Abstract
Metabolomics is one of the newer omics fields, and has enabled researchers to complement genomic and protein level analysis of disease with both semi-quantitative and quantitative metabolite levels, which are the chemical mediators that constitute a given phenotype. Over more than a decade, methodologies have advanced for both targeted (quantification of specific analytes) as well as untargeted metabolomics (biomarker discovery and global metabolite profiling). Untargeted metabolomics is especially useful when there is no a priori metabolic hypothesis. Liquid chromatography coupled to mass spectrometry (LC-MS) has been the preferred choice for untargeted metabolomics, given the versatility in metabolite coverage and sensitivity of these instruments. Resolving and profiling many hundreds to thousands of metabolites with varying chemical properties in a biological sample presents unique challenges, or pitfalls. In this review, we address the various obstacles and corrective measures available in four major aspects associated with an untargeted metabolomics experiment: (1) experimental design, (2) pre-analytical (sample collection and preparation), (3) analytical (chromatography and detection), and (4) post-analytical (data processing).
List of keywords: Metabolomics, Untargeted metabolomics, Targeted metabolomics, Techniques, Mass spectrometry, Inborn errors of metabolism
INTRODUCTION
In the ideal, a metabolomic study provides a picture of every metabolite in the organism and provides insight into metabolic response to a biological situation or experimental manipulation. The assumptions are that every metabolite will be measured, and that the measurements will be biologically informative. In reality, there are problems with these assumptions and experimental design and methodology are required to overcome them (partially, at least). The bases of the potential problems and approaches to address these issues are discussed below, first in the most general sense which applies to all experimental systems (issues inherent in drawing inference from metabolic pool measurements), and then in specific aspects of mass spectrometric (MS) measurements (pre-analytic, analytic and post-analytic processes).
At the present time, metabolomics experiments are performed with either mass spectrometry (Want et al 2007; Dunn et al 2011; Reaves and Rabinowitz 2011) or nuclear magnetic resonance (Fan and Lane 2016; Dietz et al 2017). Nuclear magnetic resonance (NMR) has the potential to measure metabolite levels in intact tissues, but sensitivity is limited (Tognarelli et al 2015; Fan and Lane 2016), and even with increased field strength (Righi et al 2012; Dietz et al 2017), it is not possible to detect low abundance compounds with currently available technology. This manuscript only discusses liquid chromatography (LC) based mass spectrometry (MS) approaches to untargeted metabolomics, with emphasis on inborn errors of metabolism (IEM).
Targeted Versus Untargeted Metabolomics
There is some confusion and ambiguity in the application of the terms “targeted” and “untargeted” in metabolomics. In targeted studies, specific compounds are quantified and compared to established reference ranges. In practice, this corresponds to setting the mass spectrometer to monitor selected transitions reflecting individual target analytes (and their internal standards) through the time course of the chromatography. This is not different from what Biochemical Genetics laboratories have traditionally done in performing amino acid, organic acid, acylcarnitine analysis, etc. Using modern day instrumentation and stable isotope dilution, target analytes can be fully quantified to clinical laboratory standards, using formal calibration, validation, and quality control (FDA 2001), though in cases where absolute quantification is not necessary, a semi-quantitative approach may be useful, and is often used instead. On the other hand, untargeted metabolomics (Want et al 2005) seeks to analyze all detectable metabolites, known and unknown, to determine if one or more is or are significantly perturbed, and then to perform identification. Untargeted metabolomics is a “discovery mode” process and it relies on differential comparison between groups (Dudzik et al 2017) of samples (for example cases versus controls); it is not applicable to individual samples. In its strictest form, untargeted metabolomics is agnostic, comparing peaks as chromatographic “features”, and then seeking to identify the compounds. The settings of the mass spectrometer would reflect that (i.e. acquisition would be in scan mode), but identification is made on review and extraction of the data collected during the chromatography. Naturally, with experience, a laboratory will accrue a library of identities of chromatographic features and spectra, so that many peaks can be immediately identified. Untargeted metabolomics however is truly intended for discovery, and is not limited to a pre-determined list of metabolites or class of compounds, with the aim to span the breadth of the metabolome.
CHALLENGES/PITFALLS AND SOLUTIONS/WORKAROUNDS
Discussion of issues, pitfalls and workarounds is organized into the phases of a metabolomics experiment: Experimental Planning/Conceptualization, Pre-Analytical, Analytical, and Post-Analytical.
Experimental Planning/Conceptualization
There are realities which raise challenges to the basic assumptions of metabolomics; in some cases, there is nothing that the experimentalist can do to overcome the challenges, but in others there are solutions or at least methods to minimize the problem. LC-MS methodology involves extraction of body fluids or tissues. The source of the material will determine which analytes are present, so in any given sample there may be groups of compounds which will never be seen at more than trace amounts. For example, certain sugar phosphates and nucleotides will not be expected in extracellular fluids, and hydrophobic compounds such as fatty acids may be seen in blood, but not in urine or CSF. The metabolomic picture will differ greatly depending upon the fluid studied and so it is imperative to choose the most relevant sample type that will demonstrate the metabolic perturbation.
It is possible that the key event and most informative biological event in metabolism will take place as a trigger or nucleation event and will not be evident at any time later. That may be the case in transient niacin deficiency which could cause defects in embryogenesis (Shi et al 2017) but might not be evident in the mother at a later time. It also may be true that through cascade effects or biochemical amplification, that a widespread change may result from a small perturbation in a key regulator, creating a sort of “butterfly effect,” as for example with microRNA species (Dorn 2013) or the trace concentration of cAMP initiating the cascade of glycogenolysis (Fischer 2013). That regulator may either be inaccessible in the study, or present at such a low concentration that it would never be measured when the experiment is performed. Instrumental dynamic range or interference from much more abundant analytes may make it impossible to monitor changes on both the regulatory and the bulk substrates. Performing longitudinal studies when possible can help detect transient changes which might not be observed in a static timepoint. In other scenarios, the levels of observed intermediates may not reveal a regulatory change, particularly when metabolite pools are defended by side reactions (such as anaplerosis), but measurement of flux could be mechanistically informative. In the last decade, initially pioneered through microbial metabolism studies (Blank et al 2005; Kummel et al 2006), researchers have used 13C (and/or 15N) labeled nutrients to follow the utilization of substrates such as glucose or amino acids, not only in cell culture, but in live mammals as well (Fan et al 2009). A bolus of stable iostopically labeled material can reveal altered ratios of labeled to unlabeled intermediates and isotopologues (containing both labeled an unlabeled atoms), and MS/MS fragmentation patterns can reveal changes in isotopomers (varying in the location of labeled atoms). This can provide insight into changes in metabolic flux in disease and allow construction of metabolic network models, revealing linkage among pathways otherwise not obviously related.
