Abstract
The GABAergic neurons in the parafacial zone (PZ) play an important role in sleep-wake regulation and have been identified as part of a sleep-promoting center in the brainstem, but the long-range connections mediating this function remain poorly characterized. Here, we performed whole-brain mapping of both the inputs and outputs of the GABAergic neurons in the PZ of the mouse brain. We used the modified rabies virus EnvA-ΔG-DsRed combined with a Cre/loxP gene-expression strategy to map the direct monosynaptic inputs to the GABAergic neurons in the PZ, and found that they receive inputs mainly from the hypothalamic area, zona incerta, and parasubthalamic nucleus in the hypothalamus; the substantia nigra, pars reticulata and deep mesencephalic nucleus in the midbrain; and the intermediate reticular nucleus and medial vestibular nucleus (parvocellular part) in the pons and medulla. We also mapped the axonal projections of the PZ GABAergic neurons with adeno-associated virus, and defined the reciprocal connections of the PZ GABAergic neurons with their input and output nuclei. The newly-found inputs and outputs of the PZ were also listed compared with the literature. This cell-type-specific neuronal whole-brain mapping of the PZ GABAergic neurons may reveal the circuits underlying various functions such as sleep-wake regulation.
Keywords: Parafacial zone, Parvocellular reticular formation, GABAergic neurons, Trans-synaptic tracing
Introduction
The parafacial zone (PZ) is located within the medulla oblongata, lateral and dorsal to the facial nerve [1]. It overlaps with the alpha part of the parvocellular reticular formation (PCRt).
Although significant progress has been made in clarifying the neuronal circuitry regulating behavioral states, including sleep and wakefulness, some fundamental gaps in our knowledge remain, and this is particularly true with respect to the nature and location of the circuitry regulating sleep [2–4]. GABAergic neurons are usually involved in the generation of brain waves [5]. Sleep-promoting GABAergic neurons in the PZ were first reported in 2012 [1]. Recently, the PZ has been further reported to be involved mainly in slow-wave sleep [6–8]. But the monosynaptic inputs or axonal projections of the GABAergic neurons in the PZ are still unknown.
Previous studies have revealed afferent projections to the PCRt (which overlaps with the PZ) using horseradish peroxidase and phytohaemagglutinin-L tracing in rats [9]. The PCRt is known to contain large populations of both inhibitory (expressing the vesicular GABA transporter VGAT) and excitatory (expressing the vesicular glutamate transporter VGlut2) craniofacial premotor neurons [10, 11]. Accordingly, Cre-dependent transfection of both VGAT and VGlut2 neurons in the PCRt with the retrograde pseudotyped rabies virus SADDG-GFP (EnvA) [12] results in neuronal labeling in the medial part of the central amygdaloid nucleus (CeA), and this newly-found CeA/PCRt projection induces “fictive feeding” behavior [13].
An important challenge in understanding the functions of the PZ circuits is the neuronal heterogeneity, so it is crucial to map its inputs and outputs with cell-type specificity. Thus, in the present study, the inputs and outputs of the GABAergic neurons in the PZ were investigated. Most previous studies of the long-range connections of the PCRt were not cell-specific [9, 14]. A recent study of these connections focused on specific regions connected to the PCRt, making it difficult to assess their whole-brain distribution [13]. Recent advances in virus-assisted circuit tracing [15, 16] and high-throughput imaging have greatly facilitated the whole-brain mapping of long-range connectivity in a cell-type-specific manner [17, 18]. In this study, we traced the long-range inputs and outputs of the GABAergic neurons in the PZ. Our quantitative analysis of the whole-brain distributions of inputs and outputs for the GABAergic neurons in the PZ serves as an anatomical blueprint for future studies of inter-regional pathways mediating the functions of the PZ.
Materials and Methods
Animals
Adult VGAT-Cre mice (2–4 months old) of the C57BL/6J strain and C57BL/6J mice were used in experiments. The VGAT-Cre mice were generated by Jackson Laboratory, Maine, USA. Mice were housed at 23 ± 1°C, 55% ± 5% humidity, and under a 12-h light/dark cycle (starting at 07:00) with food and water ad libitum. Animal experiments were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of Zhejiang University, and were approved by Committee of Laboratory Animal Center of Zhejiang University (ZJU201501501).
Trans-Synaptic Retrograde Tracing
AAV-CAG-DIO-TVA-EGFP, AAV-CAG-DIO-RvG, and EnvA-pseudotyped, glycoprotein (RG)-deleted, and DsRed-expressing rabies virus (RV-EnvA-ΔG-DsRed, RV) were provided by F. Xu (Wuhan, China). Two Cre-dependent AAVs were generated as helper viruses for retrograde monosynaptic tracing and were microinjected into the unilateral PZ area of VGAT-Cre mice. Two to three weeks later, RV-EnvA-ΔG-DsRed was injected into the same location.
All AAV vectors were packaged into 2/9 serotypes with tilters estimated at ~1012 genomic copies/mL. The rabies virus was packaged with tilters estimated at ~108 genomic copies/mL.
Anterograde Axon Tracing
In the axonal projection experiments we used a Cre-dependent AAV reporter construct, AAV-EF1α-DIO-EGFP, from Hanbio, Shanghai, China. All AAV vectors were packaged into 2/9 serotypes with titers estimated at ~1012 genomic copies/mL.
