Significance
Many patients with breast and ovarian cancer carry inherited cancer-predisposing mutations in BRCA1. However, virtually no patients have two inherited mutations in BRCA1 because the DNA repair function of BRCA1 is essential for embryonic development. We discovered that patients with two nonsense mutations from a specific region of BRCA1 may survive as the result of naturally occurring alternative splicing that yields a short but partially functional BRCA1 protein. These patients are extremely rare, and are characterized by severe chromosomal fragility, congenital anomalies, and predisposition to childhood cancers.
Keywords: BRCA1, RNA splicing, DNA repair, chromosomal fragility
Abstract
BRCA1 is essential for repair of DNA double-strand breaks by homologous recombination, and hence for survival. Complete loss of its function is lethal during early embryonic development. Patients who are compound heterozygous for BRCA1 truncating mutations and missense alleles that retain some DNA repair capacity may survive, albeit with very high risk of early onset breast or ovarian cancer and features of Fanconi anemia. However, a mechanism enabling survival of patients homozygous for BRCA1 truncating mutations has not been described. We studied two unrelated families in which four children presented with multiple congenital anomalies and severe chromosomal fragility. One child developed T cell acute lymphocytic leukemia (ALL), and a second child developed neuroblastoma. Each of the four children was homozygous for a nonsense mutation in BRCA1 exon 11. Homozygosity for the nonsense mutations was viable thanks to the presence of a naturally occurring alternative splice donor in BRCA1 exon 11 that lies 5′ of the mutations. The mutations did not affect the alternative splice site, but transcription from it produced an in-frame BRCA1 message with deletion of 3,309 bp. The translated BRCA1 protein was only 40% of normal length, but with intact N- and C-terminal sequences. These patients extend the range of BRCA1-related phenotypes and illustrate how naturally occurring alternative splicing can enable survival, albeit with severe consequences, of otherwise lethal genotypes of an essential gene.
The BRCA/Fanconi DNA repair pathway is responsible for repair of DNA double-strand breaks via homologous recombination (1). Among the components of the pathway are BRCA1, BRCA2, and PALB2, each of which harbors mutations predisposing to breast, ovarian, and prostate cancer (2–5). Homozygosity or compound heterozygosity for mutations in several of the genes in the pathway causes Fanconi anemia, presenting with congenital anomalies, bone marrow failure, and predisposition to both solid tumors and leukemia (1). However, despite the existence of relatively common cancer-predisposing BRCA1 alleles in several populations, homozygotes for these founder alleles have not been reported. Indeed, patients homozygous for BRCA1 truncating alleles have not been expected, because evidence from mouse models has shown that at least one copy of BRCA1 is necessary for embryonic development (6–8). Consistent with this observation, patients with BRCA1-related Fanconi anemia and very young onset breast or ovarian cancer have been shown to be compound heterozygous for a BRCA1 truncating mutation and a BRCA1 missense allele retaining partial DNA repair activity (9, 10).
In the course of genomic analysis of consanguineous families with severe childhood illnesses, we evaluated two families with congenital abnormalities and childhood cancers. In each family, the affected children were homozygous for an inherited BRCA1 nonsense mutation. Because we had believed inherited homozygous pathogenic mutations in BRCA1 to be incompatible with life, we explored the molecular mechanism enabling survival of these children. We discovered that these mutant alleles exploited an alternate, naturally occurring donor splice site in BRCA1 exon 11, yielding an in-frame short BRCA1 protein. This short BRCA1 isoform is known to retain partial capacity for DNA repair (11), thus enabling embryonic survival but not fully normal development.
Results
Patients and Clinical Analysis.
