Abstract
Major depressive disorder (MDD) is the most common psychiatric illness worldwide, and it displays a striking sex-dependent difference in incidence, with two thirds of MDD patients being women. Ketamine treatment can produce rapid antidepressant effects in MDD patients, effects that are mediated—at least partially—through glutamatergic neurotransmission. Two active metabolites of ketamine, (2R,6R)-hydroxynorketamine (HNK) and (2S,6S)-HNK, also appear to play a key role in ketamine’s rapid antidepressant effects through the activation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate receptors. In the present study, we demonstrated that estrogen plus ketamine or estrogen plus active ketamine metabolites displayed additive effects on the induction of the expression of AMPA receptor subunits. In parallel, the expression of estrogen receptor alpha (ERα) was also significantly upregulated. Even more striking, radioligand binding assays demonstrated that [3H]-ketamine can directly bind to ERα (KD: 344.5 ± 13 nM). Furthermore, ketamine and its (2R,6R)-HNK and (2S,6S)-HNK metabolites displayed similar affinity for ERα (IC50: 2.31 ± 0.1, 3.40 ± 0.2, and 3.53 ± 0.2 μM, respectively) as determined by [3H]-ketamine displacement assays. Finally, induction of AMPA receptors by either estrogens or ketamine and its metabolites was lost when ERα was knocked down or silenced pharmacologically. These results suggest a positive feedback loop by which estrogens can augment the effects of ketamine and its (2R,6R)-HNK and (2S,6S)-HNK metabolites on the ERα-induced transcription of CYP2A6 and CYP2B6, estrogen inducible enzymes that catalyze ketamine’s biotransformation to form the two active metabolites. These observations provide novel insight into ketamine’s molecular mechanism(s) of action and have potential implications for the treatment of MDD.
Keywords: Ketamine, Ketamine metabolites, Estrogens, CYP2A6, CYP2B6, AMPA receptors
1. Introduction
Major depressive disorder (MDD) displays a striking difference in incidence between the sexes, with two thirds of patients being women, which might be due—at least partially—to biological and hormonal factors unique to women [1, 2]. MDD is the most common psychiatric illness worldwide, with an estimated prevalence of approximately 7% in the adult population [3]. The World Health Organization has estimated that depression will be the leading cause of medical disability worldwide by 2020 [4]. Estrogens have both neuroprotective and neuroexcitatory actions and have been recognized as a factor that can influence dopaminergic, serotoninergic, cholinergic and glutamatergic neurotransmission [5]. It has been well-documented that a striking sex difference in the prevalence of MDD begins at age 13 and continues through middle-to-late adulthood [6–9]. As a result, the incidence of depression increases sharply after puberty [8], and tragically suicide is one of the leading causes of death in adolescents as a result of depression, with more than 50% of adolescent suicide victims suffering from depression at the time of death [8]. The etiology of depression remains unclear, but a recent study suggested that sex-specific patterns of glutamate receptor gene expression in MDD patients might contribute to the sex-related pathophysiology of MDD [10]. Specifically, women with MDD displayed higher levels of glutamate receptor expression in the dorsolateral prefrontal cortex as compared to non-depressed women, a pattern that was not seen in men [10].
Glutamate receptors are included among druggable targets for the treatment of depressive symptoms [11]. For example, ketamine, an antagonist of the N-methyl-D-aspartate (NMDA) receptor, has a very rapid onset of action and is increasingly entering clinical practice for use in the treatment of MDD, especially in patients with treatment resistant depression. Since the first double blind clinical trial designed to assess the antidepressant effects of ketamine in depressed patients was reported in 2000 [12], a growing body of clinical and preclinical studies has provided evidence in support of ketamine’s rapid antidepressant effects [13, 14]. Specifically, ketamine administration, in addition to blocking NMDA receptors, also results in enhanced α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor activity as a result of the effect of ketamine and/or its metabolites (2S,6S;2R,6R)-hydroxynorketamine (HNK), actions that might play a role in its rapid antidepressant effects [15]. Furthermore, sex-dependent antidepressant effects have been observed during ketamine treatment, and estrogens have been shown to mediate increased sensitivity to ketamine therapy in rodent models of depression [16–18]. In addition, (2S,6S;2R,6R)-HNK concentrations were three fold higher in the brains of female than in male mice, which might represent an important factor in the reported sex-dependent difference in the antidepressant effects of ketamine [15]. Similarly, in clinical trials, female subjects had significantly higher plasma levels of (2S,6S;2E,6R)-HNK than did male subjects [19]. A trend toward differences between the sexes in MDD ketamine response has also been reported, but those differences did not reach statistical significance, perhaps as a result of small sample sizes [20–22].
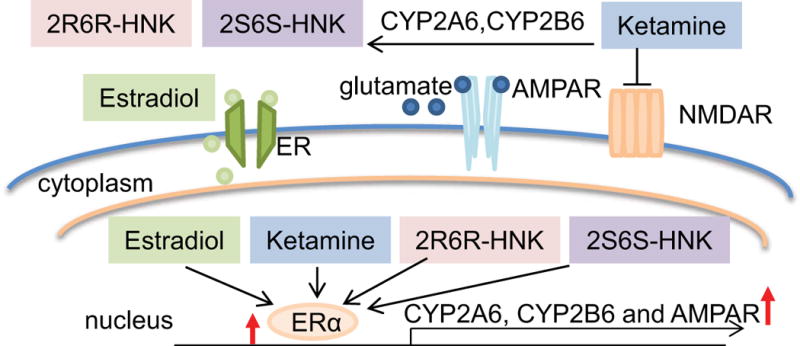
CYP2A6 and CYP2B6 are the major enzymes that catalyze ketamine metabolism [23]. Women have higher levels of gene expression and enzyme activity for CYP2A6 and CYP2B6 [24, 25], both of which are known to be estrogen inducible [26–28]. Included among the mechanisms by which ketamine might influence MDD is activation of the mTOR signaling pathway, which ultimately increases synaptic protein translation and glutamate ionotropic receptor AMPA subunit 1 (GRIA1) trafficking to the cell membrane [17, 29, 30] (see Figure 1).
Figure 1.
