Abstract
A previous study has indicated that Krüppel-like factor 7 (KLF7), a transcription factor that stimulates Schwann cell (SC) proliferation and axonal regeneration after peripheral nerve injury, is a promising therapeutic transcription factor in nerve injury. We aimed to identify whether inhibition of microRNA-146b (miR-146b) affected SC proliferation, migration, and myelinated axon regeneration following sciatic nerve injury by regulating its direct target KLF7. SCs were transfected with miRNA lentivirus, miRNA inhibitor lentivirus, or KLF7 siRNA lentivirus in vitro. The expression of miR146b and KLF7, as well as SC proliferation and migration, were subsequently evaluated. In vivo, an acellular nerve allograft (ANA) followed by injection of GFP control vector or a lentiviral vector encoding an miR-146b inhibitor was used to assess the repair potential in a model of sciatic nerve gap. miR-146b directly targeted KLF7 by binding to the 3′-UTR, suppressing KLF7. Up-regulation of miR-146b and KLF7 knockdown significantly reduced the proliferation and migration of SCs, whereas silencing miR-146b resulted in increased proliferation and migration. KLF7 protein was localized in SCs in which miR-146b was expressed in vivo. Similarly, 4 weeks after the ANA, anti-miR-146b increased KLF7 and its target gene nerve growth factor cascade, promoting axonal outgrowth. Closer analysis revealed improved nerve conduction and sciatic function index score, and enhanced expression of neurofilaments, P0 (anti-peripheral myelin), and myelinated axon regeneration. Our findings provide new insight into the regulation of KLF7 by miR-146b during peripheral nerve regeneration and suggest a potential therapeutic strategy for peripheral nerve injury.
Electronic supplementary material
The online version of this article (10.1007/s12264-018-0206-x) contains supplementary material, which is available to authorized users.
Keywords: miR-146b, KLF7, Schwann cells, Sciatic nerve injury
Introduction
Schwann cells (SCs) are the major peripheral nerve glial cells that mediate myelination, and are critical for proper nerve conduction in the peripheral nervous system. Although much is known about the regulatory transcriptional networks which confer oligodendrocyte and SC identity, the post-transcriptional regulation of these networks is not well understood. MicroRNAs (miRNAs) are an emerging mechanism by which genes can be targeted. Small, non-coding RNAs may contribute to gene silencing through translational inhibition or degradation, which may be a critical component of SC specification and function. To date, some studies have identified miRNAs 219 and 338 as critical for oligodendrogenesis through the targeted inhibition of negative regulators [1]. One of these, miR-338, has been further implicated in the positive regulation of Sox10, an important factor in the transition from SC precursors to myelinating SCs [2]. Further evidence on the essential nature of miRNAs in SC specification can be found across studies in which the expression of Dicer, a critical processing factor of miRNAs, is inhibited. In SCs particularly, loss of Dicer has been linked to delayed development and, importantly, compromises the expression of myelinating genes [3–5].
It has been reported that miRNAs regulate the signaling pathways of cellular responses to nerve injury [6, 7]. In the peripheral nerve injury response, various SC miRNAs are dynamically regulated. Several miRNAs display patterns similar to myelination gene networks during injury: they are initially inhibited and subsequently upregulated [8]. Injury-induced SC reprogramming is regulated transcriptionally by mechanisms that include transcription factors and miRNAs, which mediate the reprogramming of myelin and non-myelin SCs to myelin cells [9]. In models of sciatic nerve injury, various correlates of SC phenotypic modulation have been identified, including the miR-221/222 cluster and miR-182 [10, 11]. These and other miRNAs (miR-1, miR-132, and miR-9) have been reported to regulate post-injury proliferation and migration [12–14]. In addition, miR-340 has been implicated in debris clearance and axonal regeneration through targeting tissue plasminogen activator following sciatic nerve damage [15]. Collectively, the involvement of SC regulatory cues in the post-injury response is widely interwoven with miRNA post-transcriptional mechanisms.
An important transcriptional factor, Kruppel like factor 7 (KLF7), has been described as a regulator of axon outgrowth [16], likely through the stimulation of nerve growth factor (NGF) and its receptors Trk (tyrosine kinases) A and B, and the neuronal growth cone plasticity protein GAP43 [17–21]. In CNS and peripheral nerve injury, KLF7 upregulation promotes growth, axonal regeneration, and sprouting in injured nerves [18, 22]. In a recent study, overexpressing KLF7 in SCs during sciatic nerve injury improved SC survival, proliferation, and myelin production, as well as general myelinated fiber regeneration and function [23]. These findings collectively suggest that KLF7 and its targets play crucial roles in conferring therapeutic properties on SCs and axonal regeneration during the injury response. Despite the evidence describing the prominent role of miRNAs in SC specification and axonal regeneration, the targeting of KLF7 by miRNAs in SCs has not been investigated.
Previously, endogenous miR-146b [24], one of the most abundant miRNAs in neural tissue, was reported to directly target KLF7. MicroR-146b plays an essential role in neuronal/glial differentiation and inhibits glioma migration. Importantly, in mature neurons, miR-146b also guides axonal growth [25–28]. Previous studies and search engines such as TargetScan have identified that KLF7 is targeted by endogenous miR-146b and that KLF7 expression opposes that of miR-146b [24]. KLF7 itself has demonstrated a capacity for axonal regeneration and nerve repair [18, 23]. Together, miR-146b may negatively regulate KLF7 expression as well as its therapeutic role in peripheral nerve regeneration. Therefore, miRNA-targeting techniques could be used as an alternative therapeutic approach to the exogenous application of KLF7, which has been previously reported [18, 22, 23, 29].
In this study, we set out to determine whether KLF7 is directly targeted by miR-146b and whether inhibition of miR-146b affects SC proliferation, migration, and myelinated axon regeneration after peripheral nerve injury.
Materials and Methods
Animal
Eighteen Sprague-Dawley (SD) rats (9 males, 9 females, 180–220 g) and 72 Wistar rats (36 males, 36 females, 180–220 g) were obtained from the Experimental Animal Center of China Medical University (Certification No. SCXK Liao 2003-0009). All animal experimental protocols were reviewed and approved by the Animal Care and Use Committee of China Medical University. The animals were placed in a temperature- and humidity-controlled room under a 12-h/12-h light/dark cycle with free access to standard rat chow and water.
Study Groups and Surgical Procedures In Vivo
All 18 adult SD rats received sciatic nerve transection as described previously [30]. A 10-mm segment of the distal sciatic nerve was resected and used as a donor nerve for acellular nerve allografts (ANAs). Each donor provided two segments and they were prepared for ANAs [31]. Briefly, the two ends were fixed and incubated with Tris-HCl buffer containing protease inhibitors (0.5 μg/mL leupeptin, 0.6 g/mL pepstatin A, and 0.1 μg/mL aprotinin) for 4 days at 4 °C. Next, RNase A (1 U/mL) and DNase I (5 ng/mL) were added to digest the segments in 3% Triton X-100 Tris-HCl buffer (pH 7.4). The segments were washed several times, and these ANAs were immersed in phosphate-buffered saline (PBS) containing antibiotic and stored at 4 °C. Donor rats were sacrificed by cervical dislocation. A total of 3 μL Lenti-anti-miR-146b virus or a control Lenti-GFP virus (6.5 × 109 viral particles/mL) was injected into the ANA using a micro-injector and Hamilton syringe, incubated for 30 min, and the ANA was used for in vivo experiments [15].
Male Wistar rats were used as graft recipients, and were randomly separated into three groups. Group I: Autograft group (n = 18), the excised nerve segment was rotated 180° and orthotopically transplanted; Group II: ANA+Con group, ANA infected with control Lenti-GFP virus (n = 18); Group III: ANA+anti-miR-146b group, ANA infected with Lenti-anti-miR-146b virus (n = 18). The procedures have been described previously [32]. Briefly, rats were anesthetized via an intraperitoneal (i.p.) injection of a ketamine (87 mg/kg)/xylazine (12 mg/kg) cocktail and the right sciatic nerve was exposed. A segment (7 mm long) was excised under a surgical microscope, immediately followed by implantation of a prepared ANA to bridge the nerve gap. Then, the implant was sutured to the proximal and distal nerve stumps, and muscles and skin were closed. Pain-related behavior was monitored throughout the procedure. Four weeks later, the proximal stump of the sciatic nerve (10 mm) was harvested.
The miR-146b expression in the proximal nerve stump following sciatic nerve injury was assessed by qRT-PCR as described previously [33]. Eighteen adult Wistar rats were randomly divided into six groups: normal group, 1 day group, 1 week group, 2 week group, 3 week group, 4 week group. The right sciatic nerve was exposed and a segment was then resected at the site just proximal to the division of tibial and common peroneal nerves. The proximal stumps of the sciatic nerves (5 mm) were collected 1 day, and 1, 2, 3, and 4 weeks later.
Primary SC Culture
Primary SCs were harvested from the sciatic nerves of adult SD rats as previously described [23]. The nerves were excised and digested by type I collagenase (625 U/mL; Sigma Aldrich, St Louis, MO) and 0.25% trypsin for 60 min at 37 °C. Cell culture medium with forskolin (2 μmol/L; Sigma Aldrich), 10% FBS, and 10 ng/mL fibroblast growth factor 2 (FGF-2; Sigma Aldrich) was supplemented with cytosine arabinoside (10−5 μmol/L) to remove fibroblasts.
The final purity of cultured SCs was 98%, as determined by rabbit anti-S100 and mouse anti-GFAP immunostaining. SC cultures were passaged no more than three times before conducting experiments.
