Significance
Negative feedback regulation of ribonucleotide reductase (RNR) activity by dATP is important for maintaining balanced intracellular 2ʹ-deoxynucleoside triphosphate (dNTP) pools essential for the high fidelity of DNA replication and repair. To date, this type of allostery has been nearly universally associated with dATP binding to the N-terminal ATP-cone domain of the class Ia RNR large subunit (canonical α2), resulting in an altered quaternary structure that is unable to productively bind the second subunit (β2). Here, we report our studies on activity inhibition by dATP of the Bacillus subtilis class Ib RNR, which lacks a traditional ATP-cone domain. This unprecedented allostery involves deoxyadenosine 5′-monophosphate (dAMP) binding to a newly identified site in a partial N-terminal cone domain, forming an unprecedented noncanonical α2.
Keywords: nucleotide metabolism, allostery, ribonucleotide reductase, ATP-cone, dAMP
Abstract
The high fidelity of DNA replication and repair is attributable, in part, to the allosteric regulation of ribonucleotide reductases (RNRs) that maintains proper deoxynucleotide pool sizes and ratios in vivo. In class Ia RNRs, ATP (stimulatory) and dATP (inhibitory) regulate activity by binding to the ATP-cone domain at the N terminus of the large α subunit and altering the enzyme’s quaternary structure. Class Ib RNRs, in contrast, have a partial cone domain and have generally been found to be insensitive to dATP inhibition. An exception is the Bacillus subtilis Ib RNR, which we recently reported to be inhibited by physiological concentrations of dATP. Here, we demonstrate that the α subunit of this RNR contains tightly bound deoxyadenosine 5′-monophosphate (dAMP) in its N-terminal domain and that dATP inhibition of CDP reduction is enhanced by its presence. X-ray crystallography reveals a previously unobserved (noncanonical) α2 dimer with its entire interface composed of the partial N-terminal cone domains, each binding a dAMP molecule. Using small-angle X-ray scattering (SAXS), we show that this noncanonical α2 dimer is the predominant form of the dAMP-bound α in solution and further show that addition of dATP leads to the formation of larger oligomers. Based on this information, we propose a model to describe the mechanism by which the noncanonical α2 inhibits the activity of the B. subtilis Ib RNR in a dATP- and dAMP-dependent manner.
Class I ribonucleotide reductases (RNRs) reduce ribonucleoside diphosphates (NDPs) to supply de novo the 2ʹ-deoxyribonucleotides (dNTPs) required for DNA replication and repair in all eukaryotes and most bacteria. These enzymes are composed of two subunits termed “α,” which houses the catalytic site where the reduction occurs (C-site), and “β,” which houses a dimetallo-tyrosyl radical (Y•) cofactor responsible for transiently generating a catalytically essential cysteine thiyl radical in the C-site of α [C382 in the modeled canonical dimer (1, 2) in Fig. 1A]. During turnover, the subunits of class I RNRs form an α2β2 complex, allowing the oxidation of the C-site Cys by the dimetallo-Y• cofactor in β to occur via a long-range (>35-Å) radical transfer (RT) pathway involving redox-active tyrosines in both subunits (3). The dimetallo-Y• cofactor composition serves as the basis for the subclassification of class I RNRs, with most falling into either class Ia (α = NrdA, β = NrdB), which use a diferric-Y•, or class Ib (α = NrdE, β = NrdF), which use a dimanganic-Y• (4).
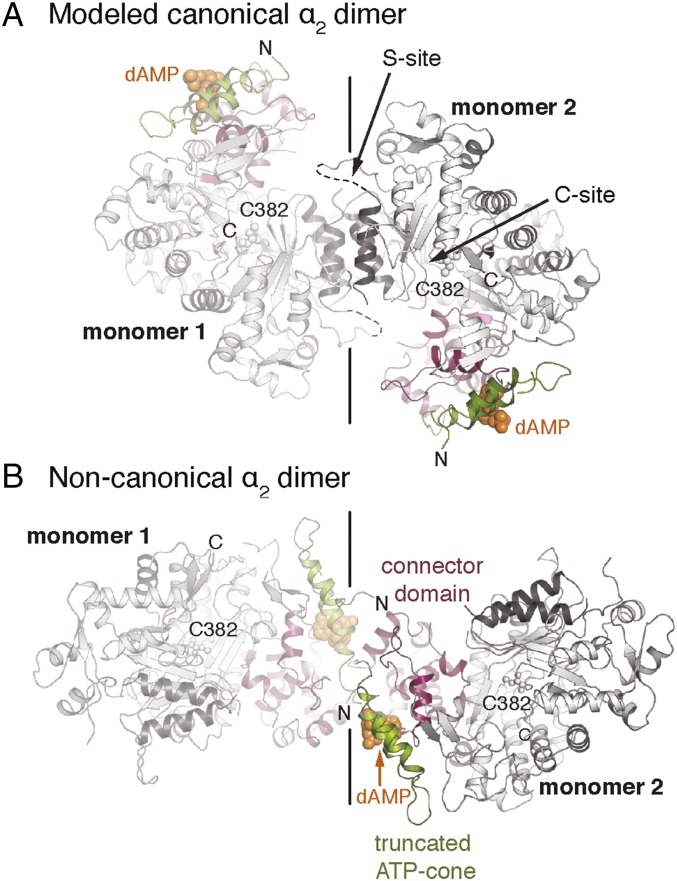
Fig. 1.
Comparison of the modeled canonical and noncanonical dimeric forms of B. subtilis NrdE. (A) A model of the canonical dimer was generated via secondary-structure matching structural superposition (1) of the dAMP-bound B. subtilis NrdE monomer structure against the Salmonella typhimurium NrdE α2 dimer (PDB ID code 1PEM) (2). Disordered S-site residues are shown as dashed lines. (B) Crystal structure of the noncanonical dimer of dAMP-bound B. subtilis NrdE at pH 4. In both panels, the dimer interface is indicated by a solid black line. The N-terminal partial cone (green), the connector region (pink), and the two helices at the S-site in the canonical NrdE dimer (dark gray) are colored by domain. These colors are used throughout this report.
RNRs contribute to the high fidelity of DNA replication and repair by being subjected to many levels of regulation (5) to control the intracellular dNTP pool sizes and ratios. All RNRs employ a sophisticated system of allosteric regulation that allows a single enzyme to reduce four different substrates (CDP, UDP, ADP, and GDP). In addition to having a conserved C-site, all class I α subunits have specificity sites (S-sites) located at the interface of a canonical α2 dimer (Fig. 1A). Each protomer contributes two helices and flexible loops to which dNTP and ATP effectors bind. These structures couple the C- and S-sites and control substrate preference (6). The substrate/effector (S/E) pairs are universally conserved in all RNRs (5): ATP and dATP stimulate the reduction of CDP and UDP, whereas dGTP and TTP stimulate the reduction of ADP and GDP, respectively.
A regulatory mechanism which is distinct between class Ia and Ib RNRs is allostery controlling the enzyme’s overall activity. NrdAs have a second effector binding site (A-site) to which ATP and dATP competitively bind and alter the enzyme’s quaternary structure between active and inactive states, respectively (7–9). The A-site is located within the evolutionarily mobile ATP-cone domain fused to α’s N terminus (10). ATP-cone domains are thought to have evolved independently on different NrdAs via horizontal gene transfer, resulting in the ability of Ia RNRs to form at least three distinct inhibited complexes (α4, α6, and α4β4) with subunit interfaces that are entirely or partially composed of this domain (11). In each of these structures, RNR activity is inhibited by disrupting the RT pathway between the α and β subunits and by prohibiting the subunits from forming an active α2β2 complex. In contrast, NrdEs lack a complete ATP-cone domain, and, until recently, the activity of previously studied class Ib RNRs has been reported to be insensitive to inhibition by dATP (12–19). These results have led to the belief that ATP-cones are modular and uniquely responsible for overall activity allosteric regulation in RNRs (5).
Contrary to this belief, we recently reported that physiologically relevant dATP concentrations (5–10 μM) are able to inhibit the activity of the Bacillus subtilis Ib RNR (20). This result is an example of RNR inhibition occurring independently of an ATP-cone and suggests that nature has evolved other methods for negative feedback regulation of elevated and/or imbalanced dNTP pools. Here, we report the results of further biochemical and biophysical studies undertaken to understand the allosteric regulation of the B. subtilis RNR and to gain insight into the structural basis for dATP inhibition. Surprisingly, we discovered that the inhibitory potency of dATP is dependent on an equivalent of deoxyadenosine 5′-monophosphate (dAMP) tightly bound to recombinantly expressed NrdE (dAMP/α). Furthermore, isolation of endogenous NrdE as the NrdE⋅NrdF (NrdEF) complex from B. subtilis and analysis by high-resolution mass spectrometry also reveals bound dAMP, providing support for its physiological importance. Structural characterization of NrdE by analytical ultracentrifugation (AUC), X-ray crystallography, and anion exchange (AEX) chromatography-coupled small-angle X-ray scattering (SAXS) reveals a previously uncharacterized NrdE dimer (Fig. 1B). The interface of this noncanonical α2 dimer is formed by a partial N-terminal cone domain, which houses one dAMP-binding site per α. Using AUC and SAXS, we further show that the addition of dATP to dAMP-bound NrdE leads to the formation of higher-order oligomers in a manner that is distinct from but reminiscent of dATP inhibition in class Ia RNRs. Based on these results, a model for the basis of overall activity allosteric regulation of the B. subtilis Ib RNR is proposed.
Results
Substrate Specificity Allosteric Regulation of the Mn(III)2–Y•–Dependent Enzyme Is Similar to Other Class I RNRs.