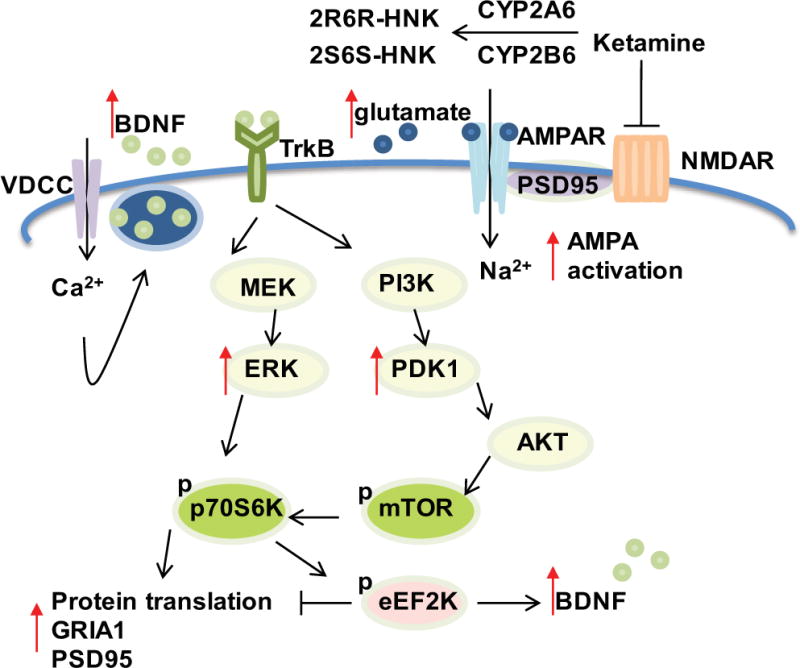
Schematic diagram illustrating ketamine mechanism(s) of action. CYP2A6 and CYP2B6 are major enzymes responsible for ketamine metabolism. Ketamine is an NMDA receptor (NMDAR) antagonist and the active ketamine metabolites (2R,6R-HNK and 2S,6S-HNK) also reportedly contribute to antidepressant effects through the activation of AMPA receptors (AMPRs). Ketamine induces glutamate release. Glutamate in turn binds to AMPARs to induce depolarization as well as sodium (Na2+) and calcium (Ca2+) influx through the AMPA receptors and L-type voltage gated calcium channels (VDCCs), respectively. This results in increased brain-derived neurotropic factor (BDNF) release from synaptic vesicles. BDNF binds to tyrosine kinase receptor B (TrkB) with high affinity, which leads to activation of both the ERK and AKT/mTOR pathways, increasing synaptic protein translation and GRIA1 trafficking to the cell membrane.
In the present study, we have performed a series of experiments designed to help increase our understanding of molecular mechanisms responsible for variation in ketamine’s therapeutic response. Those experiments focused on estrogen receptor alpha (ERα) and the role of ERα as a transcription factor. The first set of experiments was designed to determine whether estrogen might influence the expression of AMPA receptors, the glutamate receptor type reported to be up-regulated in the brains of depressed women [10]. We also pursued underlying mechanism(s) by which ketamine might be able to interact with ERα, a ligand-activated transcription factor, and, as a result, might influence the transcriptional regulation of genes encoding AMPA receptors as well as CYP2A6 and CYP2B6, genes encoding the major enzymes that catalyze ketamine metabolism. Finally, we tested the possibility that ketamine itself might be an ER ligand—and found that it can bind to ER. As a result, the series of observations reported subsequently provides novel insight into ketamine’s molecular mechanisms of action. They also have potential implications for the treatment of MDD with ketamine and/or its active metabolites.
2. Materials and methods
2.1. Cell culture and drug treatment
Human iPSC-derived astrocyte progenitors were cultured and differentiated to mature astrocytes according to the supplier’s instructions (Axol Bioscience, Cambridgeshire, UK). U251-MG cells were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (Cellgro, Manassas, VA, USA) supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA, USA). HepaRG cells were purchased from Biopredic International (Saint Grégoire, France) and were maintained according to the supplier’s instructions. Ketamine hydrochloride was purchased from Pfizer (New York, NY, USA). Both (2R,6R)-2-amino-2-(2-chlorophenyl)-6-hydroxycyclohexanone hydrochloride (2R,6R-HNK) (lot number: NCGC00378227-18) and (2S,6S)-2-amino-2-(2-chlorophenyl)-6-hydroxycyclohexanone hydrochloride (2S,6S-HNK) (lot number: NCGC00373033-12) were synthesized and characterized in Dr. Craig Thomas’s laboratory at the National Center for Advancing Translational Sciences (Rockville, MD, USA). Cells were treated with ketamine or with the ketamine metabolites (2R,6R)-HNK or (2S,6S)-HNK for 24 hours in serum and phenol red-free medium. In some experiments, cells were exposed to estradiol (E2) (Sigma, St. Louis, MO, USA) or fulvestant (Sigma, St. Louis, MO, USA) together with ketamine, (2R,6R)-HNK or (2S,6S)-HNK, at concentrations similar to plasma concentrations observed during ketamine therapy [19].
2.2. RNA isolation and quantitative real time PCR (qRT-PCR)
Total RNA was extracted using the Quick-RNA™ MiniPrep kit (Zymo, Irvine, CA, USA). The PCR reaction contained 100 ng of total RNA, 5 μl of VeriQuest 2X SYBR Green One-Step qRT-PCR master mix (Affymetrix, Santa Clara, CA USA), 1 μl of gene specific primers (Table 1), 0.1 μl of VeriQuest 100X RT enzyme mix and distilled water up to 10 μl per reaction. The expression of ESR1, CYP2A6 and CYP2B6 was quantified using TaqMan gene expression assays (Life Technologies, Carlsbad, CA, USA). qRT-PCR reactions were performed in duplicate using the Applied Biosystems ViiA 7™ Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). The 2−ΔΔCt method was employed for statistical analysis.
Table 1.
