Abstract
Diffusion-weighted magnetic resonance imaging (dMRI) is widely used to study white and gray matter (GM) micro-organization and structural connectivity in the brain. Super-resolution track-density imaging (TDI) is an image reconstruction method for dMRI data, which is capable of providing spatial resolution beyond the acquired data, as well as novel and meaningful anatomical contrast that cannot be obtained with conventional reconstruction methods. TDI has been used to reveal anatomical features in human and animal brains. In this study, we used short track TDI (stTDI), a variation of TDI with enhanced contrast for GM structures, to reconstruct direction-encoded color maps of fixed tree shrew brain. The results were compared with those obtained with the traditional diffusion tensor imaging (DTI) method. We demonstrated that fine microstructures in the tree shrew brain, such as Baillarger bands in the primary visual cortex and the longitudinal component of the mossy fibers within the hippocampal CA3 subfield, were observable with stTDI, but not with DTI reconstructions from the same dMRI data. The possible mechanisms underlying the enhanced GM contrast are discussed.
Keywords: Track-density imaging, Diffusion tensor imaging, Tree shrew, Primary visual cortex, Hippocampus
Introduction
Diffusion-weighted magnetic resonance imaging (dMRI) uses the diffusion of water molecules as an endogenous probe to characterize tissue microstructure [1]. Diffusion tensor imaging (DTI) is one of the most widely used dMRI techniques to characterize fiber orientation and integrity [2, 3]. DTI has been used extensively to study not only the white matter (WM) anatomy [4–6], but also the micro-organization of gray matter (GM) structures. For instance, DTI has been used to reveal the laminar organization of the hippocampal formation (Hipp) in humans [7], rats [8], and mice [9], and to reveal the microstructure of human cortical areas [10, 11].
More recently, a post-processing method for dMRI data, super-resolution track-density imaging (TDI), has been proposed, which is capable of providing spatial resolution beyond the acquired data, as well as novel and meaningful anatomical contrast that cannot be obtained with conventional MRI [12–14]. These are achieved by incorporating into image reconstruction the information of a large number of streamlines generated from probabilistic fiber-tracking across the whole brain [13, 14]. Short-track TDI (stTDI) is a variation of TDI, in which the contrast among GM structures is enhanced by constraining the length of the streamlines generated during fiber-tracking [13]. Previous studies have demonstrated that super-resolution stTDI can be used to delineate substructures within the human thalamus, brainstem, and cortex [10, 14–16], and to characterize GM structures in mouse [12, 13, 17], zebrafish [18], and zebra finch [19] brains. The capability of super-resolution TDI in detecting abnormal microanatomical fiber structures in the GM of human patients has also been demonstrated [20–22].
Tree shrews are squirrel-like mammals, which have a closer relationship with primates than with rodents on the evolutionary tree [23, 24]. Tree shrews have a high brain-to-body mass ratio [25]. Comparative studies have demonstrated that the neuroanatomy of this species shares many features with that of primates, and differs significantly from that of rodents [6, 26–28]. It has been proposed that tree shrews may be a better model animal than rodents, and an alternative/adjunct to non-human primates for studying human neurological/psychiatric diseases [29]. Tree shrew models of myopia [30, 31], psychosocial stress/depression [32, 33], senile diseases [34, 35], Parkinson’s disease [36], and drug addiction [37] have been developed.
DTI has been used to study the WM anatomy of tree shrews [6]. It has been demonstrated that tree shrews possess a primitive primate-like WM configuration, especially in frontotemporal connections [6]. In addition, it has been shown that DTI contrast can be used to study the microstructural organization of certain GM structures in the tree shew brain, such as the visual cortex and superior colliculus [6]. In this study, we used super-resolution stTDI to further investigate the GM micro-organization in the tree shrew brain. We demonstrated that super-resolution stTDI can reveal meaningful fine anatomical features in the primary visual cortex (V1) and Hipp of tree shrew, which could not be identified unequivocally otherwise.
Materials and Methods
Brain Samples and dMRI Data Acquisition
All animal handling protocols were approved by the Institutional Animal Care Committee. The raw ex vivo dMRI data used for stTDI analysis were the same as those used in a previous DTI WM tractography study [6]. Briefly, two adult male tree shrews, weighing ~120 g and 6 months old, were transcardially perfused first with 0.9% saline and subsequently with 4% paraformaldehyde (PFA). The brains were then extracted from the skull and stored in 4% PFA at 4 °C until use. To acquire the dMRI data, the brains were rehydrated in 0.1 mol/L phosphate buffered saline (pH 7.4), which was freshly-prepared daily, at 4 °C for 3 consecutive days. The rehydrated brains were then separately transferred into two plastic sample tubes filled with Fomblin YL VAC 06/6 oil (Y06/6 grade, Solvay, Italy), a perfluorocarbon which was used to limit tissue dehydration and minimize susceptibility to artifacts [38].
dMRI data were acquired on a 400 MHz (i.e., 9.4 T)/89 mm vertical-bore Bruker Biospin micro-imaging spectrometer using a 25 mm-diameter quadrature resonator for both radiofrequency transmission and signal reception. A three-dimensional (3D) diffusion-weighted spin-echo sequence was used for acquisition with repetition time 400 ms, echo time 23.5 ms, field of view 20 mm × 20 mm × 30 mm to cover the whole brain, matrix size 100 × 100 × 100, five b = 0 s/mm2 images, 30 diffusion-weighted images with b = 5000 s/mm2, δ 5 ms, Δ 13 ms, a gradient-encoding scheme of 30 directions homogenously distributed on a spherical surface, and 1 average. The total acquisition time was 38 h and 53 min per sample.
dMRI Data Processing
All dMRI image volumes were first corrected for eddy current distortion and possible drift using the Diffusion Toolbox in the FMRIB Software Library from the Oxford Centre for Functional MRI of the Brain (http://www.fmrib.ox.ac.uk/fsl/) [39]. Then, the diffusion tensor (DT) of each voxel was fitted with a multivariate linear fitting algorithm provided by the MRtrix software (http://www.brain.org.au/software/) [40], followed by generation of DTI parametric maps, including the apparent diffusion coefficient (ADC) map, fractional anisotropy (FA) map, and direction-encoded color (DEC) map. The DEC-DTI map is a two-dimensional visualization approach in which the image brightness of each pixel represents the FA value, and the color codes the orientation of the major eigenvector of the DT, which is assumed to represent the principal diffusion direction that coincides with the orientation of the fibers occupying this pixel [41]. A red–green–blue color encoding scheme for DEC-DTI was used [41], with red representing medial-lateral; green representing dorsal-ventral, and blue representing rostral-caudal.