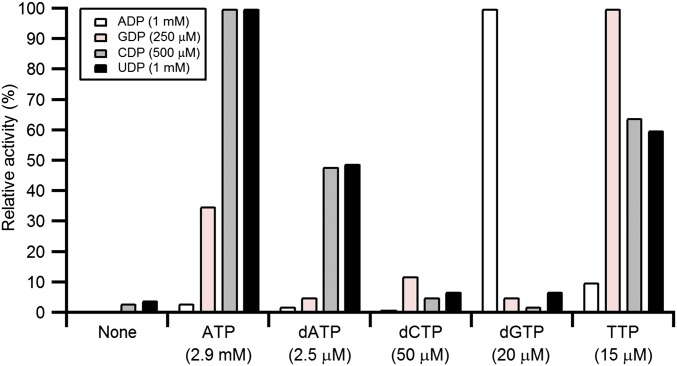
Given the surprising susceptibility of the B. subtilis Ib RNR to dATP inhibition (20), we examined its allosteric regulation in more detail. The steady-state kinetics using as-isolated NrdE (aiNrdE) with Mn(III)2–Y•–loaded NrdF were measured spectrophotometrically with endogenous reductants. The results, summarized in Fig. 2 and SI Appendix, Table S1, show that its regulation of substrate specificity conforms to the general scheme observed previously with Ia and Fe(III)2–Y•–loaded Ib RNRs (5). ATP and low dATP (<5 μM) stimulate pyrimidine reduction, TTP stimulates GDP reduction, dGTP stimulates ADP reduction, and dCTP has no effect on activity. The previously reported inhibitory effect of dATP at ≥10 μM on B. subtilis RNR is also observed with UDP reduction (SI Appendix, Fig. S1A). The kinetics follows Michaelis–Menten behavior with pyrimidine substrates but not with purine substrates, ADP (SI Appendix, Fig. S1B), or GDP (SI Appendix, Fig. S1C). These data fit best to the Hill equation and revealed negative (nH = 0.7 ± 0.1) and positive (nH = 1.8 ± 0.3) cooperative binding for ADP and GDP, respectively. The former result may explain the unusually large apparent affinity () for ADP relative to other class I RNRs (SI Appendix, Table S1). The apparent affinity of B. subtilis RNR for its other substrates (SI Appendix, Table S1) and effectors (SI Appendix, Table S2) showed characteristics similar to those of other class I enzymes: GDP is the tightest-binding substrate, UDP the weakest, and the S-site exhibits an ∼200-fold greater affinity for dNTPs relative to ATP. Finally, kcat values (1–2⋅s−1) are on the low side but are comparable to those measured with the Escherichia coli Ia RNR in the absence of effectors (2⋅s−1) vs. in the presence of effectors (5–10⋅s−1) (21). The kcat⋅Km−1 values, on the other hand, are within the range previously observed for other class I RNRs (103–105⋅M−1⋅s−1). The observation of patterns of allosteric regulation (Fig. 2 and SI Appendix, Table S1) similar to class Ia RNRs and Ib RNRs with diferric-Y•s is consistent with the formation of a canonical α2 structure for NrdE (Fig. 1A) and the assembly of an active α2β2 complex during turnover. The kinetic parameters also suggest that the presumed conformational change(s) gating RT with the Mn(III)2–Y• cluster in NrdFs might be slower than and distinct from that with the Fe(III)2–Y• cluster in NrdBs, resulting in lower turnover numbers.
Fig. 2.
Allosteric regulation of B. subtilis RNR substrate specificity using Mn(III)2–Y• NrdF and aiNrdE (described subsequently). Activities are expressed as a percentage relative to the maximum activity observed for a given substrate. Maximum activities for S (E) in α2β2 are ADP (dGTP) = 420 ± 20 nmol⋅min−1⋅mg−1; GDP (TTP) = 245 ± 10 nmol⋅min−1⋅mg−1; CDP (ATP) = 385 ± 20 nmol⋅min−1⋅mg−1; and UDP (ATP) = 435 ± 12 nmol⋅min−1⋅mg−1. Assays (500 μL) were conducted at 37 °C and contained a 1:1 mixture of 0.5-μM His6-tagged subunits, the endogenous reducing system (40-μM TrxA, 0.4-μM TrxB, and 0.2-mM NADPH), and S and E nucleotides at the concentrations indicated in the figure.
dATP and TTP Have Contrasting Effects on NrdE’s Quaternary Structure.
We next utilized sedimentation velocity analytical ultracentrifugation (SV-AUC) to compare the effects of dATP and TTP on the quaternary structure of NrdE. Previously, we showed that, in the absence of nucleotides, aiNrdE at physiological concentrations (1-μM α) sediments with a sedimentation coefficient (s20.w) value of 5.6 S, consistent with that predicted for a monomer (5.4 S) (20). Similar results (5.5 ± 0.1 S) are observed here using 1.1 ± 0.1-μM samples (SI Appendix, Fig. S2, top curves). Addition of dATP leads to the appearance of a second peak (SI Appendix, Fig. S2A). At 25-μM dATP, for example, ∼30% of aiNrdE appears to sediment as a monomer (5.6 S), while the remainder of the sample sediments with an apparent s20.w of 7.5 S. The position of this second peak is dependent on dATP concentration and shifts from ∼6.5 S at 1-μM dATP to 9.3 S at 100-μM dATP (SI Appendix, Fig. S2A). Interestingly, the peak position does not asymptotically approach the expected s20,w for a canonical NrdE dimer (8.0 S) and instead continues to shift to higher s20,w with increasing dATP. This result, along with the simultaneous shift of the monomer peak to higher s20,w values, is indicative of a higher-order species that is exchanging on a faster time-scale than sedimentation. Additionally, SV-AUC data at 100-μM dATP showed the presence of even larger species that completed sedimentation within the first 10–15 scans (details are given in SI Appendix). In contrast, the specificity effector TTP had little effect on the quaternary structure of NrdE at concentrations up to 10-fold its (0.12–12 μM) (SI Appendix, Fig. S2B). Thus, while our steady-state kinetics studies indicate that B. subtilis RNR conforms to the canonical scheme of specificity regulation, these AUC results suggest an unanticipated oligomerization-dependent mechanism for dATP inhibition.
NrdE Contains Tightly Bound dAMP.
Our preliminary biophysical studies of His6-aiNrdE with AUC and AEX chromatography revealed minor quaternary structural heterogeneity independent of protein concentration. Given these results and our previous experience with the purine biosynthetic enzyme formylglycineamide ribonucleotide synthetase, which requires tightly bound, noncatalytic Mg2+–ADP to assemble the active form (22), we examined the possibility of a bound nucleotide as a potential source of heterogeneity in aiNrdE. Two denaturation methods were pursued: aiNrdE was either dialyzed against water and then heat denatured, or it was buffer exchanged to remove glycerol and DTT and treated with 1% HClO4. With the acid protocol the supernatant was neutralized (KOH). The precipitated protein in the former case and the KClO4 salt in the latter case were then removed by centrifugation, and the supernatants were analyzed by UV-visible (UV-vis) spectroscopy. Spectra with a maximum absorbance (λmax) = 259–260 nm, minimum absorbance (λmin) = 227–229 nm, and an A280:A260 ratio = 0.16–0.19 were observed, consistent with the presence of (deoxy)adenosine analog(s) (23). To establish the phosphorylation state of the compound(s), the supernatant from heat-denatured aiNrdE was analyzed by polyethyleneimine (PEI)-cellulose TLC with several development conditions and compared with mono-, di-, and triphosphorylated adenosine (deoxyadenosine) nucleotide standards (24, 25). Unexpectedly, with His6-tagged protein we observed a mixture of mono-, di-, and triphosphate-containing species (dAXP, X = M, D, T) with Rf values similar to the authentic standards, whereas with tagless NrdE, we only observed a monophosphate (26).
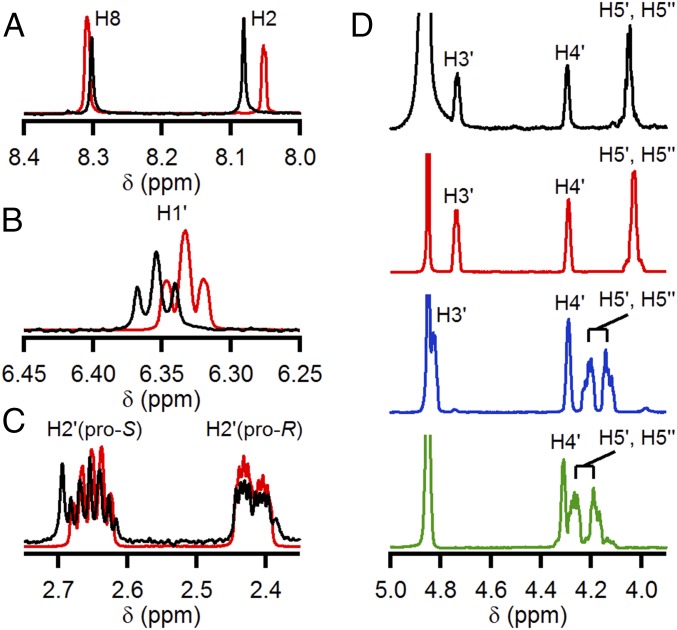
Molecular identification was performed by 1H-NMR spectroscopy. Compounds isolated by different workups from His6-tagged NrdE were chromatographed on a DEAE A25 anion exchange column with a triethylammonium bicarbonate (TEAB) gradient from 50–600 mM. For the compounds recovered with the heat denaturation procedure, the majority of the loaded material eluted at 200-mM TEAB, suggesting that the monophosphate was the major species associated with aiNrdE. Subsequent to TEAB removal, the compound was analyzed by 1H-NMR, which revealed a single species with a spectrum almost identical to dAMP (Fig. 3); the small differences in chemical shifts are due to small differences in the pH between standards and sample, salts associated with the sample preparation, and concentration differences. UV and TLC analyses also revealed variable amounts of nucleoside di- and triphosphates eluting from the column at 400- and 600-mM TEAB. NMR analysis of these species revealed chemical shifts in the base and anomeric proton spectral regions consistent with purine-like nucleotides (26). However, the amount of dAMP was always >50%. With the HClO4 protocol on tagless-NrdE (see SI Appendix for details), only dAMP (∼0.7 equiv per monomer) was recovered. Importantly, control experiments revealed that nonspecific breakdown of dATP during NrdE denaturation by heat or acid treatment was not occurring. Thus, the results establish that dAMP is the major nucleotide associated with recombinant aiNrdE.
