Significance
Mycobacteria, including the notorious pathogen Mycobacterium tuberculosis, possess a mycolic acid membrane that is a barrier to antibiotics. Although key enzymes that generate this structure are known, a full understanding of cell envelope assembly is lacking. We synthesized a fluorogenic analog of trehalose monomycolate, the building block used by mycolyltransferase enzymes to construct the mycolic acid membrane. When this analog, termed QTF, is processed in cells by mycolyltransferases, fluorescence is generated. Thus, mycolic acid membrane biosynthesis can be monitored in real time over several generations. Although mycolyltransferases are the most abundantly secreted proteins, we found that mycolyltransferase activity is localized. Finally, we show QTF has diagnostic features in that its processing by mycolyltransferases allows it to selectively detect mycobacteria.
Keywords: tuberculosis, cell wall, lipid, mycolic acid, Ag85
Abstract
Control and manipulation of bacterial populations requires an understanding of the factors that govern growth, division, and antibiotic action. Fluorescent and chemically reactive small molecule probes of cell envelope components can visualize these processes and advance our knowledge of cell envelope biosynthesis (e.g., peptidoglycan production). Still, fundamental gaps remain in our understanding of the spatial and temporal dynamics of cell envelope assembly. Previously described reporters require steps that limit their use to static imaging. Probes that can be used for real-time imaging would advance our understanding of cell envelope construction. To this end, we synthesized a fluorogenic probe that enables continuous live cell imaging in mycobacteria and related genera. This probe reports on the mycolyltransferases that assemble the mycolic acid membrane. This peptidoglycan-anchored bilayer-like assembly functions to protect these cells from antibiotics and host defenses. Our probe, quencher-trehalose-fluorophore (QTF), is an analog of the natural mycolyltransferase substrate. Mycolyltransferases process QTF by diverting their normal transesterification activity to hydrolysis, a process that unleashes fluorescence. QTF enables high contrast continuous imaging and the visualization of mycolyltransferase activity in cells. QTF revealed that mycolyltransferase activity is augmented before cell division and localized to the septa and cell poles, especially at the old pole. This observed localization suggests that mycolyltransferases are components of extracellular cell envelope assemblies, in analogy to the intracellular divisomes and polar elongation complexes. We anticipate QTF can be exploited to detect and monitor mycobacteria in physiologically relevant environments.
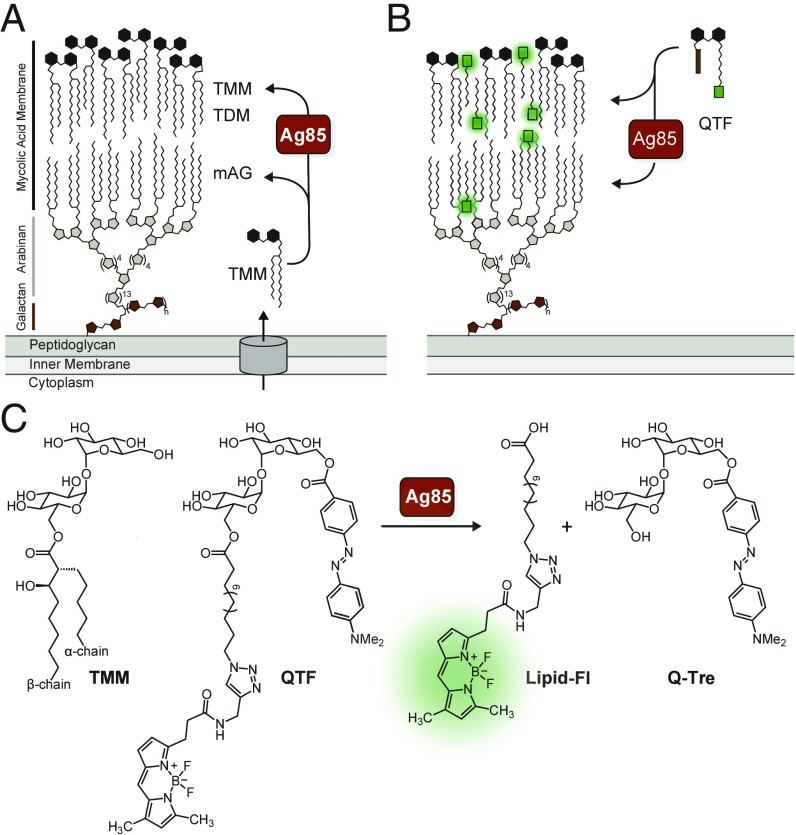
The Corynebacterineae suborder includes human pathogens that cause devastating diseases, such as Corynebacterium diphtheriae and Mycobacterium tuberculosis. These bacteria are distinguished from other prokaryotes by the unique composition of their cell envelope, whose major constituent is the mycolyl-arabinogalactan (mAG) (Fig. 1A). The mAG is assembled from an eclectic set of building blocks, including a branched polysaccharide constructed from galactofuranose and arabinofuranose, which is anchored to the peptidoglycan. Appended to this heteropolysaccharide are long chain (up to C90) α-branched, β-hydroxylated lipids, known as mycolic acids, arranged in a pseudobilayer orientation. Cell envelope mycolic acids are found as mono- and diesters of the disaccharide trehalose [trehalose monomycolate (TMM) and trehalose dimycolate (TDM)], or covalently linked to the cell wall as mAG (1–3). The mycolic acid membrane forms the interface of the cell with its environment, functions as a permeability barrier to antibiotics, and facilitates survival in host environments (4). Bacteria with this shared cell wall architecture display diversity in growth rates, cell size, and patterns of elongation and division (5–7). Modulation of these features enables pathogens to evade antibiotics and host defenses (8, 9). The importance of cell elongation and division in antibiotic function, disease latency, and microbe survival highlights the need to understand mAG biosynthesis and maintenance. Many of the enzymes that generate this physical barrier have been identified, but when, where, and how they function in live cells to construct and remodel the mAG is not known.
