed statistical significance (Z-score > 2). This resulted in 15 motifs of C-terminal extensions (for a total of 396 unique 10-mer peptides) and no motif of N-terminal extensions (Fig. 2A and SI Appendix, Fig. S1). Seven motifs corresponded to C-terminal extensions associated to HLA-A03:01 across different samples, two motifs to HLA-A68:01 and one motif to each of the other alleles (i.e., HLA-A02:03, HLA-A02:07, HLA-A31:01, HLA-A68:02, HLA-B27:05, and HLA-B54:01; Fig. 2A, SI Appendix, Fig. S2, and Datasets S2 and S3). We first note that motifs describing C-terminal extensions associated to the same allele (i.e., HLA-A03:01 or HLA-A68:01) in different datasets displayed a very high similarity among each other (rectangles in Fig. 2A). This highlights the remarkable reproducibility of our predictions across our very heterogeneous set of HLA peptidomics studies. The average frequency of C-terminal extensions among 10-mer ligands is shown in Fig. 2B.
Fig. 2.
(A) Predicted 10-mer C-terminal extension motifs found for different HLA-I alleles in the different datasets (see also Datasets S2 and S3). Parentheses indicate the number of peptides associated to each motif. (B) Estimates of the frequency of C-terminal extensions among 10-mers for alleles shown in A. (C) C-terminal extensions motifs comprising two residues after the second anchor residue in 11-mers ligands. (D) Estimates of the frequency of C-terminal extensions among 11-mers for alleles shown in C.
To investigate whether C-terminal extensions may extend for more than one amino acid, we applied our approach to longer peptides (SI Appendix) and found statistically significant evidences of C-terminal extensions for HLA-A02:07 and HLA-B27:05 for a total of 46 unique 11-mer peptides (Fig. 2 C and D and Datasets S2 and S3). The motifs are consistent with the 10-mer C-terminal extensions (Fig. 2A). A trend was also observed in some samples for HLA-A02:01, HLA-A02:03, HLA-A03:01, HLA-A68:02, and HLA-B54:01 (Dataset S2), although it did not pass our thresholds on the number of ligands or the Z-score. When analyzing even longer ligands (12-mers), we did not find anything statistically significant, but the number of such ligands identified by MS is much smaller so that it may be difficult to confidently identify C-terminal extensions with our approach.
Robustness to Noise.
To explore the robustness of our findings with respect to noise in HLA peptidomics data, we reran our whole pipeline, adding 5% of randomly selected peptides from the human proteome to all datasets. Remarkably, the predicted C-terminal extensions remained basically unchanged (SI Appendix, Fig. S3). In particular, we did not observe any new C-terminal extension motifs that would arise from the random peptides. This clearly suggests that C-terminal extensions predicted in this work are not resulting from contaminations in HLA peptidomics data.
In Vitro Validation.
To experimentally test our predictions, we selected 10-mer ligands predicted to follow the C-terminal extension binding mode for three alleles (HLA-A03:01, HLA-A31:01, HLA-A68:01). We mutated either the last or second-to-last residue and experimentally measured the stability of the wild-type and the two mutated peptides (SI Appendix). 9-mer peptides without the predicted C-terminal extensions were used as positive controls. As expected, mutating the last residue had little effect on their binding stability, while mutating the second-to-last residue significantly decreased the stability (Fig. 3). These data strongly suggest that, for these peptides, the second-to-last residue is playing the role of the anchor residue and the last residue is extending beyond the canonical C terminus of HLA-I ligands. We also tested other P10 mutants that did not match the motif predicted by our analysis. In general, the stability of these mutants was lower than for peptides seen in MS data, especially for HLA-A03:01 and HLA-A68:01 (SI Appendix, Fig. S4). Of note, peptides with R or K at P10 could also form bulges, which likely explains their higher stability. To test whether longer C-terminal extensions may bind to HLA-A03:01, we added all amino acids not compatible with the bulge model at the C terminus of the 10-mer HLA-A03:01 ligand KLAYTLLNKL and measured the stability of these peptides (SI Appendix, Fig. S5). Our results indicate that most of the 11-mers did bind, although with lower stability compared with the 10-mer peptide. This suggests that C-terminal extensions can extend for more than one residue in HLA-A03:01 ligands, although this likely corresponds to a small fraction of the actual HLA peptidome, as suggested by their low frequency in MS data.