Sample Size
A bad outcome for a metabolomics experiment would be finding no meaningful associations, and worse would be reaching spurious conclusions. Since untargeted metabolomics depends inherently on statistical comparison between or among experimental groups (controls vs. cases, treatment A vs. treatment B, etc.), meaningful results require an adequate number of samples in each group. In general, the objective is to predict the number of samples needed to generate a given power (e.g., 0.8) and a given degree of confidence (e.g. an adjusted p-value ≤0.05), given the experimental variability between replicate runs. One approach is to use data sets from pilot studies or from related samples in public data repositories. The power for a given false discovery rate (FDR) may be estimated for a given set of pilot data by a number of methods, including a module of the publically-available MetaboAnalyst package (Xia and Wishart 2016). The larger the number of samples, the less work in the post-analytical phase and the more definitive the results. As a rule of thumb, it is not practical to perform untargeted analysis with groups of less than 5–10 individual samples per group, and it is not realistic to consider running single samples for untargeted metabolomics, The metabolomics standards initiative (MSI) recommends a minimum of 5 biological replicates in their minimum reporting standards (Sumner et al 2007), but of course the true number required depends heavily on the intrinsic variation in the biological samples as well as the magnitude of the observed perturbation, all factors incorporated into power analysis. It is possible that a pathognomonic metabolite will by chance be seen in a single sample from a patient with a given disease, and Miller et al. (Miller et al 2015) recently demonstrated the ability to identify such elevations in metabolomic studies of various inborn errors of metabolism by comparison to previously established reference ranges. However, only known metabolites were evaluated in this single sample fashion, while for biomarker search, multiple patient samples were processed in cohorts, a key aspect of untargeted metabolomics. Novel biomarker search poses a specific challenge with low sample numbers, as various analytical, environmental, and even dietary factors may result in aberrant levels of certain features from any single sample/run, normally evaluated by rigorous false discovery analysis in untargeted experiments. The variation posed by these factors are discussed in detail throughout this review, but it is worth considering that colleagues’ requests to “run untargeted metabolomics on a single sample” or small sample cohorts should be handled with discussion about experimental design, and redirected to either run exhaustive targeted analysis (similar to extended Biochemical Genetics assays) or to extend the study population to provide appropriate statistical power.
Pre-Analytical
Sample Preparation
When blood is sampled, there are advantages to using plasma over serum, since the specimen can be immediately placed on ice prior to separation. It is possible to use dried blood spot (and urine) cards for some applications (Barri and Dragsted 2013), but there is some uncertainty about extraction efficiency, depending upon the compound’s polarity. There is controversy regarding the choice of anticoagulant for plasma preparation. There may be interferences and serious matrix effects depending on the particular experimental setup, the specific anticoagulant (EDTA or heparin), the counter-ion (Na, K2, K3, Li), and the type (glass versus polypropylene) or brand of the tube. Some investigators favor heparin for plasma samples and state that EDTA should be avoided (Barri and Dragsted 2013), whereas others favor EDTA (Yin et al 2015; Metabolon 2017). Citrate should be avoided when studying central metabolism. There may also be artefactual features from surfactants and detergents used to treat the subject’s skin (Denery et al 2011). The best advice is to perform pretesting, and above all, to be consistent throughout the sample acquisition phase of the experiment, so that all samples are handled identically. Urine samples, which do not require special collection tubes (and should generally not include additives), must also be considered carefully. Metabolite concentrations may vary significantly in an individual throughout the course of the day based on hydration and diet. Often this is managed by normalization to creatinine levels, but that process may be compromised in kidney dysfunction. Alternative methods for normalization that have been used include use of osmolality and “total useful signal” from MS-data (MSTUS), a process by which many (hundreds or thousands) of common ions among all samples are used for scaling (Warrack et al 2009). Other factors to take into account when acquiring any animal or human samples include control of diet (or fasting time) to prevent exogenous metabolite interferences and to minimize variation, in addition to variables associated with sample storage and repeated freeze/thawing (Alvarez-Sanchez et al 2010).
Tissue/cell harvesting, metabolite extraction, and quenching of metabolism
Extracting and quenching metabolism is a critical factor for any metabolomics experiment. The need to effectively deproteinize biological sample while solubilizing the metabolome is of course important, but if additional metabolism or compound degradation occurs during this process, the readout by LC-MS may no longer be biologically valid. Certain compound classes are especially labile and are represented in many of the primary energy pathways. These include sugar phosphates (glycolysis and pentose phosphate pathways), nucleotides (ATP, GTP, etc.), coenzymes and cofactors whose stability, especially in terms of phosphorylation state, are greatly influenced by factors such as pH and temperature (Sellick et al 2011; Vuckovic 2012; Leon et al 2013). These are intracellular metabolites for the most part and are rarely considered when extracting extracellular material such as plasma (or serum), CSF, or urine for example. Researchers interested in bacterial metabolism and flux analysis have increasingly considered such issues, often employing filtration systems that avoid perturbation from centrifugation allowing for quick washing and sampling (Aragon et al 2006; McCloskey et al 2014). There may be advantages to bloodspots in limiting ex vivo metabolism (Hill et al 2017), but that approach may entail differences in recovery and stability of different classes of metabolites (Koulman et al 2014). Adherent cell lines face a unique set of challenges in order to limit artifactual metabolic perturbation. In general practice, adherent mammalian cell cultures are washed with PBS, trypsinized, harvested, and centrifuged for further media washing, a process that has been implicated to be poorly compatible with preservation of the metabolome (Teng et al 2009). This has recently led to alternative, creative strategies to allow for quick harvesting and quenching of cellular material (Lorenz et al 2011; Martano et al 2015), where the intracellular energy metabolites mentioned above may be critical to the study. The commonality among the methods is that trypsinization and centrifugation steps are avoided, and cells are quenched quickly directly on the surface that they are grown on. They are then scraped off manually, often after freezing, before final preparation for LC-MS analysis. Validation of proper quenching can be performed by calculating ratios of the intact to degraded forms of labile metabolites such as nucleotides. For example, concentrations of ATP, ADP, and AMP can be incorporated in the equation for energy charge (([ATP]+0.5[ADP])/([ATP]+[ADP]+[AMP)), and then compared to established ranges in various cell types, generally centered near 0.9 in normal conditions (Chapman et al 1976).
A variety of extraction/quenching methodologies have been compared for tissue that has been excised or biopsied from animals. Issues that have warranted extensive investigation include the need to cryo-freeze tissue, the use of freeze clamping, as well as variables associated with animal anesthesia and euthanasia methods (Belanger et al 2002; Want et al 2013; Overmyer et al 2015). In addition, the extraction solution used can have a major influence on the scope of the metabolome observed. For an untargeted metabolomics experiment that assumes many compound classes will be represented, it is critical to test extraction efficiency of both highly polar metabolites such as organic and amino acids, as well various lipid classes with varying hydrophobicity. For extraction methods that solubilize both polar and hydrophobic compounds, biphasic strategies such as the Bligh Dyer (Bligh and Dyer 1959) or Folch (Folch et al 1957) method, or several variations (Rose and Oklander 1965; Jensen 2008) are commonly used. These primarily use a combination of chloroform, methanol, water, and in some cases acid, resulting in a separation of the aqueous and organic layers of solvent with a protein/DNA layer in between. More recently, a new method that utilizes methyl-tert-butyl-ether (MTBE) instead of chloroform has improved two important aspects of biphasic extraction (Chen et al 2013): 1) MTBE is less toxic than chloroform and safer to handle, and 2) the DNA/protein pellet from extraction is localized to the bottom of the tube following centrifugation. This allows for a simple removal of the two phases without contaminating the lower phase with the insoluble material. A variety of monophasic methods are also widely used and include solvents such as methanol, acetonitrile, ethanol, perchloric acid, as well as others, either in cold or boiling conditions, and are preferred for certain classes of compounds (Kolarovic and Fournier 1986; Canelas et al 2009; Dietmair et al 2010; Yanes et al 2011). It is important to note that there are significant differences in the coverage of the metabolome when comparing the various extraction methods, muddying the true definition of “untargeted” metabolomics.