Fig. 1.
Features of QTF, a fluorogenic probe of Ag85-mediated cell wall biogenesis. (A) Components of the mycobacterial cell envelope include the peptidoglycan-anchored mycolyl-arabinogalactan (mAG) complex. The mycolyltransferases, including the antigen 85 complex (Ag85) of M. tuberculosis, construct the mycolic acid membrane by processing endogenous trehalose monomycolate (TMM) to afford trehalose dimycolate (TDM) or mycolyl-arabinogalactan (mAG). (B) QTF is a TMM mimic bearing a fluorophore and quencher. QTF processing by mycolyltransferases generates a fluorescent signal. (C) Chemical structure of TMM, QTF, and QTF products Q-Tre and lipid-FL generated by Ag85 cleavage.
Fluorescent reporters that can visualize cell envelope biosynthetic enzymes or their products are providing insight into cell growth and division. For example, protein and small molecule reporters have revealed that peptidoglycan (PG) biosynthesis in mycobacteria is asymmetric, with increased synthesis occurring at the older inherited pole. This growth mechanism has been invoked to explain the observation of drug resistance in subpopulations of M. tuberculosis (9, 10). Additionally, cytosolic enzymes that mediate galactan and peptidoglycan biosynthesis (i.e., the glycosyltransferases GlfT2 or MurG) localize to a subpolar region proximal to sites of newly emerging PG. The spatial localization of these intracellular enzymes has led to a model in which they form a mycobacterial polar elongation complex (10, 11). In contrast, information is lacking regarding the activity and localization of extracellular enzymes responsible for cell envelope assembly and maintenance.
Several groups have used the metabolic incorporation of labeled derivatives of trehalose to visualize the Corynebacterineae outer membrane (12–16). These probes exploit the action of metabolic proteins, including those that import and process trehalose. Mycolic acids originate in the cytoplasm, where they are synthesized, incorporated into TMM, and exported by a suite of proteins (Fig. 1A) (17–22). On the cell’s exterior, TMM is processed by mycolyltransferases to construct the mycolic acid membrane. All bacteria known to generate a mycolic acid membrane produce multiple mycolyltransferases (up to 11); M. tuberculosis possesses three, Ag85A–C mycolyltransferases (17). Mycolyltransferases carry out transesterification of TMM through an acyl-enzyme intermediate. This mycolyl-enzyme adduct persists until transfer to a mycolyl acceptor (e.g., trehalose, TMM, or AG), a reaction that can afford three different products: TDM, mAG, or TMM (SI Appendix, Fig. S1A). Trehalose analogs bearing fluorophores (12, 13) or possessing intermediary reactive handles (e.g., an azide or alkyne group) can be used to label mycobacteria (14–16). Such fluorescent or chemically reactive trehalose analogs are of limited use for real-time monitoring of bacterial growth. Fixing and washing steps are required because the trehalose analog precursor must be removed before visualization of the labeled metabolic products. The conversion of these probes can require the action of intracellular enzymes of the mycolic acid production pathway; thus, they do not report directly on Ag85 activity. In all cases, the timing and location of mycolyltransferase activity is lost. As a result, fluorescent probes have been lacking that report on mycolyltransferase activity during cell growth.
To address this deficiency, we developed quencher-trehalose-fluorophore (QTF), a fluorogenic probe for imaging mycolyltransferase activity. QTF, an analog of the mycolyltransferase donor TMM, is hydrolyzed by mycolyltransferases to provide a real-time fluorescent readout of mycolyltransferase activity and mycolic acid membrane assembly (Fig. 1 B and C). Additionally, QTF specifically detects bacteria with mycolic acid membranes, even in mixed cultures. In studies of single cells of Mycobacterium smegmatis, we visualized cells dividing over several generations. Although mycolyltransferases are the most abundantly secreted proteins by mycobacteria, our data reveal their activity is spatially and temporally localized.
Results
Fluorogenic Probe Design.
To create a fluorogenic reporter of Ag85 activity, we modified the natural donor TMM with a fluorophore-quencher pair using the available structural data and postulated mycolyl transfer mechanism. The substrate TMM is composed of the disaccharide trehalose whose 6-hydroxyl bears a mycolate substituent, which is a branched lipid with an α-chain and a longer β-chain (Fig. 1C). The structure of Ag85B bound to trehalose determined by X-ray crystallography [Protein Data Bank (PDB) ID code 1F0P] indicates the 6′-hydroxyl of the disaccharide points out of the sugar binding pocket and should tolerate chemical modification (SI Appendix, Fig. S1B) (12, 23). Similarly, TMM is thought to be oriented with the shorter α-branch of the mycolic acid residing in a hydrophobic tunnel, while the longer β-chain is outside of the active site (SI Appendix, Fig. S1C). We reasoned that mycolyltransferases would process substrates bearing a range of nonnatural lipids, by placing any bulky lipid groups in the binding groove occupied by the substrate β-chain. Accordingly, we designed a reporter in which the hydrophobic fluorescent dye BODIPY-FL was appended to the terminus of a simplified lipid chain and the fluorescence quencher DABCYL linked to the 6′-hydroxyl (SI Appendix, Fig. S1D). We reasoned that this potential Ag85 substrate would be dark, as DABCYL efficiently quenches the fluorescence emission of BODIPY-FL (24, 25). If QTF undergoes processing with nucleophilic attack by the catalytic serine residue, the release of DABCYL–6′-trehalose would separate the FRET pair and trigger fluorescence emission (26–29).