Fig. 3.
In vitro binding stability assays for peptides predicted to follow the C-terminal extension binding mode for HLA-A03:01, HLA-A31:01, and HLA-A68:01.
We then analyzed peptides that displayed similar scores for the bulge and the C-terminal extension model (red circles in SI Appendix, Fig. S1). Most of them displayed the same or very similar amino acids at P9 and P10 (Dataset S4). To investigate whether they are more likely to adopt a bulge or a C-terminal extension binding mode, we measured the stability of RYIEIFPSRR with HLA-A31:01. R(9)S did not affect the binding, while R(10)S decreased the stability (SI Appendix, Fig. S6). This suggests that peptides displaying similar scores for the two models may preferentially adopt a bulged conformation.
Crystal Structures of C-Terminally Extended HLA-I Ligands.
To investigate the structural mechanisms underlying C-terminal extensions uncovered in this work, we generated a high-resolution (1.6 Å) crystal structure of HLA-A68:01 in complex with a 10-mer peptide (ETSPLTAEKL; Fig. 3) predicted to follow the C-terminal extension binding mode (Fig. 4A and SI Appendix, Fig. S7). As expected, the lysine at P9 (yellow sidechain) filled the F pocket and superimposed nicely with the last residue (K9) of canonical 9-mer ligands of HLA-A68:01 (blue sidechain in Fig. 4A). More importantly, to accommodate the C-terminal extension (L10), the Y84 side chain was flipped by 90° and the two alpha helices around the F pocket moved away from each other (Fig. 4A), as measured by the distance between C-alpha atoms of residues 80 and 143 (D80–143 = 11.1 Å versus D80–143 = 10.1 Å for the complex with a 9-mer peptide). Interestingly, the same flip had been observed in one of the HLA-A02:01 structures in complex with C-terminally extended ligands (9) (Fig. 4B). However, in contrast to HLA-A02:01 where the side chain of the first residue of the C-terminal extension points toward the solvent [K12 Fig. 4B (9), see also L10 in PDB ID code 5FA4 and D10 in PDB ID code 5F7D (10)], in the case of HLA-A68:01, the C-terminal extension side chain filled the pocket created by the flip of Y84 side chain (SI Appendix, Fig. S8). This result is fully consistent with the specificity for hydrophobic residues in the C-terminal extension motif of HLA-A68:01 (Fig. 2A). It is also interesting to note that the flip of Y84 side chain was recently observed in a crystal structure of peptide-MHC-β2m in complex with TAPBPR, suggesting that loading of C-terminal extensions may be favored in vivo (20).
Fig. 4.
(A) New crystal structure of HLA-A68:01 (white ribbon) in complex with a C-terminally extended ligand (yellow) superimposed with HLA-A68:01 (green ribbon) in complex with a canonical 9-mer ligand (blue, PDB ID code 4WHZ) (29). (B) Comparison of HLA-A68:01 (white ribbon) in complex with a C-terminally extended ligand (yellow) and the complex of HLA-A02:01 (pink ribbon) bound to a C-terminal extended 12-mer peptide (FVLELEPEWTVK, magenta, PDB ID code 5DDH) (9). (C) HLA-I residues surrounding the C terminus of canonical ligands and displaying the largest Jensen–Shannon divergence between alleles with C-terminal extensions (Top Left) and alleles without C-terminal extensions (Bottom Left). For comparison, the sequence logos of HLA-A (Top Right) and HLA-B/C (Bottom Right) alleles at the same positions are displayed. (D) Analysis of the distance D80–143 for alleles with available crystal structures (SI Appendix, Table S1). Green lines correspond to alleles displaying C-terminal extensions. The orange line represents HLA-A02:01.
