Significance
IFN regulatory factor 3 (IRF3) is one of the most well-characterized transcription factors involved in the regulation of innate immune responses. Many studies have used conventional Irf3-deficient mice to analyze the functions of IRF3 in immunity and other biological systems; however, these mice were found to carry another null mutation in the Bcl2l12 gene, which has confounded the analyses of these mice. For this study, we generated conditional Irf3-deficient mice while leaving the Bcl2l12 gene intact. Conditional ablation of IRF3 in myeloid cells showed increased survival when challenged with endotoxin-induced shock, indicating a myeloid cell-specific role of IRF3. The IRF3 conditional knockout mouse may be a useful tool to identify cell type-specific functions of this transcription factor.
Keywords: IRF3, infection, Bcl2l12, interferon, inflammation
Abstract
IFN regulatory factor 3 (IRF3) is a transcription regulator of cellular responses in many cell types that is known to be essential for innate immunity. To confirm IRF3’s broad role in immunity and to more fully discern its role in various cellular subsets, we engineered Irf3-floxed mice to allow for the cell type-specific ablation of Irf3. Analysis of these mice confirmed the general requirement of IRF3 for the evocation of type I IFN responses in vitro and in vivo. Furthermore, immune cell ontogeny and frequencies of immune cell types were unaffected when Irf3 was selectively inactivated in either T cells or B cells in the mice. Interestingly, in a model of lipopolysaccharide-induced septic shock, selective Irf3 deficiency in myeloid cells led to reduced levels of type I IFN in the sera and increased survival of these mice, indicating the myeloid-specific, pathogenic role of the Toll-like receptor 4–IRF3 type I IFN axis in this model of sepsis. Thus, Irf3-floxed mice can serve as useful tool for further exploring the cell type-specific functions of this transcription factor.
A hallmark of the innate immune response is the induction of the type I IFN (IFN-α/β) that occurs on activation of the signal-transducing pattern recognition receptors (PRRs) of the innate immune system (1–3). The transcription factor IFN regulatory factor 3 (IRF3) plays an essential role in this process (3, 4). It was also reported that IRF3 is required to protect the host from viral infections, such as encephalomyocarditis virus (EMCV) and herpes simplex virus type 1 (HSV-1) (4, 5). Furthermore, IRF3 is also critical to the suppression of dextran sodium sulfate-induced colitis (6). On the other hand, IRF3 may also be involved in unfavorable immune responses, such as proinflammatory diseases and autoimmune diseases (7). Finally, IRF3 participates in the induction of septic shock syndrome (8).
IRF3 is constitutively expressed in various types of cells, where it resides in an inactive form with the cytoplasm. On activation of PRR by pathogen infection or stimulation with a synthetic ligands, IRF3 is phosphorylated on specific serine residues by the kinases TANK-binding kinase 1 (TBK1) or inhibitor of NF-κB kinase ε/i (IKKε/i) (9, 10), resulting in translocation of IRF3 into the nucleus, where it induces the transcription of type I IFN genes on binding to conserved sequences known as IFN stimulated response elements (ISREs) (3, 11). Several PRRs, including the cytosolic RNA helicase retinoic acid-inducible gene-I (RIG-I)-like receptor (RLR) family members; cytosolic DNA sensors, including cGMP-AMP synthase (cGAS), stimulator of IFN genes (STING), and DNA-dependent activator of IRFs (DAI); and the transmembrane Toll-like receptor (TLR) 3 and 4, are identified as inducers of type I IFN gene expression through the phosphorylation of IRF3 (1–3, 11, 12). IRF3 directly targets several cytokine genes, including CXCL10, RANTES, IFN-stimulated gene 56, IL-12p35, IL-15, and arginase II, as well as type I IFN genes (8, 13, 14). IRF3 also functions as a negative regulator of gene expression, where, for example, IRF3 is directly recruited to the Il12b gene promoter and enhancer on RLR or cytosolic DNA sensor stimulation, where it suppresses Il12b mRNA induction by competing with IRF5, the bona fide transcriptional activator for the gene induction (15, 16). Furthermore, IRF3 activated by RLR signaling interacts with a transcription factor SMAD3 to interfere with transforming growth factor-β (TGF-β)-induced gene expression (17).
It should be noted that much of the foregoing data were obtained from Irf3-deficient mice (Irf3−/−) generated in our laboratory (3, 4). However, subsequent reanalysis revealed that this line of mice was also carrying a null mutation for the Bcl2l12 gene located adjacent to the Irf3 gene (18). Thus, these mutant mice are renamed Irf3−/−Bcl2l12−/− mice (6, 18). The Bcl2l12 gene encodes Bcl2-like-12 (Bcl2L12), a protein that functions as an antiapoptotic factor that suppresses DNA damage-induced apoptosis by inhibiting caspase-7 activity or p53-mediated gene transcription (19, 20). In fact, mouse embryonic fibroblasts (MEFs) from Irf3−/−Bcl2l12−/− mice show resistance to apoptosis induced by UV irradiation or doxorubicin treatment (18). As a result, analysis of IRF3 in these mice has been complicated by loss of the Bcl2l12 gene.
In this study, we constructed Irf3-floxed (Irf3f/f) mice, which retains an intact Bcl2l12 gene, to achieve the conditional deletion of the gene in a cell- and tissue-specific manner affected by crossing these mice with a corresponding Cre recombinase transgenic strain. In fact, there is no mouse model with which to study cell type- or tissue-specific function of IRF3 in vivo. The contribution of IRF3 in the innate immune responses was reexamined without the influence of Bcl2l12 nullizygosity. As exemplified by our results showing the myeloid cell-specific contribution of IRF3 in LPS-induced shock, the newly generated Irf3f/f mice offers a useful tool with which to further study the cell- and tissue-specific function of this transcription factor.
Results
Generation of Irf3-Floxed (Irf3f/f) Mice.