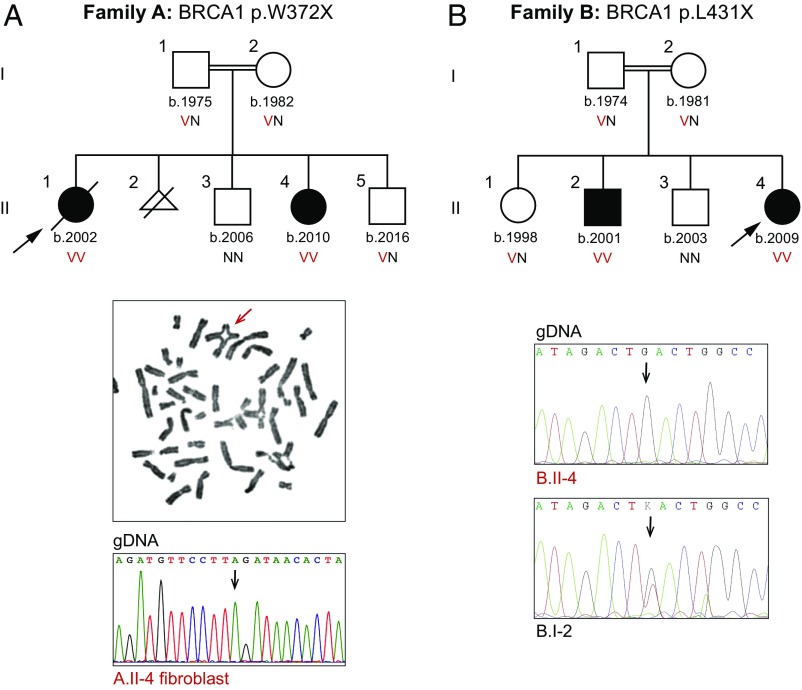
Clinical features of the four children in the two families in the study are summarized in Table 1. Family A, consanguineous and of Arab ancestry, has two affected daughters (Fig. 1A). Family B, consanguineous and of Turkish ancestry, has an affected daughter and an affected son (Fig. 1B). All four children had failure to thrive, manifesting from birth or postnatally. All had microcephaly, intellectual disability, microphthalmia, and skin pigmentation lesions (hyperpigmented and hypopigmented). Limb defects (small hands or feet and hypoplastic thumb) were present in three. Dysmorphic features were noted in both affected children in family A, and brain gliosis was observed in both affected children in family B. Other manifestations (e.g., congenital heart defects, optic nerve hypoplasia) were present in only one patient. Chromosomal breakage was substantially increased in all cases (Table 1). Proportions of cells with radial chromosomes (e.g., Fig. 1A) were very similar to those of patients with other forms of Fanconi anemia; proportions of cells with other forms of chromosome breaks were somewhat lower than among patients with Fanconi anemia generally (12). Notably, except during periods of active malignant disease (discussed below), none of the children had anemia or bone marrow failure.
Table 1.
Clinical characteristics of patients with two BRCA1 mutations
| Features | A.II.1 | A.II.4 | B.II.2 | B.II.4 | Ref. 9 | Ref. 10 |
| BRCA1 genotype | p.W372X homozygote | p.W372X homozygote | p.L431X homozygote | p.L431X homozygote | p.846X/p.V1736A | p.233X/p.R1699W |
| Sex and karyotype | Female; 46;XX | Female; 46;XX | Male; 46;XY | Female; 46;XX | Female; 46;XX | Female; 46;XX |
| Age last follow-up | 5 y (deceased) | 6 y | 15.5 y | 7 y | 28 y (deceased) | 25 y (deceased) |
| Growth | ||||||
| At birth | SGA: 33 wks, 1.47 kg | AGA: 38 wks, 2.79 kg | SGA: term, 1.6 kg | AGA: 2.9 kg | nd | SGA: 1.99 kg |
| Postnatal | 5 y: 12 kg, 92 cm | FTT | Height < 3%ile | Height < 3%ile | Short stature | 25 y: 40 kg,135 cm |
| Intellectual disability | Developmental delay | Mild developmental delay | Yes (IQ: 50–69) | Yes (IQ: 62) | Developmental delay | Yes |
| Microcephaly | Yes (39 cm at 5 y) | Yes | Yes | Yes | Yes | Yes (microsomia) |
| Microphthalmia | Yes | Yes | Yes | Yes | nd | Yes (microsomia) |
| Skin pigment lesions | ||||||
| Hypopigmented | Yes | Yes | Yes | Yes | nd | Yes |
| Hyperpigmented | Yes | Yes | Generalized | No | nd | Yes |
| Café-au-lait spots | Yes | Yes | Yes | Yes | nd | No |
| Limb defects | Small edematous palms, soles | Small palms | Hypoplastic thumb (Blauth 2) | None | None | Proximally inserted thumbs, second digit captodactyly, second/third toe syndactyly |