Virus Production and Transfer to SCs
We packaged lentiviruses that contained the vectors Lenti rno-mir-146b lentivirus (mr45059, pLenti-III-mir; ABM Inc. Richmond, BC, Canada), blank control (m004, pLenti-III-mir-Blank; ABM Inc.), inhibitor rno-miR-146b Lentivector (mr35103, pLenti-III-miR-Off; ABM Inc.), anti-blank control (m004, pLenti-III-mir-Off-Blank; ABM Inc.), Klf7 siRNA1 lentivirus (rat) (iV053669a, piLenti-siRNA; ABM Inc.), Klf7 siRNA2 lentivirus (rat) (iV053669b, piLenti-siRNA; ABM Inc.), and scrambled siRNA GFP lentivirus (LVP015-G, piLenti-siRNA-GFP; ABM Inc,) (Table S1). We then used Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA) to infect the primary cultured SCs with these lentiviruses (final concentration, 6.5 × 109 viral particles/mL) for 12 h at a multiplicity of infection of four (150 μL/well) to generate the miR-146b, negative control (NC), anti-miR146b, anti-NC, KLF7 siRNA1, KLF7 siRNA2, and siRNA NC groups,
SC Proliferation Assay
SCs were seeded at 3 × 105 cells/mL on 96-well plates coated with poly-L-lysine for 36 h. 5-ethynyl-2-deoxyuridine (EdU) was applied 4 h before cell fixation. Next, EdU immunostaining was performed using the Cell-Light™ EdU DNA Cell Proliferation Kit (Ribobio, USA). To determine the SC proliferation level, five fields with equal areas were randomly selected. The EdU labeling index was calculated as the number of EdU-labeled nuclei divided by the number of DAPI-labeled nuclei. The assays were repeated three times.
SC Migration Assay
Migration of SCs was assessed using Transwell chambers (Costar, Cambridge, MA) [34]. Each membrane was coated with 10 mg/mL fibronectin. Next, A 100 μL sample of DMEM containing resuspended SCs (106 cells/mL) was transferred to the top of each chamber for migration into the lower chambers. Cells adhering to the bottom surface of each membrane were subsequently stained with 0.1% crystal violet and counted. Assays were performed three times.
Luciferase Reporter Assay
The 3′-UTR sequence of the KLF7 gene was amplified from genomic DNA and subcloned into the region downstream of the stop codon of the luciferase gene in the luciferase reporter vector. PCR amplification of the 3′-UTR sequence of KLF7 generated different p-Luc-UTR luciferase reporter vectors. The sequencing confirmed the sequences of wild-type and mutant 3′-UTRs. HEK293T cells were next seeded in 96-well plates at 3–4 × 105 cells/well, and transfected with a mixture of 30 ng p-Luc-UTR, 5 μL mirR-146b lentivirus, and 5 ng Renilla. After 48 h incubation, firefly and Renilla luciferase activity was measured (Promega, Madison, WI).
DRG Explant Cultures
To evaluate the effect of anti-miR-146b on neurite outgrowth, DRGs were transfected with Lenti-anti-miR-146b or Lenti-anti-NC and then seeded onto Aclar coverslips [33]. Three days later, the DRGs were fixed and immunostained with mouse anti-β-III-tubulin (a neuronal marker) (1:200; Sigma Aldrich). The average neurite length was measured using a Neurolucida system (MicroBrightField, Williston, VT) and statistically compared [35].
Fluorescent in situ Hybridization (FISH) and Immunofluorescence Staining
MiR-146b in situ hybridization and immunofluorescence staining for S100 (labels SCs) were performed as previously described [36]. One week after injury, the proximal sciatic nerve stump was removed and cut into 15 μm sections. For FISH, a double-digoxigenin-labeled miR-146b detection probe (Exiqon, Vedbaek, Denmark) was used. The procedure comprised the following steps: 30% H2O2 with pure methanol at a 1:50 ratio was mixed and dropped onto each section, which was incubated for 30 min at room temperature (RT) and washed thrice with distilled water. Then 3% citric acid was added to the sections to expose mRNA for 2 min at RT, and were then washed thrice with PBS and once with distilled water, post-fixed with 1% paraformaldehyde/0.1 mol/L PBS at RT for 10 min, and washed again. This was followed by pre-hybridization using hybridization buffer (50% formamide, 500 μg/mL yeast tRNA, 1×Denhardt’s solution, 0.75 mol/L NaCl, 0.075 mol/L Na citrate, and diethyl pyrocarbonate-treated water) for 2–4 h at 37 °C in a humidified box. Then the sections were hybridized with the digoxin-labeled miR-146b probe (5′-AGCCTATGGAATTCAGTTCTCA-3′) overnight at 42 °C. After stringent washes (2×, 0.5×, and 0.2× standard saline citrate for 5 min at 37 °C, sections were blocked with blocking buffer for 30 min at 37 °C, and incubated with biotin-conjugated mouse anti-digoxin antibody (1:200, Sigma Aldrich) for 60 min at 37 °C. After washes, the sections were incubated with SABC-CY3 (SABC-CY3 kit, Boster, Wuhan, China) for 30 min at 37 °C for amplification. To identify the co-expression of S100 and miR-146b, the sections under FISH were further incubated overnight with rabbit anti-S100 (1:200; Sigma Aldrich) at 4 °C. Then, the sections were washed four times with PBS, and incubated for 1–2 h with anti-rabbit IgG (FITC) (1:200; Jackson Immuno Research, West Grove, PA). The sections were again washed four times with PBS and visualized using a fluorescence microscope (Olympus Optical Co, Tokyo, Japan).
Immunohistochemistry
The nerve graft (~7 mm long) was harvested at the proximal site, fixed in 4% paraformaldehyde for 3 days, and transferred to sucrose phosphate buffer (30% w/v, pH 7.4) for a day until longitudinal sections. (25 μm thick) were cut. Tissues were stained with rabbit anti-peripheral myelin (P0, 1:200; Sigma Aldrich), mouse anti-neurofilament (NF 200, 1:200; Sigma Aldrich), mouse anti-KLF7 (1:500; Novus Biologicals, Littleton, CO), or rabbit anti-S100 (1:200; Sigma Aldrich). The immunoreactive signals were visualized by goat anti-rabbit IgG (FITC) or goat anti-mouse IgG (TRITC) (1:200; Jackson Immuno Research, West Grove, PA). Images were captured by the MetaMorph/DP10/BX41 analytical imaging system to analyze the integrated optical density of positive immunological reactions.
Histopathological Analysis
Regeneration of myelinated nerve, axon diameter, and myelin sheath thickness were evaluated by transmission electron microscopy [37]. Briefly, the middle of the ANA (4–6 mm long) at the proximal site was fixed, dehydrated, and embedded in plastic. Semi-thin transverse sections were stained with 1% toluidine blue for light microscopy. High-power fields were randomly selected to detect regenerated axons, axon diameter, number of myelinated axons, and the myelin sheath thickness from all groups using an analytical imaging system (MetaMorph, Universal Imaging Corporation, Dowington, PA; DP10/Bx41; Olympus Corp, Tokyo, Japan). Systematic random sampling and two-dimensional dissector rules method were applied to circumvent sampling bias due to fiber location or discrepancies in fiber size.
qRT-PCR
A prime-Script RT reagent kit (TaKaRa, Dalian, China) was used to synthesize reverse-transcribed complementary DNA and PCR was performed using the SYBR Premix Ex Taq system (TaKaRa). To detect miRNA, mature miR146b and KLF7 were reverse-transcribed, quantified with a TaqMan probe, and normalized with RNU6B mature miRNA using TaqMan miRNA assays (Applied Biosystems, Foster City, CA). The sequences of the miR-146b primers were 5′-UGAGAACUGAAUUCCAUAGGCUGU-3′ and 5′-ACUCUUGACUUAAGGUAUCCGACA-3′. The KLF7 primers used were 5′-TTTCCTGGCAGTCATCTGCAC-3′ and 5′-GGGTCTGTTTGTTTGTCAGTCTGTC-3′. The comparative 2−△Ct method was used to calculate the relative expression levels. All reactions were run in triplicate.
Western Blot
Protein was extracted from cultured cells or the proximal stumps of nerve grafts. Protein samples (20 μg) were electrophoresed and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). The blots were incubated overnight with the primary antibodies mouse anti-KLF7 (1:500; Novus Biologicals), mouse anti-NGF (1:500; Sigma Aldrich), mouse anti-TrkA (1:1000; Sigma Aldrich), or mouse anti-TrkB (1:1000; Sigma Aldrich) followed by incubation with an HRP-conjugated goat anti-mouse secondary antibody for 1 h at room temperature (1:5000). The enhanced chemiluminescence-plus detection system (GE Healthcare, Little Chalfont, UK) was used to visualize the blots. GAPDH was used as an internal control.
Analysis of Sciatic Function Index (SFI)
Functional regeneration of the sciatic nerve was evaluated based on the SFI [38]. A technician blinded to the experimental grouping evaluated sciatic nerve function every week after surgery. In hindlimb footprint analysis, the distances between the third toe and heel (PL), first and fifth toes (TS), and second and fourth toes (ITS) were measured on the experimental side (EPL, ETS, and EITS, respectively) and the contralateral normal side (NPL, NTS, and NITS). The SFI was calculated as follows: SFI = − 38.3 × (EPL−NPL) / NPL + 109.5 × (ETS−NTS) / NTS + 13.3 × (EITS−NITS) / NITS−8.8. An SFI value around 0 indicated normal function and a value around − 100 indicated total dysfunction (n = 8/group).
Measurements of Axonal Conduction
Transcranial magnetic motor-evoked potentials were used to assess nerve conduction [39]. Action potentials were elicited by activation of subcortical structures, descended in the ventral spinal cord, synapsed onto motoneuron pools, and finally output signals were recorded from both gastrocnemius muscles.
Muscle Wet Weight Ratio and Motor Endplate Analysis
At 28 days post-injury, anterior tibial (TA) muscles on the injured and uninjured sides were collected and weighed to determine the wet weight ratio (wet weight on the injured side/wet weight on the uninjured side). The muscles were then fixed overnight in 4% paraformaldehyde, transferred to sucrose phosphate buffer, rinsed in distilled water, blocked into proximal and distal segments, flash-frozen in 2-methylbutane, and longitudinally sectioned. The Roots-Karnovsky method was used to assess the density of motor endplate acetylcholinesterase staining. A grid (1 mm × 1 mm) was randomly placed on the muscle section (1 sample field per section, 5 sections per animal). The number of muscle fibers and endplates in this grid was counted to estimate the number of motor endplates per muscle fiber.
Statistical Analysis
SPSS software (version 13.0; SPSS, Chicago, IL) was used for statistical analysis. The values are expressed as the mean with the corresponding standard deviation. Sample sizes were calculated to determine the minimum number of animals or assays required. All reported groups were larger than the minimum calculated sample size.
The Kolmogorov-Smirnov test was used to distinguish between parametric or nonparametric data. A two-tailed Student’s t tests or one-way ANOVA, and Tukey’s post hoc test were used to analyze all parametric values. The Wilcoxon Mann-Whitney test, or the Kruskal-Wallis and Dunnett’s method was used to analyze nonparametric data. A P value of < 0.05 was considered statistically significant.