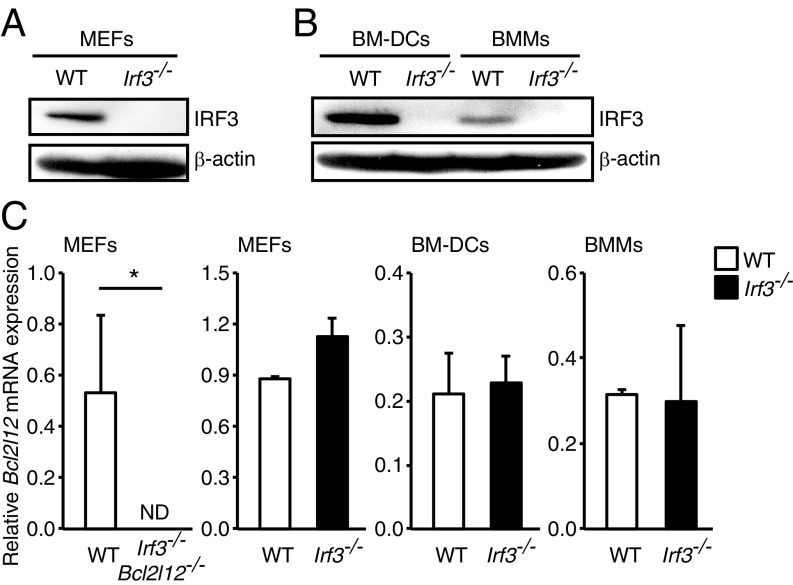
To generate Irf3f/f mice, a vector targeting exons 2, 3, and 4 flanked by loxP (floxed) sites designed to delete these exons on expression of Cre recombinase was constructed (SI Appendix, Fig. S1A). TT2 mouse embryonic stem cells (ES cells) (21) were electroporated with this vector, and the clones with the homologously recombined allele were identified by PCR and Southern blot analysis. The resulting ES clones, carrying the floxed Irf3 gene, were used to generate chimeric mice with germ line transmission (SI Appendix, Fig. S1B). The FRT-flanked neomycin cassette was then removed from the chimeric mice by crossing the mice with Flippase-expressing transgenic mice. Mice homozygous for the floxed Irf3 gene (Irf3f/f) were born at the expected Mendelian ratios with no obvious abnormality. Irf3f/f mice were crossed with transgenic mice with a CAG promoter-driven Cre (CAG-Cre) gene, and then further crossed with wild-type (WT) mice to eliminate the CAG-Cre gene to obtain systemic Irf3-deficient mice (Irf3−/−). IRF3 deletion was confirmed in both the genome (SI Appendix, Fig. S1C) and at the protein expression level in MEFs derived from Irf3−/− mice (Fig. 1A); it was also confirmed in bone marrow-derived dendritic cells (BM-DCs) and macrophages (BMMs) (Fig. 1B).
Fig. 1.
Bcl2l12 mRNA expression in WT and Irf3−/− cells. (A) Whole-cell extracts were prepared from WT and Irf3−/− MEFs and subjected to immunoblot analysis with anti-IRF3 and β-actin antibodies. (B) Whole-cell extracts were prepared from WT and Irf3−/− GM-CSF–cultured BM-DCs and macrophage-CSF–cultured BMMs and subjected to immunoblot analysis. (C) MEFs were prepared from WT, Irf3−/−Bcl2l12−/−, or Irf3−/− mice. Total RNA was extracted, and Bcl2l12 mRNA expression was determined by qRT-PCR analysis. ND, not detected. *P < 0.05. Results shown are mean ± SD of three independent experiments.
We next examined the status of the Bcl2l12 gene in these mice. Since an antibody reactive to mouse Bcl2L12 protein is not available, Bcl2l12 mRNA expression levels were monitored by quantitative reverse-transcription PCR (qRT-PCR) analysis. As shown in Fig. 1C, Bcl2l12 mRNA expression levels in Irf3−/− MEFs were comparable to those in WT MEFs. We further examined DNA damage-induced apoptosis in the Irf3−/− MEFs. As expected, MEFs from Irf3−/−Bcl2l12−/− mice showed resistance to UV-induced apoptosis (SI Appendix, Fig. S1D), whereas Irf3−/− MEFs did not (SI Appendix, Fig. S1E). Collectively, these results indicate that the newly generated Irf3f/f mice retain the Bcl2l12 gene.
Impairment of Type I IFN Gene Expression Evoked by Stimulation of PRRs in Irf3−/− Cells.
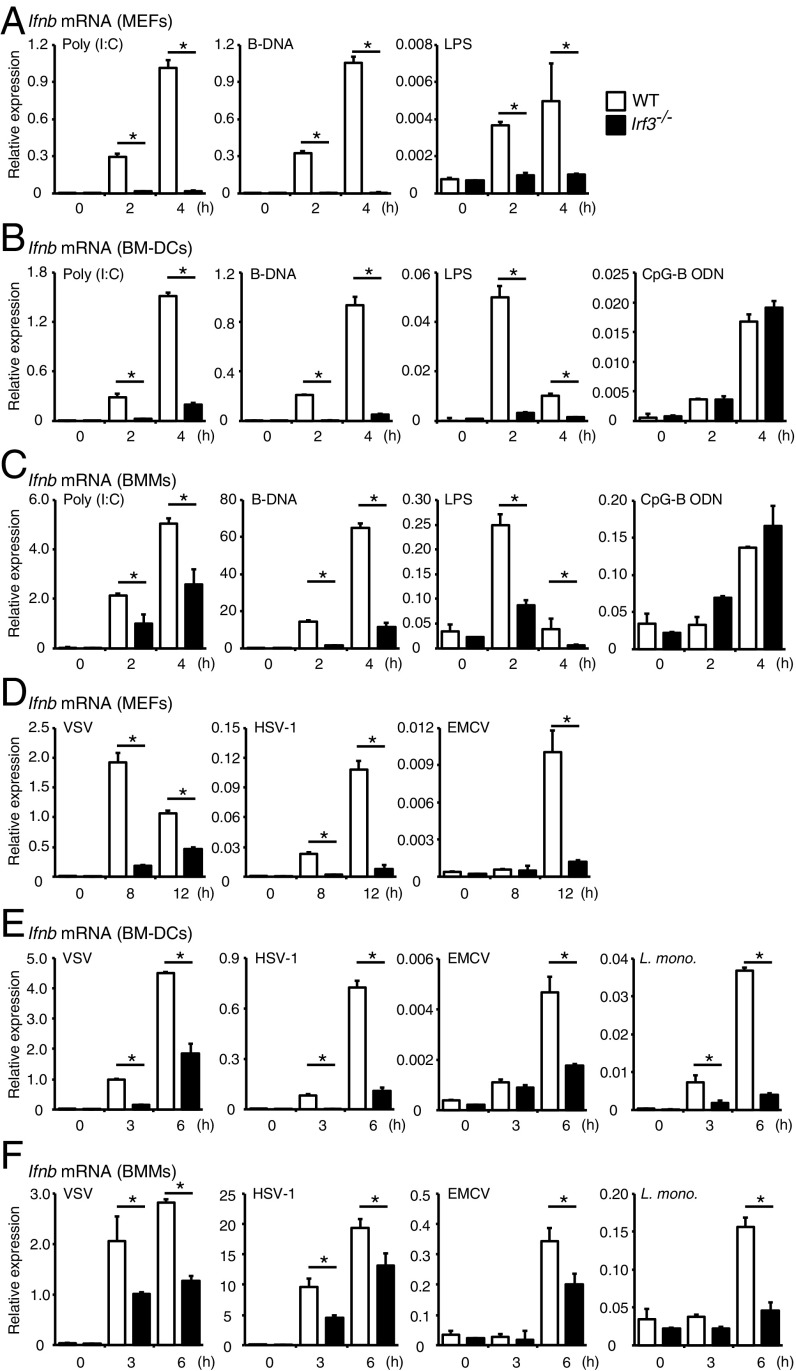
We next examined the role of IRF3 in the induction of type I IFN gene expression in response to PRR ligands. MEFs from WT or Irf3−/− mice stimulated with double-stranded RNA (polyinosinic-polycytidylic acid [poly (I:C)]), double-stranded DNA [poly(deoxyadenylic-deoxythymidylic) acid (B-DNA)], or lipopolysaccharide (LPS) to activate RLRs, cytosolic DNA sensors, or TLR4 (1–3, 11, 12), respectively, and then Ifnb mRNA expression levels examined by qRT-PCR analysis. As shown in Fig. 2A, Ifnb mRNA expression induced by stimulation of these ligands was impaired in Irf3−/− MEFs, confirming the sine qua non role of IRF3.