Primer sequences
| Gene name | Accession number | Forward primer 5′-3′ | Reverse primer 5′-3′ | PCR size (bp) | |
|---|---|---|---|---|---|
| qPCR | GRIA1 | NM_000827.3 | CGAGCTTTCCCGTTGATACAT | CTGTATCCAGGACTTTCTGCAGG | 162 |
| qPCR | GRIA2 | NM_000826.3 | CATCATTTTGCGGAACACTC | GGTATGCAAACTTGTCCCATTGA | 150 |
| qPCR | GRIA3 | NM_007325.4 | TATTGTGTAGACCTAGCCTATG | ATAGACAAGTTCCCCAACCATG | 138 |
| qPCR | GRIA4 | NM_000829.3 | GTGCAAATAGGT GGTCTCTTCA | GCATTGGGGCTGGTGTTATGA | 92 |
| qPCR | ESR1 | NM_000125.3 | TACATCACGGGGGAGGCAGA | CCGGAGTGTATGCAGGAGAC | 140 |
| qPCR | GAPDH | NM_002046.5 | AGGTCGGAGTCAACGGATTTG | TGTAAACCATGTAGTTGAGGTCA | 123 |
| Reporter gene | CYP2A6 | NC_000019.10 | CTAGAGTTCTCCCAACCTGT | ATGGTGGTAGTGGGATGATA | 2481 |
| Reporter gene | CYP2B6 | NC_000019.10 | GGTACCGGAGAACAGAAAAGAACCTA | GAGCTCCTTTTATCCTGACCCTGCC | 2325 |
2.3. Cloning of promoter reporter gene constructs for CYP2A6 and CYP2B6
The promoter sequences of CYP2A6 and CYP2B6 were PCR amplified using genomic DNA extracted from human lymphoblastoid cell lines. The amplified PCR products (2481 bp and 1845 bp) for the CYP2A6 and CYP2B6 promoters, respectively, were cloned into the Nhel and Sal1 sites of the pGL3 basic plasmid to create pGL3-promoter constructs for CYP2A6 and CYP2B6. PCR primer sequences for the CYP2A6 and CYP2B6 promoters are listed in Table 1. The sequences of the constructs were verified by Sanger sequencing.
2.4. Luciferase reporter gene assays
U251-MG cells were used to perform luciferase reporter gene assays. Cells (1×105) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with 1 μg of the plasmid DNA and 100 ng of pRL-SV40 Renilla luciferase reporter as an internal control (Promega, Madion, WI, USA) in a 6-well plate. After 24 hours, the cells were treated with increasing concentrations of ketamine, (2R,6R)-HNK or (2S,6S)-HNK for 24 hours. Luciferase assays were performed using a dual-luciferase reporter assay system (Promega, Madison, WI, USA). Renilla activity was used to correct for possible variation in transfection efficiency. The pGL3 basic vector without insert was used as a negative control. Expression was calculated as the relative firefly luciferase activity normalized by use of the activity of the transfection control, Renilla luciferase. Results are presented as fold change in relative luciferase units as compared to the pGL3 basic vector. All experiments were repeated three times in triplicate.
2.5. ERE luciferase reporter assays
U251-MG cells (1×105) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with ERE responsive firefly luciferase reporter constructs (Qiagen, Valenicia, CA, USA). Twenty four hours after transfection, the cells were treated for an additional 24 hours with increasing concentrations of ketamine, (2R,6R)-HNK or (2S,6S)-HNK. Luciferase assays were then performed with the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA). Relative luciferase activities were normalized to Renilla luciferase activities. Results were presented as fold change in relative luciferase units compared with vehicle treatment. All experiments were performed at least three times in triplicate.
2.6. ERα ligand binding assays
Recombinant full length human estrogen receptor alpha (ESR1-1033H, Creative BioMart, NY, USA) was used to perform radioligand binding assays and surface plasmon resonance (SPR). For the radioligand binding assays, protein was diluted in ice-cold assay buffer (10 mM Tris-HCl; 1 mM EDTA; 1 mM EGTA; 1 mM NaVO3; 1% glycerol; 0.25 mM leupeptin; 1% BSA; 1 mM DTT). Estradiol-[2,4,6,7- 3H(N)], 9.25 MBq (specific activity 70-115 Ci/mmol, lot number 2117185) was purchased from Perkin Elmer Life Sciences (Boston, MA, USA). Unlabeled 17β-estradiol and components of assay buffer were obtained from Sigma-Aldrich (St. Louis, MO, USA). (±)-Ketamine [N-methyl-3H] hydrochloride, 2.96-3.145 TBq/mmol (specific activity 80 Ci/mmol, lot number 17115) was purchased from American Radiolabeled Chemicals (St. Louis, MO, USA). Unlabeled ketamine, and the two ketamine metabolites, (2R,6R)-HNK and (2S,6S)-HNK, were obtained as described above. ER saturation binding assays were performed to measure total binding (TB) and non-specific binding (NSB). To determine nonspecific binding, a 500-fold excess of non-radioactive ligand, i.e. 500 nM E2, was incubated overnight together with 0.75 nM ER protein and radioactive ligand, i.e. 1 nM Estradiol-[6,7-3H(N)] at 4°C. Bound and unbound estradiol were separated by incubation with ice-cold dextran-coated charcoal (DCC) on ice for 10 minutes. After centrifugation for 15 mins at 8000rpm, 100 μl of DCC suspension was gently transferred into LSC vials containing 1 ml Ultima Gold scintillation cocktail (Perkin Elmer Life Sciences, Boston, MA, USA). Radioactivity was then measured using a Beckmann LS 6500 liquid scintillation counter (Ramsey, MN, USA). Specific binding (SB) was calculated by subtracting the value for NSB from that for TB. The equilibrium dissociation constant for the radioligand (KD) was calculated using non-linear regression analysis (GraphPad Prism Software v7, San Diego, CA, USA). [3H]-estradiol displacement binding assays were performed with recombinant ER protein, 1 nM [3H]-estradiol and increasing concentrations of non-radioactive ligand, i.e. non-radioactive E2 (0.002 nM to 20 μM), or ketamine and the two active ketamine metabolites: (2R,6R)-HNK and (2S,6S)-HNK (4 nM - 40 mM). [3H]-ketamine displacement assays were performed in a similar fashion using ESR1 protein, 100 nM [3H]-ketamine and increasing concentrations of non-radioactive ligand, i.e. non-radioactive E2 or ketamine and the two active metabolites: (2R,6R)-HNK and (2S,6S)-HNK (0.04 nM – 400 μM). Separation of bound and unbound radioligands was performed as described above. Competition curves were plotted as the percentage of SB of radioactive ligand versus increasing concentrations of non-radioactive ligand. %SB=[(bound-NSB)]×100/(TB-NSB). The IC50 value was the competitor concentration that caused 50% displacement of non-radioactive ligand. IC50 values were calculated using nonlinear regression analysis (GraphPad Prism Software v7, San Diego, CA, USA). The radioligand binding assays were performed in duplicate with at least three independent experiments.