Analytical
Once sample acquisition and extraction has been achieved, the analytical aspects associated with LC-MS analysis are the next key part of a successful experiment, and though seemingly straightforward, it contains a number of permutations that the experimenter must choose. As with the extraction steps described above, none of these will be perfect for all subsets of metabolites. The analytical choices, which include sample resuspension, chromatography and instrumentation, will determine the breadth of the metabolome covered and the degree of reliability in the collected data. The following section will highlight some of the major areas where major consideration must be applied.
Importance of Chromatography
Though several groups have published methods that utilize direct injection into mass spectrometers for analysis of metabolites (Madalinski et al 2008; Fuhrer et al 2011), the vast number of researchers utilize inline chromatography in their platforms to minimize ionic suppression and increase both sensitivity and specificity of the analytes they report. Added complexity, be it in the form of non-volatile salts, buffers, or even metabolites can greatly influence the ionization efficiency of any given compound and cause interfering compounds that will convolute the accurate reporting of data, issues that can be greatly alleviated with successful chromatographic methods. From the early days of untargeted LC-MS based metabolomics dating back a little over a decade ago, C18 reverse phase columns have been a stalwart of many platforms. There are many iterations of C18 columns and nearly every manufacturer sells a version of these, though with sometimes distinguishing features that result in varying degrees of performance. Differences in particle technology, particle size, uniformity, column dimensions and other factors will affect binding, separation, and elution properties, as well as back pressure. Smaller particle sizes result in increased column efficiency but cause an increase of back pressure that necessitates ultra high performance LC (UHPLC) systems and fast acquisition mass spectrometers to match narrow elution profiles (Guillarme et al 2010). Ultimately though, their frequent use throughout the LC-metabolomics era is based on their high reproducibility, which is a necessity for accurate run-to-run alignment, their versatility in retaining many non-polar and hydrophobic compound classes, and the simple mobile phase compositions (often acetonitrile/water or methanol/water gradients with small amounts of additives such as formic acid) required for their use. The latter factor ensures ideal compatibility with electrospray (ESI) and atmospheric pressure chemical ionization, the two primary LC-MS ionization techniques. The weakness of these columns is in the polar regime of the metabolome, and many such compounds will have poor retention, eluting near the solvent front of a run where the greatest amount of ionic suppression and potential interferences reside. Unfortunately, many of the metabolites of interest, especially in the realm of primary energy metabolism (e.g. organic acids and amino acids) related to both human disease as well as intracellular studies are highly polar. An example of this was an early study involving our group to demonstrate the utility of untargeted metabolomics to detect known biomarkers of IEM (Wikoff et al 2007). In this study of a small group of patients with propionic acidemia, methylmalonic acidemia and controls, the controls were of course distinguished from propionic and methylmalonic acidemia by an elevation of propionyl-carnitine and related acylcarnitines The distinction between methylmalonic and propionic acidemia, however, was less clear, because methylmalonyl-carnitine was not detected (presumably attributable to a lack of suitable stationary phase ideal for such a highly polar compound). That study demonstrated feasibility, but also limitations: no single chromatography will permit “global” untargeted metabolomics.
Normal phase and HILIC columns, with stationary phases containing polar groups such as amino, cyano and silica among others, are now frequently employed for additional runs to analyze the polar chemical realm (Jandera and Janas 2017; McCalley 2017). In the past, these columns were more difficult to use reproducibly, as they generally required longer re-equilibration times and more complex mobile phases that also incorporated buffers and higher ionic strength for efficient metabolite elution. A more recent alternative to using normal phase is the use of reverse phase stationary phases containing polar groups such as pentafluorophenyl (PFP) columns (Csató et al 1990), which we have previously validated for use in a combined targeted/untargeted metabolomics platform (Gertsman et al 2014). Various versions of these exist, including with a propyl (PFPP) linker (manufacturers include Phenomenex, Resetek, ES industries, and UCT) or even combined with a C18 stationary phase for a mixed mode effect (Mac-Mode). Mixed mode columns, which generally utilize both non-polar and polar stationary phases to extend versatility in metabolite selection, can often be used in standard reverse phase conditions and have been a preferred choice in some untargeted studies (Yanes et al 2011; Gertsman et al 2015). This is not to say that drawbacks do not exist in these columns as well, and weaknesses can include poor elution of polar lipids or other compound classes that carry both polar and hydrophobic moieties.
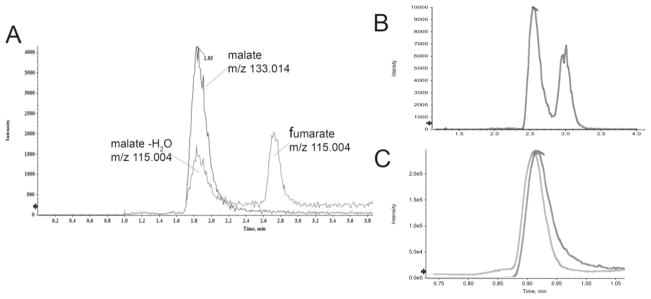
Yet another important aspect of chromatography lies in the ability to separate isomers, isobaric compounds, and other interferences. An example of an unexpected interference often ignored is a co-eluting compound that actually has a different parent mass, but undergoes an in-source fragmentation that contributes to the signal of the other. This can occur for the organic acids fumarate and malate for example, where a water loss from malate (m/z 133.014) during electrospray ionization in negative ion mode will cause a m/z 115.004 ion to appear that is indistinguishable in MS and even MS/MS profiles from fumarate (Fig. 1a). If chromatography cannot distinguish these two, fumarate, a very critical TCA cycle intermediate, will be falsely reported. An example of necessitating chromatographic distinction of isomers can be seen in Fig. 1c, where a certain C18 column was unable to resolve 2- and 3-hydroxybuturate under a typical reverse phase gradient, while a C18-PFP column successfully could (Fig. 1b). Though it will be nearly impossible to qualify the separation all such possible pairs or isomers from each other, it is worthwhile to qualify a platform for the critical metabolites that are routinely measured and reported (e.g. major energy pathways). In the above example, 2- and 3-hydroxybutyrate stem from completely different metabolic pathways (threonine/methionine metabolism and fatty acid metabolism respectively), and their combined signal will obscure potentially significant results from either of these.