Fig. 2.
Ifnb mRNA induction in WT and Irf3−/− cells in response to PRR ligands or virus infection. (A–C) MEFs (A), BM-DCs (B), and BMMs (C) obtained from WT and littermate Irf3−/− mice were stimulated with poly (I:C) (10 μg/mL), B-DNA (10 μg/mL), LPS (200 ng/mL), or CpG-B ODN (1 μM) for the indicated times. Total RNA was extracted, and Ifnb mRNA expression was determined by qRT-PCR analysis. *P < 0.05. Results shown are mean ± SD from three independent experiments. (D–F) WT and littermate Irf3−/− MEFs (D), BM-DCs (E), and BMMs (F) were infected with VSV, HSV-1, EMCV, or L. monocytogenes (L. mono) at a multiplicity of infection of 1 for the indicated times. Total RNA was extracted, and Ifnb mRNA expression was determined by qRT-PCR analysis. *P < 0.05. Results are mean ± SD of three independent experiments.
We further examined the induction of mRNA for the activation of type I IFN and other cytokines in myeloid-derived BM-DCs and BMMs. BM-DCs and BMMs were stimulated with poly (I:C), B-DNA, LPS, or CpG-B oligodeoxynucleotide (ODN), a TLR9 agonist (2, 11), and cytokine mRNA induction levels were measured by qRT-PCR. We found that Ifnb mRNA induction by poly (I:C), B-DNA, or LPS stimulation was significantly reduced in both BM-DCs and BMMs from Irf3−/− mice (Fig. 2 B and C). Of note, Il6 and Rantes mRNA induction levels were also decreased in the Irf3−/−, BM-DCs, and BMMs, indicating the involvement of IRF3 (SI Appendix, Fig. S2 A–D). On the other hand, the induction of these cytokine mRNAs remained unaffected when the cells were stimulated by a TLR9 agonist, CpG-B ODN (Fig. 2 B and C and SI Appendix, Fig. S2 A–D). The data are consistent with the previous observations that other IRFs are involved in gene induction by the TLR-myeloid differentiation primary response gene 88 (MyD88) signaling pathway (2, 3).
Consistent with our previous study (15, 16) in which IRF3 suppressed the Il12b promoter on stimulation by cytosolic nucleic acid receptors (15, 16), Il12b mRNA expression was enhanced when reanalyzed in Irf3−/− BM-DCs and BMMs (SI Appendix, Fig. S2 E and F). Since M1-type macrophages secrete larger amounts of proinflammatory cytokines, such as IL-12p40, compared with M2-type macrophages (22), we next analyzed whether IRF3 skews macrophage polarization toward the M2-type phenotype, but found no significant differences in the in vitro polarization of M1 and M2 macrophages in Irf3−/− BMMs (SI Appendix, Fig. S2G).
We next evaluated the regulation of type I IFN on virus infections. In Irf3-deficient MEFs, BM-DCs, and BMMs infected with vesicular stomatitis virus (VSV) and EMCV, which activate RLR family members RIG-I and the melanoma differentiation-associated gene (MDA5), respectively, we observed a reduction in type I IFN mRNA expression compared with WT cells. A similar observation was made in MEFs infected by HSV-1 and Listeria monocytogenes, which are activated mainly by cytosolic DNA sensors (2, 12, 23, 24) (Fig. 2 D–F).
Involvement of IRF3 in Antiviral Response and Tumor Growth in Vivo.