Surgery and Viral Injections
VGAT-Cre mice were anesthetized with sodium pentobarbital (80 mg/kg) and mounted on a stereotaxic frame (Stoelting Corp., Wood Dale, IL). After exposing the skull and drilling a small hole, a glass micropipette was placed above the PZ [antero-posterior (AP), −5.5 mm from bregma; medio-lateral (ML), 1.4 mm; dorso-ventral (DV), −4.5 mm]. Two helper AAVs (AAV-CAG-DIO-TVA-EGFP and AAV-CAG-DIO-RvG mixed at a 1:1 ratio) in 100 nL were first injected into the PZ at 0.01 μL/min using a microsyringe pump controller (WPI, Sarasota, FL). To allow diffusion of the virus, the micropipette was not retracted until 10–15 min after the end of injection. After three weeks, 200 nL RV-EnvA-ΔG-DsRed was injected into the same site. One week after spread of the rabies virus, the mice were sacrificed to collect brain sections.
Histology and Immunostaining
Mice were deeply anesthetized with sodium pentobarbital and perfused with 0.9% saline followed by 4% paraformaldehyde in 0.1 mol/L phosphate-buffered saline (PBS, pH 7.4). Brains were removed on ice and placed in 4% paraformaldehyde buffer at 4°C for 6-h post-fixation. Then the fixed brains were cryoprotected in 30% sucrose/PBS (w/v) at 4°C for two days. After that, the whole brain was embedded in optimal cutting temperature compound, and cut into 30-μm coronal sections on a freezing cryostat (Leica CM1950, Nussloch, Germany). The sections were preserved in anti-freeze solution (0.1 mol/L PBS and glycerol mixed at 1:1) at −20°C. For further imaging, the sections were stuck onto gelatin-coated slides, dried for 2–3 h at room temperature, and washed three times for 10 min in 0.01 mol/L PBS. After that, the sections were covered with DAPI for 10 min to stain nuclei, and washed three times. Finally, the stained sections were coverslipped with 90% glycerol.
Imaging and Data Analysis
Images of whole-brain sections were captured using a 10×/20× objective in an inverted confocal microscope (Olympus FV-1200, Tokyo, Japan). We used this microscope to adjust the brightness and contrast of the images, and then manually defined the outlines of nuclei based on the mouse brain atlas [19]. Also, we manually counted the number of DsRed+ neurons on image projections. Only data from the mice with precise injection sites are shown in the figures and were included in the statistics.
Results
In the present study, VGAT-Cre mice were used to target the PZ GABAergic neurons for virus-mediated circuit tracing.
Monosynaptic Inputs to the PZ GABAergic Neurons Using the Rabies Virus System
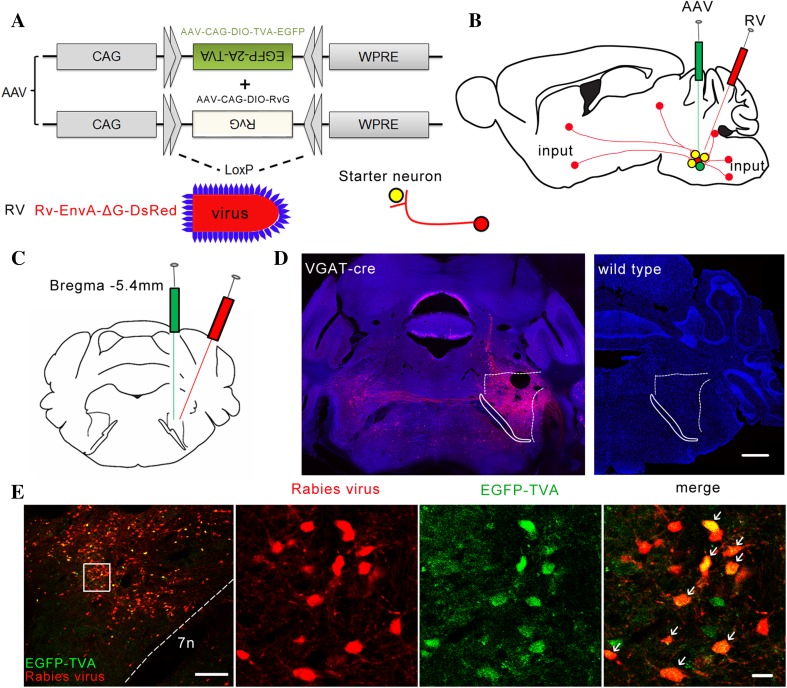
To identify the long-range inputs to the PZ GABAergic neurons, we used rabies virus-mediated trans-synaptic retrograde tracing, which has been shown to label monosynaptic inputs to selected starter cells with high specificity [20–23]. We used the modified rabies virus EnvA-ΔG-DsRed combined with a Cre/loxP gene-expression strategy (Fig. 1A). First, two Cre-dependent AAVs, AAV-CAG-DIO-TVA-EGFP and AAV-CAG-DIO-RvG, were microinjected unilaterally into the PZ of VGAT-Cre mice. Two to three weeks later, RV-EnvA-ΔG-DsRed was injected into the same location (Fig. 1B). After 1 week, starter neurons were characterized by the co-expression of RV-EnvA-ΔG-DsRed and AAV-CAG-DIO-TVA-EGFP, which was restricted to the PZ, ipsilateral to the injection site (Fig. 1E). There were also numerous neurons that were DsRed-positive but did not express EGFP in the PZ, demonstrating the presence of direct monosynaptic input from the PZ neurons to the PZ GABAergic neurons (Fig. 1E). In contrast, we did not detect any DsRed-positive neurons in wild-type mice that were investigated using the same strategy (Fig. 1D). These results demonstrated the specificity of our strategy.
Fig. 1.
Experimental procedures for retrograde monosynaptic tracing to identify direct inputs to PZ GABAergic neurons. A Design of viral vectors for RV-mediated trans-synaptic retrograde tracing. B Schematic of the PZ injection procedure for AAV-CAG-DIO-TVA-EGFP, AAV-CAG-DIO-RvG, and RV-EnvA-ΔG-DsRed in VGAT-Cre mice. C Schematic showing unilateral injection site of virus in the PZ in VGAT-Cre mice. D Representative images showing RV-labeled neurons (red) in VGAT-Cre mice but not in wild-type mice (blue, nuclei stained with DAPI). Scale bar, 550 μm. E Representative immunostaining images of the PZ from VGAT-Cre mice (left-most) showing that virus co-expression was restricted to the PZ. Scale bar, 200 μm. Starter cells (yellow, expressing both EGFP and DsRed) indicated by arrows in enlarged view of the PZ (right). Scale bar, 20 μm. 7n, facial nerve or its root.