The pre-processed dMRI data for DTI analysis were subsequently used to reconstruct super-resolution track density maps with the MRtrix software. To overcome the well-known limitation of DT in modeling crossing fibers, the constrained spherical deconvolution (CSD) algorithm provided in the MRtrix package [42] was applied to model the intravoxel fiber orientation distribution (FOD) with a maximum harmonic order of 6. Probabilistic fiber-tracking [43] was then carried out to generate a large number of tracks within the whole brain for track density map reconstruction. The track number was selected according to the maximum track length used to avoid saturated DEC track density maps [13, 18]. All the other tracking parameters were optimized empirically (step size 0.01 mm–0.04 mm, maximum turning angle 25°–65°, FOD amplitude cutoff value 0.05–0.3, and maximum track length 1 mm–7 mm for short-track reconstruction). By visual inspection of the reconstructed track density maps, step size 0.02, maximum turning angle 45°, and FOD amplitude cutoff 0.1 were taken as the parameters of choice. A maximum track length of 4 mm was considered optimal for short-track reconstruction. Track numbers of 80 million and 8 million were used for short-track and long-track (i.e., no constraint on track length) reconstruction respectively, corresponding to roughly 250 and 25 tracks per voxel. The TDI/stTDI versions of the DEC maps [13] were reconstructed at either the native resolution (the same as the source data) or an isotropic resolution of 40 μm (Fig. 1D–I), corresponding to an about 190-fold reduction in voxel volume. For each pixel in the DEC-TDI and DEC-stTDI maps, the color and saturation corresponded to the averaged direction and the number of traversing streamlines, respectively [13]. The color encoding scheme for the DEC-TDI and DEC-stTDI maps were the same as that for the DEC-DTI maps.
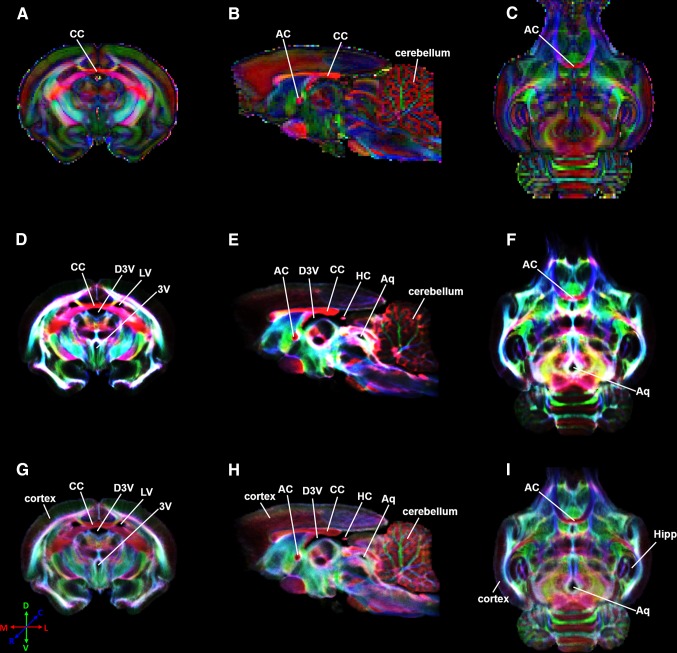
Fig. 1.
DEC-DTI (A–C) and super-resolution DEC-TDI (D–F)/DEC-stTDI (G–I) maps of selected coronal (A, D, G), sagittal (B, E, H), and transverse (C, F, I) slices of a fixed tree shrew brain. All reconstructions used the same dMRI dataset. CC corpus callosum, AC anterior commissure, D3V dorsal third ventricle, LV lateral ventricle, 3V third ventricle, HC habenular commissure, Aq aqueduct, Hipp hippocampus.
Results
All dMRI results displayed in Figs. 1, 2, 3 and 4 are from the same fixed brain sample used previously to construct the tree shrew brain WM atlas [6]. Figure 1 shows the DEC-DTI, DEC-TDI, and DEC-stTDI maps of the tree shrew brain. These maps showed largely similar contrast for major WM structures, including the corpus callosum and anterior commissure. Because of the improved spatial resolution, depiction of the complex WM/GM configurations in the cerebellum and small WM structures such as the habenular commissure (Fig. 1E, H) was better in the super-resolution DEC-TDI and DEC-stTDI maps than in the corresponding DEC-DTI maps. The DEC-TDI and DEC-stTDI maps also showed enhanced contrast for the ventricular system, including the lateral ventricle (Fig. 1D, G), the third ventricle (Fig. 1D, G), the dorsal third ventricle (Fig. 1D, E, G, H), and aqueduct (Fig. 1E, F, H, I), for the lack of fiber-tracks originating from the cerebrospinal fluid [14]. With reduced track density dynamic range [13], the DEC-stTDI maps showed enhanced contrast for neocortex, Hipp, and cerebellum (Fig. 1D–F) relative to the DEC-TDI maps (Fig. 1G–I).
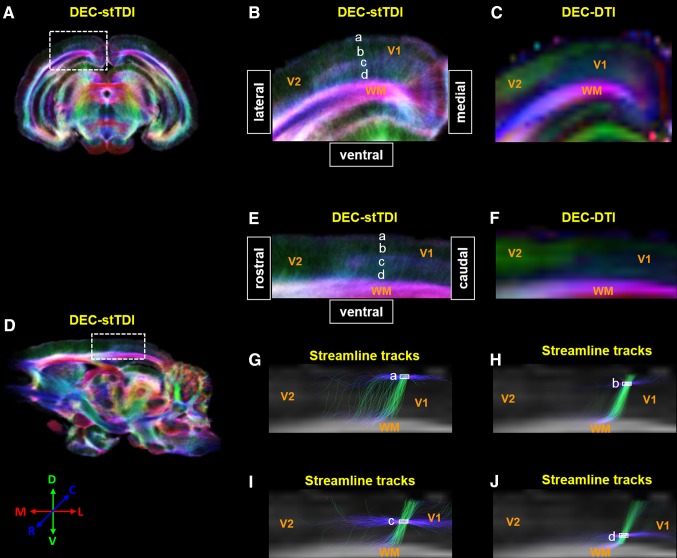
Fig. 2.