Fig. 3.
Comparison of 1H-NMR spectra of the deoxynucleoside monophosphate recovered from aiNrdE (black trace) with authentic dAMP (red trace), dADP (blue trace), and dATP (green trace) standards. The panels show regions of the spectra containing peaks that correspond to protons associated with the adenine base (A), the anomeric carbon (C1′) (B), the 2′ carbon (C), and carbons C3′, C4′, and C5′ (D). Note that H3′ for dATP is masked by the H2O/DOH solvent peak.
dAMP Copurifies with Endogenous RNR.
The tight association of dAMP with recombinant NrdE was an unexpected result. We therefore examined whether dAMP is also associated with NrdE isolated from B. subtilis. Molecular identification and quantitation were performed with high-resolution LC/MS as endogenous RNR levels are low in all organisms, including B. subtilis. To further facilitate measurement, RNR was isolated from the B. subtilis strain 1B-UP that we previously engineered to increase its levels ∼17-fold relative to wild type (27). The RNR as isolated from two different preparations was composed of an ∼1:1 mixture of NrdE and Mn-loaded NrdF, and the purity, as judged by SDS/PAGE, was ∼80%. In each case, NrdEF was precipitated with HClO4 and separated from the supernatant by centrifugation. UV analysis of the neutralized, desalted supernatant revealed an absorption feature with a λmax of 260 nm, corresponding to ∼0.4 dAXP/NrdEF (see SI Appendix for details). High-resolution electrospray ionization (ESI)-MS of this material yielded a major [M + H]− ion with a mass of 331.0609 and a MS/MS fragmentation pattern identical to that of a dAMP standard. dAMP thus copurifies with NrdEF. Consistent with the UV analysis, the amount of dAMP recovered from NrdEF was determined to be 0.3/α by generating a standard curve using known amounts of dAMP that was subjected to the same treatment as the nucleotide isolated from NrdEF. Precise quantitation is not possible, as endogenously isolated NrdEF is not pure and the subunit ratio is determined by Western analysis. However, despite these limitations, the data strongly support dAMP copurification with NrdEF and hence its physiological importance.
Recombinant aiNrdE Can Be Separated into Nucleotide-Free (Apo) and dAMP-Bound (Holo) Protein by AEX Chromatography.
An average loading of 0.7 ± 0.1 dAXP/α was measured in seven different preparations of recombinant aiNrdE using the heat-denaturation method. In an effort to investigate the effects of dAMP on RNR activity and structure, we developed a method capable of separating apo- and holo-NrdE by FPLC using MonoQ AEX chromatography and elution with a 50- to 500-mM linear NaCl gradient. The initial separation produced a complex chromatogram (SI Appendix, Fig. S3A) with multiple unresolved species, perhaps reflecting the heterogeneity of the bound nucleotides and/or the quaternary structure of NrdE. Quantitation of dAXP, however, revealed that the protein eluting at 250- to 270-mM NaCl contained little to no nucleotide and that eluting at 330- to 350-mM NaCl had close to one equiv. By pooling the respective fractions and rechromatographing them, we obtained nearly homogenous preparations of apo-NrdE (SI Appendix, Fig. S3B, 0.02 ± 0.02 dAXP/α, average of five preparations) and holo-NrdE (SI Appendix, Fig. S3C, 1.1 ± 0.2 dAMP/α, average of eight preparations). Subsequent PEI-cellulose TLC analysis revealed that only a monophosphate was associated with holo-NrdE. Any nucleoside di- and triphosphates that could be associated with aiNrdE are thus lost during this purification procedure.
Sensitivity of Apo- and Holo-NrdE to dATP Inhibition.
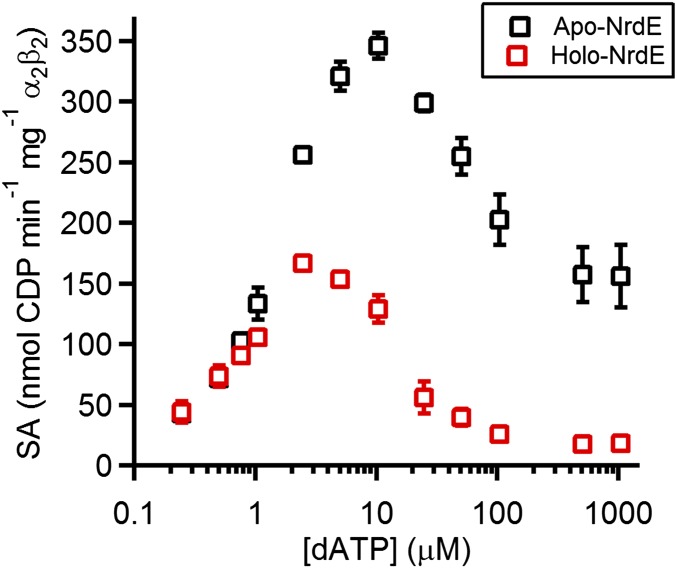
Spectrophotometric assays using apo- and holo-NrdE revealed that the presence of dAMP has a pronounced effect on the inhibition of RNR activity by dATP (Fig. 4). The stimulatory effect of dATP on CDP reduction from 0.25 to 1 μM is very similar for both forms of NrdE, consistent with S-site binding. Higher dATP concentrations result in potent inhibition of holo-NrdE, with only ∼15% activity remaining at 25 μM. In contrast, apo-NrdE activity continues to increase until dATP concentrations reach 5–10 μM. Milder inhibition sets in above 10-μM dATP, with 60% activity still remaining at 100 μM. Furthermore, the activity of apo-NrdE remains significantly higher (∼7.5-fold on average) relative to holo-protein, even up to 0.5-mM dATP. The data thus reveal that dAMP enhances RNR’s susceptibility to dATP inhibition.
Fig. 4.
Tightly bound dAMP associated with NrdE enhances inhibition of the B. subtilis Ib RNR by dATP. Spectrophotometric assays (500 μL) were conducted at 37 °C using 1-mM CDP, the indicated concentrations of dATP, the endogenous reducing system (40-μM TrxA, 0.4-μM TrxB, 0.2-mM NADPH), a 1:1 ratio of Mn(III)2–Y• NrdF (0.9 Y•/β2) and either apo-NrdE (black squares, 0.05 dAMP/α) or holo-NrdE (red squares, 1.2 dAMP/α). Each data point is the average of two replicates, and in most cases the error bars (±1 SD) are smaller than the marker.
dAMP Binds to an N-Terminal Effector Site to Facilitate Assembly of a Noncanonical α2 Dimer.
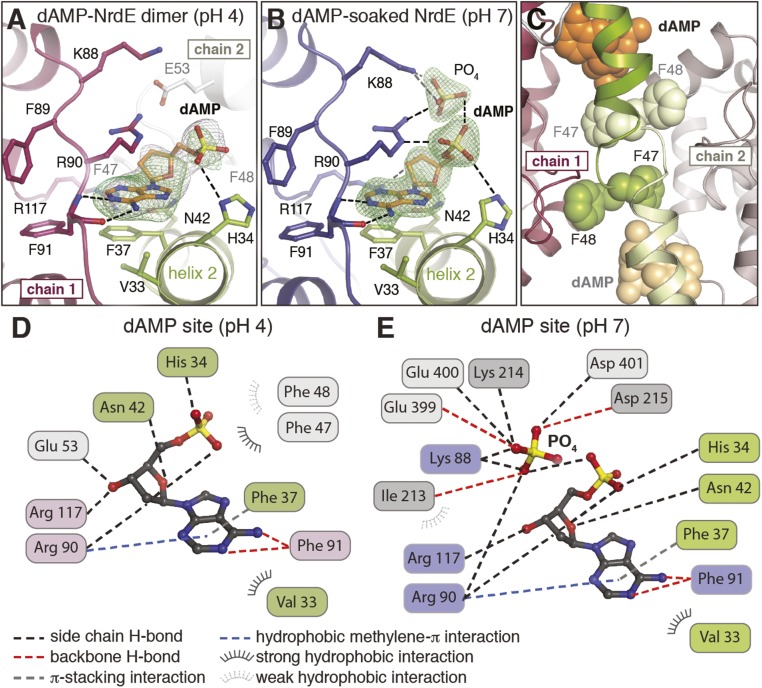
To identify the location of the dAMP-binding site, holo-NrdE was characterized by X-ray crystallography. Initial crystallization conditions [0.1 M citric acid (pH 4.0) and 1.6 M ammonium sulfate] yielded a 3.20-Å resolution structure (SI Appendix, Table S3) solved by molecular replacement using the coordinates of Salmonella typhimurium NrdE [Protein Data Bank (PDB) ID code 1PEM] as the initial search model (2). Subsequent refinements revealed a surface-accessible pocket at the N terminus of the protein containing extra electron density that could be reasonably modeled as dAMP (Fig. 5 A and D). The asymmetric unit (ASU) of the crystal structure contains two NrdE monomers that interact by a long carboxylate-rich loop (D396–E402) distal to the dAMP-binding site (SI Appendix, Fig. S4A). Analysis of this quaternary interface indicates that it is likely a crystallographic artifact. Inspection of symmetry-related monomers in the crystal lattice reveals a second interface near the N terminus involving the dAMP-binding sites of each protomer. The interaction constitutes a symmetric dimer interface that buries ∼620 Å2 of protein surface area (Fig. 1B) (28) and includes two consecutive Phe residues (F47 and F48) (Fig. 5 C and D) that buttress the dAMP site in the adjacent monomer. The new dimer surface is fully distinct from the canonical α2 dimer interface (Fig. 1A) observed in class Ia RNRs (29) and in the other class Ib NrdE structure (2).
