The next new disease
Emerging infectious diseases are a major threat to health: AIDS, SARS, drug-resistant bacteria and Ebola virus are among the more recent examples. By identifying emerging disease 'hotspots', the thinking goes, it should be possible to spot health risks at an early stage and prepare containment strategies. An analysis of over 300 examples of disease emerging between 1940 and 2004 suggests that these hotspots can be accurately mapped based on socio-economic, environmental and ecological factors. The data show that the surveillance effort, and much current research spending, is concentrated in developed economies, yet the risk maps point to developing countries as the more likely source of new diseases.
Supplementary information
The online version of this article (doi:10.1038/nature06536) contains supplementary material, which is available to authorized users.
The origins of emerging infections diseases are significantly correlated with socio-economic, environmental and ecological factors.
Supplementary information
The online version of this article (doi:10.1038/nature06536) contains supplementary material, which is available to authorized users.
Abstract
Emerging infectious diseases (EIDs) are a significant burden on global economies and public health1,2,3. Their emergence is thought to be driven largely by socio-economic, environmental and ecological factors1,2,3,4,5,6,7,8,9, but no comparative study has explicitly analysed these linkages to understand global temporal and spatial patterns of EIDs. Here we analyse a database of 335 EID ‘events’ (origins of EIDs) between 1940 and 2004, and demonstrate non-random global patterns. EID events have risen significantly over time after controlling for reporting bias, with their peak incidence (in the 1980s) concomitant with the HIV pandemic. EID events are dominated by zoonoses (60.3% of EIDs): the majority of these (71.8%) originate in wildlife (for example, severe acute respiratory virus, Ebola virus), and are increasing significantly over time. We find that 54.3% of EID events are caused by bacteria or rickettsia, reflecting a large number of drug-resistant microbes in our database. Our results confirm that EID origins are significantly correlated with socio-economic, environmental and ecological factors, and provide a basis for identifying regions where new EIDs are most likely to originate (emerging disease ‘hotspots’). They also reveal a substantial risk of wildlife zoonotic and vector-borne EIDs originating at lower latitudes where reporting effort is low. We conclude that global resources to counter disease emergence are poorly allocated, with the majority of the scientific and surveillance effort focused on countries from where the next important EID is least likely to originate.
Supplementary information
The online version of this article (doi:10.1038/nature06536) contains supplementary material, which is available to authorized users.
Main
In the global human population, we report the emergence of 335 infectious diseases between 1940 and 2004. Here we define the first temporal origination of an EID (that is, the original case or cluster of cases representing an infectious disease emerging in human populations for the first time—see Methods and Supplementary Table 1) as an EID ‘event’. Our database includes EID events caused by newly evolved strains of pathogens (for example, multi-drug-resistant tuberculosis and chloroquine-resistant malaria), pathogens that have recently entered human populations for the first time (for example, HIV-1, severe acute respiratory syndrome (SARS) coronavirus), and pathogens that have probably been present in humans historically, but which have recently increased in incidence (for example, Lyme disease). The emergence of these pathogens and their subsequent spread have caused an extremely significant impact on global health and economies1,2,3. Previous efforts to understand patterns of EID emergence have highlighted viral pathogens (especially RNA viruses) as a major threat, owing to their often high rates of nucleotide substitution, poor mutation error-correction ability and therefore higher capacity to adapt to new hosts, including humans5,8,10,11. However, we find that the majority of pathogens involved in EID events are bacterial or rickettsial (54.3%). This group is typically represented by the emergence of drug-resistant bacterial strains (for example, vancomycin-resistant Staphylococcus aureus). Viral or prion pathogens constitute only 25.4% of EID events, in contrast to previous analyses which suggest that 37–44% of emerging pathogens are viruses or prions and 10–30% bacteria or rickettsia5,8,11. This follows our classification of each individual drug-resistant microbial strain as a separate pathogen in our database, and reflects more accurately the true significance of antimicrobial drug resistance for global health, in which different pathogen strains can cause separate significant outbreaks12. In broad concurrence with previous studies on the characteristics of emerging human pathogens5,8,11, we find the percentages of EID events caused by other pathogen types to be 10.7% for protozoa, 6.3% for fungi and 3.3% for helminths (see Supplementary Data and Supplementary Table 2 for a detailed comparison to previous studies).