We sought a fluorogenic probe that mycolyltransferases would hydrolyze and thereby avoid covalent cell envelope modification. A reported kinetic study using the artificial substrate 6,6′-dihexanoyl trehalose revealed Ag85 exhibits substantial acylhydrolase activity when acting on simple lipid linkages, a reactivity orthogonal to its primary role as an acyltransferase (23). We therefore replaced the native α-branched, β-hydroxylated lipid head group of TMM with an unbranched lipid in QTF to promote hydrolysis of the acyl-enzyme intermediate. The active site homology evident in the X-ray crystal structures of Ag85A–C and in alignments of mycolyltransferase sequences suggest the design of QTF should allow broad utilization by mycolyltransferases across genera (30–32). Thus, our modified mycolyltransferase substrate should avoid covalent modification of the cell wall with a nonnatural lipid, minimize any perturbation of Ag85 function, and improve substrate turnover. In this way, QTF’s features should simplify the interpretation of fluorescence imaging data.
QTF Synthesis and Chemical Properties.
We synthesized QTF from a hexasilylated trehalose derivative by adapting a known route to maradolipid, a naturally occurring substituted trehalose derivative (SI Appendix, Fig. S2A) (33). Similarly, we prepared the predicted cleavage products (SI Appendix, Scheme S2B) as standards for analysis. QTF is freely soluble on dilution into aqueous buffer at all tested concentrations and maximally soluble up to 100 µM (in 2% DMSO). As expected, an equimolar mixture of the cleavage standards gave rise to a 1,500-fold increase in relative fluorescence emission corresponding to BODIPY-FL [λmax (excitation = 503 nm), λmax (emission = 515 nm)], while QTF lacks a measurable emission across these wavelengths (SI Appendix, Fig. S3). Solutions of QTF retained quenched fluorescence emission for extended periods at elevated temperatures (SI Appendix, Fig. S4). Lack of QTF fluorescence is presumably due to intramolecular fluorescence quenching by DABCYL, and we calculated a quenching efficiency of 99.9% between 10 nM and 10 µM (SI Appendix, Fig. S5). These observations confirm QTF has the spectral properties required for a “turn-on” probe.
QTF Processing by Mycolyltransferases.
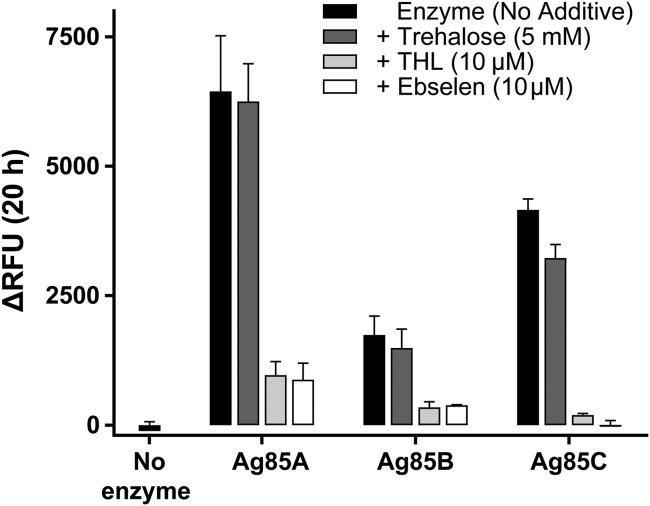
A robust increase in fluorescence occurred upon QTF exposure to Ag85A, Ag85B, or Ag8C (Fig. 2, black bars), the three catalytically active mycolyltransferases of M. tuberculosis. This result reveals that QTF can be cleaved by all M. tuberculosis mycolyltransferases. QTF does not undergo spontaneous hydrolysis, as no change in fluorescence emission was observed after 20 h at 37 °C. Probe turnover without a mycolyl acceptor present to intercept the acyl-enzyme intermediate suggested that the mycolyltransferases cleave QTF by hydrolysis. QTF processing could be accelerated in the presence of a mycolyl acceptor; therefore, we assessed fluorescence upon mixing QTF, an Ag85 homolog, and trehalose (Fig. 2, dark gray bars). No increase in fluorescence was observed with trehalose addition, a finding consistent with the proposed hydrolytic mechanism. To assess whether known Ag85 inhibitors block QTF turnover, we monitored fluorescence in the presence of tetrahydrolipstatin (THL) or ebselen. THL is a nonspecific lipase inhibitor that covalently modifies catalytic serine residues, whereas ebselen is reported to disrupt the Ag85 catalytic triad through allosteric covalent modification of a nonactive site cysteine (34). Addition of either THL (Fig. 2, light gray bars) or ebselen (Fig. 2, white bars) inhibited QTF processing, consistent with QTF hydrolysis mediated by the catalytic triad of Ag85.
Fig. 2.
Fluorescence resulting from QTF exposure to the native M. tuberculosis mycolyltransferases Ag85A, Ag85B, and Ag85C. Fluorescence emission was measured after incubation of QTF (1 μM) with purified M. tuberculosis Ag85A, Ag85B, or Ag85C (20 nM) for 20 h at 37 °C in 20 mM phosphate buffer (pH = 7.2) (black bars). Turnover was also assessed in the presence of a mycolyl acceptor (5 mM trehalose, dark gray bars) and generic mycolyltransferase inhibitors tetrahydrolipstatin (THL) (10 μM light gray) or ebselen (10 μM white). RFU, relative fluorescence units. Error bars depict SD.