Laminar organization of tree shrew primary visual cortex (V1) revealed by stTDI. DEC-stDTI maps of a coronal slice and a sagittal slice are shown in A and D, respectively. The visual cortex areas covered by the white dashed rectangles in A and D are enlarged in B and E, along with the corresponding native-resolution DEC-DTI maps in C and F. A four-layer organization of V1 was discernible in B and E, which are labeled as layers a–d. Panels G–J show the streamline tracks passing through regions of interest in the four layers (white solid rectangles) selected from whole-brain fiber-tracking results. The backgrounds in these panels were fractional anisotropic maps corresponding to the regions in E and F. The color coding scheme of the streamline tracks is the same in the DEC-DTI and DEC-stTDI maps. V2 secondary visual cortex, WM white matter.
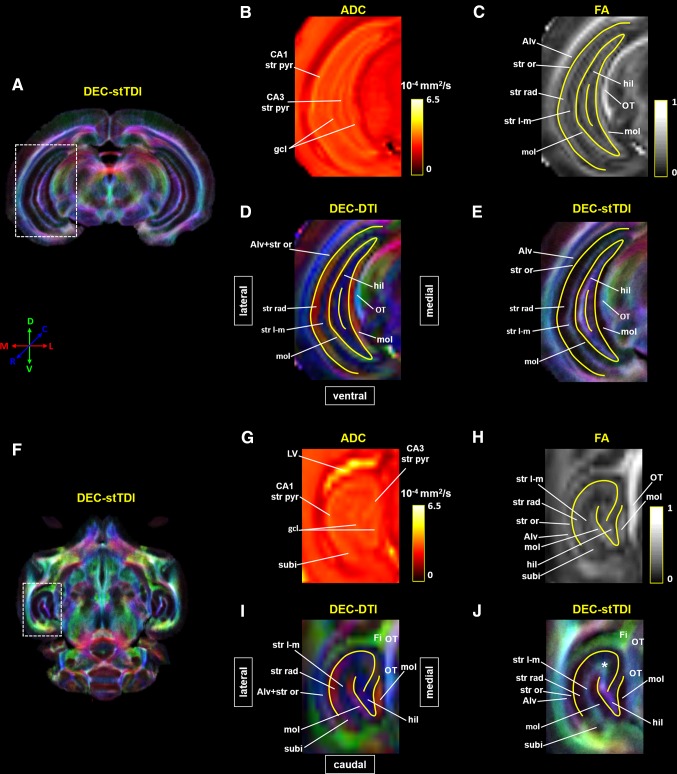
Fig. 3.
Laminar organization of tree shrew hippocampal formation (Hipp) revealed by DTI and stTDI. A, F Super-resolution DEC-stTDI maps of a coronal slice and a transverse slice at the level of the Hipp. The hippocampal areas (dashed white rectangles) are enlarged in E and J. Native-resolution apparent diffusion coefficient (ADC) (B, G), fractional anisotropic (FA) (C, H), and DEC-DTI (D, I) maps of the corresponding areas are also shown. Yellow solid lines in C–E and H–J were manually drawn according to the ADC contrast to show the location of the granule cell layer (gcl) of the dentate gyrus and the stratum pyramidale (str pyr) of CA1 and CA3. The white asterisk in J indicates a structure revealed by the stTDI reconstruction only. Alv alveus, Fi fimbria of hippocampal formation, gcl granule cell layer of the dentate gyrus, hil hilar region of the dentate gyrus, LV lateral ventricle, mol molecular layer of the dentate gyrus, OT optic tract, str l-m stratum lacunosum-moleculare, str or stratum oriens, str pyr stratum pyramidale, str rad stratum radiatum, subi subiculum.
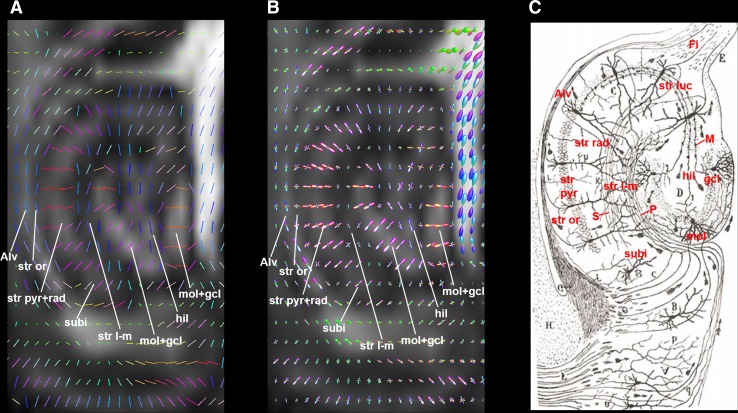
Fig. 4.
DTI principal direction (A) and CSD FODs (B) maps of tree shrew hippocampal formation. For comparison, the rodent hippocampal anatomy and connectivity depicted by Cajal [46], based on the silver staining method, is shown in C. The background images in A and B are fractional anisotropy maps. The color coding scheme in these maps are the same as that used for DEC-DTI maps. Alv alveus, CSD constrained spherical deconvolution, Fi fimbria of hippocampal formation, FODs fiber orientation distributions, gcl granule cell layer of dentate gyrus, hil hilar region of the dentate gyrus, M mossy fibers, mol molecular layer of the dentate gyrus, P performant pathway axons, S Schaffer collateral axons, str l-m stratum lacunosum-moleculare; str luc stratum lucidum of CA3, str or stratum oriens, str pyr stratum pyramidale, str rad stratum radiatum, subi subiculum.