2.7. Surface Plasmon Resonance (SPR)
We performed NTA mixed capture of His6-tagged hER (ESR1-1033H, Creative BioMart, NY, USA) using a Biacore T200 surface plasmon resonance (SPR) analyzer (GE Healthcare). Binding of ketamine to ERα was detectable by this method, which an equilibrium dissociation constant (KD) of 215 ± 100 nM. Briefly, the NTA chip (BR-1000-34) was conditioned with 350 mM EDTA (pH 8.3) for 1 min followed by washing with immobilization buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.01 % (w/w) Polysorbate) for 5 min at a flow 30 μL/min. After a 0.5 mM NiCl2 injection (60 s), the His6-tagged hER protein (0.2 mg/ml) was captured at a flow of 5 μl/min. Additionally, after capture, the ligand was cross-linked using amine coupling with a 1 min pulse of N-ethyl-N′-[dimethylaminopropyl]carbodiimide/N-hydroxysuccinimide) and ethanolamine, and reached a level of 11000-12000 resonance units (RU). Ketamine in calcium- and magnesium-free Dulbecco’s phosphate buffered saline containing 0.01% (w/w) Polysorbate was run over the chip with a flow of 50 μl/min for 30 s and allowed to dissociate for 60 s. Kinetic analysis of SPR data was performed using BiaEvaluation (GE). Sensograms were subtracted for background contributions, and affinity constants were derived using a steady state affinity fitting 1:1 interaction model.
2.8. Immunofluorescence staining and confocal imaging analysis
U251-MG cells were grown on glass coverslips and treated with E2 0.1 nM or ketamine or the two active ketamine metabolites (400 nM) for 24 hours. Cells were then fixed in 4% paraformaldehyde at room temperature for 10 min. Cells were washed in cold PBS and permeablized with 0.2% Triton X-100 in PBS. After blocking for one hour with 3% BSA, cells were incubated with mouse anti-ESR1 antibody (Santa Cruz Biotechnology, Dallas, Texas, USA) overnight at 4°C. The secondary antibody (red) was ab150119 Alexa Fluor® 647 goat anti-mouse IgG (H+L) used at 1:1000 dilution for an hour. DAPI was used to stain the cell nuclei (blue) at a concentration of 1.43 μM. Slides were visualized using fluorescence microscopy (Olympus, FV1200).
2.9. Statistics
GraphPad Prism Software v7 (San Diego, CA, USA) was used for data analysis. Results were presented as mean ± S.E.M. Gene expression and luciferase activities were analyzed using One-way or two-way ANOVA, followed by Tukey’s multiple comparison tests for individual comparisons when significant effects were detected. Differences were considered significant at p<0.05.
3. Results
3.1. Estrogens and ketamine act additively to induce AMPA receptor mRNA expression
As a first step in this series of experiments, the mRNA expression of AMPA receptors in U251-MG cells was determined after exposure of the cells to estradiol (E2), ketamine, (2R,6R)-HNK or (2S,6S)-HNK—either alone or together. The concentrations of E2, ketamine and ketamine metabolites used to perform these experiments were selected to fall within the physiological range for E2 and within the range of concentrations of ketamine and ketamine metabolites observed during ketamine infusion therapy for patients with depression [19, 31]. As shown graphically in Figure 2A, E2 induced the expression of the GRIA1, GRIA2 and GRIA4 AMPA receptor subunits by approximately 2 fold in a dose-dependent fashion. In similar fashion, ketamine exposure induced mRNA expression of the same AMPA receptor subunit genes, GRIA1, GRIA2 and GRIA4, approximately 1.5-2 fold in these cells (Figure 2B). However, GR1A3 mRNA expression was not altered significantly in response to either E2 or ketamine treatment. The expression of GRIA1, GRIA2 and GRIA4 responded in a fashion very similar to the response to E2 when the cells were exposed to ketamine and to the ketamine metabolites (2R,6R)-HNK and (2S,6S)-HNK (Figure 2B)—at concentrations that are observed in vivo during ketamine infusion therapy [19]. Strikingly, the addition of E2 at physiologic concentrations was additive to the effect of ketamine on GRIA1, GRIA2 and GRIA4 mRNA expression (Figure 2B). However, this additive effect was only observed at 0.1 nM E2 together with 400 nM ketamine. Similarly, E2 exposure together with the ketamine metabolites (2R,6R)-HNK or (2S,6S)-HNK also displayed an additive effect on the induction of AMPA receptor subunits, once again with the exception of GRIA3 (Figure 2B). Similar results with regard to E2 plus ketamine or ketamine metabolites resulting in the induction of AMPA receptor subunit mRNA expression were observed when we conducted experiments using primary human astrocytes (Figure 3).
Figure 2.
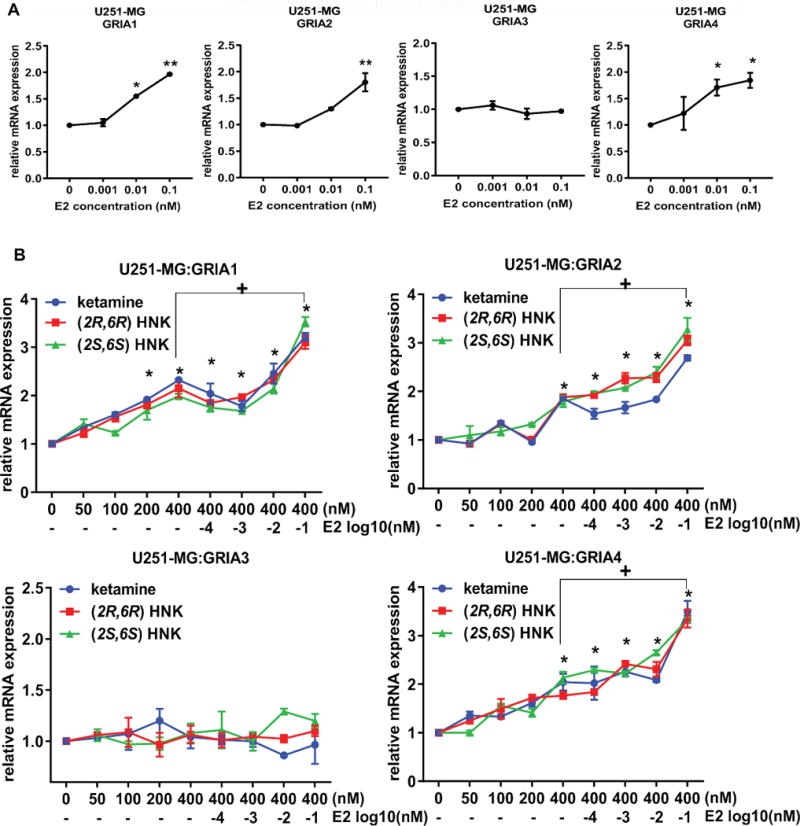
Effects on the expression of GRIA1, GRIA2, GRIA3 and GRIA4 in U251-MG cells in response to various treatment conditions, including exposure to increasing concentrations of (A) E2 (0.001-0.1 nM) and (B) ketamine or the active ketamine metabolites (2R,6R)-HNK or (2S,6S)-HNK (0-400 nM) alone or ketamine (400 nM) together with increasing concentration of E2 (0.0001-0.1 nM). ANOVA was performed to compare gene expression, followed by Tukey’s multiple comparisons when significant effects were detected. *p ≤0.05, **p ≤0.005 as compared to vehicle treatment. + p ≤0.05 as compared to the same treatment with or without E2.