Input Patterns of PZ GABAergic Neurons
To investigate the monosynaptic input areas to the PZ GABAergic neurons, we used coronal whole-brain sections. In VGAT-Cre mice, we injected 3 viruses into the PZ, and those labeled with DsRed were the monosynaptic inputs to the PZ GABAergic neurons. The DsRed-labeled presynaptic neurons spanned almost the entire brain, including the cortex, hypothalamus, midbrain, pons, and medulla.
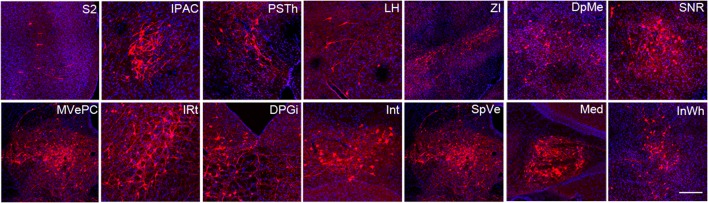
To show the whole-brain distribution of DsRed-labeled presynaptic neurons, we selected representative coronal images from the secondary somatosensory cortex (S2), interstitial nucleus of the posterior limb of the anterior commissure (IPAC), parasubthalamic nucleus (PSTh), lateral hypothalamic area (LH), zona incerta (ZI), deep mesencephalic nucleus (DpMe), substantia nigra, pars reticulata (SNR), medial vestibular nucleus, parvocellular part (MVePC), intermediate reticular nucleus (IRt), dorsal paragigantocellular nucleus (DPGi), interposed cerebellar nucleus (Int), spinal vestibular nucleus (SpVe), medial cerebellar fastigial nucleus (Med) and intermediate white layer of the superior colliculus (InWh) (Fig. 2).
Fig. 2.
Representative images of selected regions with monosynaptic inputs to PZ GABAergic neurons. RV-labeled cells identified by red dots in a series of coronal sections to show the whole-brain input pattern. Scale bar, 200 μm. DPGi, dorsal paragigantocellular nucleus; DpMe, deep mesencephalic nucleus; Int, interposed cerebellar nucleus; InWh, intermediate white layer of the superior colliculus; IPAC, interstitial nucleus of the posterior limb of the anterior commissure; IRt, intermediate reticular nucleus; LH, lateral hypothalamic area; Med, medial (fastigial) cerebellar nucleus; MVePC, medial vestibular nucleus, parvocellular part; PSTh, parasubthalamic nucleus; S2, secondary somatosensory cortex; SNR, substantia nigra, pars reticulata; SpVe, spinal vestibular nucleus; ZI, zona incerta. Regions were identified based on the mouse brain atlas [19].
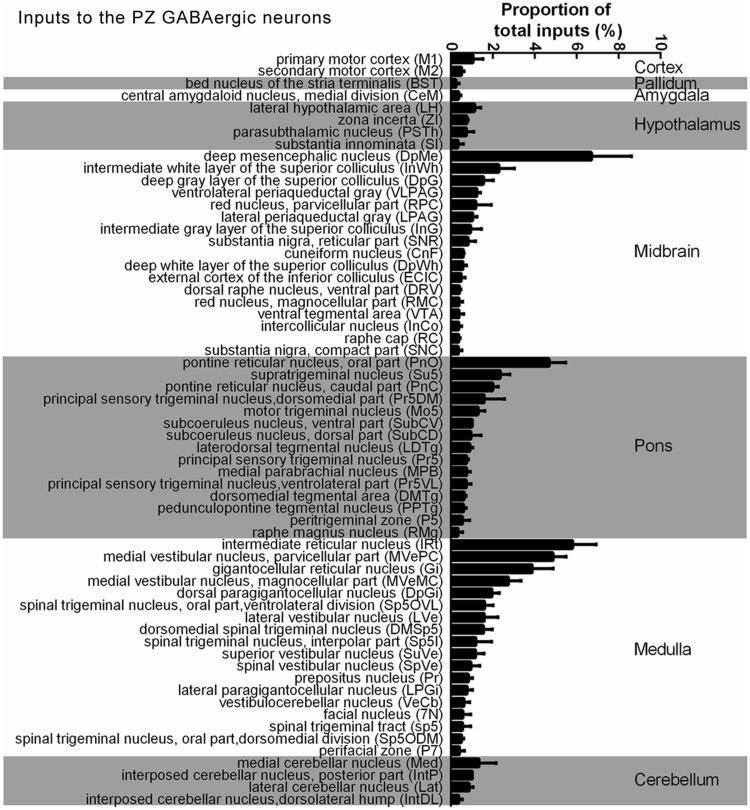
We constructed a list of the whole-brain inputs to the PZ GABAergic neurons based on the number of DsRed-labeled neurons (Fig. 3), and then normalized the number of labeled neurons in each nucleus by the total number of labeled neurons in each mouse brain. This identified 62 nuclei that contained an average of >0.2% of the total number of DsRed-labeled neurons in the whole brain. The largest number of neurons providing input to the PZ GABAergic neurons was found in the DpMe (6.70% ± 1.90%, n = 3 mice) in the midbrain (Fig. 3). The pontine reticular nucleus, oral part (PnO) (4.69% ± 0.78%, n = 3 mice) in the pons, and the IRt (5.80% ± 1.12%, n = 3 mice) in the medulla also had strong projections to the PZ GABAergic neurons (Fig. 3). In summary, this whole-brain mapping of all DsRed-labeled input neurons allowed us to identify specific regions for further research (Figs. 3 and 4).
