Significance
Pattern recognition receptors control the cellular entry of pathogenic as well as symbiotic microbes. While ligand-induced changes in receptor mobility at the plasma membrane and their localization in membrane nanodomains are general features, the molecular mechanism and the biological relevance of this phenomenon have remained unknown. Here we show that immobilization of the symbiotic cell entry receptor LYK3 in nanodomains requires the presence of actin and two molecular scaffold proteins, FLOT4 and SYMREM1. While FLOT4 forms the initial core structure, infection-induced expression and subsequent physical interaction of SYMREM1 with LYK3 stabilize the activated receptors in membrane nanodomains. This recruitment prevents stimulus-dependent endocytosis and ensures progression of the primary infection thread into root cortical cells.
Keywords: membrane nanodomain, symbiosis, scaffold proteins, receptor, remorin
Abstract
Plant cell infection is tightly controlled by cell surface receptor-like kinases (RLKs). Like other RLKs, the Medicago truncatula entry receptor LYK3 laterally segregates into membrane nanodomains in a stimulus-dependent manner. Although nanodomain localization arises as a generic feature of plant membrane proteins, the molecular mechanisms underlying such dynamic transitions and their functional relevance have remained poorly understood. Here we demonstrate that actin and the flotillin protein FLOT4 form the primary and indispensable core of a specific nanodomain. Infection-dependent induction of the remorin protein and secondary molecular scaffold SYMREM1 results in subsequent recruitment of ligand-activated LYK3 and its stabilization within these membrane subcompartments. Reciprocally, the majority of this LYK3 receptor pool is destabilized at the plasma membrane and undergoes rapid endocytosis in symrem1 mutants on rhizobial inoculation, resulting in premature abortion of host cell infections. These data reveal that receptor recruitment into nanodomains is indispensable for their function during host cell infection.
Physical recruitment of membrane-resident ligand-binding receptors into membrane nanodomains is as a common phenomenon within the plant kingdom (1, 2). It represents a possible generic theme for the activation and maintenance of receptor-controlled signal transduction. Indeed, a number of plant receptor-like kinases (RLKs), as well as receptor-associated and immunity-related proteins, localize to nanodomains (1–3). Given the diversity of coexisting nanodomains (4), these membrane compartments might serve as active hubs that allow a spatially confined assembly of pathway-specific and functionally related signaling complexes (3).
The ability of multivalent and nanodomain-localized molecular scaffold proteins to associate and/or colocalize with RLKs and other signaling proteins (5–8) makes them potent candidates to act as central integrators of these membrane subcompartments. In plants, plasma membrane-resident scaffolding functions have been proposed for flotillins and remorins (9). These soluble proteins associate with the inner leaflet of the plasma membrane and interact with different partners (6, 8, 10–15), contributing to phytohormone signaling (5), developmental processes (13, 16), abiotic stress responses (17, 18), plant–virus interactions (14, 15, 19, 20) and root nodule symbiosis (6, 7, 11). The latter mutualistic interaction between rhizobia and legumes is initiated at the epidermis by a highly specific molecular dialog between the two partners (21).
In Medicago truncatula, the primary perception of rhizobial lipo-chitooligosaccharides, the so-called “Nod factors” (NFs), and subsequent colonization of host root hair cells by symbiotic bacteria is tightly controlled by LysM-type RLKs, such as the ligand-binding receptor NOD FACTOR PERCEPTION (NFP) and the coreceptor LYSINE MOTIF KINASE 3 (LYK3) (22, 23). Rhizobial infection is preceded by NF- and microbe-triggered changes in root hair morphology (i.e., root hair deformation and root hair curling) (24). This results in inclusion of rhizobia and formation of an infection chamber inside the root hair curl, which is followed by an invagination of the host cell plasma membrane and the subsequent formation of an infection thread (IT) (25). Molecularly, infection-induced activation of LYK3 results in a transition of its membrane-partitioning including restricted lateral mobility and accumulation in FLOTILLIN 4 (FLOT4)-labeled nanodomains (26). Along with FLOT4, two other scaffolds, FLOT2 and the remorin SYMREM1, contribute to host-driven infection control (6, 11) in a so-far elusive molecular manner. Here we used rhizobial infections as an appropriate biological system to unravel the spatiotemporal sequence of molecular events that control protein patterning at the host cell surface. In addition, we genetically tested whether receptor recruitment into nanodomains is functionally relevant.