Figure 3.
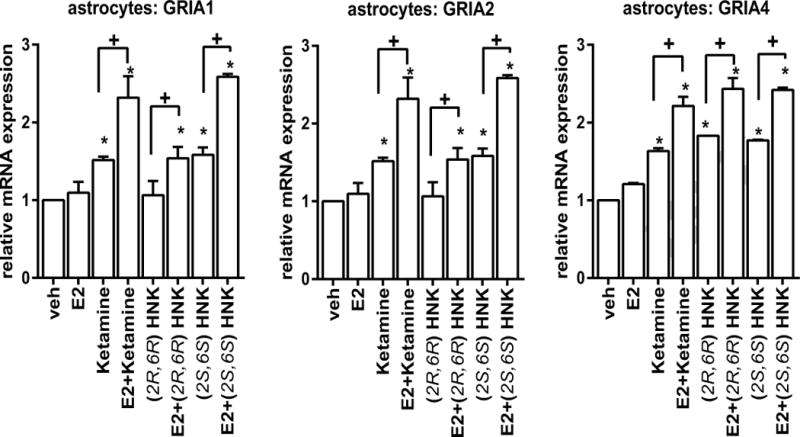
Effects on the expression of AMPA receptor subunits GRIA1, GRIA2, and GRIA4 in primary human astrocytes. ANOVA was performed to compare gene expression, followed by Tukey’s multiple comparisons when significant effects were detected. *p ≤0.05, as compared to vehicle treatment. + p ≤0.05 as compared to the same treatment with or without E2.
The majority of MDD patients are women, so our results raised the possibility that these E2-based effects might contribute to ketamine treatment response in women [19]. As the next step in this series of studies, we set out to determine whether ERα—a ligand-activated transcription factor—might play a role in response to ketamine treatment. We found that ketamine and its active metabolites significantly increased ERα expression in a concentration-dependent fashion (Figure 4A) and, even more striking, that E2 acted additively with ketamine to further induce ERα expression (Figure 4A). These observations were paralleled by the results of estrogen response element (ERE) luciferase reporter assays (Figure 4B). This series of findings led us to ask whether ketamine and its metabolites might induce the expression of the two CYP isoforms that are known to catalyze the formation of the ketamine metabolites, CYP2A6 and CY2B6 [23], both of which are known to be estrogen inducible as a consequence of functional EREs located in their promoters [26–28].
Figure 4.
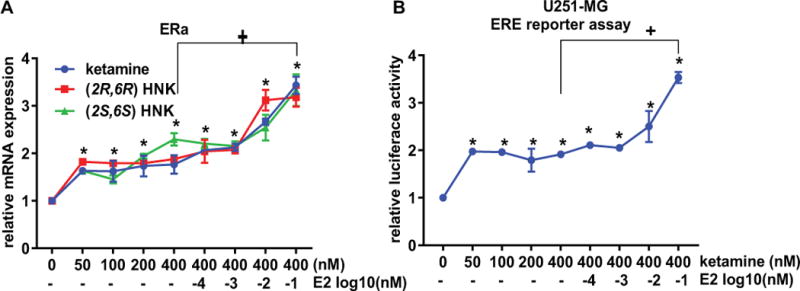
(A) ERα mRNA expression was determined by qPCR 24 hours after U251-MG cells were exposed to increasing concentrations of ketamine, (2R,6R)-HNK, or (2S,6S)-HNK. E2 plus ketamine or E2 plus the active metabolites resulted in additive effects on the induction of ERα expression. (B), U251-MG cells were transfected with the ERE dual luciferase reporter constructs, and were exposed to increasing concentrations of ketamine, (2R,6R)-HNK, or (2S,6S)-HNK alone or in combination with increasing concentrations of E2. ANOVA was performed to compare gene expression, followed by Tukey’s multiple comparisons. *p ≤0.05, **p ≤0.005 as compared to vehicle treatment. + p ≤0.05 as compared to the same with or without E2.
3.2. CYP2A6 and CYP2B6 induction by ketamine and its metabolites
Based on observations described in preceding paragraphs, we considered the possibility that the expression of CYP2A6 and CYP2B6 might also be induced by ketamine and its metabolites. CYP2A6 and CYP2B6 are not highly expressed in the brain. Therefore, we created reporter constructs for these two genes that included the functional EREs present in their promoters, as shown schematically in Figure 5A. Consistent with previous reports [26–28], E2 induced transcriptional activity for both the CYP2A6 and CYP2B6 constructs (Figure 5B). In addition ketamine, (2R,6R)-HNK and (2S,6S)-HNK could also induce transcriptional activity for both the CYP2A6 and CYP2B6 promoter constructs. Even more striking, this induction could be further enhanced by E2 exposure after expression in both U251-MG and HepaRG cells (Figure 5C and 5D).
Figure 5.