Fig. 3.
Whole-brain distribution of inputs to PZ GABAergic neurons. Percentages of retrogradely-labeled input neurons from 62 regions in VGAT-Cre mice. Brain areas are grouped into 8 general structures (cortex, pallidum, amygdala, hypothalamus, midbrain, pons, medulla, and cerebellum).
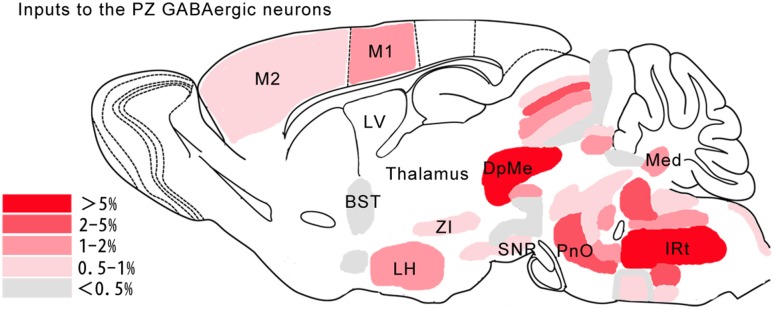
Fig. 4.
Schematic of the major presynaptic inputs to PZ GABAergic neurons. Color scale represents the percentage of total inputs. BST, bed nucleus of the stria terminalis; DpMe, deep mesencephalic nucleus; IRt, intermediate reticular nucleus; LH, lateral hypothalamic area; LV, lateral ventricle; M1, primary motor cortex; M2, secondary motor cortex; Med, medial (fastigial) cerebellar nucleus; PnO, pontine reticular nucleus, oral part; SNR, substantia nigra, pars reticulata; ZI, zona incerta.
Axonal Projection Patterns of PZ GABAergic Neurons
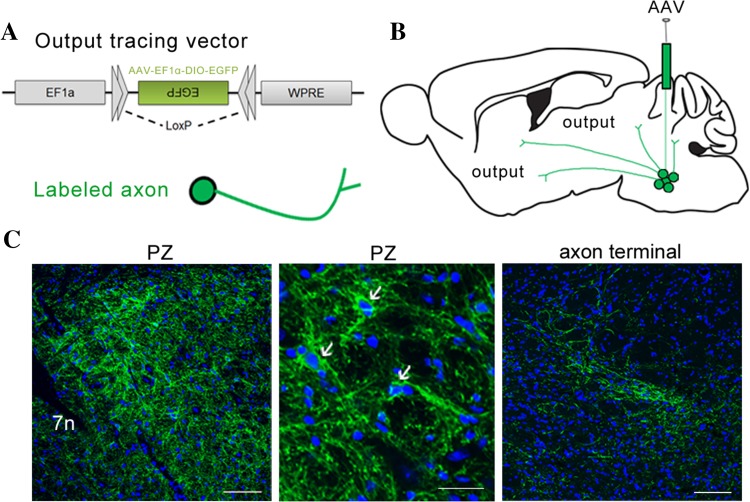
In order to define the output of the PZ GABAergic neurons, we mapped their axonal projections using the Cre-dependent AAV reporter construct AAV-EF1α-DIO-EGFP (Fig. 5A). This virus was stereotaxically infused into the PZ in VGAT-Cre mice (Fig. 5B). When they expressed EGFP, the PZ GABAergic neurons and their axons exhibited green fluorescence (Fig. 5C) and we imaged coronal whole-brain sections to show the distribution of their labeled output areas (Fig. 6).
Fig. 5.
Experimental procedures for tracing whole-brain efferents of PZ GABAergic neurons. A Design of the AAV construct expressing a loxP site and EGFP for Cre-dependent cell tracing. B Schematic of injection procedure of the output tracing virus, AAV-EF1α-DIO-EGFP, in VGAT-Cre mice. C Example image of virus expressed in the PZ (left). Scale bar, 100 μm. Arrows indicate virus-infected GABAergic neurons (middle). Green, EGFP; blue, DAPI. Scale bar, 30 μm. Axons exhibiting green fluorescence were traced by the virus (right). Scale bar, 100 μm. PZ, parafacial zone; 7n, facial nerve or its root.
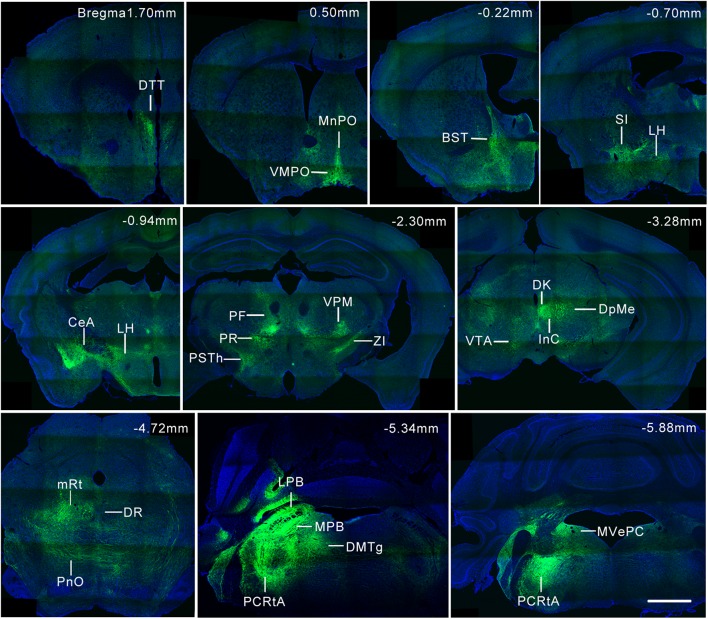
Fig. 6.