Results and Discussion
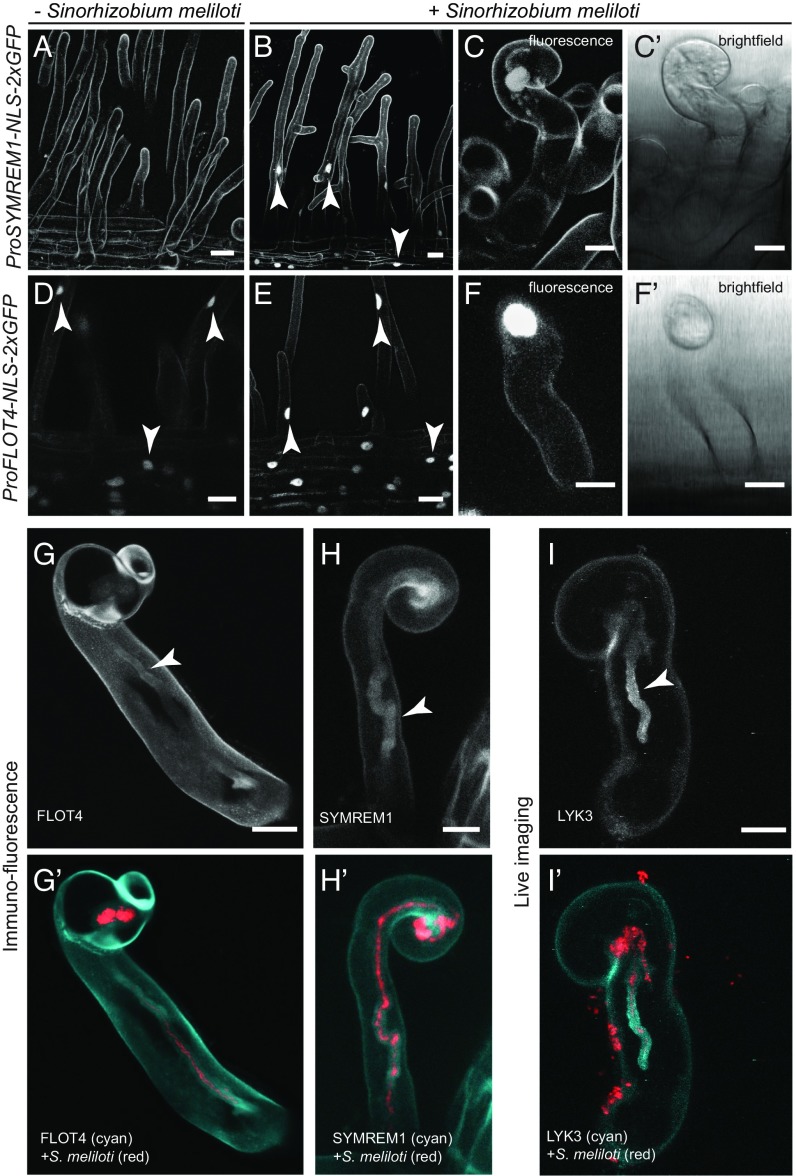
To precisely define the experimental setup, we first determined whether SYMREM1 and FLOT4 function in the same pathway during rhizobial infections of M. truncatula root hairs. Since stringent control over infection requires tightly coordinated cell type-specific regulatory networks, two transcriptional reporters expressing nuclear-targeted tandem GFPs (ProSYMREM1-NLS-2xGFP and ProFLOT4-NLS-2xGFP) were independently transformed into M. truncatula wild-type (WT) roots. For SYMREM1, no nuclear reporter activity was observed in the absence of Sinorhizobium meliloti (Fig. 1A), while nuclear fluorescence was clearly detected in curling as well as adjacent trichoblasts and atrichoblasts at 1 d after rhizobial inoculation (Fig. 1 B–C′). In contrast, the FLOT4 promoter was constitutively active at low levels in all epidermal cells of the infection zone even in the absence of rhizobia (Fig. 1D) and was further induced on inoculation (Fig. 1 E–F′).
Fig. 1.
Expression and localization analyses of SYMREM1 and FLOT4. Monitoring promoter activation of SYMREM1 (A–C′) and FLOT4 (D–F′) with cellular resolution in transgenic M. truncatula root hairs using the genetically encoded reporters ProSYMREM1:NLS:2xGFP and ProFLOT4:NLS:2xGFP, respectively. (A and B) Activation of SYMREM1 as indicated by nuclear fluorescence (arrowheads) in uninoculated (A) and S. meliloti 2011 (mCherry) inoculated (B) roots. (C, C′, F, and F′) Close-ups of individual curled root hairs with fluorescent nuclei. (A–C′) Autofluorescent contours of roots hairs and epidermal cells are visible due to ultra-sensitive imaging settings that were chosen due to low signal intensities. (D–F′) Activation of the FLOT4 promoter in uninoculated (D) and inoculated (E) conditions (arrowheads). (G–H′) Immunofluorescence targeting FLOT4-GFP (G and G′) and GFP-SYMREM1 (H and H′), both expressed from their endogenous promoters. (I and I′) Transgenic line expressing a ProLYK3::LYK3-GFP construct. Arrowheads indicate the localization of the respective protein around the ITs. (Scale bars: 20 µm in A, B, D, and E; 10 µm in C, C′, F, F′, G, H, and I.)
To precisely localize the encoded proteins during root hair infection, we independently expressed FLOT4 and SYMREM1 as GFP fusion proteins under the control of their respective endogenous promoters in the A17 WT background. Due to low signal intensities, GFP fluorescence was enhanced by immune-staining using Atto488-coupled anti-GFP-nanobodies. Both scaffold proteins localized to IT membranes and accumulated around the primary IT and around the infection chamber in curled root hairs at 7 d postinoculation (dpi) with S. meliloti (Fig. 1 G–H′). This resembles the localization pattern of LYK3 when expressed as a GFP-fusion protein driven by the LYK3 promoter (Fig. 1 I and I′) and described previously (26). Since fluorescent signals of SYMREM1 and FLOT4 around ITs were comparably low, we inoculated WT roots with mCherry-labeled rhizobia and subjected these roots to the same immunofluorescence protocol. While fluorescent signals were found along ITs on plants expressing SYMREM1 (Fig. S1 A–A″) and FLOT4 (Fig. S1 B–B″) as GFP fusion proteins, no signal was detected along ITs of control plants when being imaged with the identical microscope settings (Fig. S1 C–C″), indicating that the obtained signals were not of an autofluorescent nature.