Both the DEC-DTI maps and the super-resolution DEC-stTDI maps (Fig. 2), no matter coronal or sagittal, showed clear contrast between V1 and the secondary visual cortex (V2). The boundaries between V1 and V2 revealed by DTI/stTDI were similar to those observed by histology [28, 44, 45]. V2 appeared as a homogeneous structure on these maps, presumably containing mainly radial fibers running along the ventral-dorsal axis. In comparison, the fiber configurations in V1 were more heterogeneous. The color coding on the DEC-DTI maps (Fig. 2C, F) indicated that the dorsal-most layer of V1 contained mainly radially-oriented fibers; while the more ventral portion appeared to be abundant in tangentially-oriented fibers. The super-resolution DEC-stTDI maps revealed a more definitive and better resolved laminar organization in V1 (Fig. 2B, E). Four layers were identifiable by visual inspection, which, for convenience, are referred to as layers a–d below (Fig. 2B, E). It should be emphasized that layers a–d were defined empirically and did not necessarily correspond to the histological nomenclature of cortical layers I–IV [28]. Layer c was seen only in V1, but not in V2 (Fig. 2B, E). To better understand the fiber configurations in these empirically-defined layers, streamlines traversing the ROIs representing these layers were selected from the whole-brain tracking result and superimposed on the sagittal FA maps (Fig. 2G–J). All four layers in V1 contained both well-aligned tangentially-oriented streamlines running in parallel with the brain surface and radial streamlines extending perpendicular to the brain surface. However, the percentages of tangential streamlines in layers a and c (61.3% and 70.9%, respectively) appeared to be much higher than those in the layers b and d (26.2% and 38.5%, respectively). The superficial layer in V2 had a streamline configuration similar to that in layer a of V1, while the rest of V2 was characterized by predominantly vertical streamlines mixed with a limited number of ill-aligned tangential streamlines (data not shown).
Figure 3 shows the laminar organization of the tree shrew Hipp revealed by dMRI data. The hyperintense layers on the ADC maps (Fig. 3B, G), with an averaged ADC of ~3.2 × 10−4 mm2/s, were considered to represent the granule cell layer (gcl) of the dentate gyrus and the stratum pyramidal layer (str pyr) of CA1 and CA3 [8, 9]. The centers of these layers were then traced manually (yellow solid lines in Fig. 3C–E, H–J), and used as landmarks to determine the identities of the other layers. Combining the FA and DEC-DTI contrasts, the stratum radiatum (str rad), molecular layer (mol), and hilar region (hil) of the dentate gyrus, stratum lacunosum-moleculare (str l-m), stratum oriens (str or), and alveus (Alv) could be assigned, taking into account prior knowledge of the microstructural organization of the rodent hippocampus (Fig. 4C) [46], the histology of the tree shrew Hipp [47], and DTI contrasts in rodent and human Hipp [7–9]. The str rad was located along the medial side of the CA1 str pyr, while the mol was situated next to the gcl. Both the str rad and mol had a relatively high FA of ~0.3, mainly containing fibers running in the medial-lateral direction. The str l-m was between the str rad and mol, but had a relatively lower FA than its neighbors and a distinct fiber orientation (Fig. 3C, D, H, I). Positioned side-by-side and having similar DEC-DTI contrasts (Fig. 3D, I), the Alv and str or could be distinguished from each other by FA contrast (Fig. 3C, H). A region at the distal end of CA1 str pyr in the transverse plane was considered to be the subiculum (subi, Fig. 3G), which had a DEC-DTI contrast similar to the adjacent str pyr, but a relatively lower ADC and FA (Fig. 3G–I). As indicated by the DEC-DTI contrast, the fibers in the hil and str l-m ran mainly in the rostral-caudal direction (blue in Fig. 3D, I). The fiber orientations of different Hipp substructures could be appreciated more clearly on the DTI vector map and FODs map (Fig. 4A, B).
The laminar organization of the tree shrew Hipp revealed by DEC-stTDI contrast was largely similar to that shown on the DTI maps. However, there were also evident differences between the two modalities. First, the track-density contrast did not correlate with the FA contrast in some Hipp substructures. For example, the str l-m showed a relatively lower FA, but a relatively higher track density than the neighboring mol and str rad, while the mol had a relatively higher FA, but a relatively lower track density than its neighbors (Fig. 3E, J). Second, the fiber orientation revealed by the DEC-stTDI and DEC-DTI contrasts did not always agree with each other. For instance, the DEC-stTDI contrast indicated that the fibers in the subi extended roughly in the dorsal-ventral direction (viridescent in Fig. 3J), while a largely medial-lateral orientation was suggested by the DEC-DTI contrast (Fig. 3I). Finally, there were Hipp substructures that could be discerned unequivocally only on the stTDI maps, but not on the DTI maps, such as the region at the junction of str l-m and the hilus (white asterisk in Fig. 3J). This structure showed no distinctive ADC, FA, or DEC-DTI contrast against the adjacent areas, but a stTDI contrast from the hil and str l-m was clear. The color coding on the stTDI map indicated that the fibers in this structure ran predominately in the dorsal-ventral orientation, as compared with a rostral-caudal orientation in the hil and str l-m (Fig. 3I, J).
Discussion
In this study, super-resolution stTDI was used to study the microstructural organization of the V1 and Hipp of the tree shrew. These two anatomical structures in this species have been attracting the most interest in neuroscience research [48, 49]. V1 and the Hipp are also the two brain regions in the tree shrew with abundant histological/cytoarchitectural data available [28, 47]; these are crucial for understanding and interpreting the imaging data. With the high spatial resolution and unique anatomical contrast it offers, super-resolution stTDI improved the visualization of certain fine anatomical features in these regions, features that could not be discerned otherwise.
DTI and stTDI of Tree Shrew V1
Histological studies have demonstrated that the neuronal fibers within V1 have an apparent laminar organization [28, 50–52]. For instance, the superficial layer and central Baillarger bands contain prominently tangentially-oriented fibers, while the other layers are dominated by radially-oriented dendrites and co-ascending axons. The heavily myelinated outer band of Baillarger (the stria of Gennari) in human and monkey V1 can be observed by high resolution anatomical imaging [53–56]. High angular and spatial resolution dMRI has also been used to study the 3D layer-specific fiber architecture within V1 in human [10, 11, 57] and mouse [17].
V1 in the tree shrew is more structurally heterogeneous and has a DEC-DTI contrast distinct from the lateral-lying V2 [6]. However, no clear laminar organization of V1 could be discerned on the DEC-DTI maps, probably due to limited spatial resolution of the source dMRI data and the well-known limitation of DTI in modeling complex fiber configurations, such as fiber crossing.