Fig. 5.
(A and B) The N-terminal dAMP-binding site in the noncanonical holo-NrdE dimer at pH 4 (A) and the dAMP-soaked NrdE monomer at pH 7 (B). A 2Fo-Fc electron density map (gray mesh) is contoured at 1.0σ in A and 2.0σ in B, and Fo-Fc omit maps (green mesh) are contoured at 3.0σ. Selected amino acid side chains are shown as sticks and are colored by atom type. (C) A zoomed-in view of the symmetric N-terminal dimer interface shows a stacked pair of consecutive Phe residues (F47 and F48) that link the dAMP-binding sites in each monomer. (D and E) Ligand-interaction diagrams for the holo-NrdE pH 4 structure (D) and dAMP-soaked pH 7 structure (E). Interactions with symmetry-related monomers are shown in light gray in D and in light gray and dark gray in E.
An additional structure was obtained at pH 7 in a different space group, and it exhibits a distinct packing arrangement with one NrdE monomer in the ASU (SI Appendix, Table S3). The aforementioned carboxylate-rich loop and specificity site loop from two different symmetry-related molecules now approach the N-terminal dAMP pocket of NrdE (SI Appendix, Fig. S4 B and C). Consequently, the new diffraction datasets lack electron density for a nucleotide in this region, despite initial dAMP occupancy of the protein sample at 90–100%, and the side chains in the vicinity of the original site (F37, N42, and R117) are either disordered or in alternative conformations (SI Appendix, Fig. S4C). A soak of dAMP (10 mM at pH 7.0) into the crystals resulted in reincorporation of the nucleotide into the N-terminal binding site (SI Appendix, Fig. S4D) with a significant improvement in resolution (2.26 Å) relative to the original pH 4 structure. The interactions with dAMP can now be modeled in more detail (Fig. 5 B and E), but the quaternary structure rearrangement into a noncanonical dimer is prevented by the crystal lattice.
Although the pH 4 crystallization condition is seemingly less representative of the protein structure under physiological conditions inside the cell, in this case we believe that the low-pH condition is beneficial in minimizing the impact of disruptive lattice contacts, particularly those involving the carboxylate-rich loop (SI Appendix, Fig. S4 A and B). While the function of this loop is unknown, it is interesting to note that B. subtilis NrdF harbors a similar surface loop containing a stretch of four consecutive carboxylate side chains (E278–D281) of unknown function, and the analogous region complicates crystallographic analysis of that subunit as well (30). In the NrdE structures at both pH conditions, the surface-exposed location of the dAMP-binding site was unexpected given that the ligand copurifies with recombinant NrdE and endogenous NrdEF through multiple chromatography steps. However, the open nature of the binding site could be a consequence of either the dimeric quaternary structure or a crystallographic artifact. In the soak experiment, nucleotide binding induces ordering in neighboring loops contributed by adjacent monomers (SI Appendix, Fig. S4 C and D), suggesting that dAMP incorporation triggers local protein structural changes that might promote further burial of the nucleotide in solution.
Comparison of the N-Terminal Nucleotide-Binding Sites in Class Ia and Ib RNRs.
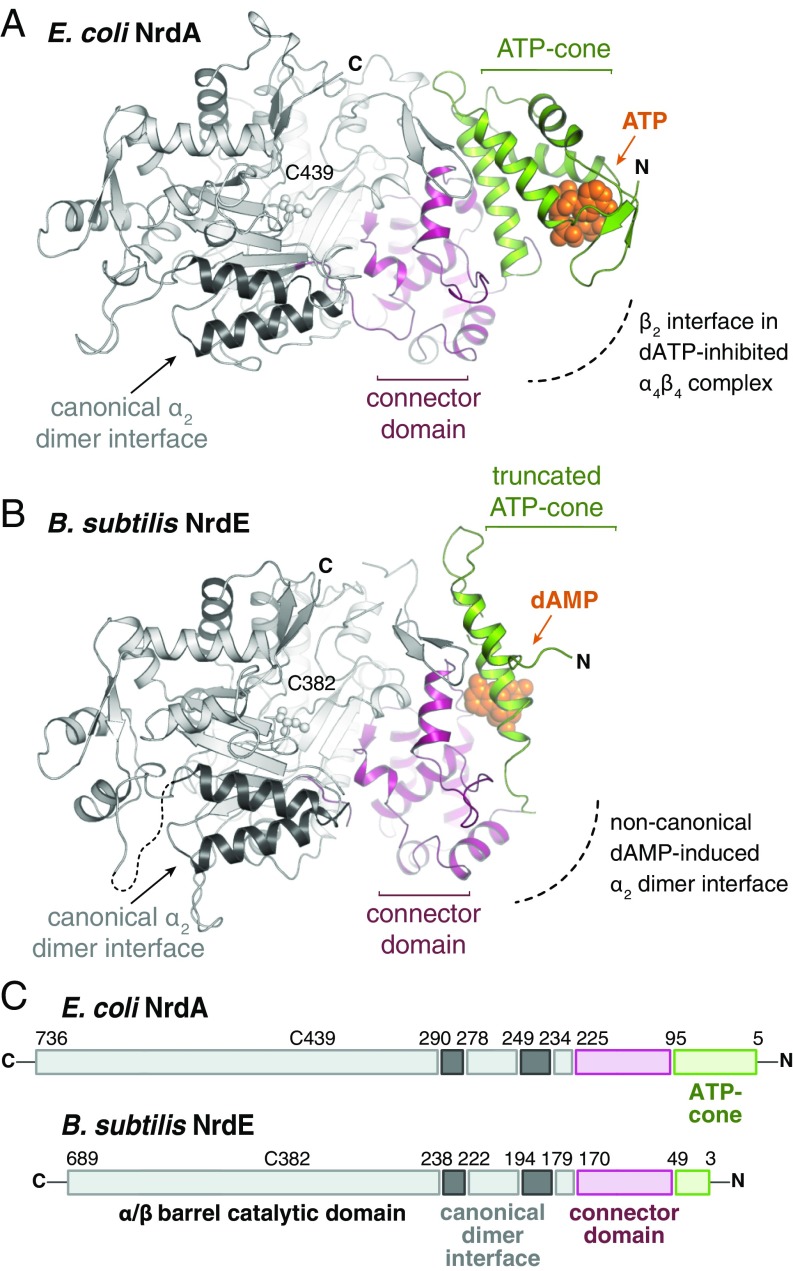
The domain architecture of a prototypical RNR α subunit from E. coli (NrdA) (Fig. 6 A and C) was originally described as a 480-residue α/β barrel housing the active site with a 220-residue α-helical N-terminal domain (29). The first ∼100 residues of this domain (residues 5–95) (Fig. 6C) constitute an ATP-cone motif that is characterized by a four-helix bundle with a conserved nucleotide-binding sequence in an N-terminal β-strand cap (10). The remainder of the N-terminal domain (residues 96–225) (Fig. 6C) forms a globular structure that connects the ATP-cone to the catalytic domain. Spatially, this connector domain is positioned between the ATP-cone (Fig. 6A, green) and the specificity site (Fig. 6A, dark gray). An explicit role for the connector domain (Fig. 6A, magenta) in RNR catalysis has not been defined, but its position between the ATP-cone (Fig. 6A, green) and the specificity site (Fig. 6A, dark gray) suggests it could facilitate communication between the two established regulatory nucleotide-binding sites in RNRs.
Fig. 6.
(A and B) A comparison of the structure of the N-terminal nucleotide-binding domains in E. coli NrdA (PDB ID code 3R1R) (A) and B. subtilis NrdE (B). (C) B. subtilis NrdE retains a shorter version of the ATP-cone motif found in E. coli NrdA and many other class I RNRs.
The sole X-ray crystal structure of a class Ib RNR catalytic subunit reported before this study, from S. typhimurium NrdE, revealed a domain architecture similar to that of E. coli NrdA (Fig. 6C and SI Appendix, Fig. S5 A and B), except the S. typhimurium NrdE structure was initially described as completely devoid of the ATP-cone domain (2). A comparison of the NrdE structures from S. typhimurium and B. subtilis with those of E. coli NrdA (29), which contains a single ATP-cone domain at the N terminus, suggests that class Ib RNRs instead retain a shorter version of this motif (residues 3–49) (Fig. 6C). The truncated N-terminal domain in the NrdE structures preserves two of the four helices that comprise the ATP-cone in NrdAs (Fig. 6B and SI Appendix, Fig. S5 A and B). In the B. subtilis NrdE structure, the second helix of the truncated cone, along with a region of the connector domain, is clearly implicated in binding and positioning of dAMP at the base of the half-cone (Fig. 6B and SI Appendix, Fig. S5).
A side-by-side comparison of the two NrdE proteins crystallographically characterized to date (Bs and St) shows that the N-terminal nucleotide-binding site is structurally conserved, and many of the residues that line the pocket are either strictly (F37, R90, F91) or moderately (H34, N42, R117) conserved (SI Appendix, Fig. S5 A and B, Insets and SI Appendix, Fig. S6A). The similarity is surprising, given the reported lack of dATP inhibition in the S. typhimurium enzyme (15). A comparable pocket lined with slightly different functional groups in S. typhimurium NrdE raises the possibility of an as-yet undiscovered capacity for overall activity modulation in other NrdE systems (SI Appendix, Fig. S5 A and B), perhaps involving priming ligands other than dAMP. Interestingly, a subset of class Ib RNRs homologous to B. subtilis NrdE (including those of mycobacterial pathogens) exhibits high sequence identity in the residues that interact directly with dAMP (SI Appendix, Fig. S6), suggesting that the dAMP-dependent overall activity regulation observed in B. subtilis NrdE is likely shared with this group.