| Dysmorphic features | Upslanting palpebral fissures, micrognathia | Triangular facies, upslanting eyes, epicanthus, cupped ears, short neck, micrognathia | None | None | Low anterior hairline, prominent nasal bridge, small alae nasi, macrognathia | Upslating palpebral fissures, sparse hair, hypertelorism, epicanthal folds, ptosis, strabismus, blepharophimosis, broad nasal bridge/nasal tip |
| Other | Optic nerve hypoplasia, bilateral hip dysplasia | Right first rib hypoplasia; Left 11 ribs | Right undescended testis, adrenal insufficiency | Heart defect (ASD and VSD), growth hormone deficiency | Duodenal stenosis, hyperextensible knees, history of hip dislocation, dental malocclusion | |
| Malignancy (age at dx) | T cell ALL (5y) | None | None | Neuroblastoma (2 y) | Ovarian cancer (28 y) | Breast cancer (25 y) |
| Chromosome breakage (case vs. control) | ||||||
| Spontaneous | 13.3% vs. 6.6% | nd | 6% vs. 0 | 2% vs. 0 | nd | 0.025 vs. 0.025 |
| Induced, DEB | 70% vs. 7% | 32% vs. 8% | 1.44 vs. 1.0 | 2.14 vs. 1.0 | nd | 2.2 vs. 0.05 |
| Induced, MMC | nd | 92% vs. 34% | nd | nd | nd | nd |
| Radials | Increased | Increased | nd | nd | nd | 30% vs. 0% |
| CBC/HbF | Normal | Normal | Normal | Normal | Normal | Normal |
| Bone marrow | t(1,16)+1q | nd | 15 y: cellular, no dysplasia | 2 y: cellular, adipocyte infiltration, no dysplasia; 7 y: normal | nd | nd |
| Brain MRI | nd | nd | Gliosis: periventricular and subcortical | Gliosis, prominent cortical sulci | nd | nd |
| Cancer family history | Intestinal and urological cancer, 2nd and 3rd degree relatives | Intestinal and urological cancer, 2nd and 3rd degree relatives | Uterine, esophageal, and lung cancer, 2nd and 3rd degree relatives | Uterine, esophageal, and lung cancer, 2nd and 3rd degree relatives | Breast, ovarian, intestinal cancer in 1st–4th degree relatives |
AGA, appropriate for gestational age; ASD, atrial septal defect; CBC, complete blood count; DEB, diepoxybutane; dx, diagnosis; FTT, failure to thrive; HbF, fetal hemoglobin; IQ, intelligence quotient; MMC, mitomycin C; MRI, magnetic resonance imaging; nd, not described; %ile, percentile; SGA, small for gestational age.
Fig. 1.
Genetic and clinical features of the families. (A, Top) Coinheritance of BRCA1 p.W372X (c.1115G > A, NM_007294.3) with chromosomal breakage and congenital anomalies. (A, Middle) Example of a radial chromosome from patient A.II.4. (A, Bottom) Sequence at BRCA1 c.1115G > A from skin fibroblasts of A.II.4, indicating no reversion of the mutant to a wild-type sequence. (B, Top) Coinheritance of BRCA1 p.L431X (c.1151T > G, NM_007294.3) chromosomal breakage and congenital anomalies. (B, Bottom) Sequence at BRCA1 p.L431X (c.1151T > G) from B.II.4 compared with his father, B.I.2, indicating no reversion of the mutant to a wild-type sequence. Arrows in sequence traces indicate variant sites. N, reference allele; V, variant allele.