The incidence of EID events has increased since 1940, reaching a maximum in the 1980s (Fig. 1). We tested whether the increase through time was largely attributable to increasing infectious disease reporting effort (that is, through more efficient diagnostic methods and more thorough surveillance2,3,13) by calculating the annual number of articles published in the Journal of Infectious Diseases (JID) since 1945 (see Methods). Controlling for reporting effort, the number of EID events still shows a highly significant relationship with time (generalized linear model with Poisson errors, offset by log(JID articles) (GLMP,JID), F = 96.4, P < 0.001, d.f. = 57). This provides the first analytical support for previous suggestions that the threat of EIDs to global health is increasing1,2,14. To further investigate the peak in EID events in the 1980s, we examined the most frequently cited driver of EID emergence during this period (see Supplementary Table 1). Increased susceptibility to infection caused the highest proportion of events during 1980–90 (25.5%), and we therefore suggest that the spike in EID events in the 1980s is due largely to the emergence of new diseases associated with the HIV/AIDS pandemic2,13.
Figure 1. Number of EID events per decade.
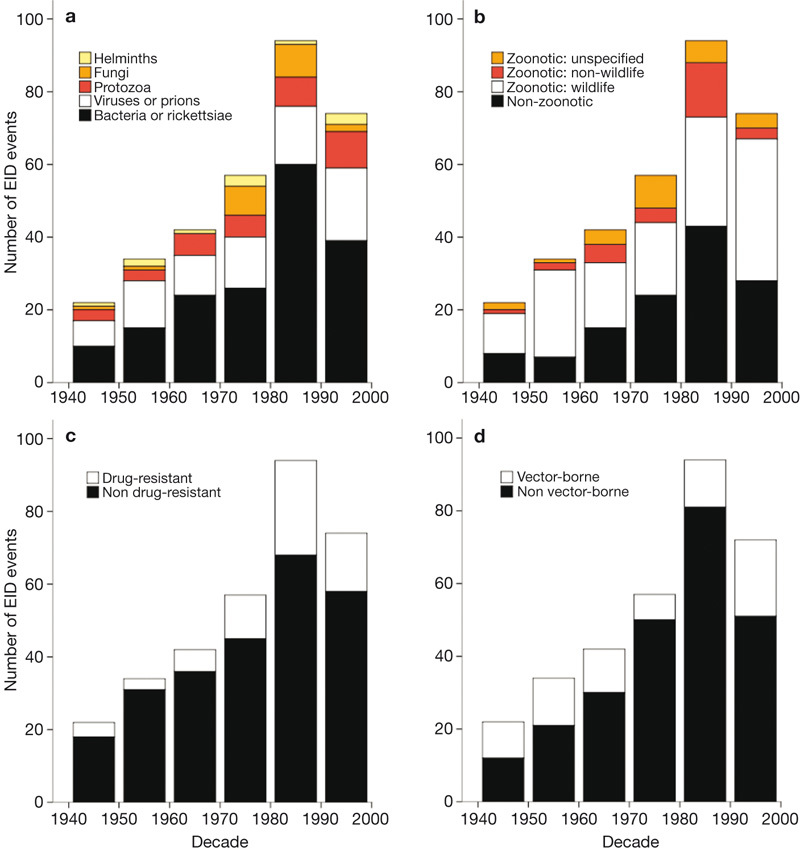
EID events (defined as the temporal origin of an EID, represented by the original case or cluster of cases that represents a disease emerging in the human population—see Methods) are plotted with respect to a, pathogen type, b, transmission type, c, drug resistance and d, transmission mode (see keys for details).
The majority (60.3%) of EID events are caused by zoonotic pathogens (defined here as those which have a non-human animal source), which is consistent with previous analyses of human EIDs5,8. Furthermore, 71.8% of these zoonotic EID events were caused by pathogens with a wildlife origin—for example, the emergence of Nipah virus in Perak, Malaysia and SARS in Guangdong Province, China. The number of EID events caused by pathogens originating in wildlife has increased significantly with time, controlling for reporting effort (GLMP,JID F = 60.7, P < 0.001, d.f. = 57), and they constituted 52.0% of EID events in the most recent decade (1990–2000) (Fig. 1). This supports the suggestion that zoonotic EIDs represent an increasing and very significant threat to global health1,2,7,13,14. It also highlights the importance of understanding the factors that increase contact between wildlife and humans in developing predictive approaches to disease emergence4,6,9,15.