Mutagenesis of the dAMP-Binding Site.
In the dAMP-bound B. subtilis NrdE structures, the adenine of the nucleotide is found inserted into a large hydrophobic pocket lined by V33 and F37 in helix 2 of the truncated ATP-cone (Fig. 5 A and B, green) and F91 and the methylene arm of R90 in the connector domain (Fig. 5A, magenta and Fig. 5B, blue). The adenine base is recognized in a base-specific fashion via H-bonding interactions between the N1 nitrogen and the N6 amino group to the backbone amide and carbonyl groups of F91 (Fig. 5 D and E) in the connector domain, enabled by a helix-strand transition at 87-KKFRFP-92. The base-specific F91 backbone interaction is an example of a reverse adenine-binding interaction (31), unusual in RNRs (6, 9, 32), although recent reports of multiple ATP molecules bound to a single ATP-cone in Pseudomonas aeruginosa class Ia RNR suggests diverse binding modes are possible (9). The motif is common to diverse classes of adenine nucleotide–binding proteins (31), implying high specificity of this site for effectors containing an adenine base. In contrast to the typical nucleotide-binding motifs found in RNRs, the dAMP site in B. subtilis NrdE exhibits more limited interaction with the sugar–phosphate moiety of the ligand and lacks a coordinating divalent cation. N42 and H34 in the truncated ATP-cone are within H-bonding distance of the furanose oxygen and one of the phosphate oxygens of dAMP, respectively (Fig. 5 D and E). N42 is additionally a possible H-bond donor/acceptor to the 3ʹ-OH of the dAMP ribose, and E53 in the noncanonical dimer partner also resides within 4 Å of this functional group.
To further validate the crystallographically observed dAMP-binding site (Fig. 5), H34 and F37 were mutated to Gln and Ile, respectively. The NrdE variants were purified in a manner similar to the wild-type enzyme and assayed for activity and dAMP incorporation. H34Q NrdE had activity higher than the wild-type protein (850–900 vs. 600 nmol⋅min−1⋅mg−1 α2), and HClO4 denaturation of the protein in the AEX fractions yielded little dAMP, as monitored by A260. Additionally, H34Q NrdE displayed a dATP-inhibition profile (SI Appendix, Fig. S7) similar to apo-NrdE (Fig. 4, black) although inhibition was observed at high dATP (0.5 mM). Thus, the H34Q mutation partially disrupts dAMP binding and renders the enzyme largely insensitive to dATP inhibition while maintaining high catalytic activity. In the case of F37I NrdE, all AEX fractions exhibited low specific activity (80–90 nmol⋅min−1⋅mg−1 α2) and no detectable A260 peak upon protein precipitation. The preliminary results are consistent with the crystallographic observations, which suggest that F37 is most important for dAMP binding because it forms part of the adenine-binding pocket. Residue H34 may be less critical because it is one of two residues within H-bonding distance of the dAMP phosphate, and the substituted Q may retain some of this functionality (Fig. 5 B and E).
Given the observed inhibition of holo-NrdE activity by dATP (Fig. 4, red), it is interesting to consider whether the dAMP site in the N terminus of the protein could accommodate deoxyadenosine 5′-diphosphate (dADP) or dATP via displacement of the endogenous dAMP ligand. The dAMP-soaked structure at pH 7 provides some insight into this question because it contains unequivocal electron density for a free phosphate positioned next to the terminal phosphate of the dAMP ligand (Fig. 5B). Although the origin of the phosphate is unknown, the structure provides a view of how this site might accommodate a deoxyadenosine di- or triphosphate. Residue K88, located near the adenine-binding motif in the helix–strand transition of the connector domain (Fig. 5B), is within H-bonding distance of the free phosphate but is too far away from the terminal dAMP phosphate to bind directly to the ligand. Interaction between the terminal phosphate of a di- or triphosphorylated nucleotide via a Lys or Arg side chain is a common pattern in other nucleotide–effector binding sites in RNR structures (6, 9, 32) and other ATP-binding proteins, including P-loop nucleotide switches (33). In the latter group of proteins, the charged residue functions in readout of the phosphorylation state of the nucleotide to communicate the status to other regions of the protein via long-range conformational changes. In B. subtilis NrdE, K88 could communicate N-terminal domain dAMP/dADP/dATP interactions to the specificity site or noncanonical dimer partner to modulate activity and/or oligomeric state. Analysis of electrostatic surface potential in the N-terminal and connector domains of B. subtilis NrdE reveals an extended positively charged region that includes the observed dAMP pocket and further supports the use of this pocket to bind nucleoside di- and triphosphates (SI Appendix, Fig. S8). The analysis indicates a second adjacent cavity that could bind another nucleotide simultaneously, as has been observed recently in one of the two cone domains of the class Ia RNR from P. aeruginosa (9).
Apo-NrdE Is a Monomer, and Holo-NrdE Is Predominantly a Noncanonical Dimer in Solution.
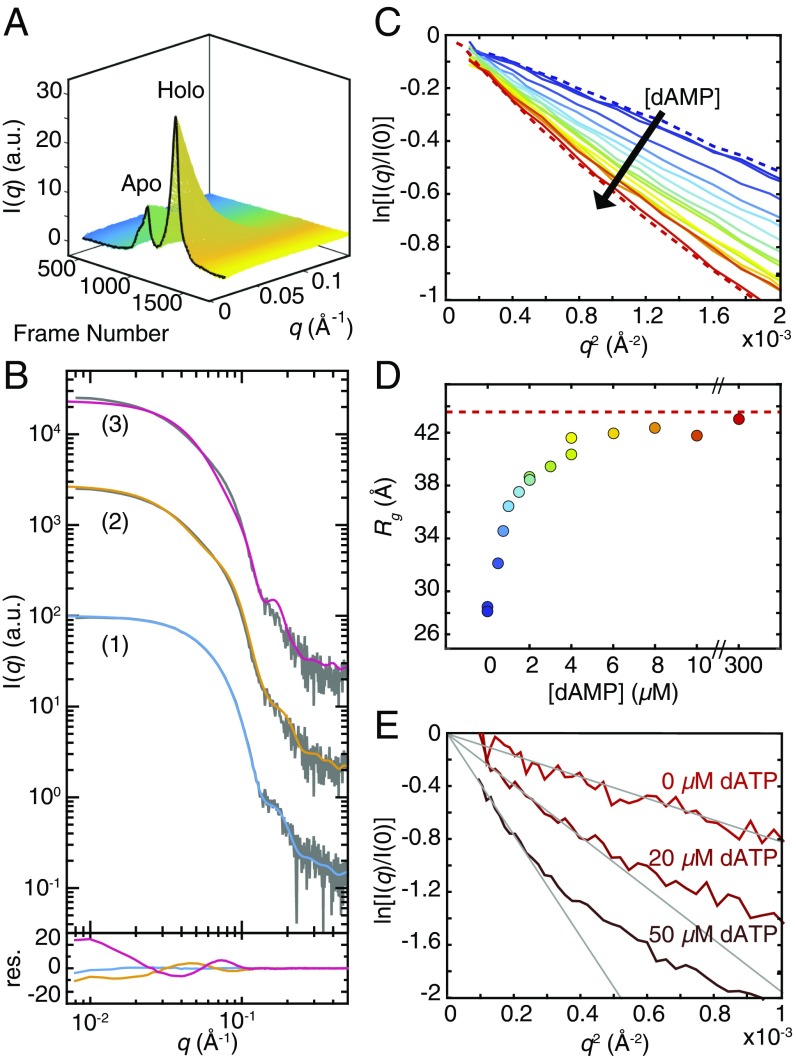
To determine if the crystallographically observed noncanonical dimer forms in solution, we employed SAXS, a technique that is particularly well suited for deconvoluting mixtures of RNR oligomers (7, 8). Because apo- and holo-NrdE were found to be separable by AEX chromatography (SI Appendix, Fig. S3), a 15-µM sample of aiNrdE was loaded onto an in-line MonoQ column and eluted directly into the X-ray path with a linear gradient of 100- to 500-mM NaCl at pH 7.6. Scattering images were collected continuously over the course of the elution to generate a 3D dataset with two major peaks corresponding to the apo- and holo-NrdE fractions (Fig. 7A), similar to those seen in the UV chromatogram described earlier (SI Appendix, Fig. S3A). The AEX–SAXS dataset was then mathematically decomposed as a linear combination of four distinct scattering components, each with an elution profile and SAXS curve, using a regularized alternating least-squares (ALS) method developed in-house (SI Appendix, Fig. S9). Mathematical decomposition by ALS was previously introduced in the context of size-exclusion chromatography (SEC) SAXS, where the sequential nature of elution affords a straightforward means to identify where individual peaks begin and end (34). In the context of AEX–SAXS, however, the background scattering varies in a complex manner throughout the chromatogram due to the NaCl gradient, complicating analysis. To decompose the AEX–SAXS data, we added smoothness regularization to the ALS method, so that background components could be forced (by selection of a large Lagrange multiplier) to vary monotonically during elution. With this approach, we found that the dataset could be represented by two background components and two partly overlapping peaks (SI Appendix, Fig. S9A). Although subtraction of the variable scattering from a salt gradient has been reported for ion exchange-coupled SAXS using a reference measurement (35), the regularized decomposition of AEX–SAXS data we present here represents a mathematical approach to background separation.