Results
Expression Profiling of miR-146b in the Proximal Nerve Stump Following Sciatic Nerve Injury
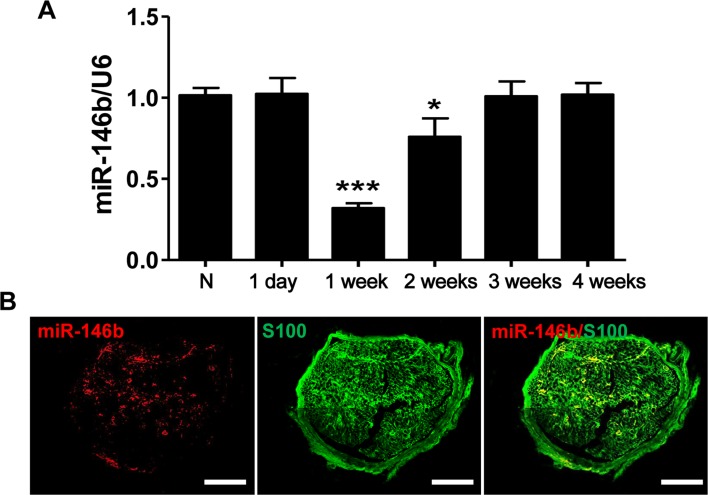
To confirm the time-dependent differential expression of miR-146b, we applied qRT-PCR to determine the expression of miR-146b mRNA in proximal nerve stumps harvested at designated time points after injury. Eighteen adult Wistar rats were randomly assigned to six times, 3 rats in each. The miR-146b expression in the stump was lower at 1 and 2 weeks post-injury than in normal, uninjured controls, and returned to baseline levels by ~3 weeks (F5, 12 = 37.73, P < 0.001) (Fig. 1A). Moreover, in situ hybridization combined with immunostaining showed that miR-146b was co-localized with S100, a marker for SCs, in the proximal stump 1 week after injury (Fig. 1B).
Fig. 1.
Expression of miR-146b in the proximal stump following sciatic nerve injury. A qRT-PCR analysis of miR-146b mRNA expression in normal (N) nerve and in the proximal stump at 1 day and 1, 2, 3, and 4 weeks after transection (n = 3 for each time point). B Representative images of in situ hybridization of miR-146b (red) and immunofluorescence staining for S100 (green, labels SCs) showing that miR-146b co-localized with SCs in the proximal stump 1 week after injury (scale bars, 100 μm).
Efficacy of miR-146b and Anti-miR146b Lentiviral Transfer to SCs In Vitro
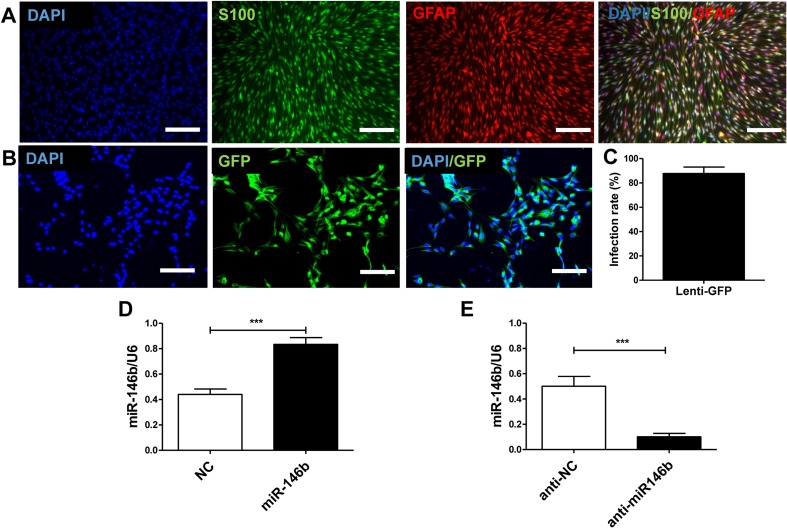
To identify SCs, cells were stained with S-100 and glial fibrillary acidic protein (GFAP). The cytoplasm of the positively-stained SC cells was visualized by double-labeling with both (Fig. 2A). The morphology of the positively-labeled cells was consistent with that of SCs observed under an inverted microscope.
Fig. 2.
Efficacy of miR-146b and anti-miR146b lentiviral transfer to SCs in vitro. A Images of SC immunohistochemical staining with S-100 (green) and GFAP (red) with nuclear counterstaining (DAPI, blue). Merged images show that SCs were double-labeled. Scale bars, 100 μm. B Images of lenti-GFP-infected SCs in vitro immunohistochemically stained with GFP (green) and nuclear staining (DAPI, blue). Scale bars, 50 μm. C Quantitation of GFP and DAPI co-labeling to assess the lenti-GFP infection rate. D qRT-PCR analysis of relative miR-146b mRNA expression in cultured SCs in the lenti-miR-146b and NC groups at 3 days in vitro (n = 6). E qRT-PCR analysis of relative miR-146b mRNA expression in cultured SCs in the lenti-anti-miR-146b and anti-NC groups at 3 days in vitro (n = 6). Error bars, standard deviation; ***P < 0.001, Student’s t test.
We then assessed the Lenti-GFP transfection of SCs in vitro (Fig. 2B). The Lenti-GFP transfection of SCs was successful, yielding a transfection efficiency of 87.71 ± 5.34% (Fig. 2C). Next, SCs were transfected with lenti-miR-146b (miR-146b), lenti-miR-146b inhibitor (anti-miR-146b), lentivirus control (NC), or inhibitor lentivirus control (anti-NC), and the transfection efficiency was assessed using qRT-PCR. We found that, compared to NCs, the mRNA expression of miR-146b was markedly increased by lenti-miR-146 infection (t10 = 5.745, P < 0.001; Fig. 2D). In contrast, lenti-anti-miR-146b transfection dramatically decreased the miR-146b mRNA expression compared to the anti-NC control (t10 = 4.938, P < 0.001) (Fig. 2E). These results showed that miR-146b and anti-miR146b lentiviruses can transfect SCs effectively and regulate miR-146b expression.
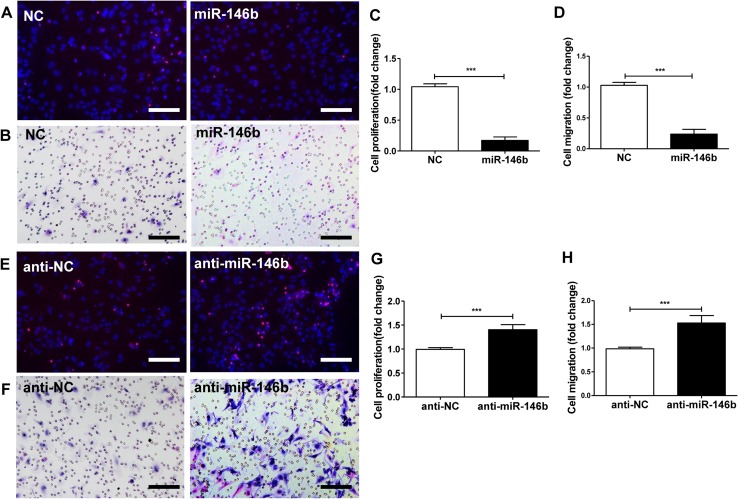
To evaluate the phenotypic effects of miR-146b manipulation on SCs, proliferation was assessed using the EdU-based proliferation assay (Fig. 3A). The proliferation rate of SCs transfected with lenti-miR-146b was lower than that of NC controls (t10 = 23.57, P < 0.001; Fig. 3C). Transwell in vitro migration assays further showed that over-expression of miR-146b decreased the migration of SCs compared with NC controls (t10 = 17.19, P < 0.001; Fig. 3B, D).
Fig. 3.
Effects of miR-146b on SC proliferation and migration. Primary SCs were transfected with lenti-miR-146b (miR-146b), lentivirus control (NC), lenti-miR-146b inhibitor (anti-miR-146b), or inhibitor lentivirus control (anti-NC) and evaluated at 3 days in vitro. A Images of all cells, stained with DAPI (blue), and proliferating SCs stained with EdU (red). B Images of migration of transfected SCs in the cell migration assay. C, D Proliferation rates and migration in the NC and miR-146b groups. E Proliferation rates of SCs transfected with anti-NC and anti-miR-146b. F Migration of SCs transfected with anti-NC and anti-miR-146b. G, H Proliferation and migration of SCs transfected with anti-NC and anti-miR-146b. ***P < 0.001, Student’s t test; scale bars, 50 μm.
On the contrary, inhibition of lenti-miR-146b with anti-miR-146b increased the proliferation rate of SCs compared to anti-NC controls (t10 = 7.534, P < 0.001; Fig. 3E, G). Similarly, the migration of SCs transfected with anti-miR-146b lentivirus was higher than that of controls (t10 = 6.808, P < 0.001; Fig. 3F, H). These data suggest that miR-146b down-regulation is sufficient to promote the proliferation and migration of SCs.
KLF7 is a Direct Target of miR-146b
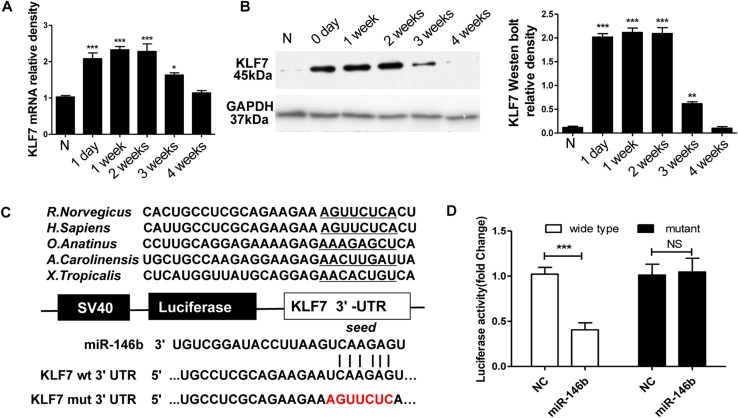
Because miRNAs function through the suppression of target genes, it is important to identify the targets of miR-146b, so we explored these using the search engine TargetScan. Although many candidates were identified in silico, the target gene KLF7 was selected for further investigation due to its demonstrated role in nerve regeneration-associated processes including SC proliferation and axonal regeneration [22, 23]. The TargetScan results notably corroborated earlier reports proposing that KLF7 is a direct target of endogenous miR-146b [24].