SI Appendix, Fig. S4 shows that the hydrophobic side chain at P10 is not strictly required, and peptides with S or D at this position can bind. In these cases, we anticipate that the P10 side chain points toward the solvent, as in previous structures of HLA-A02:01 in complex with C-terminal extensions, although we cannot exclude that additional structural changes may further modify the physical properties of the new pocket to accommodate polar or charged amino acids.
Propensity of HLA-I Alleles for C-Terminal Extensions.
To investigate the molecular determinants of C-terminal extensions, we aligned the sequences of all HLA-I alleles considered in this work and checked whether some amino acid patterns in residues surrounding the F pocket characterize alleles predicted to accommodate C-terminal extensions (SI Appendix). Clear differences were observed at specific positions (Fig. 4C) and showed overlap with properties characterizing HLA-A alleles, as expected from the higher proportion of HLA-A alleles predicted to display C-terminal extensions (Fig. 2A). Interestingly, glycine is known to destabilize alpha helices, and the presence of glycine at position 79 (i.e., exactly where the α1 helices start to be no longer aligned in Fig. 4A) in most HLA-I alleles predicted to display C-terminal extensions may endow the α1 helix with the flexibility required to accommodate such extensions. To test whether these amino acid patterns may help predict whether an allele is more likely to display C-terminal extensions, we trained a logistic regression and performed a rigorous cross-validation (SI Appendix). An average area under the ROC curve (AUC) of 0.85 could be obtained, suggesting that alleles accommodating C-terminal extensions may be reasonably well predicted from their sequence. Lower accuracy was reached in a simple model where HLA-A alleles are predicted to display C-terminal extensions and HLA-B/C alleles are not (AUC = 0.76). To further investigate molecular mechanisms allowing for C-terminal extensions, we surveyed available X-ray structures of HLA-I alleles considered in this work (SI Appendix, Table S1). As before, we computed the distance D80–143 between the two alpha helices surrounding the C terminus of canonical ligands (SI Appendix, Table S1). Interestingly, we observed that, on average, alleles predicted to display C-terminal extensions showed larger distances between these two helices already when interacting with canonical 9-mer peptides (P = 0.002, Wilcoxon rank-sum test; Fig. 4D). Using this distance to predict alleles accommodating C-terminal extensions among those with available crystal structures led to an AUC of 0.92. Of note, HLA-B51:01 is characterized by much higher frequency of 8-mer ligands compared with other HLA-I alleles (2, 3, 21). As for HLA-A01:01, we observed a trend for C-terminal extensions in some samples (e.g., the monoallelic cell line), but not in other samples (e.g., Melanoma/Mel_12; see SI Appendix, Fig. S9 and Dataset S2, although most 10-mer peptides in this sample should come from HLA-A01:01 since HLA-B08:01 and HLA-C07:01 poorly bind 10-mers). We finally point out that the clear patterns in both sequence and structural properties of HLA-I alleles predicted to display C-terminal extensions provides an additional and independent validation of our predictions in Fig. 2 based only on HLA peptidomics data.
Explicitly Incorporating C-Terminal Extensions in HLA-I Ligand Predictors.