With the same raw data, the CSD tractography-based stTDI reconstruction revealed a clear 4-layered organization in V1, but not in the adjacent V2 in the tree shrew (Fig. 2B, E). Previous stTDI studies have also observed the laminar organization of V1 in human and mouse [10, 17]. All four layers of the tree shrew V1 defined by the DEC-stTDI contrast contained both well-aligned tangential streamlines running parallel to the pial surface, and radial streamlines extending perpendicular to the pial surface (Fig. 2G–J). However, the percentages of tangential streamlines appeared to be higher in layers a and c, relative to those in layers b and d, leading to the different DEC-stTDI contrast among them. A similar 4-layer streamline configuration has also been observed in human V1 with CSD-based tractography [57]. Given the dominant fiber orientation, the superficial layer a likely corresponds to the molecular layer I defined histologically [57–59], while layer c corresponds to the Baillarger bands [28, 50]. This interpretation is consistent with the histological observation that molecular layer I is present, as well, in other cortical areas (e.g., in V2), while the Baillarger bands are present only in V1 in the tree shrew [28]. The radial streamlines in all these layers are in line with the apical dendrites of pyramidal neurons extending radially towards the pial surface [10, 50, 57].
Visualization of the laminar organization in tree shew V1 by stTDI reconstruction depended on the spatial resolution of the raw dMRI data. With the lower-spatial resolution in vivo dMRI datasets [6], we failed to observe the fine laminar organization of V1 on DEC-DTI and DEC-stTDI maps (data not shown).
DTI and stTDI of Tree Shrew Hipp
DTI has been used to characterize the microstructural organization of the Hipp in humans and rodents [7–9]. The DTI contrast of the tree shew Hipp in this study was largely similar to the results in these previous studies. It has been assumed that the gcl and str pyr layers have the highest ADC in the Hipp, due to the fact that they are rich in large and compact neuronal somata [8, 9]. Based on this assumption, we first identified the gcl and str pyr layers on the ADC maps of the tree shrew Hipp, and used them as landmarks to label the other substructures. All the other Hipp substructures appeared to have a relatively lower ADC, consistent with the fact that they are mainly composed of dendrites and axons [60].
The str rad is composed mainly of large apical dendrites of pyramidal neurons, and similarly the mol layer contains mainly apical dendrites from the granule cells in the dentate gyrus. The DEC-DTI contrast and DTI principal eigenvector map suggested that the fibers in the str rad and mol run predominately perpendicular to the surface of the str pyr and gcl, respectively, agreeing well with the known fiber orientation in these structures [46, 47, 60]. Within the str l-m, there were both the terminals of the apical dendrites of the pyramidal neurons and axons in the performant pathway axons (P, Fig. 4C) and Schaffer collaterals (S, Fig. 4C) [46, 60]. The DEC-DTI contrast of the str l-m appeared to reflect mainly the fiber orientation of the P and S. Perhaps because the two sets of fiber components in the str l-m are roughly perpendicular to each other [46, 60], it had lower FA than the str rad and mol. The str or also had two sets of orthogonally-oriented fibers: the pyramidal neuron basal dendrites and axons [46, 60]. It had a lower FA than the str rad and mol as well. The subi had a slightly lower ADC than the str pyr, as it consists of dispersed pyramidal neurons in tree shrew [47]. The dendrites of subi pyramidal neurons are also radially oriented, giving the structure a DEC-DTI contrast similar to the str rad. The Alv is a thin sheet-like WM fiber tract covering the lateral surface of the entire Hipp; this had a DEC-DTI contrast similar to, but a higher FA than the str or, likely due to the fact that it contains more well-aligned and compactly packed axons connecting the Hipp to the medial septal area, diagonal band, dorsal tenia tecta, and mammillary bodies [6]. The hil is enclosed between the blades of dentate gyrus [47, 60]. The fiber orientation in this region estimated by DTI appeared to mainly represent the horizontal portion of the mossy fibers (M, Fig. 4C), which emerge from the basal portions of the dentate gyrus granule cells and course through the hil to enter the stratum lucidum (str luc) of CA3 at the same transverse level [46, 47, 60]. The FA and DEC-DTI contrasts among the laminar structures of the tree shrew Hipp from a lower-resolution in vivo data set [6] (data not shown) were similar to the ex vivo observations in the current study, except that the Alv and str or could not be separated due to limited spatial resolution.
Both TDI and stTDI have been used to characterize the laminar organization of the Hipp in mouse [13, 17, 61]. Consistent with the results in these previous studies, the Alv, str l-m, and hilar region appeared to have a relatively higher track density, while the str or, str pyr, and str rad, as well as the mol and gcl, appeared to have a relatively lower track density. Interestingly, the FA and track density seemed to correlate negatively among the fibrous layers of the Hipp, except for the Alv. The str rad and mol had a relatively high FA, but a relatively lower track density, and vice versa for the str l-m. The explanation for this likely lies in the fact that coherently oriented fibers extend for a longer distance within the str l-m than within the str rad and mol, while all are still shorter than the maximum track length used here [46, 47]; thus the str l-m tended to be over-defined by whole-brain streamline reconstruction for a greater seeding volume [62]. The DEC-stTDI contrast also revealed novel fine anatomical features of the tree shrew Hipp, which could not be revealed by DTI. For example, the viridescent color coding on the DEC-stTDI map indicated that the streamlines in the subi extended mainly in the dorsal-ventral direction, which likely reflects the longitudinal association pathway within it [60]. Another fine anatomical feature of the tree shrew Hipp revealed by super-resolution stTDI is the region at the junction of the str l-m and hil (white asterisk in Fig. 3J), in which the fibers ran predominately in the dorsal-ventral direction, perhaps representing the longitudinal component of the mossy fibers within the CA3 str luc [60]. Perhaps due to spatial resolution, the two novel anatomical features shown on the DEC-stTDI maps of fixed brain were not evident in the DEC-stTDI reconstruction of in vivo datasets [ 6 ] (data not shown).