The KLF7 mRNA and protein levels were endogenously low in normal sciatic nerve. However, mRNA and protein expression of KLF7 in the proximal nerve segment was induced 1 day post-graft, peaked at 1–2 weeks, and returned to basal levels by ~4 weeks (mRNA: F5,30 = 22.75, P < 0.001; protein: F5,30 = 179.2, P < 0.001; Fig. 4A, B). Interestingly, the comparative expression profile of miR146b was negatively correlated with that of KLF7, providing further evidence that miR-146b negatively regulates KLF7 in injured peripheral nerve.
Fig. 4.
KLF7 is a direct target of miR-146b. A qPCR analysis of KLF7 mRNA expression in proximal sciatic nerve segments after nerve injury. B Western blot analysis of the changes in KLF7 protein expression across proximal sciatic nerve segments after nerve injury. The histogram represents the relative density of the KLF7 protein bands normalized to GAPDH at different time-points (n = 6; error bars, standard deviation; *P < 0.05, **P < 0.01, ***P < 0.001 versus normal control, one-way ANOVA and Tukey’s post hoc test. C Design of a miR-146b reporter vector containing the SV40 promoter, luciferase gene, and KLF7 3′-UTR. Sequences of miR-146b predicted binding sites in the KLF7 UTR, and the mutated KLF7 UTR. D Relative luciferase activity after the p-Luc-UTR vectors (3′-UTR of wild-type and mutant KLF7) were co-transfected into HEK293T cells with lenti-miR-146b (miR-146b) or lentivirus control (NC). Renilla luciferase vector was used as an internal control. ***P < 0.001, NS, not significant, Student’s t test.
Using miRNA target prediction software, we found that miR-146b could target the 3′-UTR of KLF7 loci. We then conducted a luciferase assay to investigate whether KLF7 is regulated by miR-146b via direct binding to its 3′-UTR. To accomplish this, wild-type and mutant 3′-UTRs of KLF7 were constructed and inserted into the downstream region of the luciferase reporter gene (Fig. 4C).
Lenti-miR-146b and p-Luc-UTR constructs were next co-transfected into HEK 293T cells to analyze the relative luciferase activity. When the UTR contained the wild-type binding site, miR-146b induced a decline in the relative luciferase activity from 100% to 40.5 ± 7.9% (t10 = 11.31, P < 0.001). In contrast, when the UTR contained the mutant binding site, miR-146b did not induce a decrease in the relative luciferase activity of KLF7 (Fig. 4D). These findings implied that binding of miR-146b to the 3′-UTR of KLF7 is sequence-specific.
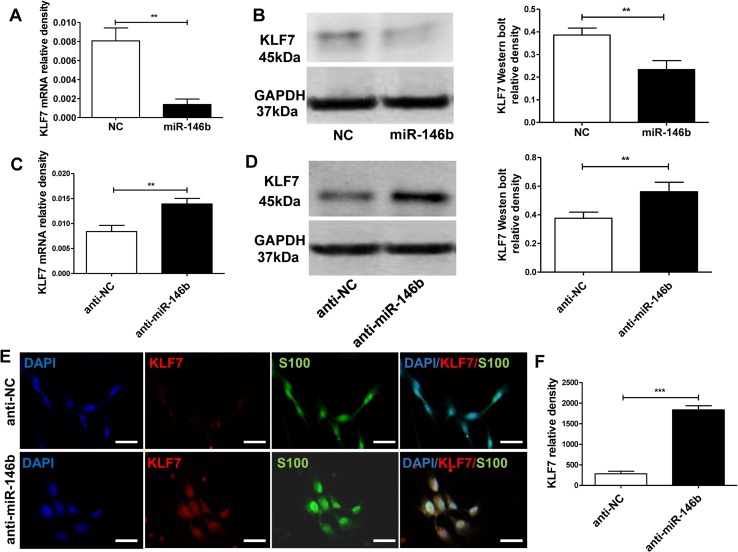
miR-146b Inhibits KLF7 Expression
To verify that miR-146b affects KLF7 expression, lenti-miR-146b or lenti-anti-miR-146b was transfected into cultured SCs. The qRT-PCR and western blot analysis showed that overexpression of miR-146b suppressed the mRNA and protein expression of KLF7 (mRNA: t10 = 7.90, P < 0.01; protein: t10 = 6.082, P < 0.001; Fig. 5A, B). Meanwhile, we found that KLF7 mRNA and protein expression was up-regulated in SCs by transfection with lenti-anti-miR-146b (anti-miR-146b), compared to the inhibitor control (anti-NC) (mRNA: t10 = 5.745, P < 0.01; protein: t10 = 7.382, P < 0.001; Fig. 5C, D).
Fig. 5.
miR-146b suppresses KLF7 expression. A mRNA expression of KLF7 in SCs transfected with lenti-miR-146b (miR-146b) or lentivirus control (NC). B KLF7 protein expression in the miR-146 and NC groups and quantitation as relative density. C KLF7 mRNA expression of SCs transfected with the lenti-miR-146b inhibitor (anti-miR-146b) or lenti-inhibitor control (anti-NC). D KLF7 protein expression in the anti-miR-146b and anti-NC groups expressed as relative density. E Images of immunohistochemical staining for KLF7 (red) and S100 (green) with nuclear counterstaining (DAPI, blue) in SCs transfected with lenti-miR-146b inhibitor or lenti-inhibitor control (scale bars, 100 μm). F Relative density of KLF7 in cultured SCs with anti-miR-146b and the anti-NC group at 3 days in vitro (**P < 0.01, ***P < 0.001, Student’s t tests).
Next, we performed immunohistochemical staining of transfected SCs with markers for SCs (S100) and KLF7 expression (Fig. 5E). We found that the expression of KLF7 was higher in the anti-miR-146b group than in the anti-NC group (t10 = 12.83, P < 0.001; Fig. 5F). These results further support the negative regulation of KLF7 by miR-146b.
Knockdown of KLF7 Compliments the Effects of miR-146b on SC Proliferation and Migration
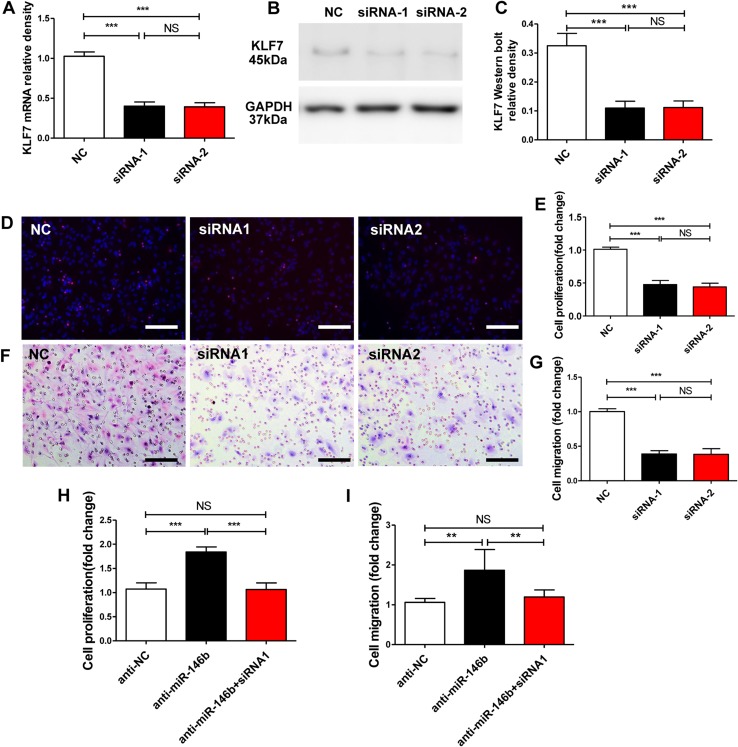
To determine the function of KLF7, two specific siRNAs against it, siRNA-1 and siRNA-2, were synthesized and delivered to SCs. KLF7 mRNA and protein expression in the exposed SCs was down-regulated by both siRNA-1 and siRNA-2 compared with siRNA control-treated SCs (mRNA: F2, 15 = 291.1, P < 0.001; protein: F2, 15 = 94.51, P < 0.001; Fig. 6A–C).
Fig. 6.
Knockdown of KLF7 compliments the effect of miR-146b on SC proliferation and migration. A Levels of KLF7 mRNA in SCs transfected with lenti-KLF7 siRNA-1 (siRNA-1), lenti-KLF7 siRNA-2 (siRNA-2), and lenti-siRNA control (NC). B, C KLF7 protein levels in SCs transfected with siRNA-1, siRNA-2, or the NC. D, E Proliferation of SCs transfected with KLF7 siRNA-1 (siRNA-1), KLF7 siRNA-2 (siRNA-2), and siRNA control (NC). F, G Migration of SCs transfected with siRNA-1, siRNA-2, and NC. H Proliferation rates of SCs after transfection with miR-146b inhibitors (anti-miR-146b) with or without KLF7-siRNA-1 (siRNA-1). I Migration of SCs after transfection with miR-146b inhibitor (anti-miR-146b) with or without KLF7 siRNA-1 (siRNA-1). Error bars, standard deviation; **P < 0.01, ***P < 0.001, NS, not significant; one-way ANOVA and Tukey’s post hoc test.
SC proliferation assays showed that both siRNA-1 and siRNA-2 inhibited SC proliferation compared with siRNA control (NC) (F2, 15 = 150.1, P < 0.001; Fig. 6D, E). Furthermore, we investigated the effect of the siRNAs targeting KLF7 on the migration of SCs. Notably, in vitro migration assays showed that both siRNA-1 and siRNA-2 inhibited SC migration compared with the NC group (F2, 15 = 141.4, P < 0.001; Fig. 6F, G). Thus, our results indicated that KLF7 inhibition parallels the SC phenotype of miR-146b transfection and further provided evidence that miR-146b acts as a functional mediator of KLF7 in the regulation of SC phenotypes.