In family A, proband A.II.1 presented at the age of 5 y with anemia (hemoglobin: 5.1 g/dL), hyperleukocytosis (156 × 109 per liter), and generalized lymphadenopathy, and was diagnosed with T cell acute lymphocytic leukemia (ALL) with chromosomal aberrations t(1,16) and +1q. She began treatment according to the ALL intercontinental Berlin–Frankfurt–Munster (IC-BFM) 2002 protocol (13), including intrathecal methotrexate and prednisone. On day 8 of treatment, her blast count was 8,000 and she received one dose of vincristine (0.75 mg/m2). Her vincristine dose was half the usual dose, given results of her chromosome breakage tests, and hence a diagnosis of possible Fanconi anemia (her genotype was not then known). Her leukemia did not respond well to chemotherapy; 2 wk later, she developed hepatitis, fulminant varicella infection, and enterococcal sepsis, and succumbed.
Patient A.II.4, the younger sister of A.II.1, also has elevated chromosomal sensitivity to diepoxybutane (DEB) and mitomycin C (MMC). She remains cancer-free at the age of 8 y. Family A also has two healthy children. Another pregnancy was terminated after in utero diagnosis of increased chromosomal breakage. The patients’ mother and father are cancer-free at the ages of 36 y and 43 y, respectively.
In family B, the proband (B.II.4) was diagnosed at the age of 2 y with stage IV neuroblastoma and completed the treatment protocol with full chemotherapy doses. At the age of 5 y, she was diagnosed with possible Fanconi anemia based on her clinical features and chromosome breakage testing. She received no transfusions except during neuroblastoma treatment; her serum ferritin was 911 ng/mL, and abdominal MRI revealed an absence of iron deposition.
Patient B.II.2, the older brother of B.II.4, had chromosome breakage testing at the same time as his sister. He remains cancer-free and with no signs of bone marrow failure at the age of 15 y and 6 mo. The genotypes of the children were not known when possible Fanconi anemia was diagnosed. Family B also has two healthy children. The patients’ mother and father are cancer-free at the ages of 37 y and 44 y, respectively.
Mutation Discovery.
Genomic analysis was carried out for family A using the MarrowSeq panel of 241 genes (14) and for family B by whole-exome sequencing. For family A, the only damaging mutation consistent with recessive inheritance of the children’s phenotype was BRCA1 p.W372X (c.1115G > A, NM_007294.3) at chr17:41,246,433 (hg19). The mutation was homozygous in both affected siblings, heterozygous in their parents, and absent from the healthy sibling (Fig. 1A). For family B, the only damaging mutation consistent with recessive inheritance was BRCA1 p.L431X (c.1292T > G, NM_007294.3) at chr17:41,246,256 (hg19). The mutation was homozygous in both affected siblings, heterozygous in their parents and one healthy sibling, and absent from the other healthy sibling (Fig. 1B).
Homozygosity for complete loss of BRCA1 function is embryonic lethal (6–8), so we first considered the possibility of somatic reversion of the nonsense allele to wild type. Somatic reversion can be evaluated by screening for mosaic sequence at the mutant site in affected individuals. From patient A.II.4, both sequencing at 250-fold coverage of genomic DNA from peripheral blood and Sanger sequencing of genomic DNA from her skin fibroblasts yielded only the mutant allele (Fig. 1A). Similarly, from patient B.II.4, Sanger sequencing of genomic DNA from peripheral blood yielded only the mutant allele and sequencing from genomic DNA of his father, B.I.2, did not reveal any loss of the mutant allele (Fig. 1B).
To assess whether truncating mutations in BRCA1 distal exon 11 might be less severe with respect to cancer risk than such mutations elsewhere in the gene, we evaluated whether cancer-predisposing mutations in BRCA1 were less likely to lie in this region. Distal exon 11 of BRCA1 includes 3,309 bp of the 5,589-bp coding sequence (59.2%). Of all known BRCA1 mutations (other than large genomic deletions) indicated as “pathogenic,” 1,318 of 2,238 mutations (58.9%) lie in distal exon 11 (https://www.ncbi.nlm.nih.gov/clinvar/). We conclude that cancer-predisposing mutations are as likely to occur in this region as elsewhere in BRCA1.
Transcription and Translation of the Mutant Alleles.