Vector-borne diseases are responsible for 22.8% of EID events in our database, and 28.8% in the last decade (Fig. 1). Our analysis reveals a significant rise in the number of EID events they have caused over time, controlling for reporting effort (GLMP,JID F = 49.8, P < 0.001, d.f. = 57). This rise corresponds to climate anomalies occurring during the 1990s16, adding support to hypotheses that climate change may drive the emergence of diseases that have vectors sensitive to changes in environmental conditions such as rainfall, temperature and severe weather events17. However, this controversial issue requires further analyses to test causal relationships between EID events and climate change18. We also report that EID events caused by drug-resistant microbes (which represent 20.9% of the EID events in our database) have significantly increased with time, controlling for reporting effort (GLMP,JID F = 5.19, P < 0.05, d.f. = 57). This is probably related to a corresponding rise in antimicrobial drug use, particularly in high-latitude developed countries2,7,12.
A recent analysis showed a latitudinal spatial gradient in human pathogen species richness increasing towards the Equator19, in common with the distributional pattern of species richness found in many other taxonomic groups20. Environmental parameters that promote pathogen transmission at lower latitudes (for example, higher temperatures and precipitation) are hypothesized to drive this pattern19. Our analyses suggest that there is no such pattern in EID events, which are concentrated in higher latitudes (Supplementary Fig. 1). The highest concentration of EID events per million square kilometres of land was found between 30 and 60 degrees north and between 30 and 40 degrees south, with the main hotspots in the northeastern United States, western Europe, Japan and southeastern Australia (Fig. 2). We hypothesize that (1) socioeconomic drivers (such as human population density, antibiotic drug use and agricultural practices) are major determinants of the spatial distribution of EID events, in addition to the ecological or environmental conditions that may affect overall (emerging and non-emerging) human pathogen distribution19, and (2) that the importance of these drivers depends on the category of EID event. In particular, we hypothesize that EID events caused by zoonotic pathogens from wildlife are significantly correlated with wildlife biodiversity, and those caused by drug-resistant pathogens are more correlated with socio-economic conditions than those caused by zoonotic pathogens.
Figure 2. Global richness map of the geographic origins of EID events from 1940 to 2004.
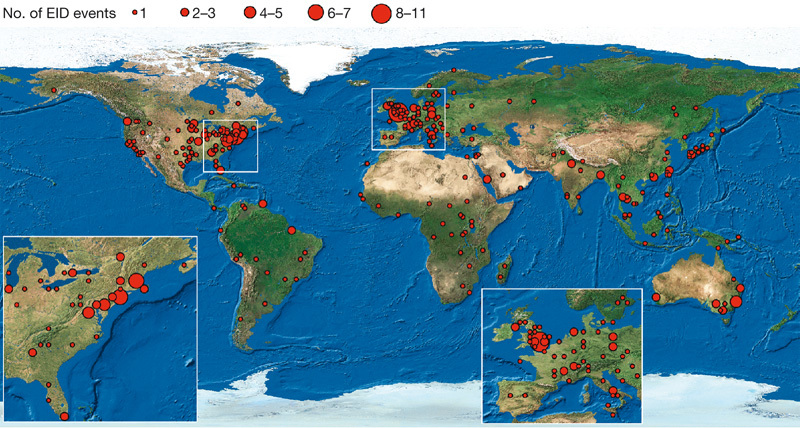
The map is derived for EID events caused by all pathogen types. Circles represent one degree grid cells, and the area of the circle is proportional to the number of events in the cell.
We tested these hypotheses by examining the relationship between the spatial pattern of the different categories of EID events (zoonotic pathogens originating in wildlife and non-wildlife, drug-resistant and vector-borne pathogens, Supplementary Fig. 2), and socio-economic variables (human population density and human population growth), environmental variables (latitude, rainfall) and an ecological variable (wildlife host species richness) (see Methods). We found that human population density was a common significant independent predictor of EID events in all categories, controlling for spatial reporting bias by country (see Methods, Table 1 and Supplementary Table 3). This supports previous hypotheses that disease emergence is largely a product of anthropogenic and demographic changes, and is a hidden ‘cost’ of human economic development2,4,7,9,13. Wildlife host species richness is a significant predictor for the emergence of zoonotic EIDs with a wildlife origin, with no role for human population growth, latitude or rainfall (Table 1). The emergence of zoonotic EIDs from non-wildlife hosts is predicted by human population density, human population growth, and latitude, and not by wildlife host species richness. EID events caused by drug-resistant microbes are affected by human population density and growth, latitude and rainfall. The pattern of EID events caused by vector-borne diseases was not correlated with any of the environmental or ecological variables we examined, although we note that the climate variable used in this analysis (rainfall) does not represent climate change phenomena.