Fig. 7.
SAXS reveals the effects of adenine nucleotides on NrdE oligomerization. (A) AEX–SAXS dataset of aiNrdE. (B) SAXS profiles of the two protein components from AEX–SAXS are shown offset with fits and corresponding residuals (res.) shown below with the same coloring. Set 1: The monomer component (gray) is well described by the calculated profile of a NrdE monomer (overlaid blue curve). Sets 2 and 3: The dimer component (gray) is well described by the calculated profile of the noncanonical dimer (overlaid orange curve), whereas that of a canonical dimer (overlaid magenta curve) shows a worse fit. (C) Titration of 0- to 300-µM dAMP into 2-µM apo-NrdE leads to a steeper slope in the Guinier region of the SAXS profiles (solid lines, blue to red) with the AEX-derived monomer (blue dashed line) and dimer (red dashed line) components shown for reference. (D) As [dAMP] increases, the Rg increases from that of a monomer and approaches that of the AEX-derived dimer (red dashed line), saturating at stoichiometric concentrations. (E) The addition of dATP to 1-µM holo-NrdE leads to the formation of increasingly large structures, as indicated by an increasingly steep Guinier region of the SAXS profile. A transformation of this plot further shows that the species becomes increasingly extended with [dATP] (SI Appendix, Fig. S11).
Decomposition of the AEX–SAXS data by regularized ALS reveals the sequential elution of two protein components (SI Appendix, Fig. S9A). The elution of apo-NrdE corresponds to that of the first component, which was found to have a radius of gyration (Rg) of 28.2 ± 0.1 Å (Guinier Rg = 27.8 ± 0.1 Å) and a scattering profile (Fig. 7B, gray curve in set 1) that is well described by the calculated scattering from a monomer model of B. subtilis NrdE having a theoretical Rg of 27.5 Å (Fig. 7B, blue curve in set 1, √χ2 = 1.1). The elution of holo-NrdE coincides with that of the second component, which was found to have an Rg of 43.6 ± 0.2 Å (Guinier Rg = 42.8 ± 0.1). The calculated scattering from a canonical dimer having an Rg of 37.5 Å shows a poor fit to the data over a wide q-range (Fig. 7B, set 3, √χ2 = 11.7). Instead, this component is better described by the calculated scattering of the noncanonical dimer (Fig. 7B, set 2, √χ2 = 6.8), which has a theoretical Rg of 45.9 Å. Although a √χ2 greater than 1 suggests that the crystal structure of the noncanonical dimer does not perfectly describe the solution conformation, it clearly provides a better fit to the experimental curve than the canonical dimer (Fig. 7B, Lower). Notably, the elongated shape of the noncanonical dimer better reproduces the larger Rg as well as the distinctive shape of the experimental data in the mid-q region (0.05 < q < 0.1 Å−1), which is sensitive to subunit arrangement. These results indicate that apo-NrdE is a monomer, whereas under the AEX conditions used here holo-NrdE elutes predominantly as a noncanonical dimer that coelutes with a small population of monomer.
To determine if apo-NrdE can be converted to holo-NrdE, a titration experiment was performed, in which 0- to 300-µM dAMP was added to a solution of 2-µM apo-NrdE and 1-mM CDP. Singular value decomposition of the titration dataset yields two major singular values (SI Appendix, Fig. S10 A and B), suggesting that dAMP induces a largely two-state transition. At 0-µM dAMP, we find that the scattering of apo-NrdE is superimposable with that of the AEX-derived monomer (blue dashed curves in Fig. 7C and SI Appendix, Fig. S10C), indicating that CDP has no effect on the oligomerization state. The addition of dAMP leads to a change in scattering that is consistent with noncanonical dimerization (Fig. 7C and SI Appendix, Fig. S10). The corresponding Rg values increase from that of a monomer to values approaching that of the AEX-derived noncanonical dimer (Fig. 7D), saturating at stoichiometric concentrations of dAMP. At 300-µM dAMP, the scattering becomes superimposable to that of the AEX-derived noncanonical dimer (SI Appendix, Fig. S10C, red dashed curve). This titration thus provides evidence that, while apo-NrdE is isolated as a dAMP-free monomer, it has a strong affinity for dAMP, such that stoichiometric concentrations are sufficient to restore a holo-like structure.
dATP Induces a Higher-Order Oligomerization of Holo-NrdE.
Having established the structure of holo-NrdE by crystallography and SAXS, we next revisited the effect of dATP on quaternary structure. To mirror the AUC experiments performed with aiNrdE, the scattering of 1-µM holo-NrdE was measured in the presence of 0-, 20-, and 50-µM dATP and 1-mM CDP. At this low protein concentration, the scattering profiles are noisier. However, addition of dATP leads to a clear change in the low-q region of the Guinier plots (Fig. 7E), corresponding to a steep increase in Rg. At 0-µM dATP, Guinier analysis yields an Rg of 49.5 ± 2.1 Å, consistent with a sample that is predominantly noncanonical dimer. At 20-µM dATP, the Rg increases to 76.5 ± 6.7 Å, and the forward scattering intensity, which is an indicator of mass, increases by a factor of 2.5. Further increasing the dATP concentration to 50-µM leads to an even greater Rg of 107.5 ± 13.3 Å and 5.7-fold increase in forward scattering relative to 0-µM dATP. Thus, we find that the size and mass of NrdE, as reported by Rg and the forward scattering intensity, do not saturate within the physiologically relevant range of dATP concentrations examined here. Instead, both our AUC data and SAXS data suggest that dATP inhibition in B. subtilis RNR involves the formation of an increasingly larger species. Additionally, the SAXS data provide evidence that this species is elongated. When replotted as a cross-sectional Guinier plot [i.e., ln(I × q) vs. q2], the addition of dATP leads to increased linearity in the low-q region, which is indicative of an extended shape (SI Appendix, Fig. S11) (8). Such a species would be possible if dATP binds the specificity sites to promote the formation of a canonical interface between two adjacent noncanonical NrdE dimers.
Discussion
Before this work, negative-feedback allosteric regulation by dATP was considered a hallmark of class Ia RNR regulation in the maintenance of balanced dNTP pools inside the cell. In these cases, inhibition occurs via an ATP-cone domain that interacts readily with other α or β chains to form ring-shaped inhibited structures (7–9). Recently, we found the B. subtilis class Ib RNR to be inhibited by dATP (20), in contrast to the previously studied unregulated members of this subclass (12–19). Sequence alignments and structural analyses indicated that all class Ib RNRs lack complete ATP-cones (36). Thus, dATP inhibition of the B. subtilis enzyme was a surprising result that implied a mechanism of overall activity regulation divergent from that of both Ia and other Ib RNRs. The work presented herein suggests that a tightly bound small-molecule modulator is a critical component of allosteric feedback regulation in B. subtilis RNR. Interaction between this ligand and the truncated cone in the N-terminal domain promotes the formation of inhibited structures distinct from those seen in previously studied class Ia systems.
The discovery of tightly bound dAMP associated with NrdE in protein obtained recombinantly from E. coli and endogenously from B. subtilis suggests that this nucleotide plays an important role in regulation and/or stability of the catalytic subunit. Although little is known about the metabolic origin(s) of dAMP in B. subtilis, the presence of ymaB in the Bs nrd operon is provocative (26, 37). While the function of YmaB has yet to be fully explored, it is homologous to members of the Nudix hydrolase superfamily reported to cleave dATP to dAMP and pyrophosphate (38–40). Thus, we speculate that YmaB might play a unique role in producing the dAMP necessary for the formation of holo-NrdE. Indeed, preliminary studies of recombinant YmaB have shown that the enzyme possesses dNTP pyrophosphohydrolase activity (26), suggesting that at the very least it may participate in controlling dATP concentrations and allosteric regulation of RNR. Further study is required to better characterize this protein and establish YmaB’s function inside the cell. We note that, of the Ib RNRs identified from other bacteria that could potentially exhibit similar allosteric regulation by dAMP and dATP (SI Appendix, Fig. S6A), only those belonging to the genus Bacillus have a homolog of YmaB encoded in the RNR operon.
Although dAMP is the primary nucleotide recovered from NrdE, other unidentified nucleoside di- and triphosphates were also detected. An outstanding issue that is related to this result is the mechanism(s) by which dATP inhibits the B. subtilis RNR. One possibility is that dATP displaces dAMP from its binding site to exert its inhibitory effects on enzymatic activity. Indeed, the observation of a phosphate in our dAMP-soaked NrdE crystal structure suggests that the dAMP-binding site may be able to accommodate dATP. A second possibility is that there is an additional binding site in the noncanonical dimer created by dAMP for dATP. Analysis of electrostatic surface potential in the N-terminal and connector domains of NrdE reveals an extended positively charged region that includes both the observed dAMP pocket and an adjacent cavity (SI Appendix, Fig. S8). Therefore, dAMP-induced formation of the noncanonical NrdE dimer observed in the pH 4 crystal structure may create a third effector site at the interface to which dATP can bind to further stabilize the noncanonical dimer. Such versatility has precedent in the ATP-cones of class Ia RNRs, which can form different types of quaternary contacts (7–9), bind nucleotides with different phosphorylation states (41, 42), and interact with multiple nucleotides at once (9). Flexibility in the overall activity allosteric sites of RNRs, whether it be the ATP-cone or the N-terminally truncated cone identified here, may give the cell the ability to finely tune enzymatic activity in response to a broader range of nucleotide (deoxynucleotide) cues that indicate imbalances in the in vivo dNTP pools.