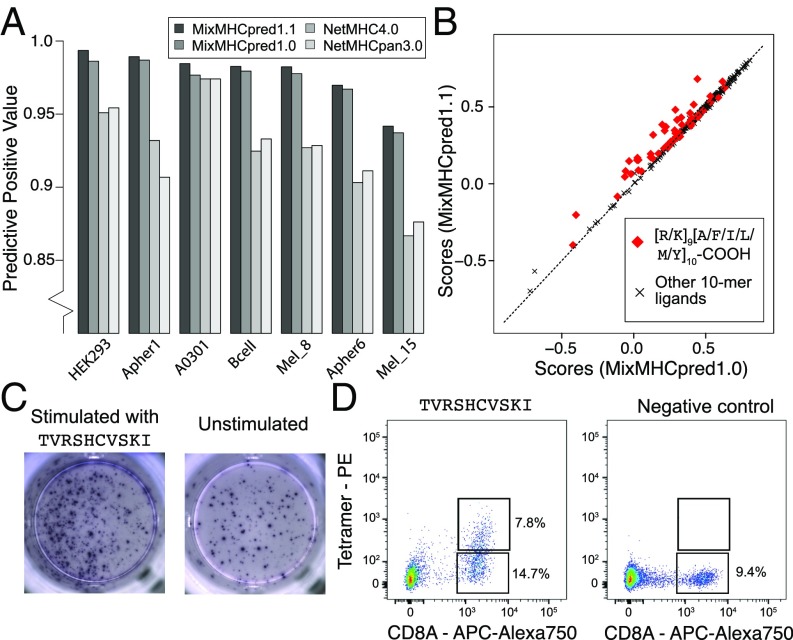
C-terminal extensions have not been routinely investigated in previous studies and the training set of most existing HLA-I ligand predictors typically does not include them, even if the recent version of NetMHC tools can mathematically handle them (5, 22, 23). We therefore retrained our predictor MixMHCpred (19) using multiple PWMs to model C-terminal extensions (24) (SI Appendix). To validate our algorithm, we took advantage of the fact that HLA-A03:01 and HLA-A68:01 alleles were present in multiple datasets and attempted to repredict all of the 10-mer peptides of these datasets, excluding data from the dataset used for testing in the training of our predictor (SI Appendix). As negative data, we included fourfold decoy (i.e., 10-mers randomly selected from the human proteome) and computed both the positive predictive value corresponding to the top 20% predictions and the AUC (SI Appendix). We observed that explicitly modeling C-terminal extensions increased the performance compared with the previous version of our predictor (MixMHCpred1.0) (19), as well as other widely used HLA-I ligand predictors that did not include unbiased MS data in their training set (22, 23) (Fig. 5A and SI Appendix, Fig. S10). The improvement came mainly from higher scores for ligands displaying C-terminal extensions, as shown in Fig. 5B for the HLA-A03:01 10-mer ligands isolated from a monoallelic cell line (2) (P = 1.0 × 10−7, Wilcoxon signed-rank test).
Fig. 5.
(A) Benchmarking of our HLA-I ligand predictor (MixMHCpred1.1). The y axis shows the positive predictive value among the top 20% of the predictions. (B) Analysis of scores when explicitly modeling C-terminal extensions (MixMHCpred1.1) or not (MixMHCpred1.0) for the 10-mer HLA-A03:01 ligands from a monoallelic cell line (2), as a function of the C-terminal amino acids (P9 and P10). (C) IFN-γ–ELISpot results obtained by stimulation with a C-terminally extended 10-mer HLA-A03:01 ligand (TVRSHCVSKI, from CMV, Left) vs. no peptide (Right) of a PBMC sample from a HLA-A03:01 and CMV seropositive healthy donor. (D) Multimer analysis of CD8 T cells from a healthy donor recognizing the HLA-A03:01 restricted C-terminally extended 10-mer peptide TVRSHCVSKI (Left) and the negative control (RVRAYFYSKV/HLA-A03:01 tetramer) for which we did not observe T cell recognition (Right).
Almost no difference between MixMHCpred1.0 and MixMHCpred1.1 could be observed when testing our algorithm on other datasets used in previous benchmarking studies. However, we anticipate that C-terminal extensions are very rare in these datasets since most of the known HLA-I ligands had been first predicted with former versions of HLA-I ligand predictors. For instance, when analyzing immune epitope database (IEDB) data (25) for HLA-A03:01 ligands coming from earlier studies than those considered in this work, we could not see any statistical evidence of C-terminal extensions, suggesting that such ligands had not been tested in binding assays, or had been filtered in MS data.
Identification of Immunogenic C-Terminally Extended Epitopes.
To investigate the immunological relevance of HLA-I ligands displaying C-terminal extensions, we used our predictor and scanned both human cancer testis antigens and viral proteomes with the motifs characterizing 10-mer C-terminal extensions for HLA-A03:01 and HLA-A68:01 (SI Appendix). This analysis revealed many putative epitopes, including peptides from the PRAME cancer testis antigen and several CMV, Epstein–Barr virus (EBV), HIV, human papillomavirus (HPV), influenza, and yellow fever peptides (Table 1). We measured the binding stability of these peptides and found values falling within the range of CD8 T cell epitopes (Table 1).