Because the KLF7 protein is strongly upregulated after sciatic nerve transection, it is likely to promote SC proliferation and migration as part of the intrinsic injury response, while miR-146b may post-transcriptionally regulate the expression of KLF7 by directly binding to its 3′-UTR. We thus hypothesized that downregulation of KLF7 is directly mediated by miR-146b, inhibiting SC proliferation and migration. To test this hypothesis, we downregulated miR-146b by transfection with an miR-146b inhibitor and dually by using siRNA-1 against KLF7. We measured a significant increase in cell proliferation and migration in groups transfected with the miR-146b inhibitors, and, in contrast, a remarkable decrease in proliferation and migration in groups co-transfected with miR-146b inhibitors and siRNA-1 (proliferation: F2, 15 = 5.745, P < 0.01; migration: F2, 15 = 7.382, P < 0.001; Fig. 6H, I). Notably, inhibited expression of KLF7 decreased the proliferation and migration induced by the miR-146b inhibitors. Taken together, these findings show that the inhibitory action of KLF7 abrogated the promotion of anti-miR-146b-induced cell proliferation and migration, suggesting that KLF7 is a functional mediator of miR-146b in SC proliferation and migration.
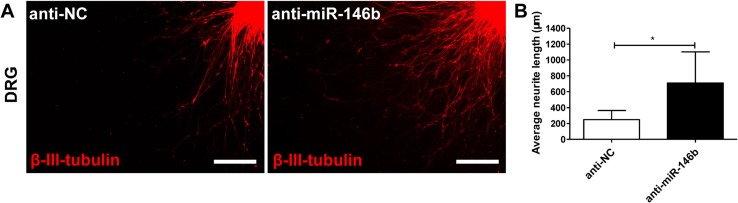
Effects of miR-146b Inhibitors on Neurite Outgrowth
To determine the effect of miR-146b on neurite outgrowth, we evaluated DRG transfection with inhibitor control (anti-NC) or an miR-146b inhibitor (anti-miR-146b). The inhibitor promoted neurite length in DRG explants (Fig. 7A) (709.4 ± 392.1 μm in the anti-miR-146b group, 246.1 ± 116.0 μm in the anti-NC group, t24 = 2.978, P < 0.05; Fig. 7B).
Fig. 7.
Effects of anti-miR-146b on neurite outgrowth. DRGs were transfected with lenti-anti-miR-146b or lenti-anti-NC and then seeded onto Aclar coverslips. A Representative immunostaining images of DRG explants in each group. Axons were labeled with β-III-tubulin (red). Scale bars, 100 μm. B Average neurite length (error bars, standard deviation; *P < 0.05 versus anti-NC group, Student’s t test).
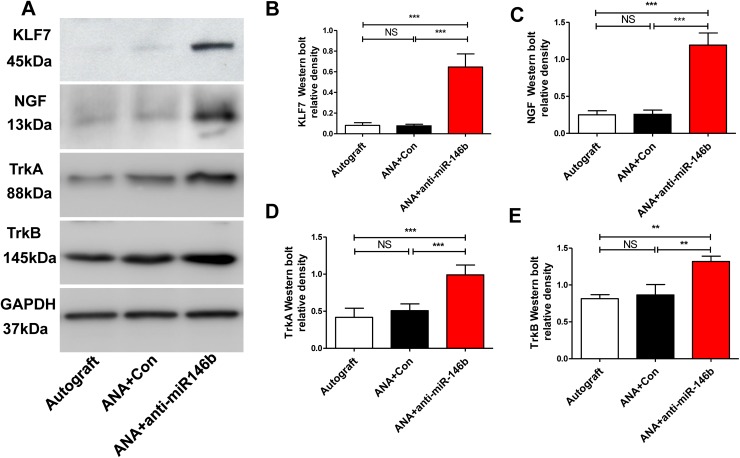
Inhibition of miR-146b Increased the Expression of KLF7, NGF, TrkA, and TrkB in Regenerating Nerves in ANAs
To verify the successful transplantation of anti-miR-146b virus, we assessed the expression of KLF7 and its targets using Western blot protein analysis of proximal ANA tissue (~7 mm long) harvested 28 days post-injury (Fig. 7A). In the Autograft and ANA+Con groups, KLF7 expression was low; however, animals receiving anti-miR-146b had higher KLF7 protein expression (F2, 15 = 56.85, P < 0.001; Fig. 8B). To test the effect of anti-miR-146b on nerve regeneration, we measured the expression of the KLF7 target genes NGF, TrkA, and TrkB in ANA tissues (Fig. 8A). The results demonstrated that ANA+anti-miR-146b promoted NGF, TrkA and TrkB expression in the graft compared to the Autograft and ANA+Con groups. (NGF: F2, 15 = 26.54, P < 0.01; TrkA: F2, 15 = 28.0, P < 0.01; TrkB: F2, 15 = 24.23, P < 0.001; Fig. 8C–E). Further, there was no difference in the NGF, TrkA, and TrkB expression between the Autograft and ANA+Con groups. These results demonstrate that anti-miR-146b promotes the expression of KLF7 and induces an increase in the expression of target proteins such as NGF, TrkA, and TrkB in regenerating ANA tissues.
Fig. 8.
Anti-miR-146b significantly increased the expression of KLF7, NGF, TrkA, and TrkB in regenerating axons in ANAs. A Representative Western blots of KLF7, NGF, TrkA, and TrkB expression in ANA tissues harvested 28 days post-injury in the Autograft, ANA+ Con, and ANA + anti-miR-146b groups. B–E The graphs show the relative protein density of KLF7 (B), NGF (C), TrkA (D), and TrkB (E) (n = 6 rats/group; error bars, standard deviation; **P < 0.01, ***P < 0.001, NS, not significant, one-way ANOVA with Tukey’s post hoc test).
Inhibition of miR-146b Promoted Myelinated Fiber Regeneration in ANAs
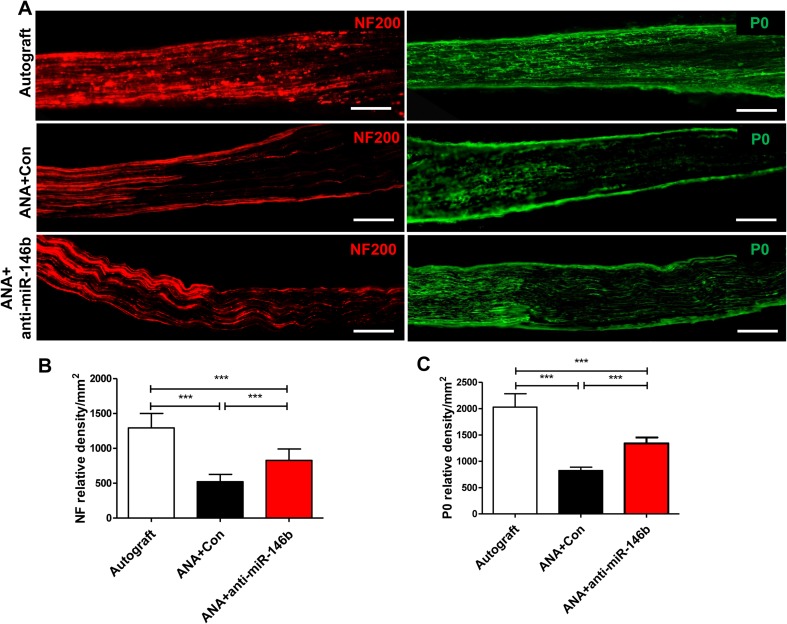
To further assess axonal regeneration in ANAs, they were sectioned longitudinally and NF-positive axons (a pan-neuronal marker) and P0 (a marker of peripheral myelin) were visualized by immunohistochemistry. In the Autograft group, NF and P0 immunoreactivity extended the full length of the ANA. In contrast, the ANA+anti-miR-146b and ANA+Con groups had shorter NF- and P0-positive axons across the ANA (Fig. 9A); moreover, the expression of NF and P0 in the ANA+anti-miR-146b group was higher than that in the ANA+Con and ANA groups (NF: F2, 15 = 56.03, P < 0.001; P0: F2, 15 = 94.0, P < 0.001; Fig. 9B, C). The findings showed that anti-miR-146b promotes the myelination of regenerated axons in ANAs.
Fig. 9.
Anti-miR-146b increased the expression of neurofilaments (NF) and P0 protein (myelination) in ANAs. A Representative immunohistochemical staining for NF (red) and P0 (green) in longitudinal sections from the Autograft, ANA+Con, and ANA+anti-miR-146b groups (n = 6 rats/group; scale bars, 100 μm. B, C Quantitative analyses of NF (B) and P0 (C) protein expression in immunohistochemical sections (error bars, standard deviation; ***P < 0.001, one-way ANOVA with Tukey’s post hoc test).
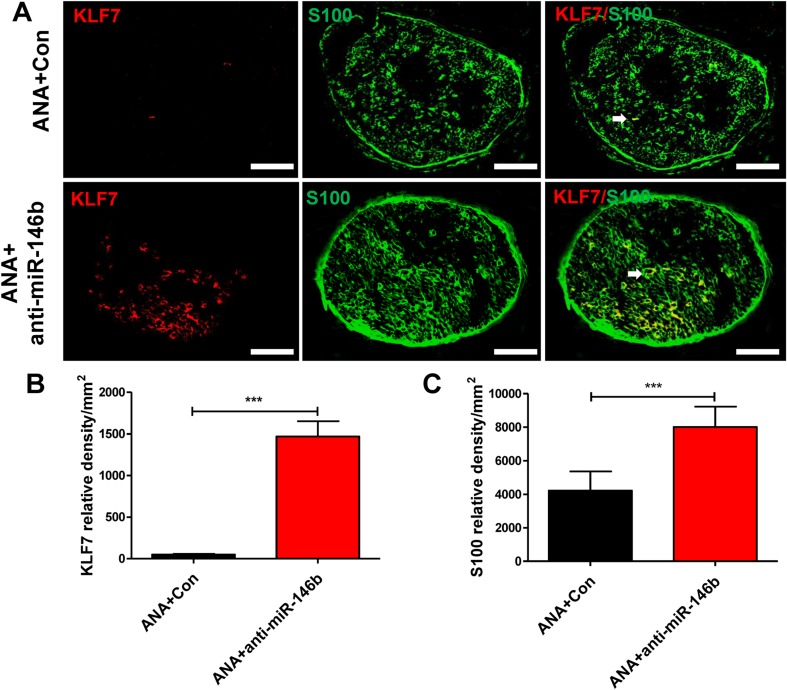
To reveal the detailed expression profiles and to further identify the cell types expressing KLF7 in the sciatic nerve in vivo, cells positive for KLF7 and S100 (a marker of SCs) in the middle of the ANA were visualized by immunohistochemistry. The results showed that KLF7 co-localized with SCs (Fig. 10A); further, the expression of KLF7 and S-100 in the ANA+anti-miR-146b group was higher than in the ANA+Con group (KLF7: t24 = 7.78, P < 0.001; S100: t24 = 5.55, P < 0.001; Fig. 10B, C). Therefore, KLF7 protein is localized in the SCs in which miR-146b is expressed, and anti-miR-146b promotes the expression of KLF7 and SC regeneration in ANAs.