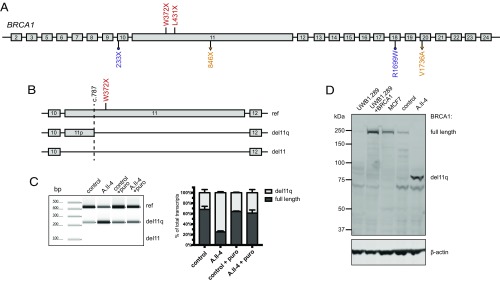
Transcripts and proteins from the patients were evaluated to determine the molecular mechanism by which homozygosity for these nonsense mutations could be viable (Fig. 2A). Wild-type BRCA1 generates multiple minor transcripts, including several lacking part or all of exon 11 (15). One such naturally occurring transcript exploits an alternate donor splice at BRCA1 c.787 at chr17:41,246,761 (Fig. 2B). Transcription from this alternate donor site leads to loss of nucleotides c.788–4,096 of exon 11, and translation yields a short BRCA1 isoform of 759 amino acids, lacking residues 263–1,365 of the normal 1,863-residue protein. The short isoform has normal 5′ and 3′ UTR sequences. We call this transcript and isoform BRCA1 del11q. Another naturally occurring minor transcript, BRCA1 del11, completely lacks BRCA1 exon 11 and yields an isoform of 760 amino acids with normal 5′ and 3′ UTR sequences (11). BRCA1 p.W372X and BRCA1 p.L431X lie 3′ of the alternate donor splice site at BRCA1 c.787, and consequently are deleted from the BRCA1 del11q mutant transcript as well as from the BRCA1 del11 mutant transcript. The BRCA1 del11q isoform from the homozygous children would therefore not be subject to premature stops but would be only 40% of the length of normal BRCA1.
Fig. 2.
Transcript and protein analysis of BRCA1 mutant alleles. (A) Homozygous mutations in families A and B (shown in red) and previously reported compound heterozygous mutations V1736A/846X (c.2457delC; shown in orange) and R1999W/233X (c.594_597del; shown in purple). (B) Schematic overview of splicing at BRCA1 exons 10–12. The dashed line at BRCA1 c.787 indicates a naturally occurring alternate splice site that yields an in-frame transcript, BRCA1 del11q, lacking 3,309 bp of exon 11. Another in-frame transcript, BRCA1 del11, lacking all 3,429 bp of exon 11 has also been reported. (C, Left) BRCA1 transcripts, measured by RT-PCR of RNA isolated from fibroblasts from a control and from patient A.II.4, and then repeated from fibroblasts treated with puromycin to suppress nonsense-mediated decay. (C, Right) Relative amounts of BRCA1 transcripts from three replicate experiments. Results suggest that both full-length and del11q transcripts are made in all cells, with full-length transcripts from the mutant allele subject to nonsense-mediated decay. Transcripts completely lacking exon 11 were not detected. (D) Western blot of nuclear fractions of protein lysate from fibroblasts of A.II.4 compared with controls. UWB1.289 is a BRCA1-null ovarian cancer cell line (1). UWB1.289+BRCA1 is UWB1.289 complemented with a full-length BRCA1 transcript. MCF7 is a breast cancer cell line with a normal BRCA1 sequence. β-Actin is an internal loading control.
Profiles of BRCA1 transcripts of A.II.4 compared with controls reflected these effects (Fig. 2C). In RNA from control fibroblasts, 66% of BRCA1 transcripts were full-length and 34% were del11q. In contrast, in RNA from fibroblasts of A.II.4, 23% of BRCA1 transcripts were full-length and 77% were del11q (P < 0.001). BRCA1 del11 was not detected in either sample. After inhibition of nonsense-mediated decay by puromycin, expression of full-length and del11q transcripts was similar in A.II.4 fibroblasts, as in controls. This result suggests that from the mutant allele, enrichment of del11q transcripts is the result primarily of nonsense-mediated decay of full-length transcripts rather than increased alternative splicing.
Analysis of proteins from fibroblasts of A.II.4 compared with controls revealed differences in BRCA1 isoforms (Fig. 2D). Cells of A.II.4 had no detectable full-length BRCA1 protein but, instead, had BRCA1 protein corresponding to the size of the del11q isoform.