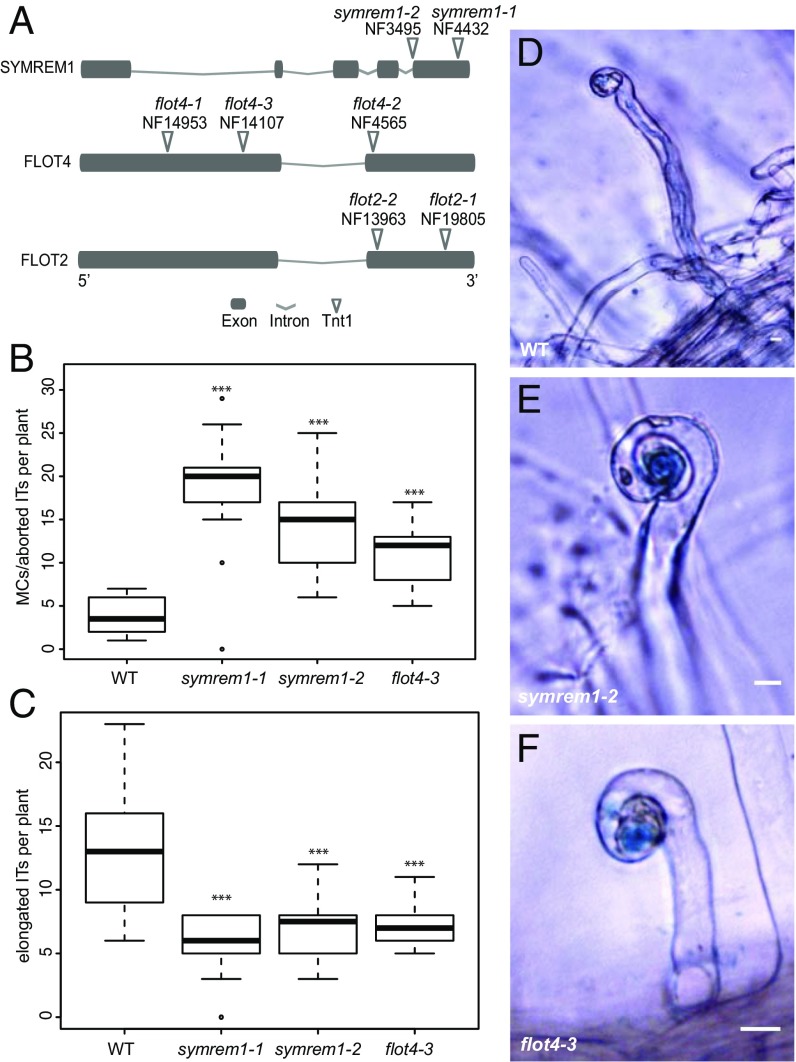
To date, the importance of flotillins and SYMREM1 during primary root hair infection has been assessed only through RNAi-mediated knockdown in transgenic roots (6, 11). To test overlapping functions genetically, we isolated three independent flot4 mutant alleles—NF14593 (flot4-1), NF4565 (flot4-2), and NF14107 (flot4-3) (Fig. 2A)—by PCR screening of the Tnt1 transposon insertion population at the Noble Foundation (27). While we were unable to recover homozygous offspring of the flot4-1 allele, transcript levels were not significantly decreased in homozygous T2 flot4-2 plants. However, FLOT4 mRNA was barely detectable in flot4-3 mutant plants, confirming that it can be considered a transcriptional null allele (Fig. S2A). For FLOT2, two independent lines were obtained (Fig. 2A). However, despite PCR-based screening of 64 individuals for flot2-1 (NF19805) and 95 individuals for flot2-2 (NF13963), no homozygous T2 plants were recovered, indicating the lethality of this mutation. A similar phenotype was reported for AtFLOT1, the closest homolog of MtFLOT2 from Arabidopsis thaliana (13). Consequently, we excluded FLOT2 from further analysis.
Fig. 2.
symrem1 and flot4 mutants are impaired in IT initiation. (A) Mapped Tnt1 transposon insertions. Phenotypical analysis of symrem1 and flot4 mutants. (B and C) Infection structures were scored at 7 d after inoculation with S. meliloti (lacZ) and classified into microcolony (MC)/aborted ITs that were arrested within trichoblasts (B) and elongated ITs that progressed into the root cortex (C). (D–F) Representative elongated ITs as found in WT (D), and infections arrested in the MC stage in symrem1-2 (E) and flot4-3 (F) mutants. In B and C, ***P < 0.001, Tukey–Kramer multiple-comparison test. (Scale bars: 10 μm.)