We further examined mycolyltransferase processing of QTF by carrying out a Michaelis–Menten analysis of initial reaction velocities (SI Appendix, Fig. S6 and Table S1). The apparent Km values for QTF (Ag85A = 107 ± 27 µM, Ag85B = 28 ± 3 µM, and Ag85C = 44 ± 3 µM) are similar to the reported Michaelis constant for the natural mycolyl donor TMM (Km = 130 µM) (35). Additionally, the apparent kcat values reflect the relative activities measured by end-point analysis (Fig. 2 and SI Appendix, Fig. S6 and Table S1). Specifically, Ag85A cleaves QTF more efficiently than Ag85C, while the activity of Ag85B is lower; these relative rankings are consistent with the known mycolyltransferase activities of these enzymes (36). Thus, the kinetic parameters of mycolyltransferase-catalyzed QTF hydrolysis are consistent with those observed for mycolyltransferase activity.
QTF Is Selectively Processed by Bacteria with Mycolic Acid Membranes.
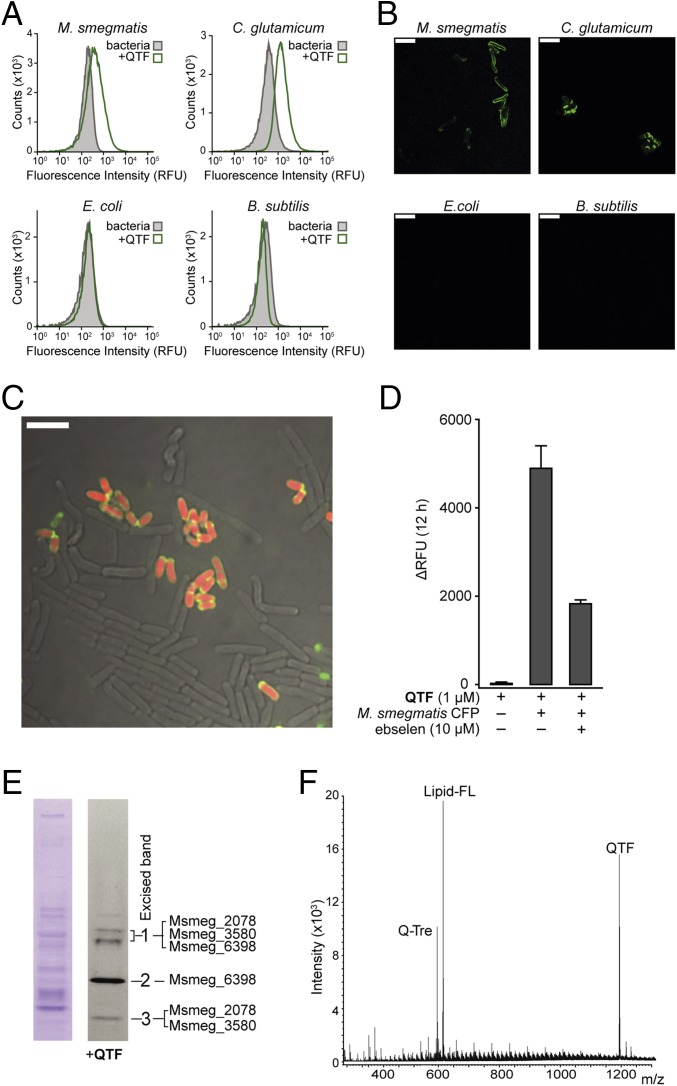
When species that contain mycolyltransferases, including M. smegmatis and Corynebacterium glutamicum, were treated with QTF (2.5 μM), bacterial growth was unaffected (SI Appendix, Fig. S7), but changes in bulk fluorescence emission of more than 10,000-fold were observed for each species (SI Appendix, Fig. S7A). We compared the fluorescence resulting from these mycolyltransferase-producing organisms to that from Escherichia coli and Bacillus subtilis, as the latter two species serve as representative Gram-negative and Gram-positive species that lack Ag85 orthologs. When cells were cultured with QTF (1 µM) for two to three doubling times and then analyzed using flow cytometry (Fig. 3A), either M. smegmatis and C. glutamicum became fluorescent. In contrast, neither E. coli nor B. subtilis cells lit up (SI Appendix, Fig. S8).
Fig. 3.
QTF processing by mycolyltransferases in live bacteria. (A) Flow cytometry analysis of M. smegmatis, C. glutamicum, E. coli, and B. subtilis with QTF. Cells were treated with QTF (1 µM) for two to three doubling times and their fluorescence was compared with that of untreated bacteria. Data are representative of two independent experiments (see SI Appendix, Fig. S8 for scatterplots and controls). (B) Confocal fluorescence microscopy images of different bacterial species exposed to QTF (1 µM) before flow cytometry analysis. (Scale bar: 5 µm.) (C) Confocal fluorescence microscopy overlay of all channels of a mixed culture [C. glutamicum (red), E. coli, and B. subtilis) exposed to QTF (1 µM). (D) Fluorescence emission from incubation of QTF (1 µM) with M. smegmatis culture filtrate protein (CFP) (200 ng) at 37 °C for 12 h. Probe turnover in the presence of ebselen (10 µM) was monitored. SE from three replicates is depicted. (E) M. smegmatis CFP (9 µg) analyzed by native-PAGE [(Left) Coomassie stain; (Right) immersion in QTF solution (2.5 µM)]. Fluorescent bands were excised and analyzed by protein digest mass spectrometry. Identified mycolyltransferase proteins are listed (see also SI Appendix, Fig. S10 and Tables S2 and S3 for protein list). (F) High-resolution mass spectrometry (HRMS) analysis following exposure of QTF (50 µM) to M. smegmatis CFP (1 µg). Signals corresponding to QTF and predicted cleavage standards Q-Tre and lipid-FL are labeled (see also Fig. 1 and SI Appendix, Fig. S11).