Limitations
In this study, the DEC-stTDI maps were reconstructed from whole-brain fiber-tracking results. A large number of streamlines (80 million) were generated to boost the signal-to-noise ratio (SNR) [63]. Using a large track number, however, was time-consuming. For example, it took ~4 h to reconstruct the DEC-stTDI maps for a single brain on an 8-core 64 GB RAM Intel-Xeon workstation. In order to reduce the computation time while retaining a high SNR, the DEC-stTDI maps of particular structures of interest can be reconstructed by constraining fiber-tracking within the relevant ROI and at neighboring voxels (partial-brain fiber-tracking) [64].
It has been suggested that, for human brain studies, a minimum of 45 diffusion weighted (DW) directions should be used to obtain optimized CSD FOD [65]. In the current study, we used only 30 DW directions such that the FOD-based tracking may not have the best possible performance. However, in practice it would be too time-consuming to acquire DWI datasets with both high spatial resolution and high angular resolution. In previous studies, datasets with 30 DW directions have been used to reconstruct TDI/stTDI maps, and the results showed well-defined anatomical features which were not revealed by DTI reconstructions from the same raw datasets [10, 12, 13, 17, 18, 64]. Our results are consistent with these previous studies. Nevertheless, we acknowledge that acquiring datasets with higher angular resolution may result in improved FOD-based tracking results and TDI/stTDI reconstructions, which could enable the visualization of even more subtle anatomical features.
The anatomical features of tree shrew V1 and Hipp revealed in this study were interpreted mainly by inference from previous dMRI studies and the micro-organization of their counterparts in humans and rodents. Although it is generally accepted that DTI and stTDI can provide meaningful anatomical information of the normal and abnormal brains [3, 8, 9, 12, 16, 20], these fine anatomical features in the tree shrew demonstrated by DTI and stTDI should be further confirmed directly by histological methods, such as silver and Golgi staining, in future studies.
It has been proposed that TDI can be used to detect structural differences quantitatively using group comparisons [14]. Although some studies have used TDI maps as scalar maps to infer brain aberrances in patients, it should be borne in mind that the locally-optimal nature of streamline propagation, such as tracking algorithm, streamline seeding, orientation selection, and streamline stop criteria, can introduce biases into TDI maps [62, 66, 67]. The stTDI, as a variation of TDI, can be further affected by the selection of maximum track length. As a result, the track-density of stTDI as a fully quantitative parameter should be used with caution.
Conclusion
DTI models the local diffusion process of water molecules directly, and can provide fully quantitative parameters (e.g., FA) of tissue. In comparison, the stTDI is reconstructed from whole-brain fiber-tracking which takes neighborhood regions with coherent fiber orientation into account. Despite the limitation in quantification, the stTDI performs better than DTI in detecting subtle structures in GM. In the current study, our results showed that super-resolution stTDI improved the visualization of certain fine anatomical features in V1 and the Hipp of the tree shrew, features that could not be discerned using DTI reconstructed from the same dMRI data. All the substructures revealed by DTI and/or stTDI can potentially be used as anatomical guides for future studies using tree shrew models of neurological/psychiatric diseases, particularly using dMRI-based methods.
Acknowledgements
This work was supported by grants from the National Basic Research Development Program of China (2011CB707800) and the National Natural Science Foundation of China (21790390, 21790392, and 61371014).
References
- 1.Taylor DG, Bushell MC. The spatial mapping of translational diffusion coefficients by the NMR imaging technique. Phys Med Biol. 1985;30:345–349. doi: 10.1088/0031-9155/30/4/009. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Jones DK. Diffusion-tensor MRI: theory, experimental design and data analysis - A technical review. NMR Biomed. 2002;15:456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- 3.Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- 5.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 6.Dai JK, Wang SX, Shan D, Niu HC, Lei H. A diffusion tensor imaging atlas of white matter in tree shrew. Brain Struct Funct. 2017;222:1733–1751. doi: 10.1007/s00429-016-1304-z. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd TM, Ozarslan E, Yachnis AT, King MA, Blackband SJ. Diffusion tensor microscopy indicates the cytoarchitectural basis for diffusion anisotropy in the human hippocampus. AJNR. 2007;28:958–964. [PMC free article] [PubMed] [Google Scholar]
- 8.Shepherd TM, Ozarslan E, King MA, Mareci TH, Blackband SJ. Structural insights from high-resolution diffusion tensor imaging and tractography of the isolated rat hippocampus. Neuroimage. 2006;32:1499–1509. doi: 10.1016/j.neuroimage.2006.04.210. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, van Zijl PC, Mori S. Three-dimensional diffusion tensor magnetic resonance microimaging of adult mouse brain and hippocampus. Neuroimage. 2002;15:892–901. doi: 10.1006/nimg.2001.1012. [DOI] [PubMed] [Google Scholar]