Discussion
Thousands of families have been identified with inherited cancer-predisposing mutations in BRCA1, yet virtually no patients with two BRCA1 mutations are known. Previous reports described two patients, each compound heterozygous for a truncating mutation and a missense mutation in BRCA1, with features of Fanconi anemia and very young onset breast or ovarian cancer (9, 10). These reports were at first surprising, because it had been known for more than 20 y that at least one copy of BRCA1 is necessary for murine embryonic development (6–8). Characterization of the BRCA1 mutations of the human patients resolved the paradox, revealing that BRCA1 compound heterozygosity was compatible with life because the missense allele of each patient retained enough function to enable embryonic survival (9, 10).
Analyses of the children in families A and B suggest that homozygosity for their nonsense mutations is compatible with life for a different reason: the existence of a naturally occurring, minor isoform of BRCA1, generated by an alternate splice donor site in BRCA1 exon 11. The short isoform created by the alternate splice is only 40% the length of normal BRCA1 protein but retains the normal reading frame. The nonsense mutations of families A and B do not create or alter the alternate splice donor but lie 3′ of it in exon 11, so that the nonsense mutations are spliced out of the short isoform. Notably, splicing at the alternate donor site is not elevated in cells of patients compared with controls, but loss of full-length mutant BRCA1 transcripts (but not full-length normal BRCA1 transcripts) by nonsense-mediated decay yields significant enrichment of alternately spliced transcripts in patients’ cells (Fig. 2C). Additional support for the importance of the alternative splice site is provided by the very recent report of another child with Fanconi-like features who is homozygous for nonsense mutation BRCA1 p.C903X (c.2709T > A) (16). This mutation also lies in exon 11 at a site 3′ of the alternate splice donor, so it likely also yields the BRCA1 del11q isoform. Despite the existence of relatively common BRCA1 truncating alleles in several populations, patients homozygous for BRCA1 founder alleles have not been encountered. The absence of such patients is likely due to the positions in BRCA1 of these founder alleles: None lie in distal exon 11.
The response to chemotherapy varied considerably among the four patients with cancer, who were homozygous or compound heterozygous for BRCA1 mutations. One of the two previously reported patients had multiple congenital anomalies; was diagnosed with ovarian cancer at the age of 28 y; and developed severe neutropenia, anemia, and thrombocytopenia during chemotherapy. Her BRCA1 missense mutation, V1736A, had a reduced, but not completely lost, capacity to repair double-strand DNA breaks (9). The other previously reported patient had multiple congenital anomalies and elevated formation of radial chromosomes on chromosome breakage assay, and she was diagnosed with breast cancer at the age of 25 y. She was compound heterozygous for a BRCA1 frameshift and for R1699W. Fibroblasts from this patient retained partial capacity for DNA repair (10). This patient tolerated chemotherapy and radiation without unusual toxicity. Of the patients in the present study, A.II.1 did not respond well to steroids. Her fatal complications (varicella and sepsis) may have been the result of both her active leukemia and her treatment. In contrast, patient B.II.2 tolerated chemotherapy for her stage IV neuroblastoma very well, which is unusual for a patient with Fanconi anemia.
The BRCA1 del11q isoform has been shown to have lost ∼50% capacity to repair DNA damage (11). This deficiency is reflected in sensitivity in chromosome breakage assays, in congenital anomalies, and in cancer predisposition of the homozygous children. Other functions of BRCA1, including interactions with PALB2, BRCA2, RAD51, and CTIP, remain intact in this isoform (11). Binding of these protein partners to the C-terminal BRCT domain is critical to BRCA1 function, and the BRCT domain is intact in BRCA1 del11q.