Fig. 10.
Anti-miR-146b promoted the expression of KLF7 and SC regeneration in ANAs. A Representative images of immunohistochemical staining for KLF7 (red) and S100 (green) in sections from the ANA+Con and ANA+anti-miR-146b groups. Arrows show that KLF7 was co-localized with S100 (a marker of SCs) (n = 6 rats/group; scale bars, 100 μm.). B, C Quantitative analyses of KLF7 (B) and S100 (C) protein expression in immunohistochemical sections (error bars, standard deviation; ***P < 0.001 versus ANA+Con, Student’s t test.
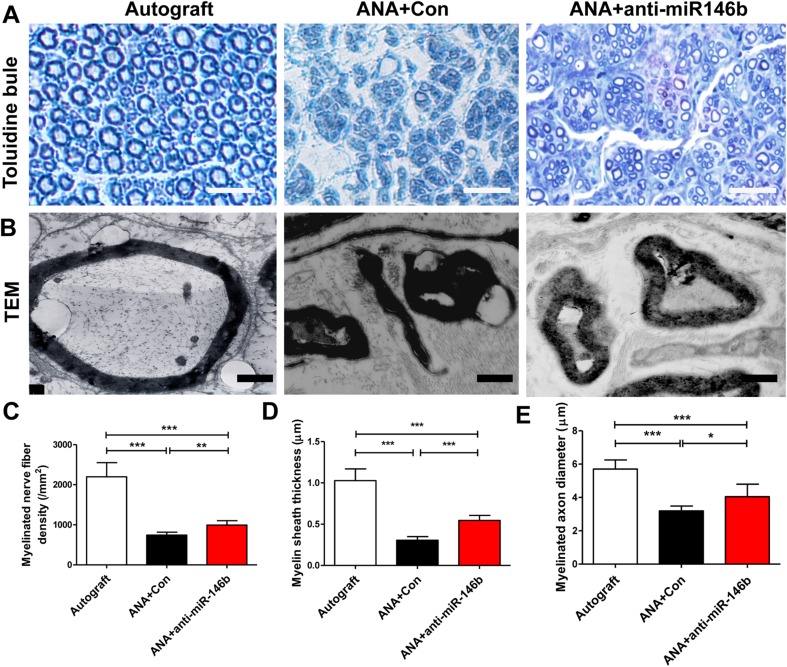
Toluidine blue staining further showed that the numbers of myelinated axons in the ANA+Con and ANA+anti-miR-146b groups were lower than in the Autograft group. Notably, the number of myelinated fibers in the ANA+anti-miR-146b group was significantly higher than that in the ANA+Con group (F2, 15 = 77.37, P < 0.001; Fig. 11A–C). Myelin sheath thickness and axon diameter demonstrated similar patterns of change across the three groups (myelin sheath thickness: F2, 15 = 92.76, P < 0.001; axon diameter: F2, 15 = 31.36, P < 0.001; Fig. 11A, B, D, E). These results demonstrate that the myelination of regenerated axons in the ANA+anti-miR-146b group was superior to that in the ANA+Con group. Collectively, these results show that ANA+anti-miR-146b treatment promotes myelinated axon regeneration in ANAs.
Fig. 11.
Anti-miR-146b promoted myelinated axon regeneration in ANAs. A, B Representative toluidine blue staining (A; scale bars, 50 μm) and transmission electron microscope images (B) of myelinated axon regeneration in the ANAs of the three groups (n = 6). C–E Quantitative analyses of the number of myelinated axons (C), myelin sheath thickness (D), and axon diameter (E) in the middle of the ANAs (error bars, standard deviation; *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA with Tukey’s post hoc test).
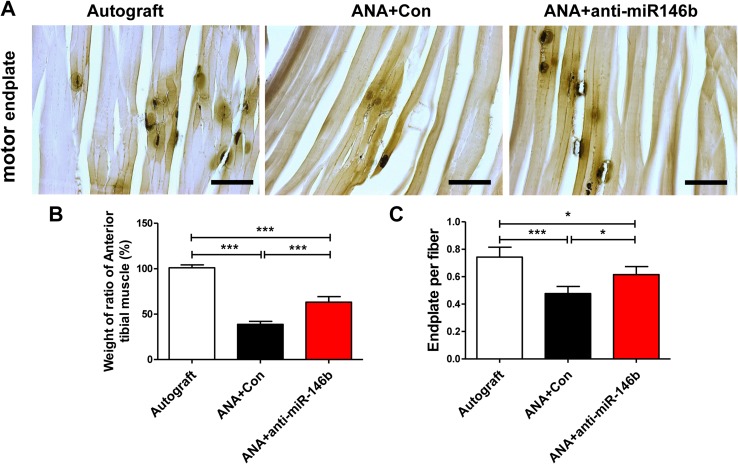
Inhibition of miR-146b Promoted Target Muscle Motor Endplate Regeneration and Wet Weight Ratio
The TA muscle began to atrophy after sciatic nerve injury. We found that the weight ratios of the TA muscles were lower in the ANA+Con and ANA+anti-miR-146b groups than in the Autograft group. The mean ratio was particularly higher in the ANA+anti-miR-146b group than in the ANA+Con group (F2, 15 = 293.4, P < 0.001; Fig. 12B).
Fig. 12.
Muscle wet weight and motor endplate analysis. A Representative images of motor endplates in longitudinal sections of the tibialis anterior (TA) on the injured side in each group. B Histogram of the wet weight ratios in each group. C Quantitative analysis of the number of motor endplates per TA muscle fiber (n = 6; error bars denote standard deviation; *P < 0.05, ***P < 0.001, one-way ANOVA with Tukey’s post hoc test).
Cholinesterase staining of the motor endplate revealed re-innervation of the target muscle cells by regenerated nerve fibers. To assess the effect of anti-miR-146b on recovery of motor function, the motor endplate density in the TA was assessed (Fig. 12A). We found that the number of motor endplates was lower in the ANA+Con and ANA+anti-miR-146b groups than in the Autograft group. Motor endplate numbers were particularly higher in the ANA+anti-miR-146b group than in the ANA+Con group (F2, 15 = 18.67, P < 0.001; Fig. 12C). The above results show that anti-miR-146b has a positive effect on the re-innervation of denervated TA muscle. In addition, they show that inhibition of miR-146b may have a protective effect on the atrophy of target muscles after injury.
Inhibition of miR-146b Significantly Enhanced Recovery of Motor Function
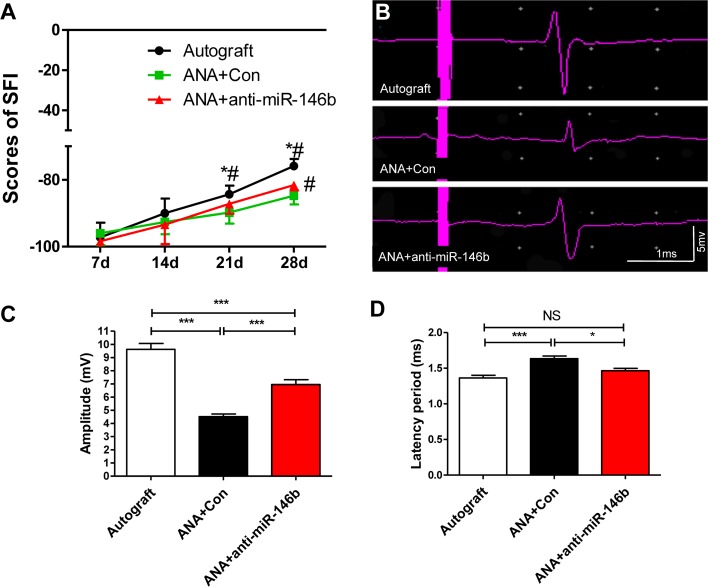
Finally, evaluation of sciatic nerve function as SFI was carried out. The results showed a significant increase in the SFI in the ANA+anti-miR-146b group compared to the ANA+Con group 28 days after ANA grafting (F2, 21 = 22.90, P < 0.001; Fig. 13A).
Fig. 13.
KLF7-SCs significantly enhanced recovery of motor function of regenerating axons in ANAs. A Quantitative analyses of the sciatic function index (SFI). B Representative electrophysiological recordings. C and D Quantitation of amplitude (mV) (C) and latency (ms) (D) recorded in ANAs from each group (error bars, standard deviation; *P < 0.05, ***P < 0.001, one-way ANOVA with Tukey’s post hoc test; n = 8/group).
We next measured the functional recovery of the ANAs 4 weeks after sciatic nerve injury (Fig. 13B) using electrophysiology. The ANA+anti-miR-146b and Autograft groups exhibited higher amplitudes than the ANA+Con group (F2, 21 = 50.87, P < 0.001; Fig. 13C). Moreover, decreased latencies were found in the Autograft and ANA+anti-miR-146b groups compared to the ANA+Con group (F2, 21 = 14.28, P < 0.001; Fig. 13D). Overall, ANA+anti-miR-146b showed a better electrophysiological response than the ANA+Con group. Taken together, these data demonstrate that inhibition of miR-146b had the most beneficial effect in the context of the recovery of motor function.
Discussion
Peripheral nerve injury initiates complex processes requiring the precise expression and quiescence of numerous gene targets for intrinsic repair. Recently, many investigators have turned to the study of miRNAs as regulators of the response to injury. Particularly interesting is their demonstrated role in the specification of SCs, the major cell type required for the myelination of regenerating axons. In previous work, the transcription factor KLF7 has shown strong post-injury induction and may play an important role in the activation of growth signals which mediate axonal regeneration [18, 22, 31]. Moreover, in SCs, KLF7 has been shown to regulate SC proliferation during peripheral nerve injury. A crucial report has proposed KLF7 as a target of miR-146b [24]. Despite this, neither the dynamics of miR-146b and KLF7 nor the therapeutic potential has been explored in peripheral nerve injury. In this study, we aimed to determine whether KLF7 is a direct target of miR-146b and whether inhibition of this miRNA plays a role in SC specification, axonal regeneration, and overall functional recovery after sciatic nerve injury.