When QTF-treated bacteria were visualized by confocal fluorescence microscopy (Fig. 3B), fluorescence was observed at the cell surface of M. smegmatis and C. glutamicum, but not E. coli or B. subtilis. To test whether QTF could label mycolyltransferase-containing organisms within a community, we added the probe to mixed cultures of B. subtilis, E. coli, and C. glutamicum expressing cytoplasmic mCherry. The colocalization of QTF-derived and mCherry fluorescence (Fig. 3C) was observed only for C. glutamicum, a mycolyltransferase-producing species. These findings indicate that QTF is a fluorogenic probe of mycolyltransferase-producing bacteria.
QTF Processing in Live Cells.
To determine whether mycolyltransferases are responsible for QTF cleavage in live cells, we harvested culture filtrate protein (CFP) from M. smegmatis, which contain secreted Ag85 proteins, and we evaluated QTF processing (Fig. 3 D–F). Exposure of the CFP to QTF afforded large increases in fluorescence emission (Fig. 3D). Treatment of M. smegmatis CFP with the Ag85 inhibitor ebselen reduced fluorescence by 63%. This reduction is consistent with mycolyltransferase-mediated probe turnover. The residual activity is likely due to inefficient inhibition of M. smegmatis mycolyltransferases by ebselen (13). To examine this possibility, we purified four M. smegmatis Ag85 homologs and measured their inhibition by ebselen (SI Appendix, Fig. S9). These data indicate ebselen is less potent against M. smegmatis mycolyltransferases (23–52% inhibition) than M. tuberculosis Ag85A–C (78–99% inhibition).
CFP from M. smegmatis was treated with QTF and the resulting mixture was analyzed by nondenaturing PAGE (Fig. 3E and SI Appendix, Fig. S10). Exposure of the gel to QTF followed by fluorescence imaging revealed only three fluorescent regions. Each band was excised and subjected to protein digest mass spectrometry analysis, which revealed the presence of at least one mycolyltransferase protein in each band (SI Appendix, Table S2). To determine the products of QTF processing, the probe (100 µM) was incubated with M. smegmatis CFP and the resulting sample was analyzed by high-resolution MS. We identified intact QTF and its predicted cleavage standards Q-Tre and lipid-FL (Fig. 3F), but no other cleavage products were detected (Fig. 3E and SI Appendix, Fig. S11). These findings are consistent with data from TLC analysis of membrane extracts and released cell wall mycolates from whole cells treated with QTF. In all cases, lipid-FL was the only fluorescent product recovered from M. smegmatis and C. glutamicum (SI Appendix, Figs. S12 and S13). These experiments establish that QTF is activated via mycolyltransferase-catalyzed hydrolysis in living cells.
QTF as a Reporter of Cell Envelope Assembly.
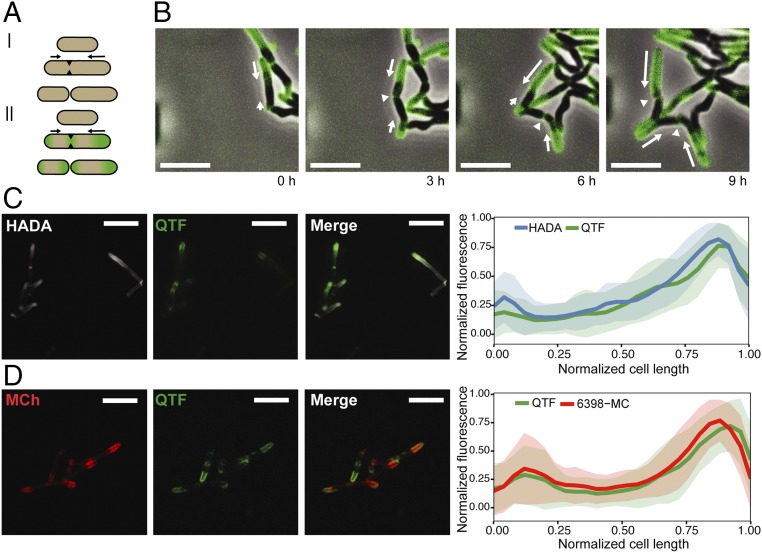
To examine the spatial and temporal dynamics of mycolyltransferase activity, we performed time-lapse microscopy of M. smegmatis exposed to QTF. We developed a microfluidic device (SI Appendix) to maintain cells in a single focal plane for imaging and to facilitate long-term growth by perfusion of growth media containing QTF. As a result, cells could be monitored for three to four generations before crowding effects precluded distinguishing specific activity of individual cells. QTF fluorescence was readily observable during growth. Moreover, because of the probe’s sensitivity, even low concentrations (i.e., 250 nM) afforded a robust signal with high contrast and low background. The fluorescence was localized to the septa and cell poles, with enhanced signal at the older poles (i.e., the poles inherited from the previous cell cycle) (Fig. 4 A and B, for videos see SI Appendix and Movie S1). To examine whether the fluorescence arising from mycolyltransferase activity is correlated with peptidoglycan biosynthesis, we incubated M. smegmatis with QTF and 7-hydroxycoumarin-3-carboxylic acid-3-amino-d-alanine (HADA), a fluorescent analog of d-alanine that is incorporated into nascent peptidoglycan (37, 38). Mycolyltransferase activity (i.e., QTF-derived fluorescence) was coincident with HADA-based fluorescence. Both signals colocalized at the faster growing, older pole (Fig. 4C).
Fig. 4.
Live-cell imaging of mycolyltransferase activity during growth and division of M. smegmatis. (A, I) Asymmetric growth model for mycobacteria. (A, II) Summary of QTF fluorescence during M. smegmatis growth. (B) M. smegmatis was exposed to QTF (250 nM) and the sample was continuously monitored in a custom microfluidic device. White arrows depict direction and magnitude of growth. (Scale bar: 5 µm.) (C) HADA (500 µM, pulse duration of 30 min, 10% of one doubling time) with 1 µM QTF. (Scale bar: 5 µm.) Colocalization analysis is reported for 71 cells. (D) M. smegmatis expressing an mCherry fusion to mycolyltransferase Msmeg_6398 treated with QTF (1 μM). (Scale bar: 5 µm.) Colocalization analysis is reported for 51 cells. Error bars reflect SDs from all measured cells.