Representative coronal sections of PZ GABAergic neurons projecting to selected brain regions. Green, axon projections stained with EGFP; blue, nuclei stained with DAPI. Scale bar, 1 mm. BST, bed nucleus of the stria terminalis; CeA, central amygdaloid nucleus; Dk, nucleus of Darkschewitsch; DMTg, dorsomedial tegmental area; DpMe, deep mesencephalic nucleus; DR, dorsal raphe nucleus; DTT, dorsal tenia tecta; InC, interstitial nucleus of Cajal; LH, lateral hypothalamic area; LPB, lateral parabrachial nucleus; MnPO, median preoptic nucleus; MPB, medial parabrachial nucleus; mRt, mesencephalic reticular formation; MVePC, medial vestibular nucleus, parvocellular part; PCRtA, alpha part of the parvocellular reticular formation; PF, parafascicular thalamic nucleus; PnO, pontine reticular nucleus, oral part; PR, prerubral field; PSTh, parasubthalamic nucleus; SI, substantia innominata; VMPO, ventromedial preoptic nucleus; VPM, ventral posteromedial thalamic nucleus; VTA, ventral tegmental area; ZI, zona incerta. Brain regions were identified based on the mouse brain atlas [19].
Reciprocal Projections Between the PZ and Other Nuclei
We compared the direct inputs and outputs of PZ GABAergic neurons, and found that some nuclei had strong reciprocal projections with the PZ. Thirty-four nuclei sent projections to and received projections from the PZ GABAergic neurons, such as the LH, ZI, central amygdaloid nucleus, medial division (CeM), and the bed nucleus of the stria terminalis (BST) (Table 1). These results showed that most of the nuclei that sent projections to the PZ also received projections from the PZ GABAergic neurons, indicating the presence of feedback control to regulate specific functions.
Table 1.
Nuclei having reciprocal connections with PZ GABAergic neurons.
| Abbreviation | Complete name |
|---|---|
| BST | Bed nucleus of the stria terminalis |
| CeM | Central amygdaloid nucleus, medial division |
| CnF | Cuneiform nucleus |
| DMSp5 | Dorsomedial spinal trigeminal nucleus |
| DpMe | Deep mesencephalic nucleus |
| DRV | Dorsal raphe nucleus, ventral part |
| ECIC | External cortex of the inferior colliculus |
| IntP | Interposed cerebellar nucleus, posterior part |
| IRt | Intermediate reticular nucleus |
| LH | Lateral hypothalamic area |
| LPAG | Lateral periaqueductal gray |
| LVe | Lateral vestibular nucleus |
| Med | Medial (fastigial) cerebellar nucleus |
| MVeMC | Medial vestibular nucleus, magnocellular part |
| MVePC | Medial vestibular nucleus, parvocellular part |
| P5 | Peritrigeminal zone |
| PnO | Pontine reticular nucleus, oral part |
| PPTg | Pedunculopontine tegmental nucleus |
| Pr | Prepositus nucleus |
| PSTh | Parasubthalamic nucleus |
| RC | Raphe cap |
| RMC | Red nucleus, magnocellular part |
| RPC | Red nucleus, parvocellular part |
| SI | Substantia innominata |
| SNC | Substantia nigra, compact part |
| SNR | Substantia nigra, pars reticulata |
| Sp5I | Spinal trigeminal nucleus, interpolar part |
| Sp5ODM | Spinal trigeminal nucleus, oral part, dorsomedial division |
| Sp5OVL | Spinal trigeminal nucleus, oral part, ventrolateral division |
| SpVe | Spinal vestibular nucleus |
| SuVe | Superior vestibular nucleus |
| VLPAG | Ventrolateral periaqueductal gray |
| VTA | Ventral tegmental area |
| ZI | Zona incerta |
Previously Unreported Inputs and Outputs of the PZ
Previous studies have revealed the afferent and efferent projections of the PCRt (which overlaps with the PZ according to the brain atlas) using HRP and PHA-L tracing in rats [9, 14]. So we compared our results with the literature and listed the newly-found inputs and outputs of the PZ for brevity.
Compared with previous studies [9], the inputs to both PZ GABAergic neurons (mice) and the PCRt (rats), inputs to the PCRt (rats) but not to the PZ GABAergic neurons (mice), and inputs to the PZ GABAergic neurons (mice) but not to the PCRt (rats) are listed in Table 2.
Table 2.
Inputs to the PZ found in our study compared with the literature [9].