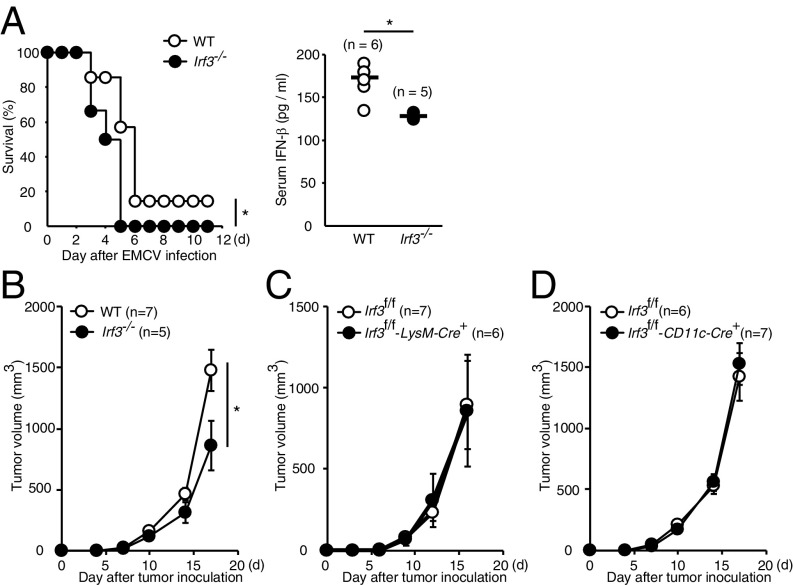
We next examined the contribution of IRF3 to the type I IFN responses induced by virus infection in vivo. WT and Irf3−/− mice were i.v. infected with EMCV, and IFN-β production levels in serum and survival rate were monitored. As shown in Fig. 3A, Irf3−/− mice were more vulnerable than WT mice to EMCV infection with reduced IFN-β production, indicating the critical role of IRF3 in antiviral response in vivo.
Fig. 3.
In vivo role of IRF3 in EMCV infection and tumor progression. (A) WT (n = 7) and Irf3−/− (n = 6) mice were i.v. infected with EMCV (105 pfu/mouse). (Left) Mouse survival was monitored every 24 h. (Right) IFN-β levels in serum were measured by ELISA at 6 h after EMCV infection. *P < 0.05. (B–D) 1 × 105 of B16F10 cells were inoculated s.c. into Irf3−/− (B), Irf3f/f-LysM-Cre+ (C), and Irf3f/f-Cd11c-Cre+ mice (D). Tumor volume was measured every 3 or 4 d.
We also examined tumor growth in Irf3−/− mice (Fig. 3B), since the STING-IRF3-type I IFN axis is implicated in antitumor immunity (25, 26). In this context, it has been shown that Sting−/− mice are more susceptible to tumor growth compared with WT mice when s.c. inoculated with B16F10 tumor cells (25, 26). Somewhat unexpectedly, tumor size was smaller in Irf3−/− mice compared with WT mice, indicating a tumor-promoting function of IRF3 (Fig. 3B). We then examined the involvement of myeloid cells or DCs in the IRF3-mediated tumor growth by crossing Irf3f/f mice with mice expressing the Cre recombinase gene under the promoter of LysM or CD11c (27). As shown in Fig. 3 C and D, the B16F10 tumor growth was not affected in Irf3f/f-LysM-Cre+ or Irf3f/f-CD11c-Cre+ mice (Fig. 3 C and D). Therefore, IRF3 may be involved in tumor progression by an as-yet unknown mechanism, but likely is not involved in STING-mediated antitumor immune responses, at least in this experimental setting. Of note, no significant difference was observed in the frequencies of tumor-infiltrating immune cells (SI Appendix, Fig. S3).
B Cell-Specific Ablation of Irf3 and Its Effect on the Differentiation of B Cells and Antibody Production.
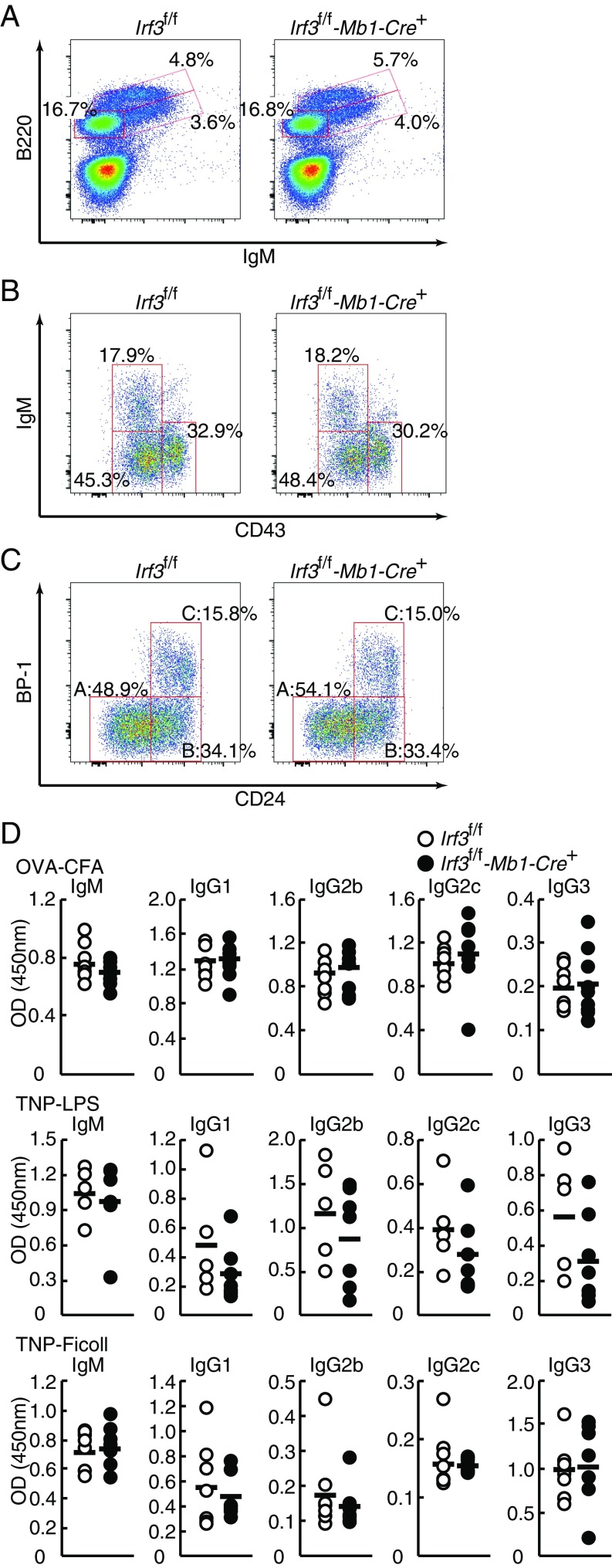
Recent reports have raised the question of whether IRF3 modulates adaptive immune responses (7, 16, 17, 28, 29). To delineate functions of IRF3 in B cells, we first generated mice with B-cell–specific IRF3 ablation by crossing Irf3f/f mice with Mb1-Cre knockin mice. The Mb1-Cre mice express Cre recombinase during early B cell development (30). We examined the distribution of bone marrow B cell lineage subsets by flow cytometry analysis. The percentages of B220+ B cells (Fig. 4A), B220loCD43–IgM+ immature B cells, B220loCD43–IgM– pre-B cells, B220loCD43+IgM– pro-B cells (Fig. 4B), or B220+CD43+ (Hardy fraction ABC) (31) pro-B cells, including B220+CD43+CD24–BP-1– (prepro-B cells; fraction A), B220+CD43+CD24+BP-1– (early pro-B cells; fraction B), and B220+CD43+CD24+BP-1+ (late pro-B and large pre-B cells; fraction C), were not affected by the IRF3 deficiency (Fig. 4C).