The recovery of dAMP associated with B. subtilis NrdEF indicates that the holo-form of NrdE occurs in vivo. The SAXS analyses at pH 7.6 show that in vitro dAMP induces a shift in equilibrium from monomer to a noncanonical dimer. However, given that full S-sites do not form within the noncanonical NrdE dimer (Fig. 1B), at least at low [dATP], we believe that this structure cannot be the active form in NDP reduction. Based on our current understanding of substrate specificity allostery in other class I RNRs, the canonical dimer interface is required to fully form the S-site (Fig. 1A). This configuration minimizes the distance between the S- and C-sites so that the correct pairing of substrate and effector nucleotides occurs (43–46). The fact that B. subtilis RNR conforms to the universal scheme of substrate specificity allostery (Fig. 2) indicates that NrdE is able to form a canonical dimer (5, 6) in addition to the noncanonical form we have described here. We propose that the dAMP-bound noncanonical dimer is an inhibited form of the protein and that canonical dimerization is favored when a monomer of NrdE binds to NrdF (β2), a second α, and effector to form the active α2β2 complex. The existence of an α2β2 structure is supported by our previous SV-AUC studies (20).
A notable feature of the canonical and noncanonical dimers is that the uncomplexed dimer interfaces are solvent-exposed and therefore are capable of interacting with other NrdE species. If dATP is able to stabilize both interfaces, we can envision higher-order association of NrdE into oligomeric structures (Fig. 8). Unlike the dATP-inhibited oligomers found in RNRs with ATP-cones (7–9), the N-terminal region of B. subtilis NrdE is likely not sufficiently long or flexible to twist the oligomer to the degree necessary to terminate in a relatively compact ring structure, leading to the increasingly large oligomers observed experimentally. As in other class I RNRs, the formation of higher-order oligomeric structures after subunit dissociation of the α2β2 complexes following turnover could attenuate RNR activity. The exact molecular basis for dATP inhibition in B. subtilis class Ib RNR requires further study. An initial analysis of the structure of two docked models of α2β2 complexes linked via the noncanonical α subunit dimer interface, for example, leads to a steric clash between the β2 components, potentially a route to inactivation via quaternary structure disruption (SI Appendix, Fig. S12).
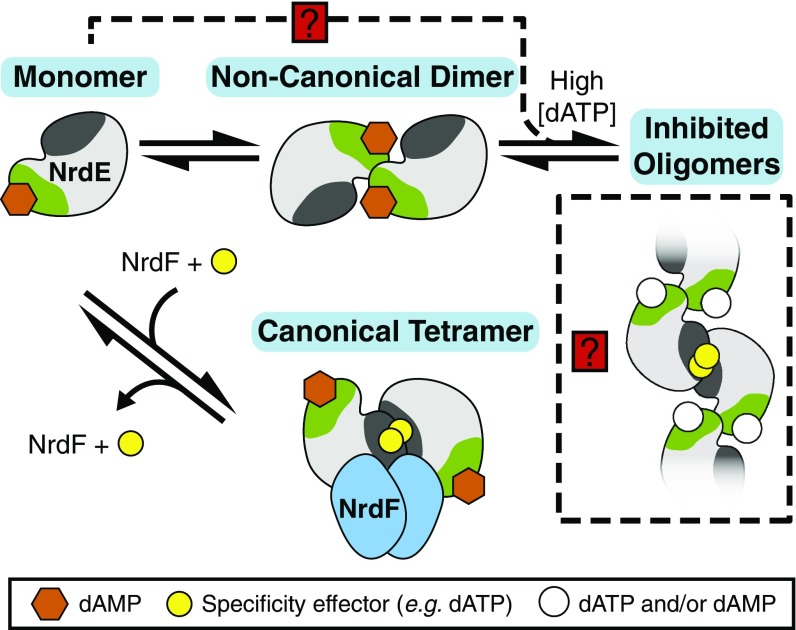
Fig. 8.
A model describing the overall activity allosteric regulation of the B. subtilis class Ib RNR. NrdE, when loaded with dAMP (orange hexagons), exists in an equilibrium between monomer and the noncanonical dimer. The monomer pool is capable of forming a canonical dimer and becoming active for NDP reduction when it binds to Mn(III)2–Y• NrdF with a second monomer, and in the presence of the specificity effector (yellow circles) it potentiates α2β2 complex formation. At high dATP concentrations, noncanonical dimers and possibly monomers (shown as a dashed line with a red box and a question mark) can associate into inhibited, extended oligomeric structures involving the canonical dimer interface, shown in dark gray. The N-terminal dATP-binding site of the oligomer is represented by an open circle suggesting two possible cases: in one, dATP displaces dAMP, and in a second, both dAMP and dATP bind.
Based on these observations, we propose the model shown in Fig. 8 to describe the overall activity allosteric regulation of the B. subtilis Ib RNR. As in enzymes with ATP-cone domains, we propose that high concentrations of dATP inhibit the enzyme by disruption of the RT pathway between the subunits via sequestration of NrdE into inhibited states, here represented by both the noncanonical dimer and higher-order oligomers. A possible route to the assembly of larger structures is indicated by the dashed line in Fig. 8. In this proposal, high [dATP] promotes addition of the NrdE monomer to the noncanonical dimer, potentially via a canonical dimer interface or a structurally similar equivalent. The relative locations of these binding sites in B. subtilis NrdE on opposite sides of the protein would result in extended structures consistent with SAXS analysis of the dATP-bound form. In this model, the key to controlling the amount of active RNR is the quaternary structural equilibrium of NrdE monomer and noncanonical dimer (Fig. 8 and SI Appendix, Fig. S2). The dATP ligand signals elevated or imbalanced dNTP pools in vivo and reduces RNR activity by shifting the equilibrium toward higher-order oligomers composed of noncanonical dimers. The absence of dATP promotes dissociation into monomeric NrdE, capable of binding NrdF dimers to form an active α2β2, thus leading to enzyme turnover. On-going studies in our groups are focused on investigating this model and other alternatives, including one in which NrdF (β) is a component of the inhibited state. The latter option has precedent in the dATP-inhibited α4β4 complex of the E. coli class Ia RNR (7, 32).
Finally, sequence comparison of B. subtilis NrdE with other class Ib RNRs reveals that only a limited subset may exhibit inhibition by dATP and/or dAMP. These include the phylogenetically close relatives of B. subtilis, Mycobacterium tuberculosis, and several other soil bacteria that are known to be opportunistic human pathogens (SI Appendix, Fig. S6A). Curiously, M. tuberculosis NrdE, which has all the dAMP-interacting residues conserved, and the Corynebacterium NrdEs, which have all but an H34 equivalent, have previously been reported to be insensitive to dATP inhibition (13, 19). The insensitivity of M. tuberculosis NrdE may be a result of its low activity measured using NrdF loaded with a diferric-Y• cofactor and DTT as the reductant (19). The results with Corynebacterium ammoniagenes RNR require further studies (13, 47). Given what we now know about the assembly of the physiologically relevant Mn(III)2–Y• cofactor and the importance of the endogenous reductant, it will be interesting to reexamine the effects of dATP and dAMP on the activity of these enzymes. Likewise, further study of other closely related enzymes is warranted, as this distinctive form of overall activity allostery may present opportunities for the development of new antibiotics.
The work reported here suggests that negative-feedback regulation of RNR activity to ensure the high fidelity of DNA replication and repair may well be more nuanced than previously thought. Traditionally, ATP-cone domains were considered to be the universal activity regulator of class Ia RNRs, operating as a discrete, indivisible unit that can be added or subtracted to imbue or remove activity regulation (9, 36). Our studies of B. subtilis Ib RNR illustrate that regulatory function can also be imparted to an ATP-cone by truncation. Additionally, the discovery of the unique role of dAMP in B. subtilis NrdE regulation suggests that RNRs may be connected in unexpected ways to nucleotide metabolism in the cell. Future examination of the allosteric regulation of other RNRs lacking ATP-cones may reveal other forms of overall activity allostery mediated by new effector molecules.
Materials and Methods
His6-tagged NrdE (720 nmol·min−1·mg−1·α2), NrdF, NrdI, B. subtilis TrxA (200 nmol·min−1·mg−1), and B. subtilis TrxB (18 µmol·min−1·mg−1) were prepared as described previously (20, 27, 48). Tagless variants of NrdE wild type (NrdE-wt, 710 nmol·min−1·mg−1 α2), NrdE-H34Q, NrdE-F37I, NrdF, and NrdI were produced using SUMO gene fusion technology (49, 50) as described in SI Appendix. NrdF was reconstituted with Mn(III)2–Y• and the holo-proteins (1,300 nmol⋅min−1⋅mg−1 β2) purified on a MonoQ column as described previously (20). Apo- and holo-NrdE were also separated by AEX chromatography on a MonoQ column as described in SI Appendix. Unless noted otherwise, NrdE and NrdF concentrations are reported relative to dimer. BsNrdEF was overexpressed in the IB-UP strain and was purified to ∼80% purity as previously described (27). Detailed descriptions of materials and methods, including cloning, purification, spectrophotometric assays, SV-AUC, nucleotide isolation, identification, and quantification, X-ray crystallography, and SAXS are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Massachusetts Institute of Technology’s Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems, supported by National Science Foundation (NSF) Grant NSF-0070319, and in particular Deborah Pheasant, for AUC instrument access and assistance in experimental setup and execution; Drs. Richard Gillilan and Jesse Hopkins for assistance with beamline setup at the Cornell High Energy Synchrotron Source (CHESS); and the Advanced Photon Source (APS), a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357, for use of its resources. The General Medical Sciences and National Cancer Institute Collaborative Access Team at the APS was funded in whole or in part with Federal funds, National Cancer Institute Grant ACB-12002 and National Institute of General Medical Sciences (NIGMS) Grant AGM-12006. The Eiger 16M detector was funded by NIH Office of Research Infrastructure Programs, High-End Instrumentation Grant 1S10OD012289-01A1. Use of the Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817. We also acknowledge the Berkeley Center for Structural Biology at the Advanced Light Source (ALS), supported in part by the NIH, NIGMS, and the Howard Hughes Medical Institute. The ALS is a DOE Office of Science User Facility under Contract DE-AC02-05CH11231. SAXS data were collected at the CHESS, which is supported by NSF Grant DMR-1332208, and at the MacCHESS facility, which is supported by NIH NIGMS Grant GM-103485. This work was supported by NIH Grants GM117757 (to S.P.M.), GM119707 (to A.K.B.), GM100008 (to N.A.), GM124847 (to N.A.), and GM081393 (to J.S.) and by start-up funds from Princeton University (N.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 6CGL, 6CGM, and 6CGN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800356115/-/DCSupplemental.