Fig. 1.
Separation of isomers and isobaric species. (A) Water loss during ionization of malate ion results in peak that is indistinguishable in mass to fumarate, necessitating their chromatographic resolution. (B) Sufficient resolution of 2- and 3-hydroxybutyrate using a PFP containing mixed-mode column, resolves isomers that were not well distinguished using a common C18 stationary phase (C).
Analytical Variation: The Case for Internal Standards and/or QC’s
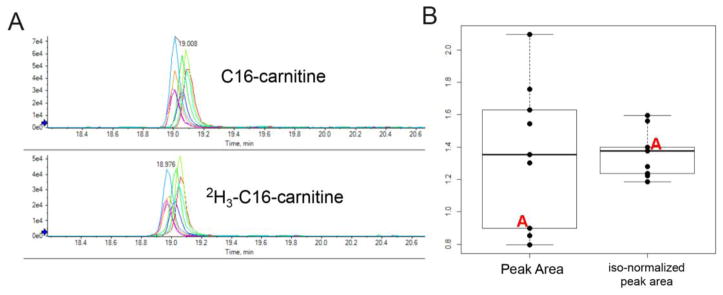
An obstacle in comparing peak area differences from one run to another is that signal variation occurs for any given compound of interest. Some of this is likely due to small but noticeable differences in signal intensity that may vary during the course of a batch, while other factors include slight differences in the matrix of one sample compared to another, resulting in differences in ionic suppression (especially with different sample types, e.g. plasma vs. urine). We show a clear example of variation in Fig. 2 (unpublished data from one of our own studies), that shows the difference between comparing non-normalized (no stable isotope) palmitoylcarnitine (C16-carnitine) to peak areas normalized to a deuterated version of the compound that was spiked in during extraction. The figure shows that one of the lower values in the un-normalized estimate (peak area) was actually one of the higher measurements for that group when normalized appropriately (Fig. 2b). Overall, omitting a stable isotope for comparison would not have changed the mean value of the metabolite in this cohort, but the concentration would have been underestimated if the single sample were studied individually. Appropriate stable isotopes can be especially useful in instances where sample numbers are low, or where compounds fall in chromatographic regions with known ionic suppression. Many groups are more commonly making use of stable isotope dilution in untargeted experiments, which not only comes in handy for the potential normalization of endogenous compounds, but can be used for assessing drift in both signal intensity and retention time. (Sysi-Aho et al 2007; Miller et al 2015). Stable isotope dilution is especially helpful in longitudinal studies acquired over years, where there may be large differences in instrument performance, different column batches, or even different operators. However care must be taken to ensure standards are adequately assessed for stability and degradation during storage times relevant to the breadth of the study. Also, if one is to use internal standards for untargeted studies, it is necessary to match the chemically diversity of the run with the standards selected, also making sure to cover the width of the chromatographic run, as intensity drift may not affect all compounds or sections of the run equally. As this can be cost prohibitive or an otherwise burden, alternatives to internal standards are used for compensating for analytical errors. These include the use of replicate samples or QC’s that can be run throughout different intervals of a batch (Dunn et al 2011; Wehrens et al 2016), and in one method, used in a serially dilute form throughout to test for signal linearity of different compounds (Kouassi Nzoughet et al 2017). A variety of processing tools have also been developed to also deal with issues of signal and retention time drift, as well as batch effects and outliers that can plague data analysis, as discussed in the post-analytical section of the review (Salerno et al 2017; Thonusin et al 2017).
Fig. 2.
Diminished variation with stable isotope dilution. (A) Comparing the endogenous and stable isotope peaks of C16-carnitine from a 9-sample (plasma) cohort. (B) Box plot depicting non-normalized peak areas of C16-carnitine compared to those normalized to 2H3-C16-carnitine in identical samples. One of the samples, labeled A, is shown to have a peak area in the lower part of the C16-carnitine range, while residing in the upper half of the range following stable isotope normalization.
Post-Analytical
Pre-processing
Following the completion of the mass spectrometry runs, a number of pre-processing and post-processing tools are available for identifying analytes of interest from untargeted data sets. Though many researchers incorporate specific target compounds in such runs that are always integrated and compared, the general processing strategy in untargeted workflows is to focus on compounds that are statistically altered. The runs must be first properly aligned, either with the aid of several freely available software packages (Lommen 2009; Tautenhahn et al 2012; Li et al 2017) or the many propriety software packages often distributed by MS vendors. Non-linear alignment is preferred in pre-processing as chromatographic shifts are often non-uniform throughout the run, and improved alignment enables more accurate peak selection and integration when unique analytes with similar m/z have small deviations in elution time. Metabolomics software packages often allow signal normalization as a pre-processing tool, either with the use of internal standards, or by other methods. Other pre-processing options prior to thorough statistical analysis include the removal of outliers and other batch effects. The following section highlights some of the intricacies and bottlenecks associated with the processing and analysis of pre-processed data.
Compound Identification
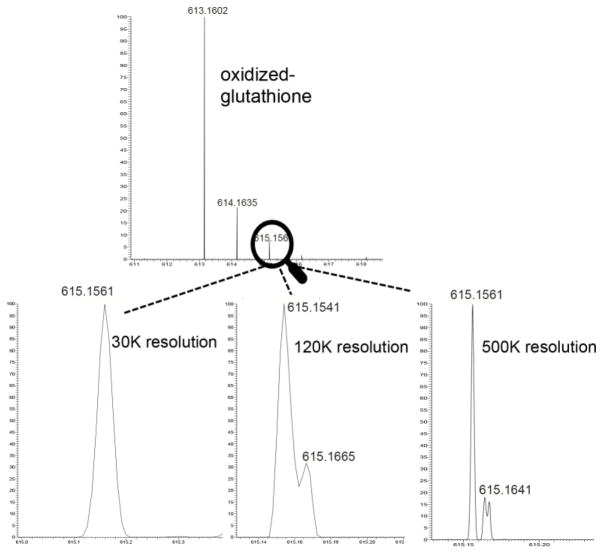
Identification of unknown compound in untargeted metabolomics is considered the greatest bottleneck of data interpretation and requires a number of tools and proper instrumentation to successfully overcome. A high resolution mass spectrometer (Q-TOF, Orbitrap, or FT-ICR instruments) using a standard reverse phase platform may lead to the observation of many thousands of peaks from a single run, the number depending on instrument sensitivity, solvent composition and purity, matrix complexity, and in-source fragmentation as possible factors. Each peak does not necessarily represent a unique metabolite though, and a single feature may be represented in a dozen or more forms that include adducts (salt or solvent complexes), dimeric or even trimeric states, and even fragments produced during ionization or transmission of ions. For compounds of interest, it is therefore important to identify the elemental composition of the ion, and some useful guidelines have been published to narrow down the possibilities for any given ion (Kind and Fiehn 2007; Watson 2013). Common considerations to reduce the number of possibilities include: 1) the nitrogen rule (better suited for masses <500 Da), which dictates that a compound with an even nominal mass will have an even number of nitrogen atoms, and with an odd mass will have an odd number of nitrogens, 2) likely hydrogen/carbon ratios and elemental probability analysis, and 3) isotopic distributions of the analyte, as atoms have different isotopic abundances. In addition, since atoms have unique mass defects due to differences in nuclear binding energy (e.g. common isotopic form of Sulfur, 32S, has mass of 31.972, while 12C Carbon is 12.000), high resolution mass spectrometry can use such properties to narrow down the possibilities. In Fig. 3, we show the parent mass of oxidized glutathione analyzed on an Orbitrap Lumos instrument collected at three different resolution: 30,000, 120,000, and 500,000. Most current Q-TOF instruments have ~30,000 resolution, and at this resolution (along with accurate mass) we demonstrate that the third peak (M+2) for oxidized glutathione (GSSG) has a lower non-integer mass than its previous two isotopic forms due to the mass defect of 34S, which is next most prominent form of sulfur after 32S. This shift to the left can be identified by Q-TOF, but the distribution of the atoms with isotopic forms in this peak are not clear. When increasing the resolution to 120K, one can see a bump next to that peak that distinguishes the Carbon and Nitrogen isotopes from the Sulfur, the latter being more predominant. When one uses ultra high resolution of 500,000 the two forms are very clearly separated and can actually be integrated accurately, enabling one to both implicate and rule out various combinations of atoms present in the analyte. In addition to resolution, high mass accuracy can help to further narrow down possible elemental compositions, a common attribute found in most instruments used for untargeted metabolomics (<~2–3 ppm mass accuracy). A number of chemical libraries can be searched for annotated compounds that match a possible elemental composition, and include: METLIN (Tautenhahn et al 2012), HMDB (Wishart et al 2009), Chemspider (Williams and Tkachenko 2014), Pubchem (Wang et al 2009)), GnPS (Wang et al 2016)), Lipidmaps (Sud et al 2012), Massbank (Horai et al 2010), Metabolomics Workbench (Sud et al 2016), and MetaCyc (Caspi et al 2014).