Table 1.
Socio-economic, environmental and ecological correlates of EID events
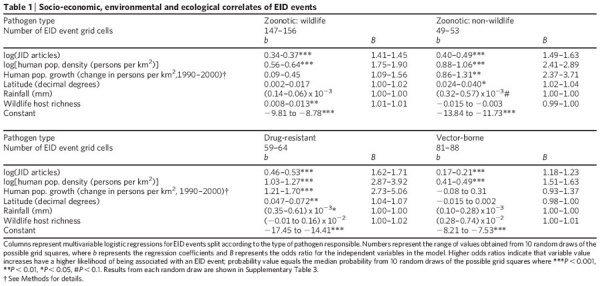
Our study examines the role of only a few drivers to understand disease emergence, whereas many other factors (for example, land use change, agriculture) have been causally linked to EIDs6,21. However, until more rigorous global data sets of these drivers become available, data on human population density and growth act as reasonable proxies for such anthropogenic changes. Other likely future improvements to the model would include a more accurate accounting for temporal and spatial ascertainment biases—for example, the development of global spatial data sets of the amount of funding per capita for infectious disease surveillance.
Our analyses provide a basis for developing a predictive model for the regions where new EIDs are most likely to originate (emerging disease ‘hotspots’). A visualization of the regression results from Table 1 for EID events from each category (Fig. 3) identifies these regions globally. This approach may be valuable for deciding where to allocate global resources to pre-empt, or combat, the first stages of disease emergence10,14,18,22. Our analysis shows that there is a high spatial reporting bias for EID events (see Methods, Supplementary Fig. 3), reflecting greater surveillance and infectious disease research effort in richer, developed countries of Europe, North America, Australia and some parts of Asia, than in developing regions. This contrasts with our risk maps (Fig. 3), which suggest that predicted emerging disease hotspots due to zoonotic pathogens from wildlife and vector-borne pathogens are more concentrated in lower-latitude developing countries. We conclude that the global effort for EID surveillance and investigation is poorly allocated, with the majority of our scientific resources focused on places from where the next important emerging pathogen is least likely to originate. We advocate re-allocation of resources for ‘smart surveillance’ of emerging disease hotspots in lower latitudes, such as tropical Africa, Latin America and Asia, including targeted surveillance of at-risk people to identify early case clusters of potentially new EIDs before their large-scale emergence. Zoonoses from wildlife represent the most significant, growing threat to global health of all EIDs (see our data in Fig. 1, and recent reviews1,2,5,8,9,13,14). Our findings highlight the critical need for health monitoring4,14,23 and identification of new, potentially zoonotic pathogens in wildlife populations, as a forecast measure for EIDs. Finally, our analysis suggests that efforts to conserve areas rich in wildlife diversity by reducing anthropogenic activity may have added value in reducing the likelihood of future zoonotic disease emergence.
Figure 3. Global distribution of relative risk of an EID event.
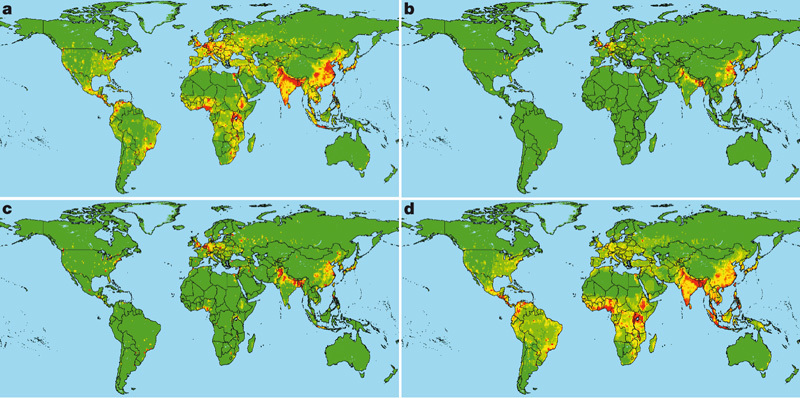
Maps are derived for EID events caused by a, zoonotic pathogens from wildlife, b, zoonotic pathogens from non-wildlife, c, drug-resistant pathogens and d, vector-borne pathogens. The relative risk is calculated from regression coefficients and variable values in Table 1 (omitting the variable measuring reporting effort), categorized by standard deviations from the mean and mapped on a linear scale from green (lower values) to red (higher values).