All patients with two mutations in BRCA1, both those previously described and our patients, had severe but not complete loss of DNA repair capacity (9, 10). Before their cancer diagnoses, all had normal complete blood cell counts and no signs of bone marrow failure (Table 1). This combination of normal blood cell counts and predisposition to early-age leukemia and solid tumors is strikingly reminiscent of the clinical features of Fanconi anemia subtype D1, caused by biallelic mutations of BRCA2 (17, 18). In the mouse, complete deletion of Brca1 specifically in the bone marrow leads to hematopoietic defects resembling Fanconi anemia (19). The children of families A and B presented with all the features of Fanconi anemia except bone marrow failure. Age at onset of bone marrow failure in other types of Fanconi anemia is highly variable, with a mean of 7.6 y (20). The oldest affected child in this study is now 15 y of age and still free of bone marrow failure. It is possible that these children may not have had bone marrow failure simply because they were too young, although the previously described patients with compound heterozygous BRCA1 mutations had no bone marrow failure at the ages of 28 y and 23 y (9, 10).
The discovery and characterization of two families with children homozygous for nonsense mutations in BRCA1 offer insights into biological and clinical features of the gene. First, homozygosity for some BRCA1 nonsense mutations can be compatible with life. Second, these patients have clinical features that include both Fanconi syndrome anomalies and predisposition to childhood cancers. Third, increased expression of an alternate in-frame BRCA1 isoform can partially compensate for loss of the full-length protein.
Finally, we suggest that evaluating other consanguineous families with Fanconi anemia, childhood cancers, or young onset breast or ovarian cancers may reveal other homozygous BRCA1 mutations that are compatible with life. Homozygosity for BRCA1 in patients with chromosomal fragility or childhood cancers may be underreported because many Fanconi anemia gene panels do not yet include BRCA1. Identifying the genotypes and phenotypes of such patients would contribute to understanding the full spectrum of clinical phenotypes associated with BRCA1 mutations. This nuanced understanding can lead to better clinical management, inform genetic counseling, and help to further reveal the role of BRCA1 in cell biology and human development.
Materials and Methods
Patients.
The study was approved by the Institutional Review Boards of the University of Washington, Seattle Children’s Hospital, Boston Children’s Hospital, Rabin Medical Center, and Hacettepe University. Informed consent was obtained from all study subjects or their parents.
Genetic Analysis.
Chromosome breakage was analyzed from peripheral blood, as previously described (21). Genomic analysis for family A was based on targeted capture and multiplexed sequencing using the MarrowSeq panel of 241 genes (14), and gene analysis for family B was performed by whole-exome sequencing. Bioinformatics analysis and variant interpretation were carried out as previously described (22). Mutations were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/): variants 54134 (BRCA1 p.W372X) and 54187 (BRCA1 p.L431X).
Transcript Analysis.
BRCA1 transcripts were evaluated by RT-PCR. RNA was extracted using TRIzol (Thermo Fisher Scientific) and then treated with DNase I (Qiagen). cDNA was transcribed using SuperScript II (Thermo Fisher Scientific). BRCA1 full-length, del11q, and del11 transcripts were amplified with the following primers (23): BRCA1ex9–10for2: 5′-CAACTTATTGCAGTGTGGGAGA-3′, BRCA1ex11FLrev: 5′-GGAGTCCGCCTATCATTACATG-3′, and BRCA1ex12rev: 5′-CCAGATGCTGCTTCACCCT-3′. PCR products were analyzed using D1000 ScreenTape and 2200 TapeStation (Agilent) and Sanger-sequenced.
Tissue Culture.
Cell line UWB1.289 (24) was grown in 50% Mammary Epithelial Cell Growth Medium (MEGM) and 50% RPMI 1640 supplemented with 3% FBS and penicillin/streptomycin (Thermo Fisher Scientific). UWB1.289+BRCA1, the same cell line transfected with wild-type BRCA1, was grown in the same medium with G418 supplementation (200 μg/mL). Breast cancer cell line MCF7 was grown in MEM supplemented with 10% FBS and penicillin/streptomycin. Human fibroblasts were grown in DMEM supplemented with 10% FBS, 2 mM glutamine, and penicillin/streptomycin. Patient fibroblasts were grown in Chang D Media (Irvine Scientific) supplemented with penicillin/streptomycin. For inhibition of nonsense-mediated decay, skin fibroblasts were cultured with 500 μg/mL puromycin (Sigma) for 4 h before cell harvesting.