Fig. 3.
High resolution for elemental composition reconstruction. Oxidized glutathione (GSSG) was collected on Orbitrap Fusion Lumos (Thermo-Fisher) mass spectrometer. The M+2 isotope is shown to have a lower non-integer mass than the previous isotope, potentially indicative of one or more sulfurs present, since 34S isotope has a smaller fractional mass than 32S. GSSG was collected at 30K, 120K, and 500K resolution to demonstrate how ultra high resolution allows one two separate the M+2 peak of GSSG into separate peaks, one reflective of 34S, and the other reflective of 13C and 15N.
An important experiment for Identification of unknown compounds involves the fragmentation of isolated ions of interest. Depending on the instrument, either collision induced dissociation (CID), electron transfer (ETD) or electron capture dissociation (ECD) is used to facilitate this experiment. Many workflows allow for an automatic selection of a number of ions during each scan cycle for fragmentation (data dependent acquisition) to provide a library of MS/MS spectra that can later be used for compound identification. For trap instruments, MSn can be useful for more thorough fragmentation and improved structure elucidation, where daughter ions are isolated and further fragmented (Rojas-Cherto et al 2012; Vaniya and Fiehn 2015). Several of the repositories mentioned above, including METLIN, HMDB, Lipid Maps and GnPS have libraries of MS/MS data for matching unknown spectra. XCMS2, an updated version of the very widely used XCMS metabolomics software package, enables fragments from MS/MS spectra to be searched against the METLIN library during data processing, and scored for their similarities to known product ion spectra to enable compound identification (Benton et al. 2008), while an online version can also be used for both analysis and spectral library searches (Tautenhahn et al 2012). GnPS, a recently established repository for natural products, allows the metabolomics community to upload data acquisition files online, which can be searched against already identified product ion spectra from previous data collections, and scored for possible matches (Wang et al 2016). The MS community can update the annotations and grade the quality of spectra submitted in this database. These automated search tools greatly help to reduce the time required to manually compare new data to existing spectral libraries. The future of quick compound identification and thorough untargeted metabolomics analysis will in large part be tied to the advancement of such spectral libraries and how they add, share, and search spectral data with fellow researchers, as this bottleneck is much too large to tackle independently.
If an unknown can be matched by some of the methods listed above, such as accurate mass, isotopic distribution, and fragmentation pattern, other factors should also be considered as well, such as: whether the elution time of the unknown likely correlates with the chemical class of the candidate compound, and whether the sample type is likely one to have such a metabolite present. At this stage purchasing an internal standard is the best way to fully confirm identity, which is often difficult or cost prohibitive if custom synthesis is required. Nonetheless, such an investment is often necessary for targeted quantitation or further study of the compound of interest. Misidentification is obviously a major pitfall for data interpretation and though compound matching using the tools described above can be very helpful, issues like isobaric or even isomeric species will often cause an additional hurdle to overcome. Having effective chromatography for the compound class of interest that can distinguish potential isomers is critical for final confirmation. Standards and guidelines for reporting identification or annotation of compounds have been authored by the metabolomics standard initiative (MSI), which have outlined criteria for reporting new compound identities in the literature (Fiehn et al 2007; Salek et al 2013). Within these reports, the MSI outlines the recommended levels of compound identification, ranging from the highest (level 1) where properties of an authentic standard are compared to experimental data, down through putatively annotated and characterized compounds (levels 2 and 3), and finally unknown (level 4). De novo identification of a compound that does not have an accessible fragmentation pattern is especially difficult, but is unfortunately the case for most analytes from a typical metabolomics study. In addition to elemental structure identification, a mass spectrometrist can also use tools to perform mock fragmentations of candidate structures, focusing especially on functional groups that are likely to fragment and ionize well, and then match these to the acquired MS/MS spectra.
Statistical Analysis
Untargeted metabolomics experiments generally use a combination of univariate and multivariate analysis to help identify compounds and pathways that are altered between cohorts. The are many different commercial as well as freely available statistical packages that can perform these functions, but in recent years several freely available online tools have been made available that carry a wide range of analysis features geared specifically towards metabolomics analysis. Two widely used online platforms include Metaboanalyst and the Metabolomics Workbench, mentioned above. Data can be uploaded to these sites, normalized and scaled as necessary, and then analyzed by tools such as: T-tests, ANOVA, Principal component analysis (PCA), as well as partial least squares (PLSDA), and orthogonal projections to latent structures (OPLSDA) determinant analysis, heatmaps, dendograms, volcano plots, and correlation analysis among other useful tools for data reduction and chemometrics.