In addition to a previously identified symrem1 insertion line (NF4432; symrem1-1) (6), we also identified a second mutant allele (NF3495; symrem1-2) (Fig. 2A). SYMREM1 transcripts were significantly reduced in both alleles when plants were inoculated for 24 h with S. meliloti (Fig. S2B). We phenotypically characterized these mutants by assessing the primary infection success of rhizobia at 7 dpi. For this, we scored infected root hairs containing microcolonies (MCs)/aborted infections or elongated ITs reaching into cortical cells. In the flot4-3 (n = 14), symrem1-1 (n = 18), and symrem1-2 (n = 18) mutants, rhizobia were trapped inside of curled root hairs mostly at the microcolonies in 60%, 76%, and 67% of the cases, respectively (Fig. 2 B, E, and F). Consequently, only 40% of all elongated ITs in flot4-3, 24% of those in symrem1-1, and 33% of those in symrem1-2 passed the epidermis/cortical cell boundary, while this was observed in 78% of all infections in the WT (Fig. 2 C and D).
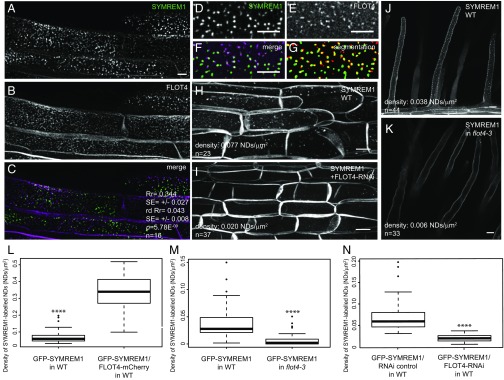
We next tested whether these proteins colocalize in the same type of membrane nanodomain. For this, we applied high-resolution live cell imaging of root epidermal cells followed by advanced quantitative image analysis of all observations. For evaluating colocalization experiments, we further refined our original quantitative image analysis workflow (3, 4). We compared the Pearson correlation coefficients (Rr) thus obtained with values generated on the corresponding image pairs in which blocks of 10 × 10 pixels in one channel were randomized (randomized Rr = rdRr). Global image analyses combined with image randomizations provide statistically sound statements on the nature of colocalizations (positive, negative, or random). Solely expressed SYMREM1 labeled nanodomains in root epidermal cells with a density of 0.077 domains/μm2 (Fig. 3 H and L). Coexpression of FLOT4 and SYMREM1 in roots in the A17 WT background revealed significant degrees of colocalization of both proteins in these cells, with an average Pearson correlation coefficient of Rr = 0.344 ± 0.027 (Fig. 3 A–G), as well as a 4.5-fold increase in SYMREM1-labeled nanodomains (Fig. 3L). Reciprocally, the density values for SYMREM1-labeled nanodomains in root hairs were decreased by more than sixfold when expressing a YFP-SYMREM1 construct in the flot4-3 mutant background compared with WT roots transformed with the same construct (Fig. 3 J, K, and M). Similar results were also obtained in epidermal cells when expressing a previously published FLOT4-RNAi construct (11) together with the YFP-SYMREM1 using the Medicago A17 accession as a genetic background (Fig. 3 H, I, and N). These data clearly demonstrate that SYMREM1 is recruited into nanodomains in a FLOT4-dependent manner, with FLOT4 serving as a central hub during primary nanodomain assembly.
Fig. 3.
SYMREM1 is recruited to nanodomains (NDs) in a FLOT4-dependent manner. (A–C) GFP-SYMREM1 (A) and FLOT4-mCherry (B) colocalized (C) in transgenic M. truncatula root epidermal cells. Quantification data are depicted within images. Rr, Pearson correlation coefficient; rdRr, Pearson correlation coefficient obtained after image randomization of the FLOT4-mCherry channel; P, confidence interval obtained from a Student t test comparing Rr and rdRr with the number of replicates (n) indicated in the panels. (D and E) Close-up views of nanodomain-localized GFP-SYMREM1 (D) and FLOT4-Cherry (E) at the cell surface. (F and G) Colocalization was quantified in merged images (F) and after image segmentation (G). (H–J) Density of GFP-SYMREM1-labeled nanodomains was greatly reduced on coexpression with a FLOT4-RNAi construct in root epidermal cells (I) and in flot4-3 mutant root hairs (K) compared with the respective controls (H and J). (L) Density of GFP-SYMREM1–labeled nanodomains was significantly increased when coexpressed with FLOT4-mCherry. (M and N) Quantification on GFP-SYMREM1–labeled nanodomains in the flot4-3 mutant (M) and FLOT4-RNAi (N) roots. ****P < 0.0001, Student t test. (Scale bars: 5 μm in A–F; 10 μm in H–K.)
Since associations of remorins with the cytoskeleton have been reported (4, 28, 29), we tested whether SYMREM1-recruitment to nanodomains requires an intact cytoskeleton. Pilot experiments in which microtubules (Fig. S3 A and B) and actin (Fig. S3 C and D) were successfully destabilized on treatment with oryzalin and cytochalasin D, respectively, confirmed the functionality of the treatment in M. truncatula roots. Indeed, application of cytochalasin D resulted in a strong and highly significant reduction in SYMREM1 nanodomain density (Fig. S3 E, F, and H), but did not block the association of SYMREM1 with the plasma membrane per se. However, at this stage, possibly indirect effects should also be considered, as application of actin destabilizing agents has a number of secondary consequences, including altered cytosolic streaming and export of transcripts from the nucleus. In contrast, destabilization of microtubules by oryzalin did not significantly affect SYMREM1 domain patterning (Fig. S3 E, G, and I).