| Inputs to both the PCRt (rats) and the PZ GABAergic neurons (mice) | Inputs to the PCRt (rats) but not to the PZ GABAergic neurons (mice) | Inputs to the PZ GABAergic neurons (mice) but not to the PCRt (rats) |
|---|---|---|
| Accumbens nucleus Anterior hypothalamic area Agranular insular cortex, dorsal part Agranular insular cortex, posterior part Agranular insular cortex, ventral part Ambiguus nucleus Bed nucleus of the stria terminalis Central amygdaloid nucleus Central gray Cuneiform nucleus Caudate putamen (striatum) Dorsal paragigantocellular nucleus Dorsal raphe nucleus Gigantocellular reticular nucleus Granular insular cortex Gigantocellular reticular nucleus, alpha part Gigantocellular reticular nucleus, ventral part Globus pallidus Interpeduncular nucleus Intermediate reticular nucleus Kolliker-Fuse nucleus Lateral (dentate) cerebellar nucleus Locus coeruleus Lateral hypothalamic area Lateral lemniscus Nucleus of the lateral olfactory tract Lateral vestibular nucleus Primary motor cortex Medial (fastigial) cerebellar nucleus Medial lemniscus Median raphe nucleus Motor trigeminal nucleus Medial vestibular nucleus Paraventricular hypothalamic nucleus Parabrachial nucleus Pontine nuclei Pontine reticular nucleus, caudal part Pontine reticular nucleus, oral part Pontine reticular nucleus, ventral part Principal sensory trigeminal nucleus, dorsomedial part Principal sensory trigeminal nucleus, ventrolateral part Red nucleus, magnocellular part Red nucleus, parvocellular part Retrorubral field Rostroventrolateral reticular nucleus Primary somatosensory cortex Secondary somatosensory cortex Superior cerebellar peduncle (brachium conjunctivum) Substantia innominata Substantia nigra, reticular part Nucleus of the solitary tract Spinal trigeminal nucleus, interpolar part Spinal trigeminal nucleus, oral part Subthalamic nucleus Supratrigeminal nucleus Subcoeruleus nucleus Superior vestibular nucleus Zona incerta |
Facial nerve or its root Hypoglossal nucleus Anterior commissure Caudoventrolateral reticular nucleus Fornix Genu of the facial nerve Internal capsule Linear nucleus of the medulla Lateral reticular nucleus Medullary reticular nucleus, dorsal part Medullary reticular nucleus, ventral part Mesencephalic trigeminal nucleus Mammillothalamic tract Optic nerve layer of the superior colliculus Parabigeminal nucleus Probst’s bundle Superior colliculus Stria medullaris of the thalamus Spinal trigeminal nucleus, caudal part Olfactory tubercle Decussation of the superior cerebellar peduncle |
Facial nucleus Dorsomedial spinal trigeminal nucleus Dorsomedial tegmental area Deep gray layer of the superior colliculus Deep mesencephalic nucleus Deep white layer of the superior colliculus Dorsal raphe nucleus, ventral part External cortex of the inferior colliculus Intercollicular nucleus Intermediate gray layer of the superior colliculus Interposed cerebellar nucleus, dorsolateral hump Interposed cerebellar nucleus, posterior part Intermediate white layer of the superior colliculus Laterodorsal tegmental nucleus Lateral periaqueductal gray Lateral paragigantocellular nucleus Secondary motor cortex Peritrigeminal zone Perifacial zone Pedunculopontine tegmental nucleus Prepositus nucleus Principal sensory trigeminal nucleus Parasubthalamic nucleus Raphe cap Raphe magnus nucleus Substantia nigra Spinal trigeminal tract Spinal trigeminal nucleus, oral part, dorsomedial division Spinal trigeminal nucleus, oral part, ventrolateral division Spinal vestibular nucleus Vestibulocerebellar nucleus Ventrolateral periaqueductal gray Ventral tegmental area |
The nuclei receiving newly-found projections from the PZ compared with the literature [14] were dorsal tenia tecta, septohippocampal nucleus, semilunar nucleus, accumbens nucleus (core), accumbens nucleus (shell), dorsal endopiriform nucleus, claustrum, lateral orbital cortex, BST, ventral pallidum, lateral preoptic area, nucleus of the horizontal limb of the diagonal band, magnocellular preoptic nucleus, medial preoptic area, median preoptic nucleus, vascular organ of the lamina terminalis, ventromedial preoptic nucleus, medial septal nucleus, anteroventral periventricular nucleus, internal capsule, lateral globus pallidus, anterior commissural nucleus, substantia innominata, medial preoptic nucleus, IPAC, paraventricular thalamic nucleus (anterior part), mediodorsal thalamic nucleus, paraventricular hypothalamic nucleus (lateral magnocellular part), paraventricular hypothalamic nucleus (medial magnocellular part), LH, CeA, nucleus of the lateral olfactory tract, reticular thalamic nucleus, ZI, rhomboid thalamic nucleus, reuniens thalamic nucleus, supraoptic nucleus, ventrolateral thalamic nucleus, parafascicular thalamic nucleus, subparafascicular thalamic nucleus, ventral posteromedial thalamic nucleus, prerubral field, fields of Forel, subthalamic nucleus, PSTh, posterior thalamic nuclear group, periventricular fiber system, posterior hypothalamic nucleus, nucleus of Darkschewitsch, interstitial nucleus of Cajal, DpMe, Edinger-Westphal nucleus, red nucleus (parvocellular part), substantia nigra (compact part), substantia nigra (lateral part), anterior pretectal nucleus, medial geniculate nucleus, posterior intralaminar thalamic nucleus, peripeduncular nucleus, ventral tegmental area, raphe cap, dorsal raphe nucleus, mesencephalic reticular formation, paratrochlear nucleus, pedunculopontine tegmental nucleus, decussation of the superior cerebellar peduncle, subpedencular tegmental nucleus, intermediate nucleus of the lateral lemniscus, paralemniscal nucleus, tectospinal tract, paramedian raphe nucleus, median raphe nucleus, epimicrocellular nucleus, microcellular tegmental nucleus, reticulotegmental nucleus of the pons, P5, middle cerebellar peduncle, red nucleus (magnocellular part), superior vestibular nucleus, dorsomedial spinal trigeminal nucleus, Med, dorsal cochlear nucleus, ventral cochlear nucleus (posterior part), interposed cerebellar nucleus (posterior part), and superficial glial zone of the cochlear nuclei.
Discussion
Using virus-mediated circuit mapping, we have characterized the whole-brain distribution of long-range PZ connections. Our experiments confirm many previously-demonstrated connections of the PCRt, which overlaps with the PZ [9, 14], but with cell-type specificity and quantitative analyses at multiple spatial scales. Furthermore, we found many new inputs and outputs of the PZ compared with those of the PCRt. These differences from the literature are probably due to our tracing method, which readily detects convincing synaptic contacts between nuclei with cell-type specificity, and the injection sites. However, to study the outputs of a nucleus, more sophisticated methods such as trans-synaptic virus tracing are still needed [24].