- 10.Aggarwal M, Nauen DW, Troncoso JC, Mori S. Probing region-specific microstructure of human cortical areas using high angular and spatial resolution diffusion MRI. Neuroimage. 2015;105:198–207. doi: 10.1016/j.neuroimage.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinnijenhuis M, Zerbi V, Kusters B, Slump CH, Barth M, van Walsum AMC. Layer-specific diffusion weighted imaging in human primary visual cortex in vitro. Cortex. 2013;49:2569–2582. doi: 10.1016/j.cortex.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Kurniawan ND, Richards KL, Yang ZY, She D, Ullmann JFP, Moldrich RX, et al. Visualization of mouse barrel cortex using ex-vivo track density imaging. Neuroimage. 2014;87:465–475. doi: 10.1016/j.neuroimage.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Calamante F, Tournier JD, Kurniawan ND, Yang Z, Gyengesi E, Galloway GJ, et al. Super-resolution track-density imaging studies of mouse brain: comparison to histology. Neuroimage. 2012;59:286–296. doi: 10.1016/j.neuroimage.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Calamante F, Tournier JD, Jackson GD, Connelly A. Track-density imaging (TDI): Super-resolution white matter imaging using whole-brain track-density mapping. Neuroimage. 2010;53:1233–1243. doi: 10.1016/j.neuroimage.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Calamante F, Oh SH, Tournier JD, Park SY, Son YD, Chung JY, et al. Super-resolution track-density imaging of thalamic substructures: comparison with high-resolution anatomical magnetic resonance imaging at 7.0T. Hum Brain Mapp. 2013;34:2538–2548. doi: 10.1002/hbm.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch MJ, Chung S, Ben-Eliezer N, Bruno MT, Fatterpekar GM, Shepherd TM. New clinically feasible 3T MRI protocol to discriminate internal brain stem anatomy. AJNR. 2016;37:1058–1065. doi: 10.3174/ajnr.A4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, Reisinger D, Xu J, Fatemi SA, van Zijl PC, Mori S, et al. Localized diffusion magnetic resonance micro-imaging of the live mouse brain. Neuroimage. 2014;91:12–20. doi: 10.1016/j.neuroimage.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullmann JFP, Calamante F, Collin SP, Reutens DC, Kurniawan ND. Enhanced characterization of the zebrafish brain as revealed by super-resolution track-density imaging. Brain Struct Funct. 2015;220:457–468. doi: 10.1007/s00429-013-0667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamaide J, De Groof G, Van Steenkiste G, Jeurissen B, Van Audekerke J, Naeyaert M, et al. Exploring sex differences in the adult zebra finch brain: In vivo diffusion tensor imaging and ex vivo super-resolution track density imaging. Neuroimage. 2017;146:789–803. doi: 10.1016/j.neuroimage.2016.09.067. [DOI] [PubMed] [Google Scholar]
- 20.Farquharson S, Tournier JD, Calamante F, Mandelstam S, Burgess R, Schneider ME, et al. Periventricular nodular heterotopia: detection of abnormal microanatomic fiber structures with whole-brain diffusion mr imaging tractography. Radiology. 2016;281:896–906. doi: 10.1148/radiol.2016150852. [DOI] [PubMed] [Google Scholar]
- 21.Ziegler E, Rouillard M, Andre E, Coolen T, Stender J, Balteau E, et al. Mapping track density changes in nigrostriatal and extranigral pathways in Parkinson’s disease. Neuroimage. 2014;99:498–508. doi: 10.1016/j.neuroimage.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barajas RF, Jr, Hess CP, Phillips JJ, Von Morze CJ, Yu JP, Chang SM, et al. Super-resolution track density imaging of glioblastoma: histopathologic correlation. AJNR. 2013;34:1319–1325. doi: 10.3174/ajnr.A3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, et al. Genome of the Chinese tree shrew. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 24.Liu FG, Miyamoto MM, Freire NP, Ong PQ, Tennant MR, Young TS, et al. Molecular and morphological supertrees for eutherian (placental) mammals. Science. 2001;291:1786–1789. doi: 10.1126/science.1056346. [DOI] [PubMed] [Google Scholar]
- 25.Peng Y, Ye Z, Zou R, Wang Y, Tian B, Ma Y, et al. Biology of Chinese Tree Shrews. Kunming: Yunnan Science and Technology Press; 1991. [Google Scholar]
- 26.Remple MS, Reed JL, Stepniewska I, Lyon DC, Kaas JH. The organization of frontoparietal cortex in the tree shrew (tupaia belangeri): II. Connectional evidence for a frontal-posterior parietal network. J Comp Neurol. 2007;501:121–149. doi: 10.1002/cne.21226. [DOI] [PubMed] [Google Scholar]
- 27.Rice MW, Roberts RC, Melendez-Ferro M, Perez-Costas E. Neurochemical characterization of the tree shrew dorsal striatum. Front Neuroanat. 2011;5:53. doi: 10.3389/fnana.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong P, Kaas JH. Architectonic subdivisions of neocortex in the tree shrew (tupaia belangeri) Anat Rec (Hoboken) 2009;292:994–1027. doi: 10.1002/ar.20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao J, Yang EB, Su JJ, Li Y, Chow P. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J Med Primatol. 2003;32:123–130. doi: 10.1034/j.1600-0684.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 30.Jobling AI, Wan R, Gentle A, Bui BV, McBrien NA. Retinal and choroidal TGF-β in the tree shrew model of myopia: isoform expression, activation and effects on function. Exp Eye Res. 2009;88:458–466. doi: 10.1016/j.exer.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Amedo AO, Norton TT. Visual guidance of recovery from lens-induced myopia in tree shrews (Tupaia glis belangeri) Ophthalmic Physiol Opt. 2012;32:89–99. doi: 10.1111/j.1475-1313.2011.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs E. Social stress in tree shrews as an animal model of depression: An example of a behavioral model of a CNS disorder. CNS Spectrums. 2005;10:182–190. doi: 10.1017/S1092852900010038. [DOI] [PubMed] [Google Scholar]
- 33.Zambello E, Fuchs E, Abumaria N, Rygula R, Domenici E, Caberlotto L. Chronic psychosocial stress alters NPY system: different effects in rat and tree shrew. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:122–130. doi: 10.1016/j.pnpbp.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Pawlik M, Fuchs E, Walker LC, Levy E. Primate-like amyloid-β sequence but no cerebral amyloidosis in aged tree shrews. Neurobiol Aging. 1999;20:47–51. doi: 10.1016/S0197-4580(99)00017-2. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita A, Fuchs E, Taira M, Hayashi M. Amyloid beta (Aβ) protein- and amyloid precursor protein (APP)-immunoreactive structures in the brains of aged tree shrews. Curr Aging Sci. 2010;3:230–238. doi: 10.2174/1874609811003030230. [DOI] [PubMed] [Google Scholar]
- 36.Ma KL, Gao JH, Huang ZQ, Zhang Y, Kuang DX, Jiang QF, et al. Motor function in MPTP-treated tree shrews (tupaia belangeri chinensis) Neurochem Res. 2013;38:1935–1940. doi: 10.1007/s11064-013-1099-8. [DOI] [PubMed] [Google Scholar]