Table 1.
Binding stability (half-lives) of C-terminally extended HLA-I ligands from cancer testis antigens and viral proteins
| Allele | Organism | Protein | Sequence | Half-life, h |
| A03:01 | HIV_B | VPU | RLIDRLIERA | 19.8 ± 2.0 |
| A03:01 | Influenza | NCAP | RMCNILKGKF | 34.2 ± 3.0 |
| A03:01 | Y. Fever | POLG | RMGERQLQKI | 30.0 ± 3.0 |
| A03:01 | HCMV | UL44 | TLLNCAVTKL | 14.7 ± 0.3 |
| A03:01 | HCMV | UL44 | TVRSHCVSKI* | 16.6 ± 3.1 |
| A03:01 | EBV | BRLF1 | RVRAYTYSKV | 51.6 ± 10.0 |
| A03:01 | EBV | BNRF1 | RTWDRMTEKL | 27.5 ± 0.8 |
| A03:01 | HIV | NEF | QVPLRPMTYKA | 48.1 ± 12.2 |
| A03:01 | HIV | NEF | QVPLRPMTYKG | 31.9 ± 7.9 |
| A03:01 | Human | PRAME | RLWGSIQSRY | 19.9 ± 12.0 |
| A68:01 | Human | PRAME | ETLSITNCRL | 14.5 ± 6.2 |
| A68:01 | HCMV | UL82 | EAASGSFGRL | 81.7 ± 14.6 |
| A68:01 | HCMV | HELI | EVVQRGLSRL | 16.6 ± 0.8 |
| A68:01 | HPV16 | E1 | ETIEKLLSKL | 12.9 ± 5.8 |
| A68:01 | EBV | BPLF1 | ETVADWKRRL | 22.3 ± 1.4 |
| A68:01 | Y. Fever | POLG | QTSRLLMRRM | 19.3 ± 2.0 |
The peptide used in the IFN-γ–ELISpot and multimer analyses of Fig. 5 C and D and SI Appendix, Fig. S11. Bold letters indicate the predicted second anchor residue.
We then focused on one CMV peptide binding to HLA-A03:01 and predicted to follow the C-terminal extension binding mode (TVRSHCVSKI, asterisk in Table 1). We stimulated a peripheral blood mononuclear cells (PBMCs) sample from a healthy donor (both HLA-A03:01 and CMV seropositive) with the 10-mer peptide for 12 d (Materials and Methods). Subsequently, we observed cytokine production by IFN-γ–ELISpot after rechallenge with the CMV 10-mer peptide for 16 h (Fig. 5C), suggesting that this epitope is potentially immunogenic in humans. To determine whether CD8 T cells could directly recognize the C-terminally extended 10-mer peptide, we constructed HLA-A03:01 tetramers loaded with the 10-mer peptide (Materials and Methods). Analyzing CD8 T cells with such tetramers revealed a population of CD8 T cells that directly bound to the 10-mer peptide/HLA-A03:01 complex (Fig. 5D). The IFN-γ–ELISpot, tetramer and refolding assays were repeated using the K(9)L and I(10)L mutants, and the truncated 9-mer (TVRSHCVSK) (SI Appendix, Fig. S11 A–C). In all cases, we could detect IFN-γ production after stimulation (SI Appendix, Fig. S11A). Tetramer analysis revealed that T cells directly interacting with each of these epitopes could be identified, although the responses were not very strong (SI Appendix, Fig. S11B). The 9-mer (TVRSHCVSK) displayed the strongest binding stability which, together with the preference of K/R of HLA-A03:01 at the second anchor position, suggests that the 10-mer CMV epitope follows the predicted C-terminal extension binding mode (SI Appendix, Fig. S11C). However, we also observed residual binding of the K(9)L mutant (similar to what was observed in Fig. 3 for the first P9 mutant). To provide further evidence that the 10-mer CMV epitope follows the C-terminal extension binding mode, we tested the cross-reactivity of T cells with the 10- and 9-mer peptides (SI Appendix). We observed that T cells recognizing the 10-mer peptide were all cross-reactive with the 9-mer peptide (SI Appendix, Fig. S11D). This level of cross-reactivity is expected with C-terminal extensions since residues in contact with the T cell receptor (TCR) are structurally conserved but is not expected with bulging 10-mers. Altogether these results suggest that TVRSHCVSKI follows the predicted C-terminal extension binding mode and that C-terminally extended peptides can form bona fide CD8 T cell epitopes.