In the present study, we identified features of the connections between the PZ and other nuclei. First, the DsRed-labeled presynaptic neurons spanned almost the entire brain, but the main inputs to the GABAergic neurons in the PZ were from the brain stem. Second, the connections to the GABAergic neurons in the PZ were strongly ipsilateral, with few contralateral projections. Third, the connections to the GABAergic neurons in the PZ were strongly reciprocal. These results reveal the long-range wiring diagram of PZ circuits with highly divergent inputs and outputs.
The PCRt has been implicated in a variety of functions such as metabolic homeostasis, suckling behavior, motor activities of the oral and forelimb body segments, and the sleep-wake mechanism [14, 25–27]. It has been reported that the glutamatergic projection from the PCRt to the motor trigeminal nucleus (Mo5) induces jaw-closer atonia during rapid eye-movement (REM) sleep [28]. The pontine sublateraldorsal nucleus sends projections to GABA/glycine neurons in the ventral gigantocellular nucleus and PCRt that show c-fos activity 90 min after a REM sleep-like state [29]. In the present study, afferents to the PZ such as M1 and M2 may reflect on inputs to the PCRt for regulation of the cranial motor system.
Although there is overlap between the PZ and the PCRt (especially the alpha part), the PZ has recently been reported to be one of several GABAergic sleep-promoting nuclei, also located in the ventrolateral preoptic area of the hypothalamus and in a compartment within the lateral hypothalamus [7, 8]. Genetic disruption of GABAergic/glycinergic transmission from the PZ in mice results in large and sustained increases in wakefulness [1].
In the newly-found brain areas which input to PZ GABAergic neurons, the DpMe, cortex (M2), and VLPAG may participate in the regulation of slow-wave sleep (SWS) [30–33]. It has been reported that Fos expression in cortical neuronal nitric oxide synthase (nNOS) neurons correlates with the amounts of NREM sleep and slow-wave activity during NREM sleep [31, 33]. Optogenetic or pharmacogenetic activation of GABAergic neurons in the VLPAG or the adjacent DpMe substantially increases NREM sleep [30, 32].
Among the newly-found outputs, the MnPO is probably involved in SWS regulation, because single-unit recordings and analysis of Fos expression showed that NREM sleep-active neurons are concentrated in the MnPO [34, 35]. In vivo application of an adenosine A2A receptor agonist increases NREM sleep and enhances c-Fos expression in the MnPO [36].
In addition, in the nuclei that connect to PZ GABAergic neurons, BF [37] and BG [38] also regulate NREM sleep. DR dopamine neurons [39], VTA [40] and LH [41] have been reported to regulate wakefulness.
But the functional connections of these sleep-regulating nuclei with GABAergic neurons located in PZ still need further studies.
Based on the neuronal circuits revealed by this tracing method, new functions have also been found [13]. Our quantitative analysis of the whole-brain distributions of inputs and outputs of the GABAergic neurons in the PZ can serve as an anatomical blueprint for future studies of the inter-regional pathways mediating the functions of the PZ.
Acknowledgements
We thank Hui-Fang Lou and Li-Ya Zhu for technical support. This work was supported by the National Natural Science Foundation of China (31571090 and 31771167), the National Key Research and Development Program (2016YFC1306700), the National High Technology Research and Development Program (863 Program) of China (2015AA020512), and the Fundamental Research Funds for the Central Universities of China (2017FZA7003).
Compliance with Ethical Standards
Conflict of interest
All authors claim that there are no conflicts of interest.
References
- 1.Anaclet C, Lin JS, Vetrivelan R, Krenzer M, Vong L, Fuller PM, et al. Identification and characterization of a sleep-active cell group in the rostral medullary brainstem. J Neurosci. 2012;32:17970–17976. doi: 10.1523/JNEUROSCI.0620-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones BE. Modulation of cortical activation and behavioral arousal by cholinergic and orexinergic systems. Ann N Y Acad Sci. 2008;1129:26–34. doi: 10.1196/annals.1417.026. [DOI] [PubMed] [Google Scholar]
- 3.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of sleep and wakefulness. Physiol Rev. 2012;92:1087–1187. doi: 10.1152/physrev.00032.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan Y. Harnessing GABAergic transmission for slow oscillations. Neurosci Bull. 2016;32:501–502. doi: 10.1007/s12264-016-0058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J, et al. The GABAergic parafacial zone is a medullary slow wave sleep–promoting center. Nat Neurosci. 2014;17:1217–1224. doi: 10.1038/nn.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz MD, Kilduff TS. The neurobiology of sleep and wakefulness. Psychiatr Clin North Am. 2015;38:615–644. doi: 10.1016/j.psc.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown RE, McKenna JT. Turning a negative into a positive: ascending GABAergic control of cortical activation and arousal. Front Neurol. 2015;6:135. doi: 10.3389/fneur.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shammah-Lagnado SJ, Costa MS, Ricardo JA. Afferent connections of the parvocellular reticular formation: a horseradish peroxidase study in the rat. Neuroscience. 1992;50:403–425. doi: 10.1016/0306-4522(92)90433-3. [DOI] [PubMed] [Google Scholar]
- 10.Travers JB, Norgren R. Afferent projections to the oral motor nuclei in the rat. J Comp Neurol. 1983;220:280–298. doi: 10.1002/cne.902200303. [DOI] [PubMed] [Google Scholar]
- 11.Stanek E, Cheng S, Takatoh J, Han BX, Wang F. Monosynaptic premotor circuit tracing reveals neural substrates for oro-motor coordination. eLife. 2014;3:e02511. doi: 10.7554/eLife.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/nmeth999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han W, Tellez LA, Rangel MJ, Motta SC, Zhang X, Perez IO, et al. Integrated control of predatory hunting by the central nucleus of the amygdala. Cell. 2017;168:311–324. doi: 10.1016/j.cell.2016.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ter Horst GJ, Copray JC, Liem RS, Van Willigen JD. Projections from the rostral parvocellular reticular formation to pontine and medullary nuclei in the rat: involvement in autonomic regulation and orofacial motor control. Neuroscience. 1991;40:735–758. doi: 10.1016/0306-4522(91)90009-D. [DOI] [PubMed] [Google Scholar]
- 15.Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 16.Callaway EM, Luo L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osten P, Margrie TW. Mapping brain circuitry with a light microscope. Nat Methods. 2013;10:515–523. doi: 10.1038/nmeth.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates: Compact second Edition. Academic Press, 2001.