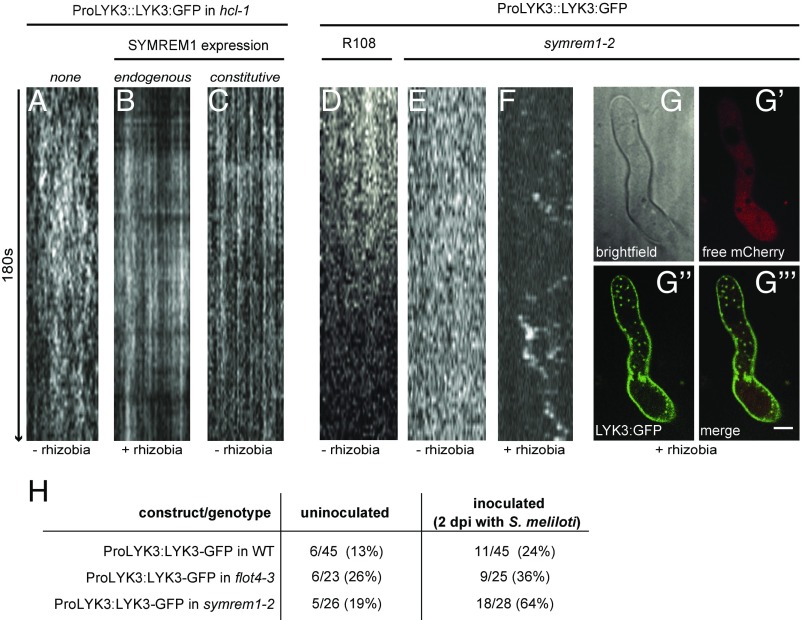
Since SYMREM1 can physically associate with LYK3, and LYK3 immobilization in FLOT4-labeled nanodomains temporally coincides with induced SYMREM1 expression (6, 26), we tested whether the presence of SYMREM1 at the onset of rhizobial infection of root hairs controls the stimulus-dependent recruitment of LYK3 into these membrane structures. For this, we first verified LYK3 mobility patterns under our laboratory conditions using the same genetic material (hcl-1 mutant complemented with a ProLYK3:LYK3-GFP construct) and imaging conditions described previously (26). Time-lapse imaging of LYK3-GFP driven by its endogenous promoter was performed over 180 s, and image sections were subjected to kymograph analysis. Vertical band and diffuse patterns indicate locally immobile protein clusters and protein mobility, respectively (Fig. 4). The receptor was indeed mobile in uninoculated roots, with dwell times of <30 s (19 of 20 roots) (Fig. 4A and Table S1). Inoculation of these roots with S. meliloti for 16 h significantly increased dwell times to >180 s in 9 of 9 roots (Fig. 4B and Table S1), confirming ligand-induced immobilization of LYK3 (26). Strikingly, this nanodomain recruitment of LYK3 was also observed when SYMREM1 was ectopically expressed in uninoculated roots, that is, in the absence of endogenous SYMREM1, with the majority of dwell times exceeding 180 s (Fig. 4C and Table S1). Thus, the presence of SYMREM1 is sufficient for the immobilization of LYK3 in nanodomains and independent of any other infection-induced genes.
Fig. 4.
SYMREM1 negatively regulates infection-triggered receptor endocytosis by stabilizing LYK3 in nanodomains. Shown are kymograph analyses of time-lapse image series taken from cell surfaces at the base of M. truncatula root hairs. LYK3 was mobile in the absence of rhizobia (A) and became immobilized at 16 h after rhizobial inoculation, i.e., at the onset of infection (B). The same effect was obtained when ectopically expressing SYMREM1 in the absence of rhizobia (i.e., in the absence of endogenous SYMREM1) (C). Analysis of uninoculated transgenic M. truncatula roots expressing LYK3-GFP under its native promoter revealed receptor mobility in WT R108 (D) and the symrem1-2 (E) mutant background. Inoculation of LYK3-GFP–expressing symrem1-2 mutant roots resulted in destabilization of the receptor (F) and its accumulation in endosome-like vesicles (G–G‴). (Scale bar in G‴: 10 μm.) (H) Frequency of LYK3 endocytosis in different genotypes (R108 WT, symrem1-2, and flot4-3) scored in independent root hairs under uninoculated conditions and at 2 dpi with. S. meliloti. (Scale bars: 10 μm.)