Discussion
MS analysis provides an unbiased view of HLA-I ligands that is not restricted by a priori assumptions on HLA-I binding specificity. Here, we capitalized on this premise to explore noncanonical binding modes of HLA-I ligands. Of the 54 alleles considered in this work, we found clear statistical evidences of C-terminal extensions for 8 of them and validated these predictions at the biochemical and structural level for some of frequent alleles in Caucasian populations.
In addition to providing evidence of C-terminal extensions, our work enabled us to characterize motifs describing this noncanonical binding mode. We observed that the C-terminal extensions are often characterized by the presence of hydrophobic residues. We also note that, for three of eight alleles (i.e., HLA-A03:01, HLA-A31:01, and HLA-A68:01), a positively charged residue is found at the second anchor. This positively charged residue interacts with D77 and D116 in our HLA-A68:01 structure, which is consistent with the fact that D is preferentially observed at these positions in alleles predicted to accommodate C-terminal extensions (Fig. 4C). The distinct binding specificity between the anchor residue and the C-terminal extension makes these cases especially amenable for the sequence-based model that we developed. However, the majority of alleles show preference for hydrophobic residues at the second anchor position. In these cases, and assuming that the preference for hydrophobic residues at the C-terminal extension is conserved, sequenced-based algorithms cannot unambiguously determine whether a 10-mer peptide with two hydrophobic amino acids at the last two positions follows the bulge or the C-terminal extension binding mode. Moreover, we anticipate that competition with high-affinity 9-mer ligands may mask cases of lower-affinity C-terminal extensions in HLA peptidomics data (e.g., peptides with only one good anchor residue). Therefore, our estimate of the number of alleles that accommodate C-terminal extensions corresponds to a lower bound, and we cannot exclude that this noncanonical binding mode may be observed in other alleles. In particular, some C-terminal extensions among HLA-A02:01 ligands may be present in our data with two hydrophobic residues at the last positions. Nevertheless, the smaller distance between the two alpha helices of HLA-A02:01 (Fig. 4D) suggests that C-terminal extensions for this allele should be rarer or involve other mechanisms like cross-presentation (9, 10). Along this line, we note that statistically significant C-terminal extensions are identified by our algorithm in the set of T. gondii HLA-A02:01 ligands (9) (SI Appendix, Table S2).
Finally, we stress that contaminations from coeluting or wrongly identified peptides, as well as challenges in aligning small peptides or in the deconvolution of pooled HLA peptidomics datasets, can easily result in cases that look like N- or C-terminal extensions. This is the reason why we developed the statistical framework described in this work and tested the robustness of our predictions with respect to noise.
Despite these inherent limitations of sequence-based approaches, it is likely that several alleles show very few, if any, C-terminal extensions. For instance, six alleles from our list showed specificity at the last anchor residue that is not restricted to hydrophobic amino acids (SI Appendix, Fig. S12), but we did not see any trend of C-terminal extensions among their 10-mer ligands (Dataset S2).
Our model also incorporated the possibility to have N-terminal extensions, but we did not find any such event, although N-terminal extensions have been recently reported (11, 12). Inspection of existing structures of HLA-I molecules in complex with 9-mer peptides shows that the C-terminal carboxyl group of 9-mer HLA-I ligands is often partly solvent exposed. Conversely, the N-terminal amide group points in general toward the binding site. This likely explains why N-terminal extensions appear to be much less frequent, although we cannot exclude that our approach may miss some N-terminal extensions if the specificity at P2 is the same as at P3 in the N-terminally extended ligands. Interestingly, this appears to be the case for N-terminal extensions reported for HLA-B57:01 and may explain why these extensions could not be easily detected with sequence-based approaches and have been first identified by X-ray crystallography (11).