Methods Summary
Biological, temporal and spatial data on human EID ‘events’ were collected from the literature from 1940 (yellow fever virus, Nuba Mountains, Sudan) until 2004 (poliovirus type 2 in Uttar Pradesh, India) (n = 335, see Supplementary Data for data and sources). Global allocation of scientific resources for disease surveillance has been focused on rich, developed countries (Supplementary Fig. 3). It is thus likely that EID discovery is biased both temporally (by increasing research effort into human pathogens over the period of the database) and spatially (by the uneven levels of surveillance across countries). We account for these biases by quantifying reporting effort in JID and including it in our temporal and spatial analyses. JID is the premier international journal (highest ISI impact factor 2006: http://portal.isiknowledge.com/) of human infectious disease research that publishes papers on both emerging and non-emerging infectious diseases without a specific geographical bias. To investigate the drivers of the spatial pattern of EID events, we compared the location of EID events to five socio-economic, environmental and ecological variables matched onto a terrestrial one degree grid of the globe. We carried out the spatial analyses using a multivariable logistic regression to control for co-variability between drivers, with the presence/absence of EID events as the dependent variable and all drivers plus our measure of spatial reporting bias by country as independent variables (n = 18,307 terrestrial grid cells). Analyses were conducted on subsets of the EID events—those caused by zoonotic pathogens (defined in our analyses as pathogens that originated in non-human animals) originating in wildlife and non-wildlife species, and those caused by drug-resistant and vector-borne pathogens.
Online Methods
EID event definition
In this paper, we are analysing the process of disease emergence, not just the pathogens that cause them. Therefore, we focus on EID ‘events’, which we define as the first temporal emergence of a pathogen in a human population which was related to the increase in distribution, increase in incidence or increase in virulence or other factor which led to that pathogen being classed as an emerging disease2,4,5,8,13,15. We chose the 1940 cut-off based on the Institute of Medicine’s2 examples of a currently or very recently emerging disease, all of which had their likely temporal origins within this time period. Single case reports of a new pathogen were not considered to represent the emergence of a disease, and emergence was normally represented by reports, in more than one peer-reviewed paper, of a cluster of cases that were identified in humans for the first time, or (for previously known diseases) considered significantly above background. Only events that had sufficient corroborating evidence for their geographic and temporal origin were included in our analysis. We based our data collection on the list of EIDs in ref. 5 updated to 2004. Unlike this previous study5, we treated different drug-resistant strains of the same microbial species as separate pathogens and the cause of separate EID events (for example, the emergence of the chloroquine-resistant strain of the malaria pathogen (Plasmodium falciparum) in Trujillo, Venezuela in 1957 and the sulphadoxine-pyrimethamine-resistant strain in Sa Kaeo, Thailand in 1981).
Variable definitions
The biological, temporal and spatial variable definitions of an EID event used are as follows: italic font indicates classes of the variables. (1) ‘Pathogen’, name of pathogen associated with the EID event. (2) ‘Year’ (the earliest year in which the first cluster of cases representing each EID event was reported to have occurred was taken where a range of years was given). (3) ‘Pathogen type’ (PathType): (i) bacterial; (ii) rickettsial; (iii) viral; (iv) prion; (v) fungal; (vi) helminth; (vii) protozoan. (4) ‘Transmission type’ (TranType): (0) non-zoonotic (disease emerged without involvement of a non-human host); (1) zoonotic (disease emerged via non-human to human transmission, not including vectors). (5) ‘Zoonotic type’ (ZooType): (0) non-zoonotic (disease emerged via human to human transmission); (1) non-wildlife (zoonotic EID event caused by a pathogen with no known wildlife origin); (2) wildlife (zoonotic EID event caused by a pathogen with a wildlife origin); (3) unspecified (zoonotic EID event caused by a pathogen with an unknown origin). (6) ‘Drug resistance’ (DrugRes): (0) event not caused by a drug-resistant microbe; and (1) event caused by a drug-resistant microbe. (7) ‘Transmission mode’ (TranMode): (0) pathogen causing the EID event not normally transmitted by a vector; and (1) pathogen causing the event transmitted by a vector. (8) ‘Driver’. We classified the most commonly cited underlying primary causal factor (or ‘driver’) associated with the EID event according to the classes listed in refs 2, 13. We re-classified ‘Economic development and land use’ and ‘Technology and industry’ to form more descriptive categories: ‘Agricultural industry changes’, ‘Medical industry changes’, ‘Food industry changes’, ‘Land use changes’ and ‘Bushmeat’. (9) ‘Location’. Description of where the first cluster of cases representing each EID event was reported to have occurred. For these descriptions, accurate spatial coordinates (point location data) were found for 51.8% of EID events (n = 220) using Global Gazetteer v.2.1 (http://www.fallingrain.com/world/) and these were assigned to corresponding one degree terrestrial spatial grids. Some EID event locations were lesser known and only described sub-regionally or regionally (for example, SARS in “Guangdong Province, China” or enterohaemorrhagic Escherichia coli in “Peru”). These locations were assigned corresponding boundaries from ESRI sub-regional or regional data24 and we randomly selected only one grid cell from the possible grid cells to represent each particular event. This treated these lesser known events equivalently to those that were assigned a specific point location.