To further elucidate whether the presence of SYMREM1 is not only sufficient but also genetically required for the observed mobility arrest of LYK3, we generated transgenic roots in the symrem1-2 mutant and the R108 WT background expressing the ProLYK3:LYK3-GFP construct. As expected and independent of the genotype, the LYK3 receptor was laterally mobile at root hair plasma membranes in the absence of rhizobia in WT (Fig. 4D) and symrem1-2 mutant plants (Fig. 4E). However, application of rhizobia to symrem1-2 mutants resulted in a dramatic change in LYK3 localization (Fig. 4F). Here the receptor was localized to mobile, endosome-like vesicles in 64% of symrem1-2 roots, while a similar structure was observed in only 24% of the control roots (Fig. 4 F–H). The plasma membrane origin of these structures was confirmed by staining membranes with the lipophilic styryl dye FM4-64 (Fig. S4). Interestingly, this effect was not observed when the same experiment was conducted in the flot4-3 mutant background (Fig. 4H), although endosomal structures were seen more frequently in flot4-3 roots (26%) compared with WT roots (13%) in the absence of rhizobia. In contrast to symrem1-2, in flot4-3 mutants and WT plants, only a modest increase in the frequency of LYK3 endocytosis was observed on inoculation with rhizobia (Fig. 4H). This indicates that infection-triggered endocytosis of LYK3 is controlled by SYMREM1 rather than by FLOT4. These data unambiguously show that, in contrast to a possible LYK3 endocytosis observed at 6 h after NF application (and in the absence of rhizobia and endogenous SYMREM1) (26), infection-induced expression and accumulation of the molecular scaffold protein SYMREM1 mediates stabilization of the activated receptor pool by receptor recruitment to membrane nanodomains. Consequently, SYMREM1 acts as a negative regulator of receptor endocytosis, a function required for successful infection, as implied by the symrem1 mutant phenotype (Fig. S5). Considering the overlapping IT phenotypes in LYK3 knockdown experiments (23) and symrem1 mutants (Fig. 2), it can be concluded that IT abortion in symrem1 mutants is caused by a depletion of LYK3 from IT membranes.
Taken together, our data genetically prove that scaffold-mediated recruitment of LYK3 to specific nanodomains is mandatory for root hair infection. This may in turn provide the host with a mechanism that allows spatiotemporal desensitization of cell surfaces toward the respective ligand, as the host might not be able to durably stabilize the activated NFP/LYK3 receptor complex at the plasma membrane. Prematurely aborted infections as observed in flot4, symrem1, and lyk3 mutants indicate that stabilization of LYK3 during host cell penetration is required for control of cellular entry along the root and during IT guidance toward the root cortex. In our present model, SYMREM1 serves as a secondary scaffold that requires FLOT4 to physically immobilize LYK3 in stable nanodomains (Fig. S5). This recruitment and phosphorylation of SYMREM1 by receptor kinase domains may result in further oligomerization of SYMREM1 and recruitment of additional proteins that function in cell entry control (Fig. S5).
Materials and Methods
Plant Growth and Phenotypic Analysis.
For phenotypical analysis, M. truncatula WT R108, symrem1-1 (NF4432), symrem1-2 (NF4395), and flot4-3 (NF14107) seeds were scarified and sterilized before being sown on 1% agar plates for germination and kept in the dark at 4 °C for 3–5 d for vernalization. Germination was allowed for up to 24 h at 24 °C before the seedlings were transferred to Fahraeus plates. Plants were grown for 4 d in the presence of 1 μM nitrate, then transferred to plates without nitrogen but containing 0.1 μM aminoethoxyvinylglycine (AVG) for 3 d. Plants were then inoculated with 1 mL of S. meliloti 2011 (lacZ) with an OD600 of 0.05. ITs were scored at 7 dpi.
Genotyping of Tnt1 Insertion Lines and Quantitative Real-Time PCR.
R0 or R1 seeds of M. truncatula R108 Tnt1 transposon insertion lines were obtained from the Samuel Roberts Noble Foundation after screening the mutant population by PCR using the primers described in Table S2.
Total RNA of control and insertion lines was extracted using a commercial kit (Promega) following the supplier’s instructions. An additional DNaseI treatment (Invitrogen) was performed. Syntheses of cDNA and qRT-PCR were conducted as described earlier using SuperScript III reverse transcriptase (Invitrogen). All data were normalized to Ct values of the housekeeping gene ubiquitin (30); the related primers are listed in Table S2.
Hairy Root Transformation and Inoculation of Rhizobia.
M. truncatula hairy root transformation was performed as described previously (31) using the Agrobacterium rhizogenes strain ARqua1. Plants were transferred weekly to fresh plates containing Fahraeus medium (pH 6.0) supplemented with 1 mM NH4NO3 and followed by 2 d of growth in nitrogen-free Fahraeus medium containing 0.1 μM AVG. Images for kymograph and immunofluorescence analyses were obtained on root hairs of plants inoculated for 2 and 6 d, respectively.
Construct Design.
The coding sequence of M. truncatula SYMREM1 (GenBank accession no. JQ061257) was recombined into the Gateway (GW)-compatible ProUBi-YFP-GW vector (12) via an LR reaction. For complementation experiments, a GW-compatible ProSYMREM1-GW vector was created by replacing the ubiquitin promoter with the functional SYMREM1 promoter (643 bp upstream of the translational start of SYMREM1) using PmeI and XbaI restriction sites. All other constructs were cloned as Golden Gate- compatible constructs. BpiI and BsaI restriction sites were removed from the following nucleotide templates before the cloning of level 2 expression vectors: SYMREM1 (GenBank accession no. JQ061257), MAP4 (GenBank accession no. M72414), the cDNA of the genomic FLOT4 (GenBank accession no. GU224281), the 2-kb FLOT4 promoter (ProFLOT4), and the SYMREM1 promoter (ProSYMREM1). A Lifeact template with flanking BsaI restriction sites were directly inserted into pUC-Bpi via blunt-end StuI (New England BioLabs) cut ligation for subsequent Golden Gate cloning. Double-stranded sequences for the FLOT4-RNAi constructs with flanking BsaI sites were also cloned via a blunt-end StuI cut ligation into pUC-BpiI. RNAi silencing vectors based on previously described sequences (11) were assembled as described previously (32). For localization of FLOT4 and SYMREM1 driven by endogenous promoters, a tandem cassette, ProUbi:NLS-2xCerulean, was inserted as the marker for selecting transgenic roots before analysis. The designed constructs are listed in Table S3.