Both univariate and multivariate analyses require special considerations to limit false interpretation from metabolomics data. Multivariate analysis is often a useful strategy for differentiating cohorts based on the covariances, or correlations of the many independent variables. Prior to using such methods, the signals of the analytes are often scaled so that high intensity ions do not overly bias the modeling. Several common scaling methods such as mean centering or Pareto scaling (Tugizimana et al 2016) are often used, depending on whether one favors treating all analytes equally, regardless of intensity (mean centering), or if one believes that high intensity analytes (compounds that either have high concentrations and/or high ionization efficiencies by ESI-LC-MS) should still have greater weight due to a higher confidence measurement (Pareto). An unbiased, or unsupervised method that is usually used as a first pass in evaluating metabolomics data is principal component analysis (PCA), which reduces the dimensionality of the many variables into primary eigenvectors that capture variance (Jolliffe and Cadima 2016). In PCA, the processing is blind to any classification in the data, and since it considers the relationships of all the independent variables simultaneously, it is generally not useful as a modeling tool in comprehensive metabolomics studies, where most of the variables are irrelevant. For generating models that are more apt to finding the independent variables that best discriminate the classifiers (dependent variables), researchers most often use partial PLSDA or OPLSDA. These are termed supervised methods, as the user inputs the classifiers (Y) along with the independent variables (X) that are projected in multi-dimensional space to enhance the variation In Y (Barker and Rayens 2003). OPLSDA differs from PLSDA in that it separates the uncorrelated variation in X from the predictive, whereas in PLSDA, variation not correlated with the Y-classifiers is still present in the data (Trygg and Wold 2002). The predictive power of both methods is thought to be the same, though (Bylesjö et al 2006).
The pitfall in theses supervised methods though are generally associated with overfitting. These methods will often show distinction of cohorts from just randomly generated data, as these tools are designed to accentuate any co-variances that differentiate the response (Y) variables, and with multicomponent testing using large numbers of independent variables with relatively low numbers of replicates, false positives are a given. From PLSDA plots for example, a VIP (variable importance parameter) describes the loadings that fit the model, and help researchers determine which analytes should be left in further iterations, and those that should be removed (not related to variation of cohorts). This process can lead to further overfitting of a model. It is therefore critical to perform validation analysis when generating these models to better ensure that false relationships between metabolites are not causing misinterpretation of the data. Permutation tests can analyze whether the assigned classes from the experiment are any more significant than randomly assigned class distinctions applied to the different samples (Golland and Fischl 2003). Cross-validation is an important process for model development and refinement, where the cohorts are split up into smaller subsets, and the model is further fitted with exclusion of various subjects (Westerhuis et al 2008; Wheelock and Wheelock 2013). Ideally, the samples can be randomly divided into a training set, validation set, and test set, where individual models can be tested and evaluated on unique sample subsets, and then applied to both other samples groups as well as the entire sample set. A cross-validated correlation (Q2) after subsequent iterations of such modeling can be assessed and compared to the R2 of the total model fitting (Wheelock and Wheelock 2013). One point often not discussed in modeling from untargeted metabolomics is the use of unidentified metabolites in the model. Though one can try to eliminate contaminants, adducts and other multiply represented features during pre-processing, a number of unknown features may persist, many of which are not biologically relevant, and which are thought to comprise the majority of features from an untargeted metabolomics run (Benton et al 2015). Some of these may be critical to the findings and are often part of the reason for choosing untargeted metabolomics in the first place. True unknowns that are significant to a data set should be attempted to be identified, but as mentioned previously, this is often a difficult task. If the unknowns cannot be identified, one is left to wonder how they should be considered in a published multivariate model.
Univariate analysis is one of the most common approaches to identify specific analytes that are significantly altered between cohorts. Assessment of normal distribution (parametric vs. non parametric) can be done prior to choosing the univariate method, most often with either student t-tests or ANOVA. Assessment of the statistical significances (i.e. p-values) of these tests are harder to interpret in untargeted metabolomics data, where thousands of individual features (or hypotheses) are being tested with comparably few unique samples tested. This multiple component problem has led to various approaches to correction in univariate testing, not just in the field of metabolomics, but in other OMICS disciplines like genomics/transcriptomics, and proteomics. The most conservative correction approach has been Bonferroni correction, where the significance level of an analyte is divided by the number of total hypotheses (Dunn 1961). Such a procedure can limit false positives in analyses (type I error), but unfortunately results in higher numbers of false negatives (type II errors) (Perneger 1998). False discovery rate (FDR) approaches have been developed to apply corrections that are more careful in limiting false negatives (Benjamini and Hochberg 1995; Genovese and Wasserman 2002). One such approach, which was made popular in genome wide studies, but is also used in metabolomics is the Q-value correction, an FDR approach that compares distributions of p-values from a data set and compares it to a distribution where all features are null (e.g. no differential between control and disease) in order to calculate the correction (q-value from a p-value) most applicable for a given dataset (Storey and Tibshirani 2003). Though there is no perfect method to remove false positives, an appropriate correction of multiple testing is nonetheless required in untargeted metabolomics reporting and interpretation.
Conclusion
Untargeted metabolomics is an exciting technology for searching for novel metabolic perturbations in various biological systems. As LC-MS metabolomics methods have developed over the last decade or two, sophisticated targeted methods have greatly expanded the breadth of metabolome that can be accurately quantified (Zhou et al 2016). Still, though, the allure of discovering novel biomarkers in disease states makes the untargeted approach remain valuable, and allows the investigator to evaluate a diverse swath of the metabolome, with less chance of missing an association, as when a particular analyte is targeted based on a single hypothesis. Unfortunately, a multitude of caveats are included when choosing untargeted metabolomics. We have attempted to address various aspects of untargeted metabolomics, including pre-analytical, analytical, and analysis aspects, all which have associated pitfalls that can jeopardize the usefulness of the data. From sample acquisition, to sample extraction and chromatographic selection, one can heavily bias the metabolites resolved, necessitating careful scrutiny and validation of each facet of the experiment. In addition, identification of novel compounds of interest presents another obstacle, but fortunately as the field grows, better tools have become available to address such issues. As these platforms further develop, we believe future untargeted studies will help to fill in the many gaps of uncharacterized metabolic perturbation in biological systems, and further benefit the clinical community by discovering novel diagnostic and therapeutic markers in disease.
Concise “Take-Home” Message.
This review discusses potential problems in the design, pre-analytical, analytical and post-analytical phases of an untargeted metabolomics experiment, and what can be done to improve the chance of a successful outcome.
Footnotes
Competing interest statement
Bruce Barshop declares that he has no conflict of interest.
Ilya Gertsman declares that he has no conflict of interest.
Details of ethics approval
Not applicable. No experimental procedures, no patient contacts.
A patient consent statement
Not applicable. No experimental procedures, no patient contacts.
Details of the contributions of individual authors:
Bruce Barshop had an equal part in the planning, conception and writing of the manuscript
Ilya Gerstman had an equal part in the planning, conception and writing of the manuscript
Details of funding
No funding.