References
- 1.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 2.Uppsten M, et al. Structure of the large subunit of class Ib ribonucleotide reductase from Salmonella typhimurium and its complexes with allosteric effectors. J Mol Biol. 2003;330:87–97. doi: 10.1016/s0022-2836(03)00538-2. [DOI] [PubMed] [Google Scholar]
- 3.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013;46:2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotruvo JA, Stubbe J. Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofer A, Crona M, Logan DT, Sjöberg BM. DNA building blocks: Keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47:50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eriksson M, et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 7.Ando N, et al. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc Natl Acad Sci USA. 2011;108:21046–21051. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ando N, et al. Allosteric inhibition of human ribonucleotide reductase by dATP entails the stabilization of a hexamer. Biochemistry. 2016;55:373–381. doi: 10.1021/acs.biochem.5b01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson R, et al. Structural mechanism of allosteric activity regulation in a ribonucleotide reductase with double ATP cones. Structure. 2016;24:906–917. doi: 10.1016/j.str.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Wolf YI, Koonin EV. The ATP-cone: An evolutionarily mobile, ATP-binding regulatory domain. J Mol Microbiol Biotechnol. 2000;2:191–194. [PubMed] [Google Scholar]
- 11.Meisburger SP, Thomas WC, Watkins MB, Ando N. X-ray scattering studies of protein structural dynamics. Chem Rev. 2017;117:7615–7672. doi: 10.1021/acs.chemrev.6b00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crona M, et al. NrdH-redoxin protein mediates high enzyme activity in manganese-reconstituted ribonucleotide reductase from Bacillus anthracis. J Biol Chem. 2011;286:33053–33060. doi: 10.1074/jbc.M111.278119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fieschi F, et al. The manganese-containing ribonucleotide reductase of Corynebacterium ammoniagenes is a class Ib enzyme. J Biol Chem. 1998;273:4329–4337. doi: 10.1074/jbc.273.8.4329. [DOI] [PubMed] [Google Scholar]
- 14.Jordan A, et al. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J Biol Chem. 1996;271:8779–8785. doi: 10.1074/jbc.271.15.8779. [DOI] [PubMed] [Google Scholar]
- 15.Jordan A, et al. A second class I ribonucleotide reductase in Enterobacteriaceae: Characterization of the Salmonella typhimurium enzyme. Proc Natl Acad Sci USA. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makhlynets O, et al. Streptococcus sanguinis class Ib ribonucleotide reductase: High activity with both iron and manganese cofactors and structural insights. J Biol Chem. 2014;289:6259–6272. doi: 10.1074/jbc.M113.533554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabinovitch I, et al. Staphylococcus aureus NrdH redoxin is a reductant of the class Ib ribonucleotide reductase. J Bacteriol. 2010;192:4963–4972. doi: 10.1128/JB.00539-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca I, Torrents E, Sahlin M, Gibert I, Sjöberg BM. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J Bacteriol. 2008;190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, et al. Characterization of two genes encoding the Mycobacterium tuberculosis ribonucleotide reductase small subunit. J Bacteriol. 1997;179:6408–6415. doi: 10.1128/jb.179.20.6408-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker MJ, Zhu X, Stubbe J. Bacillus subtilis class Ib ribonucleotide reductase: High activity and dynamic subunit interactions. Biochemistry. 2014;53:766–776. doi: 10.1021/bi401056e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge J, Yu G, Ator MA, Stubbe J. Pre-steady-state and steady-state kinetic analysis of E. coli class I ribonucleotide reductase. Biochemistry. 2003;42:10071–10083. doi: 10.1021/bi034374r. [DOI] [PubMed] [Google Scholar]
- 22.Anand R, et al. A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex. Biochemistry. 2004;43:10343–10352. doi: 10.1021/bi0491292. [DOI] [PubMed] [Google Scholar]
- 23.Characteristics of nucleic acids. Curr Protoc Mol Biol. 1996;33:A.1D.1–A.1D.11. doi: 10.1002/0471142727.mba01ds33. [DOI] [PubMed] [Google Scholar]
- 24.Randerath K, Randerath E. Ion-exchange chromatography of nucleotides on poly-(ethyleneimine)-cellulose thin layers. J Chromatogr A. 1964;16:111–125. doi: 10.1016/s0021-9673(01)82445-6. [DOI] [PubMed] [Google Scholar]
- 25.Rowley GL, Kenyon GL. PEI-cellulose thin-layer chromatography. Product studies of the creatine kinase and pyruvate kinase reactions. Anal Biochem. 1974;58:525–533. doi: 10.1016/0003-2697(74)90220-6. [DOI] [PubMed] [Google Scholar]
- 26.Parker MJ. 2017. Discovery and Investigation of the Novel Overall Activity Allosteric Regulation of the Bacillus subtilis class Ib Ribonucleotide Reductase. PhD thesis (Massachusetts Institute of Technology, Cambridge, MA)
- 27.Zhang Y, Stubbe J. Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry. 2011;50:5615–5623. doi: 10.1021/bi200348q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 30.Boal AK, Cotruvo JA, Jr, Stubbe J, Rosenzweig AC. The dimanganese(II) site of Bacillus subtilis class Ib ribonucleotide reductase. Biochemistry. 2012;51:3861–3871. doi: 10.1021/bi201925t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denessiouk KA, Rantanen VV, Johnson MS. Adenine recognition: A motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- 32.Zimanyi CM, et al. Tangled up in knots: Structures of inactivated forms of E. coli class Ia ribonucleotide reductase. Structure. 2012;20:1374–1383. doi: 10.1016/j.str.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saraste M, Sibbald PR, Wittinghofer A. The P-loop–A common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 34.Meisburger SP, et al. Domain movements upon activation of phenylalanine hydroxylase characterized by crystallography and chromatography-coupled small-angle x-ray scattering. J Am Chem Soc. 2016;138:6506–6516. doi: 10.1021/jacs.6b01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutin S, Brennich M, Maillot B, Round A. Online ion-exchange chromatography for small-angle X-ray scattering. Acta Crystallogr D Struct Biol. 2016;72:1090–1099. doi: 10.1107/S2059798316012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonna VR, et al. Diversity in overall activity regulation of ribonucleotide reductase. J Biol Chem. 2015;290:17339–17348. doi: 10.1074/jbc.M115.649624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scotti C, Valbuzzi A, Perego M, Galizzi A, Albertini AM. The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology. 1996;142:2995–3004. doi: 10.1099/13500872-142-11-2995. [DOI] [PubMed] [Google Scholar]
- 38.O’Handley SF, Frick DN, Bullions LC, Mildvan AS, Bessman MJ. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. Cloning, purification, and characterization of the enzyme. J Biol Chem. 1996;271:24649–24654. doi: 10.1074/jbc.271.40.24649. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Dunn CA, Jones CR, D’Souza G, Bessman MJ. The 26 Nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J Biol Chem. 2004;279:24861–24865. doi: 10.1074/jbc.M403272200. [DOI] [PubMed] [Google Scholar]
- 40.Xu W, Jones CR, Dunn CA, Bessman MJ. Gene ytkD of Bacillus subtilis encodes an atypical nucleoside triphosphatase member of the Nudix hydrolase superfamily. J Bacteriol. 2004;186:8380–8384. doi: 10.1128/JB.186.24.8380-8384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grinberg I, et al. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J Bacteriol. 2006;188:7635–7644. doi: 10.1128/JB.00903-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKethan BL, Spiro S. Cooperative and allosterically controlled nucleotide binding regulates the DNA binding activity of NrdR. Mol Microbiol. 2013;90:278–289. doi: 10.1111/mmi.12364. [DOI] [PubMed] [Google Scholar]
- 43.Knappenberger AJ, Ahmad MF, Viswanathan R, Dealwis CG, Harris ME. Nucleoside analogue triphosphates allosterically regulate human ribonucleotide reductase and identify chemical determinants that drive substrate specificity. Biochemistry. 2016;55:5884–5896. doi: 10.1021/acs.biochem.6b00594. [DOI] [PubMed] [Google Scholar]
- 44.Larsson KM, et al. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat Struct Mol Biol. 2004;11:1142–1149. doi: 10.1038/nsmb838. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, et al. Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc Natl Acad Sci USA. 2006;103:4022–4027. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimanyi CM, Chen PYT, Kang G, Funk MA, Drennan CL. Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli. eLife. 2016;5:e07141. doi: 10.7554/eLife.07141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stolle P, et al. Homologous expression of the nrdF gene of Corynebacterium ammoniagenes strain ATCC 6872 generates a manganese-metallocofactor (R2F) and a stable tyrosyl radical (Y˙) involved in ribonucleotide reduction. FEBS J. 2010;277:4849–4862. doi: 10.1111/j.1742-4658.2010.07885.x. [DOI] [PubMed] [Google Scholar]
- 48.Cotruvo JA, Jr, Stich TA, Britt RD, Stubbe J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: Enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J Am Chem Soc. 2013;135:4027–4039. doi: 10.1021/ja312457t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marblestone JG, et al. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.