Immunolocalization Assay.
An improved procedure for whole-mount immunolocalization was adopted (33). In brief, 0.5- to 1-cm root pieces were mounted in 2% formaldehyde in MTSB buffer supplemented with 0.1% Triton X-100. Vacuum infiltration of explants, cuticle solubilization, cell wall digestion, and membrane permeabilization were performed as described previously (33). After 1 h of blocking in BSA, samples were incubated with a GFP nano-booster (1:200 dilution) coupled to Atto488 (ChromoTek) for 1 h at room temperature, followed by three 10-min washes and then mounting in antifade medium.
Confocal Laser-Scanning Microscopy.
For cytoskeleton and NLS-GFP reporter images, confocal laser scanning microscopy was performed on a Leica TCS SP5 confocal microscope equipped with 63× and 20× HCX PL APO water immersion lenses (Leica Microsystems). GFP was excited with the argon laser line at 488 nm and the emission detected at 500–550 nm. YFP was excited with the 514 nm argon laser line and detected at 520–555 nm. mCherry fluorescence was excited using a diode-pumped solid-state laser at 561 nm, and emission was detected between 575 and 630 nm. Samples coexpressing two fluorophores were imaged in sequential mode between frames. Due to low signal intensity for the ProSYMREM1-NLS-2xGFP reporter, the corresponding fluorescence was detected using Leica HyD detectors. Images were obtained with a Leica DFC350FX digital camera.
Nanodomain patterns for FLOT4-GFP, YFP-SYMREM1, and GFP-SYMREM1 were acquired using a Leica TCS SP8 confocal microscope system with 63× HCX PL APO CS2 oil immersion and 20× HCX PL APO water immersion lenses. YFP was excited with the 514-nm argon laser line and detected at 520–555 nm. GFP was excited by white light laser at 488 nm, and emission was detected between 500 and 550 nm. The corresponding fluorescence was detected using Leica HyD detectors. mCherry fluorescence from S. meliloti was excited using white light laser at 561 nm, and emission was detected between 575 and 630 nm. To create Z-stacks of 35- to 50-µm depth, images were obtained with 0.5- to 1-µm intervals.
LYK3 mobility was assessed using a PerkinElmer UltraView Vox on an inverted Zeiss microscope stand equipped with a Yokogawa spinning-disk unit and a Hamamatsu EMCCD C9100-50 camera. GFP was excited with a 488-nm laser set to 30% with an exposure time of 900 ms. Camera binning was set to 2 during image acquisition. Time series data were collected over 90 frames taken at 2-s intervals, for a total of 180 s. The endocytosis with FM4-64 staining was imaged using a Zeiss LSM 880 with Airyscan in the fast Airyscan mode. GFP and FM4-64 were excited with 488 nm and 561 nm, respectively.
Quantitative Image Analysis.
Image analysis was performed with the open-source ImageJ/(Fiji) software (34). For illustration, images were background-subtracted according to the rolling ball algorithm, filtered with a mean filter pixel radius of 1, and then maximum Z-projected (create stack). Contrast was enhanced for visualization in figures, but not for quantification. Pixel-based colocalizations to determine Pearson correlation coefficient values were performed using the Fiji plug-ins Squassh (35) and JACoP (36). Image segmentation was performed with Squassh. Randomization was performed with the Costes automatic randomization method in JACoP, in which clusters of 10 × 10 pixels were randomly distributed in one channel and correlated to the original values. Randomization was also performed on maximum Z-projections via horizontal flip of the mCherry channel as described previously (4, 37). To quantify nanodomains, images were segmented to differentiate background from domains. For this, the background was subtracted using a rolling ball algorithm with a radius corresponding to the largest structure of interest. A mean blur with radius 1 was then applied, the slices (n = 5–12 slices, with distances of 0.25–0.7 µm) were maximum projected along the z-axis for epidermal cells, and the slice with distances of 0.5 µm was used for root hair images. A threshold was applied to the images, and the result was saved as a binary mask. The “create selection” tool was used to mark the outlines and was overlaid onto the original image to verify proper image segmentation. Domains were counted with the “particle analyzer” tool in Fiji. For mobility analysis of the LYK3-GFP nanodomain, kymographs were generated using ‘‘Reslice’’ tool in Fiji.
Cytoskeleton Depolymerization.
A 1 mM oryzalin stock solution in DMSO and a 10 mM cytochalasin D stock solution in EtOH were prepared. M. truncatula root samples (1 cm long) were incubated in final concentrations of 10 µM oryzalin or 10 µM cytochalasin D for 12 h in water. The control samples were incubated in water with an equal amount of solvent for 12 h.