First, we profiled the expression of miR-146b in the proximal nerve stump and found that expression was decreased by 1–2 weeks post-injury. In situ hybridization further revealed that this downregulation mainly occurred in the sciatic nerve 1 week after injury. These results appear to support the time course of post-injury KLF7 upregulation (peaking 1–2 weeks post-injury), confirming a negative regulatory relationship between miR-146b and KLF7, as previously suggested [24]. Indeed, in mRNA and protein analyses of the proximal nerve segment, the comparative expression profiles confirmed a negative correlation between KLF7 and miR-146b. Further, in transfected SCs, miR-146b upregulation inhibited the expression of KLF7. Collectively, these results confirm the negative regulation of the transcription factor KLF7 by miR-146b in injured nerves and transfected SCs. Finally, target prediction software and luciferase assays demonstrated that miR-146b acts on KLF7 through the direct binding of the 3′UTR, in a sequence-specific manner. These findings elaborate on the report of Chen et al. by identifying a functionally relevant binding sequence for miR-146b and its target KLF7.
After confirming the direct binding of KLF7 and miR-146b, we evaluated the effects of manipulation of the miRNA using lentiviral transfer to SCs in vitro. Lentiviral vectors have been safely and broadly used for the delivery of transgenes directly into a variety of non-dividing cells such as SCs in vitro and in vivo. Fundamentally, Lenti-GFP infection demonstrated a high efficiency of transfer (~90%). Transfection with lenti-miR-146b increased the expression of the transcript while transfection with the lenti-miR-146b inhibitor diminished the expression as assessed by qRT-PCR. These findings support the efficacious transfection of miR-146b variants in SCs in vitro and offer a potential tool for the manipulation of the SC transcriptome across injury models. First, in culture, the phenotypic characteristics of SCs were evaluated under the transfection conditions. We found that upregulation of miR-146b significantly diminished the proliferation and migration of SCs in vitro. On the contrary, transfection with an miR-146b inhibitor rescued the ability of SCs to proliferate and migrate in culture. These findings significantly implicate miR-146b targets (including KLF7) in the regulation of critical SC processes. They complement previous findings showing that miR-1 and miR-182 upregulation negatively impacts SC proliferation and migration in peripheral nerve injury models [10, 13].
Because KLF7 had previously been shown to exhibit a regulatory action on the specification of SCs, we attempted to tease out whether the effects of miR-146b on SC proliferation and migration could be specifically ascribed to diminished KLF7. Using siRNAs against KLF7, we found that treated SCs exhibited phenotypes parallel to those of miR-146b-transfected SCs. This allowed us to at least clarify that indeed miR-146b targeting of KLF7 is sufficient to drive compromised SC specification. While KLF7 silencing significantly decreased proliferation and migration (by ~50%), miR-146b transfection did appear to compromise the SCs more severely. This leaves room for the speculation that perhaps other miR-146b targets act synergistically on SC proliferation and migration, though their investigation was beyond the scope of the present study.
As the dynamics of KLF7 and miR-146b became clear, we shifted our attention to the use of miR-146b manipulators as a potential means of antagonizing the negative effects of miR-146b during injury. Given the previous finding that miR-146b inhibitors rescued SC proliferation and migration, we next attempted to evaluate a crucial component of intrinsic nerve injury repair, axonal regeneration. Using DRG explants, we found that transfection of the miR-146b inhibitor significantly increased neurite outgrowth. Our findings in culture provide early evidence that miR-146b antagonism could serve as a therapeutic strategy for peripheral nerve injury in vivo.
Using a rat model of sciatic nerve injury, we evaluated the enhancement of standard ANAs using seeding with SCs transfected with miR-146b inhibitors and controls. First, we verified the successful transplantation with the miRNA-inhibitor by confirming the expression of KLF7 and its targets. Indeed, we found that in the ANA+anti-miR-146b group, KLF7 was increased along with its target proteins NGF, TrkA, and TrkB up to four weeks after initial grafting. This suggested that miRNA inhibition specifically diminished the endogenous miR-146b and its corresponding inhibition of KLF7 and KLF7 targets. To further determine whether the miR-146b inhibitor promoted axonal regeneration in vivo as it had in vitro, we examined longitudinal sections of the experimental and control ANAs. Immunohistochemical evaluation showed that peripheral myelin and NF-positive axons were markedly increased across the graft in the anti-miR-146b group. Further supporting the increased myelination of regenerating axons were the observations that toluidine blue-stained myelinated axons, myelin sheath thickness, and axon diameter were all notably increased by the experimental treatment compared to controls. Finally, electron microscopy demonstrated that anti-miR-146b improved the myelinated axon regeneration in the regenerating nerve. Importantly, this demonstrated for the first time that antagonism of endogenous miR-146b is a viable therapeutic strategy for the regeneration of myelinated axons in peripheral nerve injury. Also, due to the upregulation of KLF7 and its targets during treatment, it is likely that the antagonist may act on nerve regeneration (and SC characteristics) via the activation of KLF7 and its networks. Indeed, previous studies have identified KLF7 and its signaling cascades as enhancers of sciatic nerve regeneration in graft transplantation models [22, 23].
Particularly, peripheral nerve injury results in motor dysfunction and many promising neuroprotectants have fallen short in this category. One of the primary signs of motor dysfunction is the muscular atrophy that follows sciatic nerve injury. To evaluate the ability of the anti-miR-146b to alleviate muscular atrophy, we measured the weight ratios of target TA muscles across groups. We found that ANA+anti-miR-146b increased these wet weight ratios, signifying diminished atrophy. Moreover, cholinesterase staining revealed that motor endplate density was increased in the target muscle following treatment. These findings extend the functional relevance of the antagonist beyond molecular cues and demonstrate that the intervention strategy has a positive effect on the re-innervation of muscle. Interestingly, motor neuron re-innervation by anti-miR-146b-induced axonal regeneration has been similarly reported with regard to miR-124 in a hypoglossal nerve injury model. Interestingly, in that study the up-regulated targets of miR-124 included KLF6 and STAT3, genes implicated (like KLF7) in axonal growth and guidance [40].
Taking one step further, we evaluated functional recovery using traditional evaluation of motor function as the SFI. Four weeks after treatment, the ANA+anti-miR-146b group outperformed the control groups by demonstrating an increased SFI score. Unsurprisingly, the electrophysiological response of the anti-miR-146b group displayed the enhanced characteristics of increased amplitude and decreased latency. These results fall in line with improved myelination and axonal conduction of regenerated nerves. Moreover, they provide a physiological basis for the improved motor functions observed in this group compared to controls. Importantly, these results demonstrate that antagonism of the endogenous miR-146b may provide a novel therapeutic strategy for the recovery of motor function following sciatic nerve injury.
MicroRNAs have in recent years become increasingly implicated in the regulation of cellular specification and function. In SCs, a critical component of the nerve injury response and principal myelin-conferring cell, miRNAs have been demonstrated to play a role in regulating SC proliferation and migration in various nerve injury models. For example, miR-1 targeting of brain-derived neurotrophic factor, miR-182 targeting of fibroblast growth factor-9 and neurotrimin, and miR9 targeting of Cthrc1 (Collagen Triple Helix Repeat Containing 1) have been reported to have an inhibitory influence on SCs during nerve injury [10, 13, 14]. Interestingly, it has been reported that miRNAs may play a positive role in SCs during nerve repair, suppressing the action of inhibitory pathways such as the action of miR-27a on the FOXO1 (Forkhead box protein O1) pathway and the targeting of LASS2 (longevity assurance homologue 2) by miR-221/222 [11, 41].
Beyond SC proliferation and migration, various miRNAs have been shown to play roles in peripheral nerve injury and regeneration including the NGF-targeting Let-7 miRNAs [15] and the PTEN (phosphatase and tensin homolog)-targeting miR-222 [42]. MicroRNAs have also demonstrated nerve injury-related pain regulation in sensory neurons, likely through the modulation of Na+ channels [36, 43]. MiR21 and miR222 have been shown to be anti-apoptotic via suppression of TIMP3 in sciatic nerve injury [44]. Finally, the role of miRNAs in nerve regeneration has been evidenced by findings in autonomic nerve remodeling, where miR-206 regulation of SOD1 has been shown to promote nerve sprouting, including sympathetic and parasympathetic nerve regeneration [45]. Collectively, these studies elucidate the range of miRNAs and their actions which contribute to the overall intrinsic nerve injury response, indicating that other miRNAs may be found in the future as additional therapeutic miRNA targets. Moreover, these studies, like our own, illuminate the molecular pathways that may be affected in peripheral nerve injury, bringing us closer to optimal therapeutic targeting.
Currently, various strategies exist for the treatment of peripheral nerve injury. Of these, nerve grafting has shown the most promising functional recovery in pre-clinical models. Despite this, there is constant need to refine treatments and optimize recovery. Lentiviral delivery of therapeutic targets encompasses an incredible range, including miRNAs, and has recently been explored as additions to traditional grafting techniques. Here, the seeding of ANAs with transfected SCs ensured the endogenous propagation of the anti-miRNA and likely minimized the invasiveness of multiple delivery required for long-term neuroprotection. Interestingly, a recent design has begun to incorporate multi-target transfection of miRNAs in collagen nerve conduits used to bridge a transected sciatic nerve. While this study did not determine whether the combined upregulation of the two targets was superior to single-target effects, it raises the potential benefits of combinatorial miRNA targeting strategies. It is plausible that future studies will indeed optimize a treatment strategy to both upregulate and downregulate injury-compromised miRNAs appropriately, synergistically acting to optimize the intrinsic nerve injury repair capacity. Though likely, much more information is required about the miRNA and gene target dynamics, particularly in disease and injury states. One important unanswered question is whether other miR-146b targets beyond KLF7 contribute to peripheral nerve regeneration. In addition, the use of general or neuron-specific miR-146b transgenic and knockout animal models is necessary to clarify the mechanisms underlying the effects of miR-146b on nerve regeneration in future studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the staff of the Experimental Animal Center of China Medical University for providing excellent animal care. This work was supported by the National Natural Science Foundation of China (81371362, 81641125, and 81500629), the Scientific Research Foundation of Heilongjiang Province, China (LC2017040), the Science Fund of Heilongjiang Provincial Health and Family Planning Commission, China (2016357 and 2016385), and the Basic Research Operating Expenses Program of Heilongjiang Provincial Universities, China (2017- KYYWFMY-0661).