Our demonstration of direct recognition by human CD8 T cells of a peptide predicted to follow the C-terminal extension binding mode indicates that these extensions are compatible with TCR binding. This did not come as a surprise since amino acids in contact with the TCR (typically positions 4–7) are structurally conserved between 9-mers and C-terminally extended 10-mers, which is consistent with the cross-reactivity observed in SI Appendix, Fig. S11D. Moreover, the vast repertoire of TCR enables recognition of many different types of epitopes (e.g., bulges, posttranslational modifications), so that the small changes in the positioning of the two alpha helices were not expected to prevent TCR recognition. C-terminal extensions may further play a role in the binding of KIR proteins, which are known to recognize amino acids surrounding the C terminus of canonical HLA-I ligands (11, 26, 27). Along these lines, we note that the HIV peptides in Table 1 represent a known escape mutant (NEF A83G), which was previously proposed to act at the level antigen processing (28). Our results suggest that this mutation may also have an effect on recognition of peptide–HLA complexes by NK receptors. As such, we anticipate that inclusion of C-terminal extensions in our HLA-I ligands predictor may help uncovering new CD8 T cell epitopes in viruses or tumors, as well as potential targets of NK cells.
Overall, our results reveal frequent C-terminal extensions in at least 8 of the 54 HLA-I alleles analyzed in this study and highlight the power of unbiased HLA peptidomics data together with new algorithms to unravel properties of HLA-I molecules. Our evidence of direct T cell recognition of such epitopes suggests that C-terminal extensions may be clinically relevant for infectious diseases or cancer immunotherapy.
Materials and Methods
Binding stability measurements were performed with standard refolding assays, and stable complexes were detected with ELISA. X-ray crystallography was carried out as described in SI Appendix. The model and structure factors have been deposited with PDB ID code 6EI2. PBMCs were peptide stimulated in vitro for 12 d in the presence of 100 U/mL IL-2. Subsequently, the Elispot was performed using the ELISpotPRO kit for Human IFN-γ from MABTECH. CD8 T cells of the CMV 10-mer peptide stimulated PBMC were stained with a PE-labeled HLA-A03:01/TVRSHCVSKI multimer and costained with anti-CD8 antibody (BC A94683). Multimer+ CD8+ T cells were analyzed at the BD ARIA III instrument equipped with the FACS Diva software.
The code for identifying C- and N-terminal extensions (MHCpExt) and the new version of the predictor (MixMHCpred1.1) are available at https://github.com/GfellerLab, and algorithmic details about the methods are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Julien Racle and Santiago Carmona for a critical reading of the manuscript, Anne Wilson and her collaborators of the flow cytometry facility, the members of our laboratories for their support, and Diamond Light Source for access to beamline I24 (proposal no. mx15433). Computations were performed at the Vital-IT (www.vital-it.ch) Center for high-performance computing of the Swiss Institute of Bioinformatics. D.G. acknowledges the financial support of Center for Advanced Modeling Science (CADMOS). S.P. and P.F. acknowledge support from Structural Genomics Consortium, a registered charity (no. 1097737) that receives funds from AbbVie; Bayer Pharma AG; Boehringer Ingelheim; Canada Foundation for Innovation; Eshelman Institute for Innovation; Genome Canada; Innovative Medicines Initiative (European Federation of Pharmaceutical Industries and Associations) (ULTRA-DD Grant 115766); Janssen, Merck, & Co.; Novartis Pharma AG; Ontario Ministry of Economic Development and Innovation; Pfizer; Sao Paulo Research Foundation; Takeda; and Wellcome Trust Grant 092809/Z/10/Z. P.F. and S.P. are supported by Wellcome Career Development Fellowship 095751/Z/11/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6EI2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717277115/-/DCSupplemental.