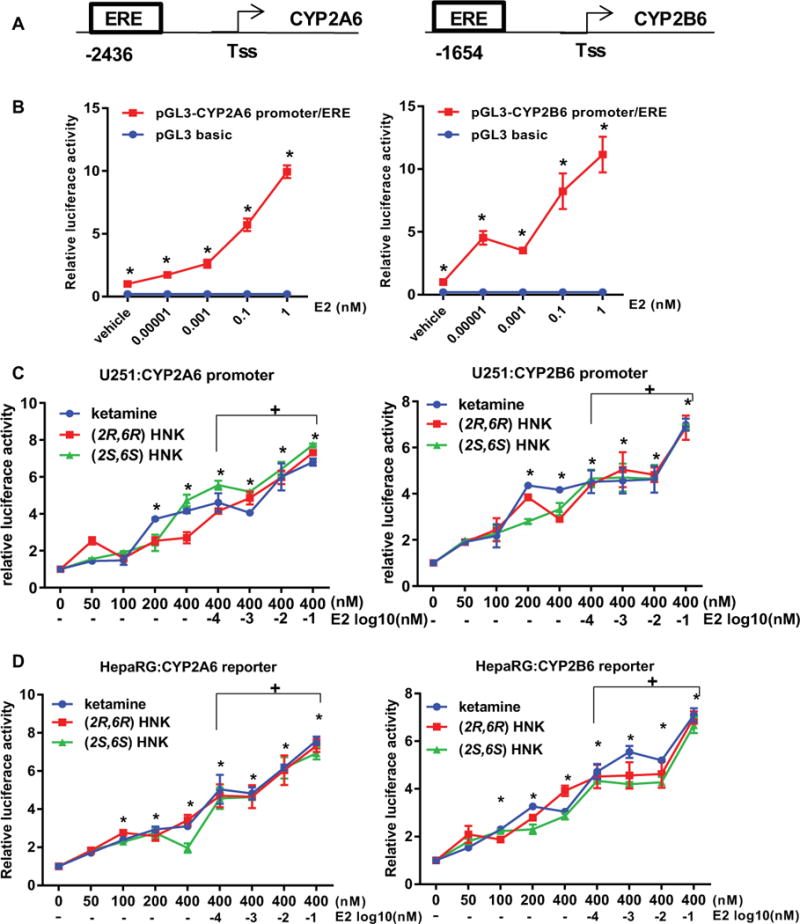
Luciferase reporter assays in U251-MG cells expressing either CYP2A6 or CYP2B6. (A) Schematic representations of the promoters for CYP2A6 and CYP2B6 with the locations of ERE motifs indicated. Tss represents the site of transcription initiation. Cells were transfected with the dual luciferase reporter constructs and treated with increasing concentrations of E2 for 24 hours. (B) Transcriptional activities for both the CYP2A6 and CYP2B6 promoter constructs could be induced by E2 in a concentration-dependent fashion. Results shown are average values for three independent experiments (*p<0.0001 when compared with the vehicle control). (C) U251-MG Cells and (D) HepaRG cells were transfected with the dual luciferase reporter constructs and were treated with increasing concentrations of ketamine, (2R,6R)-HNK or (2S,6S)-HNK and the three compounds plus increasing concentrations of E2. All three compounds significantly induced transcriptional activities for CYP2A6 or CYP2B6 reporter constructs in a dose-dependent fashion (*p<0.005, **p<0.0005 vs vehicle treatment). + p ≤0.05 as compared to the same with or without E2. Results are presented as fold change in relative luciferase units compared with the pGL3 basic vector. ANOVA was performed to compare luciferase activities, followed by Tukey’s multiple comparison tests for individual comparisons when significant effects were detected.
These observations were confirmed when gene expression studies were performed with human astrocytes (Figure 6A), and with HepaRG cells (Figure 6B), liver-derived cells that highly express CYP2A6 and CYP2B6. Surprisingly, induction of the expression of CYP2A6 and CYP2B6—both of which are known to be induced by estrogens—was also observed in the absence of estrogens in response to treatment with ketamine and the ketamine metabolites, (2R,6R)-HNK and (2S,6S)-HNK (Figure 6A and 6B).
Figure 6.
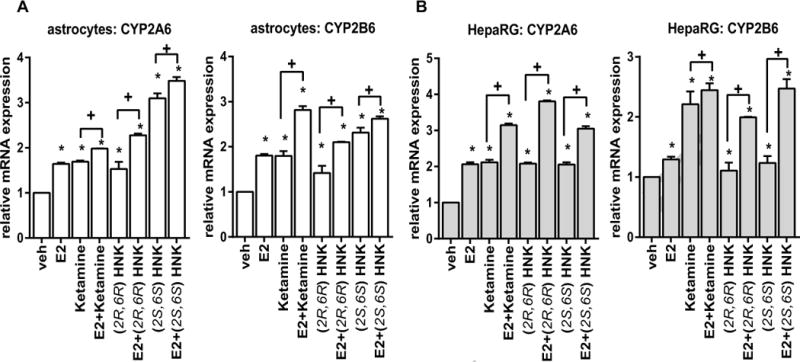
Effects on the expression of CYP2A6 and CYP2B6 in primary human astrocytes (A) or HepaRG cells (B) in response to various treatment conditions. ANOVA was performed to compare gene expression, followed by Tukey’s multiple comparisons when significant effects were detected. *p <0.05, as compared to vehicle treatment. + p ≤0.05 as compared to the same treatment with or without E2.
3.3. Induction of CYP2A6, CYP2B6 and AMPA receptors by ketamine is lost after ER blockade
To provide further evidence that the induction of CYP2A6 and CYP2B6 expression was mediated by ERα, we examined the impact of ERα knockdown on this induction. The induction of CYP2A6 and CYP2B6 mRNA expression by ketamine was lost after ERα was silenced by siRNA knockdown (Figure 7A). In addition, (2R,6R)-HNK and (2S,6S)-HNK also failed to induce the expression of CYP2A6 and CYP2B6 after ERα knockdown. Similar results were observed when ERα signaling was inhibited using fulvestant (ICI)—an ER blocker (Figure 7B). Furthermore, ketamine, (2R,6R)-HNK and (2S,6S)-HNK all failed to induce the expression of GRIA1, GRIA2 and GRIA4 when ERα was knocked down (Figure 8) or silenced pharmacologically (Figure 9). In contrast, all three of these AMPA receptor subunit genes could be induced by E2 and by ketamine and its metabolites when ERα was present, as shown in Figure 2. Taken as a whole, these results indicated that ERα can play an important role in the transcriptional effects of ketamine and the two ketamine metabolites, i.e., in the induction of the expression of CYP2A6 and CYP2B6 by ketamine and by (2R,6R)-HNK or (2S,6S)-HNK. They also raised the possibility that ketamine and its two active metabolites might either enhance ERα binding to EREs through an unknown mechanism or that they might themselves be ER ligands.