- 37.Shen F, Duan Y, Jin S, Sui N. Varied behavioral responses induced by morphine in the tree shrew: a possible model for human opiate addiction. Front Behav Neurosci. 2014;8:333. doi: 10.3389/fnbeh.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benveniste H, Einstein G, Kim KR, Hulette C, Johnson GA. Detection of neuritic plaques in Alzheimer’s disease by magnetic resonance microscopy. Proc Natl Acad Sci U S A. 1999;96:14079–14084. doi: 10.1073/pnas.96.24.14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 40.Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int J Imag Syst Tech. 2012;22:53–66. doi: 10.1002/ima.22005. [DOI] [Google Scholar]
- 41.Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med. 1999;42:526–540. doi: 10.1002/(SICI)1522-2594(199909)42:3<526::AID-MRM15>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 42.Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage. 2007;35:1459–1472. doi: 10.1016/j.neuroimage.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- 44.Chomsung RD, Wei H, Day-Brown JD, Petry HM, Bickford ME. Synaptic organization of connections between the temporal cortex and pulvinar nucleus of the tree shrew. Cereb Cortex. 2010;20:997–1011. doi: 10.1093/cercor/bhp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaas JH, Hall WC, Killackey H, Diamond IT. Visual cortex of the tree shrew (Tupaia glis): architectonic subdivisions and representations of the visual field. Brain Res. 1972;42:491–496. doi: 10.1016/0006-8993(72)90548-3. [DOI] [PubMed] [Google Scholar]
- 46.Cajal SR. Histologie Du Systeme Nerveux De L’Homme Et Des Vertebretes. Paris: A. Maloine; 1911. [Google Scholar]
- 47.Keuker JI, Rochford CD, Witter MP, Fuchs E. A cytoarchitectonic study of the hippocampal formation of the tree shrew (Tupaia belangeri) J Chem Neuroanat. 2003;26:1–15. doi: 10.1016/S0891-0618(03)00030-9. [DOI] [PubMed] [Google Scholar]
- 48.Gould E, McEwen BS, Tanapat P, Galea LAM, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17:2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzpatrick D. The functional organization of local circuits in visual cortex: insights from the study of tree shrew striate cortex. Cereb Cortex. 1996;6:329–341. doi: 10.1093/cercor/6.3.329. [DOI] [PubMed] [Google Scholar]
- 50.Nieuwenhuys R. The myeloarchitectonic studies on the human cerebral cortex of the Vogt-Vogt school, and their significance for the interpretation of functional neuroimaging data. Brain Struct Funct. 2013;218:303–352. doi: 10.1007/s00429-012-0460-z. [DOI] [PubMed] [Google Scholar]
- 51.Peters A, Sethares C. Myelinated axons and the pyramidal cell modules in monkey primary visual cortex. J Comp Neurol. 1996;365:232–255. doi: 10.1002/(SICI)1096-9861(19960205)365:2<232::AID-CNE3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 52.Balaram P, Young NA, Kaas JH. Histological features of layers and sublayers in cortical visual areas V1 and V2 of chimpanzees, macaque monkeys, and humans. Eye Brain. 2014;2014:5–18. doi: 10.2147/EB.S51814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blackwell ML, Farrar CT, Fischl B, Rosen BR. Target-specific contrast agents for magnetic resonance microscopy. Neuroimage. 2009;46:382–393. doi: 10.1016/j.neuroimage.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbier EL, Marrett S, Danek A, Vortmeyer A, van Gelderen P, Duyn J, et al. Imaging cortical anatomy by high-resolution MR at 3.0T: Detection of the stripe of Gennari in visual area 17. Magn Reson Med. 2002;48:735–738. doi: 10.1002/mrm.10255. [DOI] [PubMed] [Google Scholar]
- 55.Trampel R, Ott DV, Turner R. Do the congenitally blind have a stria of Gennari? First intracortical insights in vivo. Cereb Cortex. 2011;21:2075–2081. doi: 10.1093/cercor/bhq282. [DOI] [PubMed] [Google Scholar]
- 56.Chen G, Wang F, Gore JC, Roe AW. Identification of cortical lamination in awake monkeys by high resolution magnetic resonance imaging. Neuroimage. 2012;59:3441–3449. doi: 10.1016/j.neuroimage.2011.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leuze CW, Anwander A, Bazin PL, Dhital B, Stuber C, Reimann K, et al. Layer-specific intracortical connectivity revealed with diffusion MRI. Cereb Cortex. 2014;24:328–339. doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rockland KS, Lund JS. Intrinsic laminar lattice connections in primate visual cortex. J Comp Neurol. 1983;216:303–318. doi: 10.1002/cne.902160307. [DOI] [PubMed] [Google Scholar]
- 59.Demyanenko GP, Schachner M, Anton E, Schmid R, Feng G, Sanes J, et al. Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron. 2004;44:423–437. doi: 10.1016/j.neuron.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 60.Andersen P, Morris R, Amaral D, Bliss T, O’Keefe J. The Hippocampus Book. New York: Oxford University Press Inc; 2007. [Google Scholar]
- 61.Wu D, Zhang J. In vivo mapping of macroscopic neuronal projections in the mouse hippocampus using high-resolution diffusion MRI. Neuroimage. 2016;125:84–93. doi: 10.1016/j.neuroimage.2015.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith RE, Tournier JD, Calamante F, Connelly ASIFT. Spherical-deconvolution informed filtering of tractograms. Neuroimage. 2013;67:298–312. doi: 10.1016/j.neuroimage.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 63.Calamante F. Track-weighted imaging methods: extracting information from a streamlines tractogram. MAGMA. 2017;30:317–335. doi: 10.1007/s10334-017-0608-1. [DOI] [PubMed] [Google Scholar]
- 64.Richards K, Calamante F, Tournier JD, Kurniawan ND, Sadeghian F, Retchford AR, et al. Mapping somatosensory connectivity in adult mice using diffusion MRI tractography and super-resolution track density imaging. Neuroimage. 2014;102:381–392. doi: 10.1016/j.neuroimage.2014.07.048. [DOI] [PubMed] [Google Scholar]
- 65.Tournier JD, Calamante F, Connelly A. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed. 2013;26:1775–1786. doi: 10.1002/nbm.3017. [DOI] [PubMed] [Google Scholar]
- 66.Willats L, Raffelt D, Smith RE, Tournier JD, Connelly A, Calamante F. Quantification of track-weighted imaging (TWI): characterisation of within-subject reproducibility and between-subject variability. Neuroimage. 2014;87:18–31. doi: 10.1016/j.neuroimage.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 67.Calamante F, Smith RE, Tournier JD, Raffelt D, Connelly A. Quantification of voxel-wise total fibre density: Investigating the problems associated with track-count mapping. Neuroimage. 2015;117:284–293. doi: 10.1016/j.neuroimage.2015.05.070. [DOI] [PubMed] [Google Scholar]