Fig. 4.
Normal B-cell development in bone marrow and antibody production in B cells in IRF3-ablated mice. (A–C) Suspension of bone marrow cells were prepared from Irf3f/f and Irf3f/f-Mb1-Cre+ mice, stained with antibodies, and analyzed by flow cytometry. B220lo cells (B) and B220+CD43+ cells (C) were gated, respectively. Percentages of cells in the total or gated population are shown. Hardy fractions (fractions A, B, and C) are also shown. (D) Mice were immunized i.p. with OVA (100 μg) emulsified in complete Freund’s adjuvant (Irf3f/f, n = 7; Irf3f/f-Mb1-Cre+, n = 7; Upper), TNP-AECM-Ficoll (50 μg; Irf3f/f, n = 5; Irf3f/f-Mb1-Cre+, n = 7; Middle), or TNP-LPS (50 μg; Irf3f/f, n = 7; Irf3f/f-Mb1-Cre+, n = 7; Lower). Sera were collected at 3 wk after injections. IgM, IgG1, IgG2b, IgG2c, and IgG3 titers in sera were determined by ELISA. Thick bars represent mean values.
We next analyzed B cell distributions in peripheral tissues. CD19+ B cells were observed to be normal in the spleen, blood, mesenteric lymph nodes, and Peyer’s patches of the gut from Irf3f/f-Mb1-Cre+ mice (SI Appendix, Fig. S4A). There was no significant difference in the frequencies of CD93/AA4.1+B220+IgMhighCD23− (T1 cells), CD93/AA4.1+B220+IgMhighCD23+ (T2 cells), CD93/AA4.1+B220+IgMlowCD23+ (T3 cells), B220+CD21highCD23low (marginal zone B cells), B220+CD21lowCD23high (follicular B cells), B220lowCD138+ (plasma cells), or B220+GL7+CD95/Fas+ (germinal center B cells) subsets in the spleen (SI Appendix, Fig. S4B). Expression of IgD, Igλ, or Igκ light chain on splenic B220+ cells was also unaffected (SI Appendix, Fig. S4C). In addition, there was no significant changes in CD19+CD23low (B1 cells) or CD19+CD23+ (B2 cells) subsets in peritoneal cavity and germinal center B cells in mesenteric lymph nodes or Peyer’s patches between WT and Irf3f/f-Mb1-Cre+ mice (SI Appendix, Fig. S4D). Thus, B cells developed normally in the absence of IRF3 in B cells.
We further addressed the contribution of IRF3 in B cells during humoral immune responses. WT and Irf3f/f-Mb1-Cre+ mice were immunized with T cell-dependent antigen ovalbumin (OVA) in combination with complete Freund’s adjuvant, T cell-independent (TI)-1 antigen 2,4,6-trinitrophenyl-coupled LPS (TNP-LPS), or TI-2 antigen TNP-Ficoll, and OVA- or TNP-specific antibody subclasses in sera were examined. As shown in Fig. 4D, there were no significant differences in the levels of anti-OVA or anti-TNP IgM, IgG1, IgG2b, IgG2c, or IgG3 antibody subclasses in the sera between WT and Irf3f/f-Mb1-Cre+ mice. Thus, these data indicate that IRF3 in B cells is dispensable for B cell development and function, although it remains possible that IRF3 is involved in aspects other than those examined here.
T Cell-Specific Ablation of Irf3 Gene and Its Effect on T Cell Differentiation.
We then addressed whether IRF3 regulates T cell differentiation. Constitutive expression of IRF3 in T cells was ablated by crossing Irf3f/f mice with Lck-Cre transgenic mice. We prepared naïve CD4+ T cells from spleen of WT and Irf3f/f-Lck-Cre+ mice and performed in vitro T cell differentiation assays. IRF3 deficiency did not impact the differentiation of T helper (Th) cells, such as Th1, Th2, and Th17 cells, or induced regulatory T cells (SI Appendix, Fig. S5A).
When we examined populations of T cells in the thymus and spleen of Irf3f/f-Lck-Cre+ mice, the CD4–CD8– double-negative population was increased in the thymus of Irf3f/f-Lck-Cre+ mice (SI Appendix, Fig. S5B). In addition, CD44int-highCD62L– and CD44+CD62L+ effector/memory T cells were increased in the spleen or peripheral blood from Irf3f/f-Lck-Cre+ mice, whereas CD44–CD62L+ naïve T cells were decreased in Irf3f/f-Lck-Cre+mice (SI Appendix, Fig. S5 C and D). However, similar phenotypes have been reported with Lck-Cre transgenic mice, showing decreased numbers of naïve T cells and increased frequencies of CD44+CD62L– effector/memory T cells in the spleen and lymph nodes, as well as increased numbers of thymic CD4/CD8 double-negative cells (32). Although transgenic mice expressing Cre recombinase are widely used to generate conditional KO mice, recent publications have indicated that some models have undesirable disadvantages (33, 34).