Figure 7.
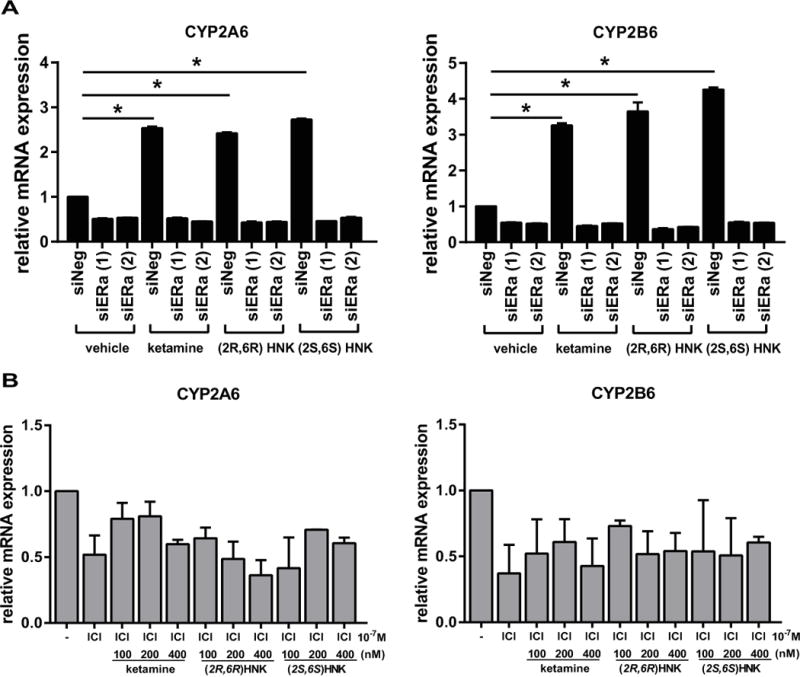
(A) Induction of both CYP2A6 and CYP2B6 expression by ketamine and the ketamine metabolites (2R,6R)-HNK, or (2S,6S)-HNK was lost when ERα was silenced by the use of siRNA. *p ≤0.005 as compared to control siRNA. (B) Induction of both CYP2A6 and CYP2B6 expression by ketamine and the ketamine metabolites (2R,6R)-HNK, or (2S,6S)-HNK was lost when ERα was blocked using ICI, an estrogen receptor blocker.
Figure 8.
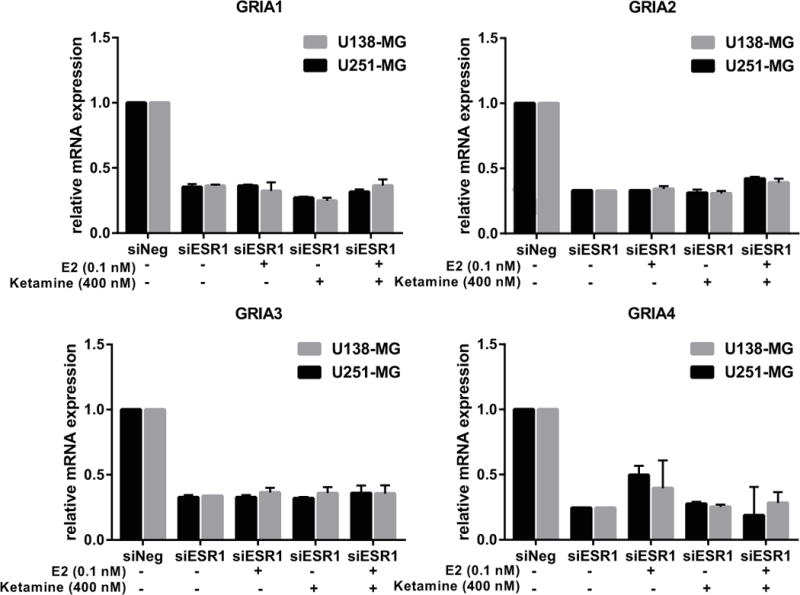
Induction of GRIA1, GRIA2, GRIA3 and GRIA4 expression by ketamine and the ketamine was lost when ERα was silenced by the use of siRNA in both U138-MG and U251-MG cells.
Figure 9.
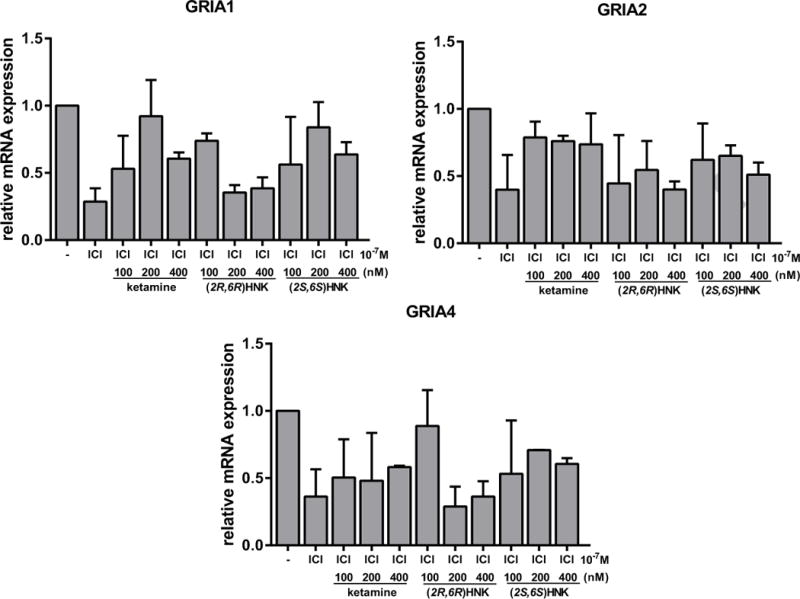
Induction of the expression of GRIA1, GRIA2 and GRIA4 by ketamine and ketamine metabolites was lost after ERα blockade by fulvestant (ICI). Specifically, the expression of AMPA receptors is known to be enhanced in response to ketamine therapy. However, we observed that GRIA1, GRIA2 and GRIA4 were not induced by increasing concentrations of ketamine or its metabolites (100, 200, 400 nM) after ER blockade. Values shown are mean +/− SEM of three independent determinations.