Our results using QTF suggested mycolyltransferases are concentrated at the growing poles. As the cellular location of these enzymes was unknown, we generated C-terminal fusions of mCherry to three M. smegmatis mycolyltransferases (Msmeg_3580, Msmeg_6398, and Msmeg_6399) and assessed the localization of each protein (SI Appendix, Figs. S14–S17). Fluorescence was localized to the cell envelope, but we observed differential subcellular localizations. Msmeg_3580 and Msmeg_6398 were concentrated at polar and septal regions, while Msmeg_6399 was preferentially located at the septum. The differences in localization indicate that the mycolyltransferases may have distinct functions. To confirm that the observed localization of the mCherry fusion proteins was derived from secreted, extracellular mycolyltransferases, we generated signal sequence variants of Msmeg_3580, Msmeg_6398, and Msmeg_6399. Secretion through the twin-arginine translocation (Tat) pathway can be tested by modifying the twin-arginine motif in the signal sequences. When these residues were substituted with lysine, a change that inhibits secretion through the Tat (39, 40) pathway, cells remained fluorescent and exhibited the same localization patterns as observed with cells producing wild-type proteins. These experiments indicate that our mCherry mycolyltransferases are not secreted through the Tat system but rather likely through the Sec translocon. Cells harboring plasmids encoding mycolyltransferases lacking signal sequences displayed broad cytoplasmic fluorescence but showed no localized fluorescence at the poles or septa (SI Appendix, Fig. S18). When the strain producing Msmeg_6398-mCherry was exposed to QTF, colocalized fluorescence emission was asymmetric and distributed at the poles (Fig. 4D). Overall, the findings support our conclusion that QTF is a reporter of mycolyltransferase activity, and that extracellular mycolyltransferase activity is coincident with sites of peptidoglycan biosynthesis.
Discussion
Small molecule probes of cell envelope biogenesis typically report on peptidoglycan assembly, an essential process in virtually all bacterial species (37, 41). Probes of distinct classes of bacteria would enable selective visualization and diagnosis. Mycobacteria are distinguished from other prokaryotes by the unique composition of their cell envelope, notably their mycolic acid-rich outer membrane. The mycolic acid layer is constructed by mycolyltransferases known as Ag85A–C in M. tuberculosis. Although mycolyltransferases are the most abundantly secreted mycobacterial proteins, whether their activity is spatially or temporally controlled was unclear (42, 43).
Chemical probes of the mycolic acid membrane lipids can label mycobacteria (12, 14–16). These agents, however, were not designed to serve as real-time reporters of mycolyltransferase activity. These labeling strategies require either multiple steps or use fluorophore-substituted trehalose substrates whose fluorescence signal is indistinguishable from their metabolic products. We envisioned complementing the available tools with a mycolyltransferase-triggered fluorogenic probe. To this end, we synthesized QTF, a selective fluorogenic probe that obviates the need for washing and reports on mycobacterial cell wall assembly in live bacteria in real time.
Fluorescence derived from QTF arises through mycolyltransferase-promoted cleavage. Our data indicate this process occurs through the formation and subsequent hydrolysis of an acyl-enzyme intermediate (23). The fluorescent lipid product produced remains at the site of its generation because of the low diffusion rates of the mycobacterial membrane (13). Mycobacteria treated with QTF exhibit fluorescence at the cell poles and cell septum, indicating mycolyltransferase activity is localized to these sites. Thus, despite the abundant secretion of mycolyltransferases, they selectively associate with the cell envelope at the poles and septa. We also observed cell cycle-dependent mycolyltransferase activity: Midcell fluorescence increased only during septal formation, suggesting the localization of Ag85 to the septum is dynamic. Cytokinesis generates incipient poles in the subsequent daughter cells, consistent with the high temporal activity of Ag85 at the septum. Following division, we observed continued mycolyltransferase activity at both poles of the resulting daughter cells; however, the fluorescence at the new pole is markedly lower than that at the older inherited pole. These observations are consistent with the known sites of peptidoglycan biosynthesis detected using fluorescent peptidoglycan probes, and they highlight QTF’s ability to reveal temporal events in the mycobacterial cell cycle.
Our findings that mycolyltransferase activity is localized led us to examine the location of individual enzyme homologs. To this end, we generated C-terminal mCherry fusions of Msmeg_3580, Msmeg_6398, and Msmeg_6399. The resulting fluorescence aligned with that afforded from QTF. Still, there were differences in the details of mycolyltransferase localization. Specifically, Msmeg_6399 was concentrated at the septa, while Msmeg_3580 and Msmeg_6398 exhibited septal and biased polar localization (SI Appendix, Figs. S15–S17). These findings suggest the expression or localization of these enzymes is cell cycle dependent and that these enzymes have distinct roles in building and maintaining the mycolic acid membrane.
Compounds that inhibit proper mycolyltransferase localization could be efficacious, as deletion of a single mycolyltransferase can profoundly affect cell viability (44). Moreover, mycolyltransferases can engage in protein–protein interactions, as Msmeg_6398 has been found to interact with LprG, a lipid transport protein; this interaction could confine the enzyme within the cell envelope (45). Alternatively, since mycolyltransferases are secreted, their mechanism of export could influence their localization. The M. smegmatis mycolyltransferases in this study are likely exported by the Sec translocase system, which can localize proteins at specific subcellular sites, including the cell poles (46, 47).