Driver definitions
Definitions of the spatial drivers used are as follows: (1) ‘Human population density’ for 200025 (persons per km2); (2) ‘Human population growth’, calculated between 1990 and 200025.We used a dummy variable to indicate grid cells that experienced rapid growth in human population. This variable was set to 1 for grid cells where the 1990–2000 human population growth exceeded 25% over the decade, and was set to 0 elsewhere; (3) ‘Latitude’ (absolute latitude of the central point of each grid cell, decimal degrees); (4) ‘Rainfall’26 (average rainfall per year, mm); (5) ‘Wildlife host species richness’. We calculated mammalian species richness as a proxy for wildlife host species richness. Richness grids were generated from geographic distribution maps for 4,219 terrestrial mammalian species27.
Controlling for sampling bias
For our temporal analysis, we included the number of JID articles per year since 1945 (nTOTAL = 17,979 articles) as an offset in our generalized linear model using a Poisson error structure. To control for bias in our spatial analysis, we calculated the frequency of the country listed as the address for every author (lead author and coauthors) in each JID article since 1973. This generated a measure of reporting effort for each country which was matched to the one degree spatial grid for analysis and was included in the multiple logistic regression models.
Regression analysis
Each logistic regression was repeated ten times using a separate random draw of the EID event grids for those events where the region reported covered more than one grid cell. The analyses are summarized in Table 1, and given in full in Supplementary Table 3. Different random draws can produce a different number of grid cells with events, even though the number of events does not change. For graphical purposes (that is, in Figs 2 and 3, and Supplementary Figs 1 and 2), we display the first random draw of the EID event grids. Human population density and number of JID articles were log-transformed before analysis. Statistical analyses were carried out using SPSS (v. 12.0)28 and R (v. 2.2.1)29. As the spatial autocorrelation (measured using Moran’s I) in the EID event occurrence spatial grids was low (0.1), the data were assumed to represent independent points in these analyses.
Supplementary information
The file contains the following: Supplementary Data; Supplementary Figures S1-S3; and Supplementary Tables S1-S3. Supplementary Data contains the EID Event Database (Table S1) along with the source details and a comparison to other variable definitions (Table S2). Figure S1 shows the Latitudinal gradient in EID events. Figure S2 shows the Global richness maps of EID events from 1940-2004 split by category and Figure S3 shows the Spatial reporting bias in EID events. Table S3 gives the results of the Socio-economic, environmental and ecological correlates of EID events for 10 random spatial draws. (PDF 4952 kb)
Acknowledgements
We thank the following for discussion, assistance and comments: K. A. Alexander, T. Blackburn, S. Cleaveland, I. R. Cooke, A. A. Cunningham, J. Davies, A. P. Dobson, P. J. Hudson, A. M. Kilpatrick, J. R. C. Pulliam, J. M. Rowcliffe, W. Sechrest, L. Seirup and M. E. J. Woolhouse, and in particular V. Mara and N. J. B. Isaac for analytical support. This project was supported by NSF (Human and Social Dynamics; Ecology), NIH/NSF (Ecology of Infectious Diseases), NIH (John E. Fogarty International Center), Eppley Foundation, The New York Community Trust, V. Kann Rasmussen Foundation and a Columbia University Earth Institute fellowship (K.E.J.).