Supplementary Material
Acknowledgments
We thank all the members of our team for fruitful discussions throughout the project and comments on the manuscript. We also thank Cara Haney (University of British Columbia) for sharing some unpublished experimental details. The complemented hcl-1 line and the ProLYK3:LYK3-GFP construct were kindly provided by Doug Cook and Brendan Riely (University of California Davis). We thank the staff of the Life Imaging Center in the Center for Biological Systems Analysis of the Albert-Ludwigs University Freiburg and the Center for Advanced Light Microscopy at the University of Munich for help with their confocal microscopy resources and the excellent support in image recording. We also greatly appreciate the methodological input of Taras Pasternak (University of Freiburg). The Medicago truncatula plants used in this research project, which are jointly owned by the Centre National De La Recherche Scientifique, were obtained from The Samuel Roberts Noble Foundation and were created through research funded in part by the National Science Foundation (Grant 703285). This work was funded by an Emmy Noether Grant from the German Research Foundation (OT423/2-1, to T.O.) and a grant from the China Scholarship Council (201506350004, to P.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721868115/-/DCSupplemental.
References
- 1.Burkart RC, Stahl Y. Dynamic complexity: Plant receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2017;40:15–21. doi: 10.1016/j.pbi.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 2.Ott T. Membrane nanodomains and microdomains in plant-microbe interactions. Curr Opin Plant Biol. 2017;40:82–88. doi: 10.1016/j.pbi.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Bücherl CA, et al. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife. 2017;6:e25114. doi: 10.7554/eLife.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarsch IK, et al. Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2014;26:1698–1711. doi: 10.1105/tpc.114.124446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gui J, et al. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev Cell. 2016;38:201–213. doi: 10.1016/j.devcel.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre B, et al. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA. 2010;107:2343–2348. doi: 10.1073/pnas.0913320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tóth K, et al. Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS One. 2012;7:e30817. doi: 10.1371/journal.pone.0030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Single-molecule fluorescence imaging to quantify membrane protein dynamics and oligomerization in living plant cells. Nat Protoc. 2015;10:2054–2063. doi: 10.1038/nprot.2015.132. [DOI] [PubMed] [Google Scholar]
- 9.Jarsch IK, Ott T. Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol Plant Microbe Interact. 2011;24:7–12. doi: 10.1094/MPMI-07-10-0166. [DOI] [PubMed] [Google Scholar]
- 10.Gronnier J, et al. Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife. 2017;6:e26404. doi: 10.7554/eLife.26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haney CH, Long SR. Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA. 2010;107:478–483. doi: 10.1073/pnas.0910081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konrad SS, et al. S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol. 2014;203:758–769. doi: 10.1111/nph.12867. [DOI] [PubMed] [Google Scholar]
- 13.Li R, et al. A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell. 2012;24:2105–2122. doi: 10.1105/tpc.112.095695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perraki A, et al. Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol. 2012;160:624–637. doi: 10.1104/pp.112.200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raffaele S, et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui J, Liu C, Shen J, Li L. Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 2014;166:1463–1478. doi: 10.1104/pp.114.246769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Checker VG, Khurana P. Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep. 2013;32:1729–1741. doi: 10.1007/s00299-013-1483-5. [DOI] [PubMed] [Google Scholar]
- 18.Yue J, Li C, Liu Y, Yu J. A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PLoS One. 2014;9:e100772. doi: 10.1371/journal.pone.0100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perraki A, et al. StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett. 2014;588:1699–1705. doi: 10.1016/j.febslet.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Son S, Oh CJ, Bae JH, Lee H, An CS. GmREM1.1 and GmREM2.1, which encode the remorin proteins in soybean, have distinct roles during root nodule development. J Plant Biol. 2015;58:17–25. [Google Scholar]
- 21.Zipfel C, Oldroyd GE. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 22.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 23.Smit P, et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray JD. Invasion by invitation: Rhizobial infection in legumes. Mol Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- 25.Fournier J, et al. Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 2015;167:1233–1242. doi: 10.1104/pp.114.253302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haney CH, et al. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell. 2011;23:2774–2787. doi: 10.1105/tpc.111.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 28.Gui J, Zheng S, Shen J, Li L. Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front Plant Sci. 2015;6:804. doi: 10.3389/fpls.2015.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanski WG, et al. Cytoskeletal components define protein location to membrane microdomains. Mol Cell Proteomics. 2015;14:2493–2509. doi: 10.1074/mcp.M114.046904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satgé C, et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat Plants. 2016;2:16166. doi: 10.1038/nplants.2016.166. [DOI] [PubMed] [Google Scholar]
- 31.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 32.Binder A, et al. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS One. 2014;9:e88218. doi: 10.1371/journal.pone.0088218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasternak T, et al. Protocol: An improved and universal procedure for whole-mount immunolocalization in plants. Plant Methods. 2015;11:50. doi: 10.1186/s13007-015-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizk A, et al. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat Protoc. 2014;9:586–596. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- 36.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 37.Jarsch IK, Ott T. Quantitative image analysis of membrane microdomains labelled by fluorescently tagged proteins in Arabidopsis thaliana and Nicotiana benthamiana. Bio Protoc. 2015;5:e1497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.