Inhibitors of Ag85 have been sought as antibiotic leads. In vitro data obtained with QTF confirmed that the Ag85 inhibitor ebselen blocks the M. tuberculosis mycolyltransferases; however, we found ebselen was poorly effective against the tested M. smegmatis orthologs. The M. smegmatis enzymes we tested possess the conserved cysteine residue ebselen targets in M. tuberculosis Ag85 proteins; however, M. smegmatis also encodes an extended linker region directly preceeding the helix that the ebselen adduct has been demonstrated to obstruct (34). M. smegmatis mycolyltransferases may be sufficiently flexible to circumvent the inhibitory effect of the ebselen adduct. A major drawback of ebselen, is its lack of selectivity; it functions as a general cysteine modification reagent (48). The ineffectiveness of ebselen in M. smegmatis, and the lack of conservation of this cysteine among mycolyltransferases from other species, highlights the need for the continued development of mycolyltransferase inhibitors, especially for those enzymes produced by pathogens.
QTF’s ability to report on Ag85 activity provides unique opportunities. The inherent sensitivity of this fluorogenic probe, together with its requisite aqueous stability and solubility, render QTF an excellent screening tool. The assays we have described could be readily adapted to high-throughput screens for mycolyltransferase inhibitors. Compounds that block Ag85 activity would augment existing antitubercular drug regimens. Moreover, this mycolyltransferase probe could be employed as a diagnostic tool to detect or selectively monitor mycobacteria in mixed bacterial populations. Our findings indicate QTF can be used in mixed populations to sensitively and rapidly detect mycolic acid-containing bacteria with no washing. In this way, the properties of QTF have the potential to enhance current microscopy-based diagnostic methods that examine patient sputum for the presence of mycobacteria. QTF has the potential to reduce the number of processing steps and duration of staining protocols by eliminating the need for washing. Additionally, the probe may afford improved accuracy and sensitivity in mycobacterial cell identification—traits that are vital for high-burden regions of M. tuberculosis with limited resources.
Materials and Methods
Synthetic details and characterization of all previously unreported compounds are provided in SI Appendix. This document also contains information regarding the origin or generation of strains, plasmids, proteins, and compounds described herein, as well as protocols for all fluorescence assays, mass spectrometric assays, and microscopic imaging.
Supplementary Material
Acknowledgments
We thank Alexander Justen and Dr. Darryl Wesener for helpful discussions; Dr. Elle Grevstad [University of Wisconsin (UW)–Madison Biochemistry Optical Core] and Dr. Dan Stevens [UW–Madison Biophysics Instrumentation Facility (BIF)] for experimental assistance. The BIF was established with support from Grants BIR-9512577 [National Science Foundation (NSF)] and S10RR13790 (NIH). We also acknowledge the help of Dr. Greg Barrett-Wilt and Grzegorz Sabat [UW–Madison Biotechnology Center Mass Spectrometry/Proteomics Facility, which was supported by the NIH (P50 GM64598 and R33 DK070297) and the NSF (DBI-0520825 and DBI-9977525). This work was also supported by the National Institute of Allergy and Infectious Diseases Grant R01-AI126592 (to L.L.K.), the National Institutes of General Medical Sciences Grant F32GM113454 (to R.A.B.), and Grant F31 GM 108408 (to H.L.H.). H.L.H. also thanks the Chemistry–Biology Interface Training Program (UW–Madison) (T32-GM008505) for support. D.B.W. was supported by the Human Frontiers Science Program (RGY0076/2013).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720996115/-/DCSupplemental.
References
- 1.Crellin PK, Luo C-Y, Morita YS. Metabolism of plasma membrane lipids in mycobacteria and corynebacteria. In: Baez RV, editor. Lipid Metabolism. InTech; Rijeka, Croatia: 2013. [Google Scholar]
- 2.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 3.Crick DC, Mahapatra S, Brennan PJ. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology. 2001;11:107R–118R. doi: 10.1093/glycob/11.9.107r. [DOI] [PubMed] [Google Scholar]
- 4.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 5.Kieser KJ, Rubin EJ. How sisters grow apart: Mycobacterial growth and division. Nat Rev Microbiol. 2014;12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyce G, et al. Cell division site placement and asymmetric growth in mycobacteria. PLoS One. 2012;7:e44582. doi: 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botella H, et al. Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. MBio. 2017;8:e01183-17. doi: 10.1128/mBio.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kester JC, Fortune SM. Persisters and beyond: Mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit Rev Biochem Mol Biol. 2014;49:91–101. doi: 10.3109/10409238.2013.869543. [DOI] [PubMed] [Google Scholar]
- 9.Aldridge BB, et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meniche X, et al. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA. 2014;111:E3243–E3251. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumgart M, Schubert K, Bramkamp M, Frunzke J. Impact of LytR-CpsA-Psr proteins on cell wall biosynthesis in Corynebacterium glutamicum. J Bacteriol. 2016;198:3045–3059. doi: 10.1128/JB.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backus KM, et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat Chem Biol. 2011;7:228–235. doi: 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Rivera FP, Zhou X, Theriot JA, Bertozzi CR. Visualization of mycobacterial membrane dynamics in live cells. J Am Chem Soc. 2017;139:3488–3495. doi: 10.1021/jacs.6b12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swarts BM, et al. Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc. 2012;134:16123–16126. doi: 10.1021/ja3062419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foley HN, Stewart JA, Kavunja HW, Rundell SR, Swarts BM. Bioorthogonal chemical reporters for selective in situ probing of mycomembrane components in mycobacteria. Angew Chem Int Ed Engl. 2016;55:2053–2057. doi: 10.1002/anie.201509216. [DOI] [PubMed] [Google Scholar]