Western Blots.
Nuclear lysates from all cell lines were extracted using the NE-PER Nuclear and Cytoplasmic Extraction Kit according to manufacturer protocols (Thermo Fisher Scientific). Nuclear lysates (40 μg) were resolved on a 4–12% Bis-Tris gel (Thermo Fisher Scientific) under denaturing conditions. Wet transfer was performed at 100 V for 2 h at 4 °C. Membranes were blocked in LI-COR Odyssey Blocking Buffer. Primary antibody incubation was performed in a cold room overnight. Antibody to detect BRCA1 was MS110 (EMD Millipore), diluted 1:500. Other antibodies were against β-actin (1:10,000; Sigma) and αMs-700nm (LI-COR). Membranes were developed using the LI-COR Odyssey Imager (LI-COR).
Acknowledgments
We thank the patients and their families for their participation; A. Bernhardy, M. Demirel, Z. Gormez, M. K. Lee, N. Rosenfeld, and M. S. Sağıroğlu for technical assistance; and J. Abkowitz, A. Koren, S. Elitzur, S. Gulsuner, N. Johnson, S. Keel, E. Levy-Lahad, E. M. Swisher, and P. Welcsh for helpful discussions. This project was supported by NIH Grants R35CA197458 (to M.-C.K.), R24DK099808 (to A. Shimamura and M.-C.K.), and F30DK103462 (to A. Seo); the Breast Cancer Research Foundation (M.-C.K.); and Turkish Ministry of Development Grant 2011K120020 (to N.A.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: New players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Marks JH, Mandell JB. New York Breast Cancer Study Group Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 3.Norquist BM, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou AC, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard CC, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 7.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 9.Domchek SM, et al. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013;3:399–405. doi: 10.1158/2159-8290.CD-12-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer SL, et al. University of Washington Centre for Mendelian Genomics FORGE Canada Consortium Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. kConFab Investigators The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 2016;76:2778–2790. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fargo JH, et al. Comparison of chromosome breakage in non-mosaic and mosaic patients with Fanconi anemia, relatives, and patients with other inherited bone marrow failure syndromes. Cytogenet Genome Res. 2014;144:15–27. doi: 10.1159/000366251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stary J, et al. Intensive chemotherapy for childhood acute lymphoblastic leukemia: Results of the randomized intercontinental trial ALL IC-BFM 2002. J Clin Oncol. 2014;32:174–184. doi: 10.1200/JCO.2013.48.6522. [DOI] [PubMed] [Google Scholar]
- 14.Zhang MY, et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica. 2015;100:42–48. doi: 10.3324/haematol.2014.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Conzen SD, Cole CN, Arrick BA. Characterization of functional messenger RNA splice variants of BRCA1 expressed in nonmalignant and tumor-derived breast cells. Cancer Res. 1996;56:4578–4581. [PubMed] [Google Scholar]
- 16.Freire BL, et al. Homozygous loss of function BRCA1 variant causing a Fanconi-anemia-like phenotype, a clinical report and review of previous patients. Eur J Med Genet. 2017;61:130–133. doi: 10.1016/j.ejmg.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch B, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- 18.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasanthakumar A, et al. Brca1 deficiency causes bone marrow failure and spontaneous hematologic malignancies in mice. Blood. 2016;127:310–313. doi: 10.1182/blood-2015-03-635599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castella M, et al. Chromosome fragility in patients with Fanconi anaemia: Diagnostic implications and clinical impact. J Med Genet. 2011;48:242–250. doi: 10.1136/jmg.2010.084210. [DOI] [PubMed] [Google Scholar]
- 22.Walsh T, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raponi M, Douglas AGL, Tammaro C, Wilson DI, Baralle D. Evolutionary constraint helps unmask a splicing regulatory region in BRCA1 exon 11. PLoS One. 2012;7:e37255. doi: 10.1371/journal.pone.0037255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DelloRusso C, et al. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res. 2007;5:35–45. doi: 10.1158/1541-7786.MCR-06-0234. [DOI] [PubMed] [Google Scholar]