References
- 1.Trolle T, et al. The length distribution of class I-restricted T cell epitopes is determined by both peptide supply and MHC allele-specific binding preference. J Immunol. 2016;196:1480–1487. doi: 10.4049/jimmunol.1501721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abelin JG, et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreatta M, Alvarez B, Nielsen M. GibbsCluster: Unsupervised clustering and alignment of peptide sequences. Nucleic Acids Res. 2017;45:W458–W463. doi: 10.1093/nar/gkx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassani-Sternberg M, Gfeller D. Unsupervised HLA peptidome deconvolution improves ligand prediction accuracy and predicts cooperative effects in peptide-HLA interactions. J Immunol. 2016;197:2492–2499. doi: 10.4049/jimmunol.1600808. [DOI] [PubMed] [Google Scholar]
- 5.Jurtz V, et al. NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo HC, et al. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992;360:364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 7.Collins EJ, Garboczi DN, Wiley DC. Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature. 1994;371:626–629. doi: 10.1038/371626a0. [DOI] [PubMed] [Google Scholar]
- 8.Tenzer S, et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 9.McMurtrey C, et al. Toxoplasma gondii peptide ligands open the gate of the HLA class I binding groove. eLife. 2016;5:246. doi: 10.7554/eLife.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remesh SG, et al. Breaking confinement: Unconventional peptide presentation by major histocompatibility (MHC) class I allele HLA-A*02:01. J Biol Chem. 2017;292:5262–5270. doi: 10.1074/jbc.M117.776542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pymm P, et al. MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat Struct Mol Biol. 2017;24:387–394. doi: 10.1038/nsmb.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Lamothe PA, Walker BD, Wang J-H. Crystal structure of HLA-B*5801 with a TW10 HIV Gag epitope reveals a novel mode of peptide presentation. Cell Mol Immunol. 2017;14:631–634. doi: 10.1038/cmi.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritz D, et al. High-sensitivity HLA class I peptidome analysis enables a precise definition of peptide motifs and the identification of peptides from cell lines and patients’ sera. Proteomics. 2016;16:1570–1580. doi: 10.1002/pmic.201500445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stryhn A, Pedersen LO, Holm A, Buus S. Longer peptide can be accommodated in the MHC class I binding site by a protrusion mechanism. Eur J Immunol. 2000;30:3089–3099. doi: 10.1002/1521-4141(200011)30:11<3089::AID-IMMU3089>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Bassani-Sternberg M, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun. 2016;7:13404. doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mommen GPM, et al. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD) Proc Natl Acad Sci USA. 2014;111:4507–4512. doi: 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilton HG, et al. The intergenic recombinant HLA-B*46:01 has a distinctive peptidome that includes KIR2DL3 ligands. Cell Rep. 2017;19:1394–1405. doi: 10.1016/j.celrep.2017.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassani-Sternberg M, et al. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput Biol. 2017;13:e1005725. doi: 10.1371/journal.pcbi.1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang J, et al. Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science. 2017;358:1064–1068. doi: 10.1126/science.aao5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guasp P, et al. The peptidome of Behçet’s disease-associated HLA-B*51:01 includes two subpeptidomes differentially shaped by endoplasmic reticulum aminopeptidase 1. Arthritis Rheumatol. 2016;68:505–515. doi: 10.1002/art.39430. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gfeller D, et al. The multiple-specificity landscape of modular peptide recognition domains. Mol Syst Biol. 2011;7:484. doi: 10.1038/msb.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vita R, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malnati MS, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 27.Vivian JP, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chassin D, et al. Dendritic cells transfected with the nef genes of HIV-1 primary isolates specifically activate cytotoxic T lymphocytes from seropositive subjects. Eur J Immunol. 1999;29:196–202. doi: 10.1002/(SICI)1521-4141(199901)29:01<196::AID-IMMU196>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Niu L, et al. Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes. Mol Immunol. 2013;55:381–392. doi: 10.1016/j.molimm.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.