- 16.Kavunja HW, et al. A chemical reporter strategy for detecting and identifying O-mycoloylated proteins in Corynebacterium. Chem Commun (Camb) 2016;52:13795–13798. doi: 10.1039/c6cc07143k. [DOI] [PubMed] [Google Scholar]
- 17.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 18.Portevin D, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci USA. 2004;101:314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavalda S, et al. The polyketide synthase Pks13 catalyzes a novel mechanism of lipid transfer in mycobacteria. Chem Biol. 2014;21:1660–1669. doi: 10.1016/j.chembiol.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Lea-Smith DJ, et al. The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J Biol Chem. 2007;282:11000–11008. doi: 10.1074/jbc.M608686200. [DOI] [PubMed] [Google Scholar]
- 21.Yamaryo-Botte Y, et al. Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem Biol. 2015;10:734–746. doi: 10.1021/cb5007689. [DOI] [PubMed] [Google Scholar]
- 22.Tahlan K, et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry CS, Backus KM, Barry CE, 3rd, Davis BG. ESI-MS assay of M. tuberculosis cell wall antigen 85 enzymes permits substrate profiling and design of a mechanism-based inhibitor. J Am Chem Soc. 2011;133:13232–13235. doi: 10.1021/ja204249p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose TM, Prestwich GD. Fluorogenic phospholipids as head group-selective reporters of phospholipase A activity. ACS Chem Biol. 2006;1:83–91. doi: 10.1021/cb5000014. [DOI] [PubMed] [Google Scholar]
- 25.Feng L, et al. A real-time fluorogenic phospholipase A(2) assay for biochemical and cellular activity measurements. Chem Biol. 2002;9:795–803. doi: 10.1016/s1074-5521(02)00168-0. [DOI] [PubMed] [Google Scholar]
- 26.Watzke A, et al. Selective activity-based probes for cysteine cathepsins. Angew Chem Int Ed Engl. 2008;47:406–409. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- 27.Folk DS, Torosian JC, Hwang S, McCafferty DG, Franz KJ. Monitoring β-secretase activity in living cells with a membrane-anchored FRET probe. Angew Chem Int Ed Engl. 2012;51:10795–10799. doi: 10.1002/anie.201206673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-López AM, Soria-Gila ML, Marsden ER, Lilienkampf A, Bradley M. Fluorogenic substrates for in situ monitoring of caspase-3 activity in live cells. PLoS One. 2016;11:e0153209. doi: 10.1371/journal.pone.0153209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav AK, et al. Fluorescence-quenched substrates for live cell imaging of human glucocerebrosidase activity. J Am Chem Soc. 2015;137:1181–1189. doi: 10.1021/ja5106738. [DOI] [PubMed] [Google Scholar]
- 30.Ronning DR, et al. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat Struct Biol. 2000;7:141–146. doi: 10.1038/72413. [DOI] [PubMed] [Google Scholar]
- 31.Anderson DH, Harth G, Horwitz MA, Eisenberg D. An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (antigen 85B), a mycolyl transferase. J Mol Biol. 2001;307:671–681. doi: 10.1006/jmbi.2001.4461. [DOI] [PubMed] [Google Scholar]
- 32.Ronning DR, Vissa V, Besra GS, Belisle JT, Sacchettini JC. Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J Biol Chem. 2004;279:36771–36777. doi: 10.1074/jbc.M400811200. [DOI] [PubMed] [Google Scholar]
- 33.Sarpe VA, Kulkarni SS. Synthesis of maradolipid. J Org Chem. 2011;76:6866–6870. doi: 10.1021/jo200979n. [DOI] [PubMed] [Google Scholar]
- 34.Favrot L, et al. Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat Commun. 2013;4:2748. doi: 10.1038/ncomms3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elamin AA, Stehr M, Oehlmann W, Singh M. The mycolyltransferase 85A, a putative drug target of Mycobacterium tuberculosis: Development of a novel assay and quantification of glycolipid-status of the mycobacterial cell wall. J Microbiol Methods. 2009;79:358–363. doi: 10.1016/j.mimet.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Belisle JT, et al. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 37.Kuru E, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuru E, Tekkam S, Hall E, Brun YV, Van Nieuwenhze MS. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 2015;10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonough JA, et al. Identification of functional Tat signal sequences in Mycobacterium tuberculosis proteins. J Bacteriol. 2008;190:6428–6438. doi: 10.1128/JB.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem. 2000;275:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 41.Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci USA. 2014;111:5456–5461. doi: 10.1073/pnas.1322727111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dautin N, et al. Mycoloyltransferases: A large and major family of enzymes shaping the cell envelope of Corynebacteriales. Biochim Biophys Acta. 2017;1861:3581–3592. doi: 10.1016/j.bbagen.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Nataraj V, et al. Mycolic acids: Deciphering and targeting the Achilles’ heel of the tubercle bacillus. Mol Microbiol. 2015;98:7–16. doi: 10.1111/mmi.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson M, et al. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999;31:1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 45.Touchette MH, et al. A screen for protein-protein interactions in live mycobacteria reveals a functional link between the virulence-associated lipid transporter LprG and the mycolyltransferase antigen 85A. ACS Infect Dis. 2017;3:336–348. doi: 10.1021/acsinfecdis.6b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsui HC, Keen SK, Sham LT, Wayne KJ, Winkler ME. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. MBio. 2011;2:e00202-11. doi: 10.1128/mBio.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose P, Fröbel J, Graumann PL, Müller M. Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS One. 2013;8:e69488. doi: 10.1371/journal.pone.0069488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azad GK, Tomar RS. Ebselen, a promising antioxidant drug: Mechanisms of action and targets of biological pathways. Mol Biol Rep. 2014;41:4865–4879. doi: 10.1007/s11033-014-3417-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.