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12264-018-0206-x) contains supplementary material, which is available to authorized users.
Contributor Information
Xiao-Feng Zhu, Email: sjkx@sohu.com.
Ying Wang, Email: yingwang2016@sina.com.
References
- 1.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gokey NG, Srinivasan R, Lopez-Anido C, Krueger C, Svaren J. Developmental regulation of microRNA expression in Schwann cells. Mol Cell Biol. 2012;32:558–568. doi: 10.1128/MCB.06270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremer J, O’Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS ONE. 2010;5:e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. J Neurosci. 2010;30:7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira JA, Baumann R, Norrmen C, Somandin C, Miehe M, Jacob C, et al. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Muheremu A, Zhang M, Gong K, Huang C, Ji Y, et al. MicroRNA-338 and microRNA-21 co-transfection for the treatment of rat sciatic nerve injury. Neurol Sci. 2016;37:883–890. doi: 10.1007/s10072-016-2500-6. [DOI] [PubMed] [Google Scholar]
- 7.Jia X, Wang F, Han Y, Geng X, Li M, Shi Y, Lu L, Chen Y. miR-137 and miR-491 negatively regulate dopamine transporter expression and function in neural cells. Neurosci Bull. 2016;32:512–522. doi: 10.1007/s12264-016-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594:3521–3531. doi: 10.1113/JP270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, et al. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding F, et al. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. J Cell Sci. 2012;125:2675–2683. doi: 10.1242/jcs.098996. [DOI] [PubMed] [Google Scholar]
- 12.Yao C, Shi X, Zhang Z, Zhou S, Qian T, Wang Y, et al. Hypoxia-induced upregulation of miR-132 promotes Schwann cell migration after sciatic nerve injury by targeting PRKAG3. Mol Neurobiol. 2016;53:5129–5139. doi: 10.1007/s12035-015-9449-y. [DOI] [PubMed] [Google Scholar]
- 13.Yi S, Yuan Y, Chen Q, Wang X, Gong L, Liu J, et al. Regulation of Schwann cell proliferation and migration by miR-1 targeting brain-derived neurotrophic factor after peripheral nerve injury. Sci Rep. 2016;6:29121. doi: 10.1038/srep29121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou S, Gao R, Hu W, Qian T, Wang N, Ding G, et al. MiR-9 inhibits Schwann cell migration by targeting Cthrc1 following sciatic nerve injury. J Cell Sci. 2014;127:967–976. doi: 10.1242/jcs.131672. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J, et al. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol Ther. 2015;23:423–433. doi: 10.1038/mt.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Caiazzo M, Colucci-D’Amato L, Esposito MT, Parisi S, Stifani S, Ramirez F, et al. Transcription factor KLF7 regulates differentiation of neuroectodermal and mesodermal cell lineages. Exp Cell Res. 2010;316:2365–2376. doi: 10.1016/j.yexcr.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Blackmore MG, Wang Z, Lerch JK, Motti D, Zhang YP, Shields CB, et al. Krüppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc Natl Acad Sci USA. 2012;109:7517–7522. doi: 10.1073/pnas.1120684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li WY, Li ZG, Guan LX, Deng LX. Transcriptional and epigenetic regulation in injury-mediated neuronal dendritic plasticity. Neurosci Bull. 2017;33:85–94. doi: 10.1007/s12264-016-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei L, Ma L, Nef S, Thai T, Parada LF. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development. 2001;128:1147–1158. doi: 10.1242/dev.128.7.1147. [DOI] [PubMed] [Google Scholar]
- 21.Li WY, Wang Y, Zhai FG, Sun P, Cheng YX, Deng LX, Wang ZY. AAV-KLF7 promotes descending propriospinal neuron axonal plasticity after spinal cord injury. Neural Plast. 2017;2017:1621629. doi: 10.1155/2017/1621629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Li WY, Sun P, Jin ZS, Liu GB, Deng LX, et al. Sciatic nerve regeneration in KLF7-transfected acellular nerve allografts. Neurol Res. 2016;38:242–254. doi: 10.1080/01616412.2015.1105584. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Li WY, Jia H, Zhai FG, Qu WR, Cheng YX, et al. KLF7-transfected Schwann cell graft transplantation promotes sciatic nerve regeneration. Neuroscience. 2017;340:319–332. doi: 10.1016/j.neuroscience.2016.10.069. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Dai Y-M, Ji C-B, Yang L, Shi C-M, Xu G-F, et al. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol Cell Endocrinol. 2014;393:65–74. doi: 10.1016/j.mce.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Stevanato L, Sinden JD. The effects of microRNAs on human neural stem cell differentiation in two- and three-dimensional cultures. Stem Cell Res Ther. 2014;5:49. doi: 10.1186/scrt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S, et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339:260–269. doi: 10.1016/j.canlet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Tang Y, Ling ZM, Fu R, Li YQ, Cheng X, Song FH, et al. Time-specific microRNA changes during spinal motoneuron degeneration in adult rats following unilateral brachial plexus root avulsion: ipsilateral vs. contralateral changes. BMC Neurosci. 2014;15:92. doi: 10.1186/1471-2202-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu DZ, Ander BP, Tian Y, Stamova B, Jickling GC, Davis RR, et al. Integrated analysis of mRNA and microRNA expression in mature neurons, neural progenitor cells and neuroblastoma cells. Gene. 2012;495:120–127. doi: 10.1016/j.gene.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Winsor K, Nienhaus C, Hess E, Blackmore MG. Combined chondroitinase and KLF7 expression reduce net retraction of sensory and CST axons from sites of spinal injury. Neurobiology of Disease. 2017;99:24–35. doi: 10.1016/j.nbd.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JS, Yao ZX. MicroRNAs: novel regulators of oligodendrocyte differentiation and potential therapeutic targets in demyelination-related diseases. Mol Neurobiol. 2012;45:200–212. doi: 10.1007/s12035-011-8231-z. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Jia H, Li WY, Tong XJ, Liu GB, Kang SW. Synergistic effects of bone mesenchymal stem cells and chondroitinase ABC on nerve regeneration after acellular nerve allograft in rats. Cell Mol Neurobiol. 2012;32:361–371. doi: 10.1007/s10571-011-9764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao R, Wang L, Sun J, Nie K, Jian H, Gao L, et al. MiR-204 promotes apoptosis in oxidative stress-induced rat Schwann cells by suppressing neuritin expression. FEBS Lett. 2014;588:3225–3232. doi: 10.1016/j.febslet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Huang L, Quan X, Liu Z, Ma T, Wu Y, Ge J, et al. c-Jun gene-modified Schwann cells: upregulating multiple neurotrophic factors and promoting neurite outgrowth. Tissue Eng Part A. 2015;21:1409–1421. doi: 10.1089/ten.tea.2014.0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantuano E, Inoue G, Li X, Takahashi K, Gaultier A, Gonias SL, et al. The hemopexin domain of matrix metalloproteinase-9 activates cell signaling and promotes migration of schwann cells by binding to low-density lipoprotein receptor-related protein. J Neurosci. 2008;28:11571–11582. doi: 10.1523/JNEUROSCI.3053-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Ma Z, Smith GM, Wen X, Pressman Y, Wood PM, et al. GDNF-enhanced axonal regeneration and myelination following spinal cord injury is mediated by primary effects on neurons. Glia. 2009;57:1178–1191. doi: 10.1002/glia.20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao J, Cao J, Wang J, Ren X, Su S, Li M, et al.. MicroRNA-30b regulates expression of the sodium channel Nav1.7 in nerve injury-induced neuropathic pain in the rat. Mol Pain 2016, 12. [DOI] [PMC free article] [PubMed]
- 37.Jia H, Wang Y, Tong XJ, Liu GB, Li Q, Zhang LX, et al. Sciatic nerve repair by acellular nerve xenografts implanted with BMSCs in rats xenograft combined with BMSCs. Synapse. 2012;66:256–269. doi: 10.1002/syn.21508. [DOI] [PubMed] [Google Scholar]
- 38.Kanaya F, Firrell JC, Breidenbach WC. Sciatic function index, nerve conduction tests, muscle contraction, and axon morphometry as indicators of regeneration. Plast Reconstr Surg 1996, 98: 1264–1271, discussion 1272–1264. [DOI] [PubMed]
- 39.Byers JS, Huguenard AL, Kuruppu D, Liu NK, Xu XM, Sengelaub DR. Neuroprotective effects of testosterone on motoneuron and muscle morphology following spinal cord injury. J Comp Neurol. 2012;520:2683–2696. doi: 10.1002/cne.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagata K, Hama I, Kiryu-Seo S, Kiyama H. microRNA-124 is down regulated in nerve-injured motor neurons and it potentially targets mRNAs for KLF6 and STAT3. Neuroscience. 2014;256:426–432. doi: 10.1016/j.neuroscience.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Zhao Y, Sun C, Hu W, Zhao J, Li G, et al. Chitosan Degradation Products Promote Nerve Regeneration by Stimulating Schwann Cell Proliferation via miR-27a/FOXO1 Axis. Mol Neurobiol. 2016;53:28–39. doi: 10.1007/s12035-014-8968-2. [DOI] [PubMed] [Google Scholar]
- 42.Zhou S, Shen D, Wang Y, Gong L, Tang X, Yu B, et al. microRNA-222 targeting PTEN promotes neurite outgrowth from adult dorsal root ganglion neurons following sciatic nerve transection. PLoS ONE. 2012;7:e44768. doi: 10.1371/journal.pone.0044768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai A, Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem Biophys Res Commun. 2013;435:176–181. doi: 10.1016/j.bbrc.2013.04.089. [DOI] [PubMed] [Google Scholar]
- 44.Zhou S, Zhang S, Wang Y, Yi S, Zhao L, Tang X, et al. MiR-21 and miR-222 inhibit apoptosis of adult dorsal root ganglion neurons by repressing TIMP3 following sciatic nerve injury. Neurosci Lett. 2015;586:43–49. doi: 10.1016/j.neulet.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Zheng S, Geng Y, Xue J, Wang Z, Xie X, et al. MicroRNA profiling of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS ONE. 2015;10:e0122674. doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.