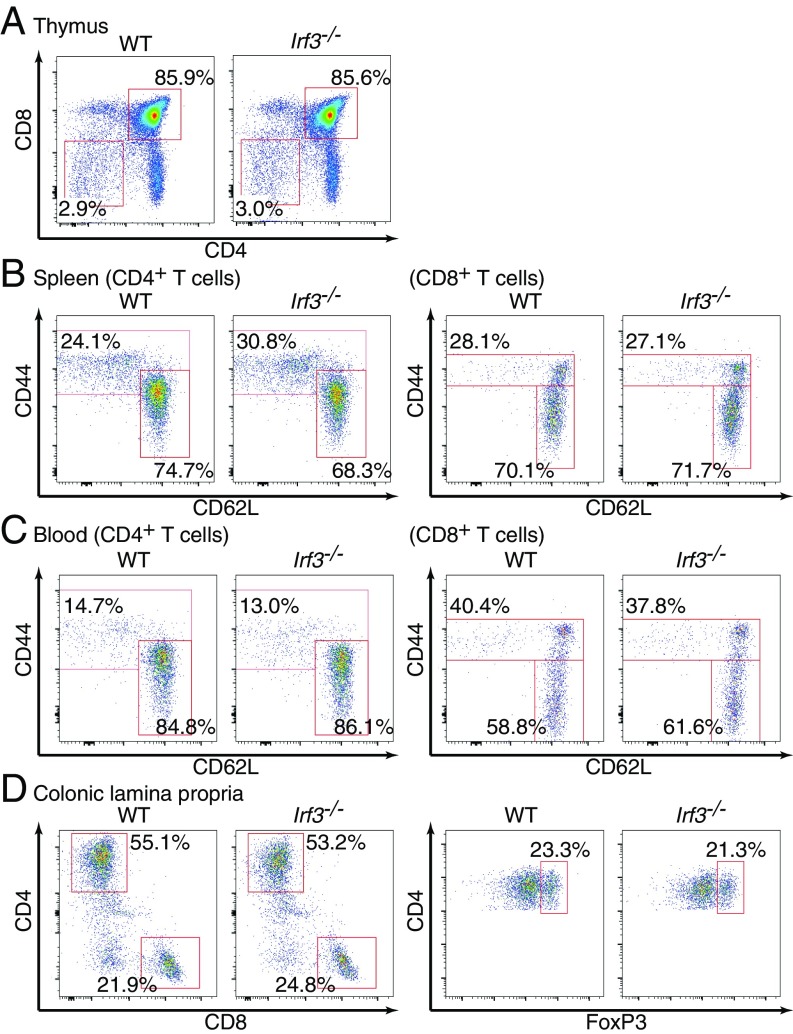
In fact, when we analyzed systemic Irf3−/− mice, the CD4/CD8 double-negative cells or double-positive cells in the thymus were found to be normal (Fig. 5A), as were frequencies of naïve and effector/memory T cells in the spleen or peripheral blood (Fig. 5 B and C). We further examined T cell subsets in colonic lamina propria, in which IRF3 is inherently activated with commensal bacteria through RLRs or cytosolic DNA sensors (6, 17); however, we found no overt abnormalities in T cell populations such as CD4+, CD8+, and Treg cells in the colonic lamina propria of Irf3−/− mice (Fig. 5D).
Fig. 5.
Normal T cell subsets in Irf3-deficient mice. (A) Thymocytes from WT or Irf3–/– mice were analyzed by flow cytometry. Percentages of CD4–CD8– and CD4+CD8+ cells in the total thymocytes are shown. (B) Splenocytes were prepared from WT or Irf3–/– mice and analyzed by flow cytometry. CD4+TCRβ+ T cells (Left) and CD8+TCRβ+ T cells (Right) were gated. Percentages of effector/memory T cells and naïve T cells in the gated population are shown. (C) Whole-blood cells were prepared from WT or Irf3–/– mice. Red blood cells were lysed, and the remaining cells were analyzed by flow cytometry as in B. (D) Colonic lamina propria cells were obtained from WT or Irf3–/– mice and subjected to flow cytometry analysis. TCRβ+ T cells (Left) and CD4+TCRβ+ T cells (Right) were gated. The results shown are representative of three independent experiments.
Pathogenic Contribution of the IRF3-Type I IFN Axis in Myeloid Cells.