Author Contributions P.D. conceived and directed the study and co-wrote the paper with K.E.J.; K.E.J. coordinated and conducted the analyses with M.A.L., A.S., N.G.P. and D.B.; N.G.P. compiled the EID event database; and J.L.G provided the mammal distribution data. All authors were involved in the design of the study, the interpretation of the results and commented on the manuscript.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smolinski MS, Hamburg MA, Lederberg J. Microbial Threats to Health: Emergence, Detection, and Response. 2003. [PubMed] [Google Scholar]
- 3.Binder S, Levitt AM, Sacks JJ, Hughes JM. Emerging infectious diseases: Public health issues for the 21st century. Science. 1999;284:1311–1313. doi: 10.1126/science.284.5418.1311. [DOI] [PubMed] [Google Scholar]
- 4.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife — threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 5.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patz JA, et al. Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004;112:1092–1098. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nature Med. 2004;10:S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerging Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morse SS. Emerging Viruses. 1993. pp. 10–28. [Google Scholar]
- 10.Burke DS. Pathology of Emerging Infections. 1998. pp. 1–12. [Google Scholar]
- 11.Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen ML. Changing patterns of infectious disease. Nature. 2000;406:762–767. doi: 10.1038/35021206. [DOI] [PubMed] [Google Scholar]
- 13.Lederberg J, Shope RE, Oakes SCJ. Emerging Infections: Microbial Threats to Health in the United States. 1992. [PubMed] [Google Scholar]
- 14.King DA, Peckham C, Waage JK, Brownlie J, Woolhouse MEJ. Infectious diseases: Preparing for the future. Science. 2006;313:1392–1393. doi: 10.1126/science.1129134. [DOI] [PubMed] [Google Scholar]
- 15.Morse SS. Factors in the emergence of infectious diseases. Emerging Infect. Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houghton JT, et al. Climate Change 2001: The Scientific Basis. 2001. [Google Scholar]
- 17.Patz JA, Campbell-Lendrum D, Holloway T, Foley JA. Impact of regional climate change on human health. Nature. 2005;438:310–317. doi: 10.1038/nature04188. [DOI] [PubMed] [Google Scholar]
- 18.Rogers DJ, Randolph SE. Studying the global distribution of infectious diseases using GIS and RS. Nature Rev. Microbiol. 2003;1:231–237. doi: 10.1038/nrmicro776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guernier V, Hochberg ME, Guegan JFO. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:740–746. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grenyer R, et al. Global distribution and conservation of rare and threatened vertebrates. Nature. 2006;444:93–96. doi: 10.1038/nature05237. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson NM, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 23.Kuiken T, et al. Public health — pathogen surveillance in animals. Science. 2005;309:1680–1681. doi: 10.1126/science.1113310. [DOI] [PubMed] [Google Scholar]
- 24.Environmental Research Systems Institute (ESRI). Data & Maps, Version 9.1 (Environmental Research Systems Institute, Inc., Redlands, California, 2005)
- 25.Center for International Earth Science Information Network (CIESIN) & Centro Internacional de Agricultura Tropical (CIAT). Gridded Population of the World, Version 3 (GPWv3): Population Grids (SEDAC, Columbia University, New York, 2005); available at 〈http://sedac.ciesin.columbia.edu/gpw〉
- 26.International Institute for Applied Systems Analysis (IIASA) & Food and Agricultural Organization (FAO). Global Agro-Ecological Zones (GAEZ) (FAO/IIASA, Rome, 2000); available at 〈http://www.fao.org/ag/agl/agll/gaez/index.htm〉
- 27.Sechrest W. Global Diversity, Endemism and Conservation of Mammals. 2003. [Google Scholar]
- 28.SPSS. SPSS for Windows, Version 12.0 (SPSS Inc., Chicago, 2006)
- 29.R Development Core Team. R: A language and environment for statistical computing, reference index, Version 2.2.1 (R Foundation for Statistical Computing, Vienna, Austria, 2005)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The file contains the following: Supplementary Data; Supplementary Figures S1-S3; and Supplementary Tables S1-S3. Supplementary Data contains the EID Event Database (Table S1) along with the source details and a comparison to other variable definitions (Table S2). Figure S1 shows the Latitudinal gradient in EID events. Figure S2 shows the Global richness maps of EID events from 1940-2004 split by category and Figure S3 shows the Spatial reporting bias in EID events. Table S3 gives the results of the Socio-economic, environmental and ecological correlates of EID events for 10 random spatial draws. (PDF 4952 kb)