References
- Alvarez-Sanchez B, Priego-Capote F, Luque de Castro MD. Metabolomics analysis I. Selection of biological samples and practical aspects preceding sample preparation. Trends Anal Chem. 2010;29:111–119. [Google Scholar]
- Aragon AD, Quinones GA, Allen C, et al. An automated, pressure-driven sampling device for harvesting from liquid cultures for genomic and biochemical analyses. J Microbiol Methods. 2006;65:357–360. doi: 10.1016/j.mimet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Barker M, Rayens W. Partial least squares for discrimination. Journal of Chemometrics. 2003;17:166–173. [Google Scholar]
- Barri T, Dragsted LO. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: effect of experimental artefacts and anticoagulant. Anal Chim Acta. 2013;768:118–128. doi: 10.1016/j.aca.2013.01.015. [DOI] [PubMed] [Google Scholar]
- Belanger MP, Askin N, Wittnich C. Multiple in vivo liver biopsies using a freeze-clamping technique. J Invest Surg. 2002;15:109–112. doi: 10.1080/08941930290085859. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Benton HP, Ivanisevic J, Mahieu NG, et al. Autonomous metabolomics for rapid metabolite identification in global profiling. Anal Chem. 2015;87:884–891. doi: 10.1021/ac5025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank LM, Kuepfer L, Sauer U. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome biology. 2005;6:R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics. 2006;20:341–351. [Google Scholar]
- Canelas AB, ten Pierick A, Ras C, et al. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Analytical Chemistry. 2009;81:7379–7389. doi: 10.1021/ac900999t. [DOI] [PubMed] [Google Scholar]
- Caspi R, Altman T, Billington R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AG, Miller AL, Atkinson DE. Role of the adenylate deaminase reaction in regulation of adenine nucleotide metabolism in Ehrlich ascites tumor cells. Cancer research. 1976;36:1144–1150. [PubMed] [Google Scholar]
- Chen S, Hoene M, Li J, et al. Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J Chromatogr A. 2013;1298:9–16. doi: 10.1016/j.chroma.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Csató E, Fülöp N, Szabó G. Preparation and comparison of a pentafluorophenyl stationary phase for reversed-phase liquid chromatography. Journal of Chromatography A. 1990;511:79–88. [Google Scholar]
- Denery JR, Nunes AA, Dickerson TJ. Characterization of differences between blood sample matrices in untargeted metabolomics. Anal Chem. 2011;83:1040–1047. doi: 10.1021/ac102806p. [DOI] [PubMed] [Google Scholar]
- Dietmair S, Timmins NE, Gray PP, Nielsen LK, Kromer JO. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Dietz C, Ehret F, Palmas F, et al. Applications of high-resolution magic angle spinning MRS in biomedical studies II-Human diseases. NMR in biomedicine. 2017 doi: 10.1002/nbm.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW., 2nd MicroRNAs and the butterfly effect. Cell cycle (Georgetown, Tex) 2013;12:707–708. doi: 10.4161/cc.23858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudzik D, Barbas-Bernardos C, Garcia A, Barbas C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. Journal of pharmaceutical and biomedical analysis. 2017 doi: 10.1016/j.jpba.2017.07.044. [DOI] [PubMed] [Google Scholar]
- Dunn OJ. Multiple Comparisons among Means. J Am Stat Assoc. 1961;56:52-&. [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN. Applications of NMR spectroscopy to systems biochemistry. Progress in nuclear magnetic resonance spectroscopy. 2016;92–93:18–53. doi: 10.1016/j.pnmrs.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Molecular cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. Guidance for Industry: Bioanalytical Method Validation (Internet at Fda.Gov/cder/guidance/index.htm) Book Guidance for Industry: Bioanalytical Method Validation. 2001 (Internet at Fda.Gov/cder/guidance/index.htm)
- Fiehn O, Robertson D, Griffin J, et al. The metabolomics standards initiative (MSI) Metabolomics. 2007;3:175–178. [Google Scholar]
- Fischer EH. Cellular regulation by protein phosphorylation. Biochemical and biophysical research communications. 2013;430:865–867. doi: 10.1016/j.bbrc.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- Genovese C, Wasserman L. Operating characteristics and extensions of the false discovery rate procedure. J Roy Stat Soc B. 2002;64:499–517. [Google Scholar]
- Gertsman I, Gangoiti J, Barshop B. Validation of a dual LC–HRMS platform for clinical metabolic diagnosis in serum, bridging quantitative analysis and untargeted metabolomics. Metabolomics. 2014;10:312–323. doi: 10.1007/s11306-013-0582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsman I, Gangoiti JA, Nyhan WL, Barshop BA. Perturbations of tyrosine metabolism promote the indolepyruvate pathway via tryptophan in host and microbiome. Mol Genet Metab. 2015;114:431–437. doi: 10.1016/j.ymgme.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Golland P, Fischl B. Permutation tests for classification: towards statistical significance in image-based studies. Inf Process Med Imaging. 2003;18:330–341. doi: 10.1007/978-3-540-45087-0_28. [DOI] [PubMed] [Google Scholar]
- Guillarme D, Ruta J, Rudaz S, Veuthey JL. New trends in fast and high-resolution liquid chromatography: a critical comparison of existing approaches. Anal Bioanal Chem. 2010;397:1069–1082. doi: 10.1007/s00216-009-3305-8. [DOI] [PubMed] [Google Scholar]
- Hill C, Drolet J, Kellogg MD, Tolstikov V, Narain NR, Kiebish MA. Blood Sampled Through Dried Blood Spots (DBS) Exhibits Diminished Ex vivo Metabolism Compared to Whole Blood Through Use of a Kinetic Isotope-Labeling Metabolomics Approach. Biochem Anal Biochem. 2017;6:325–330. [Google Scholar]
- Horai H, Arita M, Kanaya S, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Jandera P, Janas P. Recent advances in stationary phases and understanding of retention in hydrophilic interaction chromatography. A review. Anal Chim Acta. 2017;967:12–32. doi: 10.1016/j.aca.2017.01.060. [DOI] [PubMed] [Google Scholar]
- Jensen SK. Improved Bligh and Dyer extraction procedure. Lipid Technology. 2008;20:280–281. [Google Scholar]
- Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374:20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarovic L, Fournier NC. A comparison of extraction methods for the isolation of phospholipids from biological sources. Anal Biochem. 1986;156:244–250. doi: 10.1016/0003-2697(86)90179-x. [DOI] [PubMed] [Google Scholar]
- Kouassi Nzoughet J, Bocca C, Simard G, et al. A Nontargeted UHPLC-HRMS Metabolomics Pipeline for Metabolite Identification: Application to Cardiac Remote Ischemic Preconditioning. Anal Chem. 2017;89:2138–2146. doi: 10.1021/acs.analchem.6b04912. [DOI] [PubMed] [Google Scholar]
- Koulman A, Prentice P, Wong MCY, et al. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics. 2014;10:1018–1025. doi: 10.1007/s11306-014-0628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Molecular systems biology. 2006;2:2006 0034. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon Z, Garcia-Canaveras JC, Donato MT, Lahoz A. Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis. 2013;34:2762–2775. doi: 10.1002/elps.201200605. [DOI] [PubMed] [Google Scholar]
- Li L, Ren W, Kong H, et al. An alignment algorithm for LC-MS-based metabolomics dataset assisted by MS/MS information. Analytica Chimica Acta. 2017 doi: 10.1016/j.aca.2017.07.058. [DOI] [PubMed] [Google Scholar]
- Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem. 2011;83:3406–3414. doi: 10.1021/ac103313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madalinski G, Godat E, Alves S, et al. Direct introduction of biological samples into a LTQ-Orbitrap hybrid mass spectrometer as a tool for fast metabolome analysis. Anal Chem. 2008;80:3291–3303. doi: 10.1021/ac7024915. [DOI] [PubMed] [Google Scholar]
- Martano G, Delmotte N, Kiefer P, et al. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography-mass spectrometry. Nat Protoc. 2015;10:1–11. doi: 10.1038/nprot.2014.198. [DOI] [PubMed] [Google Scholar]
- McCalley DV. Understanding and manipulating the separation in hydrophilic interaction liquid chromatography-a review. J Chromatogr A. 2017 doi: 10.1016/j.chroma.2017.06.026. [DOI] [PubMed] [Google Scholar]
- McCloskey D, Gangoiti JA, King ZA, et al. A model-driven quantitative metabolomics analysis of aerobic and anaerobic metabolism in E. coli K-12 MG1655 that is biochemically and thermodynamically consistent. Biotechnol Bioeng. 2014;111:803–815. doi: 10.1002/bit.25133. [DOI] [PubMed] [Google Scholar]
- Metabolon. Book Sample Preparation and Shipping Guidelines. 2017. Sample Preparation and Shipping Guidelines. [Google Scholar]
- Miller MJ, Kennedy AD, Eckhart AD, et al. Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. Journal of inherited metabolic disease. 2015;38:1029–1039. doi: 10.1007/s10545-015-9843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Thonusin C, Qi NR, Burant CF, Evans CR. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: studies in a C57BL/6J mouse model. PLoS One. 2015;10:e0117232. doi: 10.1371/journal.pone.0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perneger TV. What’s wrong with Bonferroni adjustments. Brit Med J. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Current opinion in biotechnology. 2011;22:17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righi V, Tugnoli V, Mucci A, Bacci A, Bonora S, Schenetti L. MRS study of meningeal hemangiopericytoma and edema: a comparison with meningothelial meningioma. Oncology reports. 2012;28:1461–1467. doi: 10.3892/or.2012.1919. [DOI] [PubMed] [Google Scholar]
- Rojas-Cherto M, Peironcely JE, Kasper PT, et al. Metabolite identification using automated comparison of high-resolution multistage mass spectral trees. Anal Chem. 2012;84:5524–5534. doi: 10.1021/ac2034216. [DOI] [PubMed] [Google Scholar]
- Rose HG, Oklander M. Improved Procedure for the Extraction of Lipids from Human Erythrocytes. J Lipid Res. 1965;6:428–431. [PubMed] [Google Scholar]
- Salek RM, Steinbeck C, Viant MR, Goodacre R, Dunn WB. The role of reporting standards for metabolite annotation and identification in metabolomic studies. Gigascience. 2013;2:13. doi: 10.1186/2047-217X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno S, Jr, Mehrmohamadi M, Liberti MV, et al. RRmix: A method for simultaneous batch effect correction and analysis of metabolomics data in the absence of internal standards. PLoS One. 2017;12:e0179530. doi: 10.1371/journal.pone.0179530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Hansen R, Stephens GM, Goodacre R, Dickson AJ. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat Protoc. 2011;6:1241–1249. doi: 10.1038/nprot.2011.366. [DOI] [PubMed] [Google Scholar]
- Shi H, Enriquez A, Rapadas M, et al. NAD Deficiency, Congenital Malformations, and Niacin Supplementation. The New England journal of medicine. 2017;377:544–552. doi: 10.1056/NEJMoa1616361. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44:D463–470. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Dennis EA, Subramaniam S. LIPID MAPS-Nature Lipidomics Gateway: An Online Resource for Students and Educators Interested in Lipids. J Chem Educ. 2012;89:291–292. doi: 10.1021/ed200088u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007;8:93. doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Q, Huang W, Collette TW, Ekman DR, Tan C. A direct cell quenching method for cell-culture based metabolomics. Metabolomics. 2009;5:199–208. [Google Scholar]
- Thonusin C, IglayReger HB, Soni T, Rothberg AE, Burant CF, Evans CR. Evaluation of intensity drift correction strategies using MetaboDrift, a normalization tool for multi-batch metabolomics data. J Chromatogr A. 2017 doi: 10.1016/j.chroma.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognarelli JM, Dawood M, Shariff MI, et al. Magnetic Resonance Spectroscopy: Principles and Techniques: Lessons for Clinicians. Journal of clinical and experimental hepatology. 2015;5:320–328. doi: 10.1016/j.jceh.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) Journal of Chemometrics. 2002;16:119–128. [Google Scholar]
- Tugizimana F, Steenkamp PA, Piater LA, Dubery IA. A Conversation on Data Mining Strategies in LC-MS Untargeted Metabolomics: Pre-Processing and Pre-Treatment Steps. Metabolites. 2016:6. doi: 10.3390/metabo6040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaniya A, Fiehn O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. Trends Analyt Chem. 2015;69:52–61. doi: 10.1016/j.trac.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2012;403:1523–1548. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem: a European journal of chemical biology. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- Want EJ, Masson P, Michopoulos F, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. Journal of proteome research. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- Warrack BM, Hnatyshyn S, Ott KH, et al. Normalization strategies for metabonomic analysis of urine samples. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2009;877:547–552. doi: 10.1016/j.jchromb.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Watson DG. A rough guide to metabolite identification using high resolution liquid chromatography mass spectrometry in metabolomic profiling in metazoans. Comput Struct Biotechnol J. 2013;4:e201301005. doi: 10.5936/csbj.201301005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Hageman JA, van Eeuwijk F, et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics. 2016;12:88. doi: 10.1007/s11306-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis JA, Hoefsloot HCJ, Smit S, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–89. [Google Scholar]
- Wheelock AM, Wheelock CE. Trials and tribulations of ‘omics data analysis: assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol Biosyst. 2013;9:2589–2596. doi: 10.1039/c3mb70194h. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Gangoiti JA, Barshop BA, Siuzdak G. Metabolomics identifies perturbations in human disorders of propionate metabolism. Clinical chemistry. 2007;53:2169–2176. doi: 10.1373/clinchem.2007.089011. [DOI] [PubMed] [Google Scholar]
- Williams A, Tkachenko V. The Royal Society of Chemistry and the delivery of chemistry data repositories for the community. J Comput Aided Mol Des. 2014;28:1023–1030. doi: 10.1007/s10822-014-9784-5. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Current protocols in bioinformatics. 2016;55:14 10 11–14 10 91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Anal Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Lehmann R, Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal Bioanal Chem. 2015;407:4879–4892. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu H, Liu Y, Liu J, Zhao X, Yin Y. Development and Evaluation of a Parallel Reaction Monitoring Strategy for Large-Scale Targeted Metabolomics Quantification. Anal Chem. 2016;88:4478–4486. doi: 10.1021/acs.analchem.6b00355. [DOI] [PubMed] [Google Scholar]