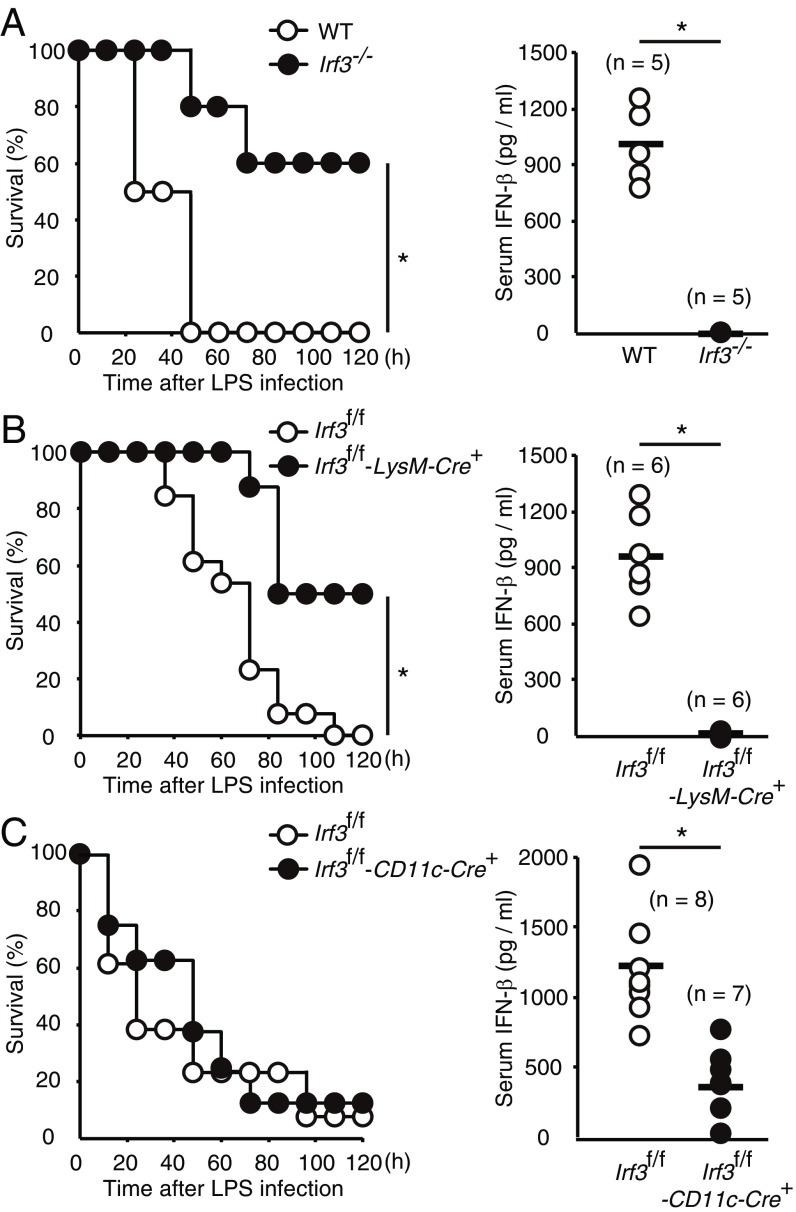
There is strong evidence that type I IFNs function as critical inflammatory mediators in high doses of LPS-induced endotoxin lethal shock (8, 35, 36). Previous reports including our own have demonstrated that Irf3−/−Bcl2l12−/− and Ifnb−/− mice are resistant to the lethal endotoxemia (8, 35). As such, we examined the susceptibility of Irf3−/− mice in the same septic shock model. We found that the production of serum IFN-β in response to i.v. LPS injection was abrogated in Irf3-deficient mice, which also had a higher survival rate than the WT mice (Fig. 6A).
Fig. 6.
The critical role of IRF3 in myeloid cells in the induction of type I IFN and mortality induced by LPS administration. IRF3 in myeloid cells, but not in DCs, plays a major role in the induction of type I IFN responses and mortality induced by high-dose LPS administration. (A) WT (n = 6) and Irf3−/− (n = 5) mice were administered i.v. LPS (8 mg/kg). Mice survival was monitored every 12 h (Left) and IFN-β levels in sera were determined by ELISA at 2 h after LPS administration (Right). (B and C) LPS was administered to Irf3f/f (n = 13) and Irf3f/f-LysM-Cre+ (n = 8) mice (B) and to Irf3f/f (n = 8) and Irf3f/f-CD11c-Cre+ (n = 6) mice (C) as in A. Mouse survival was monitored (Left), and IFN-β levels in serum were measured (Right). *P < 0.05.
The availability of Irf3f/f mice allowed us to further examine the cell type-specific contribution of IRF3 to LPS-induced lethality. TLR4 is highly expressed in antigen-presenting cells, such as macrophages and DCs. Interestingly, Irf3f/f-LysM-Cre+ mice showed prolonged survival accompanied by impaired IFN-β expression in serum in the LPS-induced shock model (Fig. 6B), whereas Irf3f/f-CD11c-Cre+ mice did not display such phenotypic abnormalities (Fig. 6C). Thus, IRF3 in myeloid cells, but not in DCs, plays a major role in the induction of type I IFN responses and mortality induced by high-dose LPS administration.
Discussion
In recent years, IRF3 has gained recognition as a key transcriptional regulator of type I IFN gene expression induced in response to pathogenic infection. Many of the discoveries on the role of IRF3 in immunity and other biological systems were made using conventional Irf3-null mice. However, these mice were recently discovered to also carry a null mutation in the Bcl2l12 gene, and hence have been renamed Irf3−/−Bcl2l12−/− (18). In this study, we successfully engineered Irf3f/f mice in which the Bcl2l12 gene remains functionally intact (Fig. 1C). This strain has enabled us to study the function of IRF3 without considering the effect of loss of function of Bcl2L12 (SI Appendix, Fig. S1 D and E). The present results, taken together with our previous reports (4, 18), confirm that IRF3 is critical for the activation of the type I IFN responses induced by innate PRRs or pathogen infections (Figs. 2, 3A, and 6).
It is interesting that the growth of s.c. transplanted B16F10 melanoma cells is significantly suppressed in Irf3−/− mice, suggesting a tumor-promoting function of IRF3 (Fig. 3B). Our results indicate that IRF3 in myeloid cells and DCs is not involved in this process (Fig. 3 C and D); therefore, further work is needed to clarify cell type(s) and mechanism(s), wherein IRF3 is functioning for the promotion of tumor growth. It is interesting that the STING-IRF3–type I IFN axis is known to inhibit tumor growth (25, 26); thus, we infer that IRF3 is bifunctional, in that it contributes to tumor suppression via the STING pathway and tumor progression by as-yet unknown mechanism(s).
The Irf3f/f mice also allow us to analyze the function of IRF3 in a cell type-specific manner. Indeed, we have demonstrated that IRF3 in myeloid cells has a profound impact on LPS-induced IFN-β production and survival (Fig. 6B). Since several reports have indicated the involvement of IRF3 in the regulation of adaptive immune responses (7, 16, 17, 28, 29), we also analyzed the potential role of IRF3 in the development of and antibody production by cells of the B cell lineage using Irf3f/f-Mb1-Cre+ mice. However, no significant differences in B cell subset frequency or antibody production were observed, at least in these experimental settings (Fig. 4 and SI Appendix, Fig. S4). T cell subsets in tissues were also normal in Irf3−/− mice (Fig. 5). Despite such observations, additional studies may yet elucidate a function of IRF3 in B cell- and T cell-mediated immune responses.
In conclusion, the conditional deletion of the Irf3 gene using the Irf3f/f mice described here will allow for more detailed and extensive investigation for the cell- and tissue-specific roles of IRF3 in the regulation of the immune and other biological processes.
Materials and Methods
Mice.