- 20.Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Wall NR, De La Parra M, Callaway EM, Kreitzer AC. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao F, Jiang HF, Zeng WB, Shu Y, Luo MH, Duan S. Anterograde trans-synaptic tagging mediated by adeno-associated virus. Neurosci Bull. 2017;33:348–350. doi: 10.1007/s12264-017-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minkels RF, Jüch PJ, Ter Horst GJ, Van Willigen JD. Projections of the parvocellular reticular formation to the contralateral mesencephalic trigeminal nucleus in the rat. Brain Res. 1991;547:13–21. doi: 10.1016/0006-8993(91)90569-H. [DOI] [PubMed] [Google Scholar]
- 26.Sahara Y, Hashimoto N, Nakamura Y. Hypoglossal premotor neurons in the rostral medullary parvocellular reticular formation participate in cortically-induced rhythmical tongue movements. Neurosci Res. 1996;26:119–131. doi: 10.1016/S0168-0102(96)01080-2. [DOI] [PubMed] [Google Scholar]
- 27.Parenti R, Cicirata F, Pantò MR, Serapide MF. The projections of the lateral reticular nucleus to the deep cerebellar nuclei. An experimental analysis in the rat. Eur J Neurosci. 1996;8:2157–2167. doi: 10.1111/j.1460-9568.1996.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 28.Anaclet C, Pedersen NP, Fuller PM, Lu J. Brainstem circuitry regulating phasic activation of trigeminal motoneurons during REM sleep. PLoS One. 2010;5:e8788. doi: 10.1371/journal.pone.0008788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boissard R, Gervasoni D, Schmidt MH, Barbagli B, Fort P, Luppi PH. The rat ponto-medullary network responsible for paradoxical sleep onset and maintenance: a combined microinjection and functional neuroanatomical study. Eur J Neurosci. 2002;16:1959–1973. doi: 10.1046/j.1460-9568.2002.02257.x. [DOI] [PubMed] [Google Scholar]
- 30.Weber F, Chung S, Beier KT, Xu M, Luo L, Dan Y. Control of REM sleep by ventral medulla GABAergic neurons. Nature. 2015;526:435–438. doi: 10.1038/nature14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morairty SR, Dittrich L, Pasumarthi RK, Valladao D, Heiss JE, Gerashchenko D, et al. A role for cortical nNOS/NK1 neurons in coupling homeostatic sleep drive to EEG slow wave activity. Proc Natl Acad Sci U S A. 2013;110:20272–20277. doi: 10.1073/pnas.1314762110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi Y, Kashiwagi M, Yasuda K, Ando R, Kanuka M, Sakai K, et al. Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science. 2015;350:957–961. doi: 10.1126/science.aad1023. [DOI] [PubMed] [Google Scholar]
- 33.Gerashchenko D, Wisor JP, Burns D, Reh RK, Shiromani PJ, Sakurai T, et al. Identification of a population of sleep-active cerebral cortex neurons. Proc Natl Acad Sci U S A. 2008;105:10227–10232. doi: 10.1073/pnas.0803125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong H, McGinty D, Guzman-Marin R, Chew KT, Stewart D, Szymusiak R. Activation of c-fos in GABAergic neurones in the preoptic area during sleep and in response to sleep deprivation. J Physiol. 2004;556:935–946. doi: 10.1113/jphysiol.2003.056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam MA, Kumar S, McGinty D, Alam MN, Szymusiak R. Neuronal activity in the preoptic hypothalamus during sleep deprivation and recovery sleep. J Neurophysiol. 2014;111:287–299. doi: 10.1152/jn.00504.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Rai S, Hsieh KC, McGinty D, Alam MN, Szymusiak R. Adenosine A(2A) receptors regulate the activity of sleep regulatory GABAergic neurons in the preoptic hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2013;305:R31–41. doi: 10.1152/ajpregu.00402.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang WC, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci. 2015;18:1641–1647. doi: 10.1038/nn.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu MH, Vetrivelan R, Fuller PM, Lu J. Basal ganglia control of sleep-wake behavior and cortical activation. Eur J Neurosci. 2010;31:499–507. doi: 10.1111/j.1460-9568.2009.07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho JR, Treweek JB, Robinson JE, Xiao C, Bremner LR, Greenbaum A, et al. Dorsal raphe dopamine neurons modulate arousal and promote wakefulness by salient stimuli. Neuron. 2017;94:1205–1219. doi: 10.1016/j.neuron.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Sun HX, Wang DR, Ye CB, Hu ZZ, Wang CY, Huang ZL, et al. Activation of the ventral tegmental area increased wakefulness in mice. Sleep Biol Rhythms. 2017;15:107–115. doi: 10.1007/s41105-017-0094-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerri M, Del Vecchio F, Mastrotto M, Luppi M, Martelli D, Perez E, et al. Enhanced slow-wave EEG activity and thermoregulatory impairment following the inhibition of the lateral hypothalamus in the rat. PLoS One. 2014;9:e112849. doi: 10.1371/journal.pone.0112849. [DOI] [PMC free article] [PubMed] [Google Scholar]