The Irf3−/−Bcl2l12−/− mice have been described previously (4, 18). C57BL/6 mice were purchased from CLEA Japan. CAG-Cre transgenic mice, LysM-Cre knockin mice, CD11c-Cre transgenic mice, and Lck-Cre transgenic mice were used (27). Mb1-Cre knockin mice were kindly provided by Michael Reth, Max Planck Institute of Immunobiology and Epigenetics, Freiburg im Breisgau, Germany. FLPe transgenic mice (RBRC01834) were provided by the RIKEN BioResource Research Center through the National BioResource Project of the Ministry of Education, Culture, Sports, and Technology of Japan. All animal experiments were done in accordance with guidelines of The University of Tokyo and RIKEN Kobe Branch.
Reagents and Cells.
CpG-B ODN (ODN 1668) and other oligodeoxynucleotides were purchased from FASMAC. LPS O55:B4 was purchased from Sigma-Aldrich. B-DNA (Sigma-Aldrich) or poly (I:C) (GE Healthcare Biosciences) was mixed with Lipofectamine 2000 (Invitrogen), and cells were stimulated with the mixture as described previously (16, 37). Bone marrow cells, splenocytes, thymocytes, colonic lamina propria cells, B16F10 melanoma cells, MEFs, BM-DCs, and BMMs were prepared as described previously (4, 6, 27, 37).
Additional information is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Reth (Max Planck Institute of Immunobiology and Epigenetics) and T. Kurosaki (RIKEN Center for Integrative Medical Sciences) for providing the Mb1-Cre mice. We also thank M. Sugahara, M. Yasui, and C.-Y. Chang for their technical assistance. This work was supported in part by Grant-In-Aid for Scientific Research (S) 15638461 from the Ministry of Education, Culture, Sports, Science of Japan; Grant AMED-PRIME 1005352 from the Japan Agency for Medical Research and Development, the Uehara Memorial Foundation, and the Princess Takamatsu Cancer Research Fund. The Department of Molecular Immunology is supported by BONAC Corporation and Kyowa Hakko Kirin Co., Ltd.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803936115/-/DCSupplemental.
References
- 1.Medzhitov R, Janeway C., Jr Innate immune recognition: Mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 4.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 5.Menachery VD, Pasieka TJ, Leib DA. Interferon regulatory factor 3-dependent pathways are critical for control of herpes simplex virus type 1 central nervous system infection. J Virol. 2010;84:9685–9694. doi: 10.1128/JVI.00706-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negishi H, et al. Essential contribution of IRF3 to intestinal homeostasis and microbiota-mediated Tslp gene induction. Proc Natl Acad Sci USA. 2012;109:21016–21021. doi: 10.1073/pnas.1219482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald DC, et al. Interferon regulatory factor (IRF) 3 is critical for the development of experimental autoimmune encephalomyelitis. J Neuroinflammation. 2014;11:130. doi: 10.1186/1742-2094-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi S, et al. Essential role of IRF-3 in lipopolysaccharide-induced interferon-β gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 11.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 13.Goriely S, et al. Interferon regulatory factor 3 is involved in Toll-like receptor 4 (TLR4)- and TLR3-induced IL-12p35 gene activation. Blood. 2006;107:1078–1084. doi: 10.1182/blood-2005-06-2416. [DOI] [PubMed] [Google Scholar]
- 14.Grandvaux N, et al. Regulation of arginase II by interferon regulatory factor 3 and the involvement of polyamines in the antiviral response. FEBS J. 2005;272:3120–3131. doi: 10.1111/j.1742-4658.2005.04726.x. [DOI] [PubMed] [Google Scholar]
- 15.Koshiba R, et al. Regulation of cooperative function of the Il12b enhancer and promoter by the interferon regulatory factors 3 and 5. Biochem Biophys Res Commun. 2013;430:95–100. doi: 10.1016/j.bbrc.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Negishi H, et al. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat Immunol. 2012;13:659–666. doi: 10.1038/ni.2307. [DOI] [PubMed] [Google Scholar]
- 17.Xu P, et al. Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol Cell. 2014;56:723–737. doi: 10.1016/j.molcel.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima A, et al. Cell type-dependent proapoptotic role of Bcl2L12 revealed by a mutation concomitant with the disruption of the juxtaposed Irf3 gene. Proc Natl Acad Sci USA. 2009;106:12448–12452. doi: 10.1073/pnas.0905702106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stegh AH, et al. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24:2194–2204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegh AH, et al. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev. 2007;21:98–111. doi: 10.1101/gad.1480007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagi T, et al. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- 22.Luo C, Chen M, Madden A, Xu H. Expression of complement components and regulators by different subtypes of bone marrow-derived macrophages. Inflammation. 2012;35:1448–1461. doi: 10.1007/s10753-012-9458-1. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 25.Corrales L, et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Reports. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demaria O, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci USA. 2015;112:15408–15413. doi: 10.1073/pnas.1512832112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanai H, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci USA. 2013;110:20699–20704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oganesyan G, et al. IRF3-dependent type I interferon response in B cells regulates CpG-mediated antibody production. J Biol Chem. 2008;283:802–808. doi: 10.1074/jbc.M704755200. [DOI] [PubMed] [Google Scholar]
- 29.Ysebrant de Lendonck L, Martinet V, Goriely S. Interferon regulatory factor 3 in adaptive immune responses. Cell Mol Life Sci. 2014;71:3873–3883. doi: 10.1007/s00018-014-1653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carow B, Gao Y, Coquet J, Reilly M, Rottenberg ME. lck-driven cre expression alters T cell development in the thymus and the frequencies and functions of peripheral T cell subsets. J Immunol. 2016;197:2261–2268. doi: 10.4049/jimmunol.1600827. [DOI] [PubMed] [Google Scholar]
- 33.Jeannotte L, et al. Unsuspected effects of a lung-specific Cre deleter mouse line. Genesis. 2011;49:152–159. doi: 10.1002/dvg.20720. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 35.Karaghiosoff M, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 36.Mahieu T, Libert C. Should we inhibit type I interferons in sepsis? Infect Immun. 2007;75:22–29. doi: 10.1128/IAI.00829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanai H, et al. HMGB proteins function as universal sentinels for nucleic acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.