3.4. Ketamine, (2R,6R)-HNK and (2R,6R)-HNK as novel ERα ligands
We next set out to test the hypothesis that ketamine might bind directly to ERα. In initial experiments, we examined the ability of ketamine to bind to ERα using SPR, a biophysical technique that detects interactions between an immobilized protein and a ligand in solution. As indicated in Figure 10, binding of ketamine to ERα was readily detectable by this method, with an equilibrium dissociation constant (KD) of 215 ± 100 nM. Using a more sensitive and more physiological approach, we also performed radioligand binding assays. When the amount of radioactive ligand bound to ERα protein was assessed at nine different concentrations of [3H]-ketamine (range 4 nM to 400 nM) [3H]-ketamine bound to ERα with a KD= 344 ± 13 nM based on non-linear regression analysis of the saturation binding assay (Figure 11A). We also performed [3H]-ketamine displacement assays and found that both of the ketamine active metabolites, (2R,6R)-HNK and (2S,6S)-HNK, could displace [3H]-ketamine binding with similar affinity (IC50 values= 3.40 ± 0.2, and 3.53 ± 0.2 μM, respectively) (Figure 11B). These observations suggested that ketamine and its’ two active metabolites were most likely binding to the same site on ERα. However, E2 was unable to displace bound [3H]-ketamine, indicating that ketamine might bind to ERα at a novel site different from the estradiol binding site and, as a result, E2 was ineffective in displacing [3H]-ketamine binding (Figure 11B). To further explore that possibility, we also performed [3H]-estradiol displacement assays to confirm that estradiol and ketamine might have different binding sites at ERα. Specially, [3H]-estradiol saturation binding assays (range of concentrations tested from 0.156 nM to 20 nM), demonstrated that estradiol had a much higher affinity for ERα (KD= 3.67 ± 0.56 nM), as compared to that for ketamine (Figure 12A). [3H]-Estradiol displacement assays confirmed that non-radioactive estradiol displaced bound [3H]-estradiol in a concentration-dependent fashion (IC50= 64.4 ± 0.56 nM). However, ketamine and the two active ketamine metabolites failed to displace [3H]-estradiol binding (Figure 12B). Finally, to complement and extend our studies of the potential functional effects of ketamine on ERα as demonstrated by the luciferase reporter gene assay results shown graphically in Figure 5, we performed immunofluorescence studies of U251-MG cells after exposure to E2, to ketamine or to the two ketamine active metabolites. Just as exposure to E2 resulted in nuclear translocation of ERα, so too did exposure to ketamine or its active metabolites (Figure 13).
Figure 10.
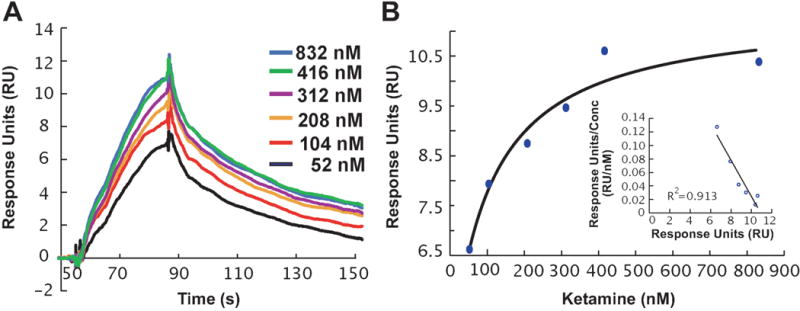
(A) SPR sensorgrams showing the dose-dependent binding of ketamine (52 – 823 nM) to ERα, as indicated by relative response units (RUs) after background subtraction. (B) Plot of RUmax versus ketamine concentration. Inset, Scatchard plot of ketamine binding.
Figure 11.
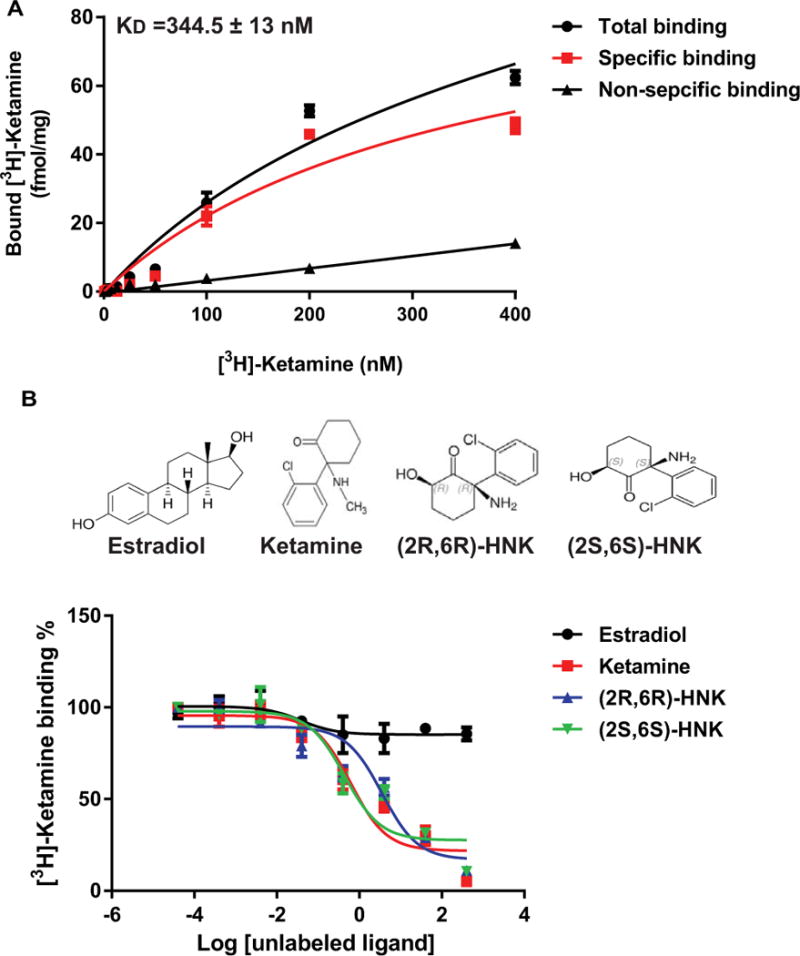
(A) Saturation curves for the binding of increasing concentrations of [3H]-ketamine to ERα (KD=344.5 ± 13 nM). (B) [3H]-Ketamine competition binding curves. Data are expressed as percentages of specific binding of [3H]-ketamine vs. log of the competitor concentration (non-radioactive drug concentrations ranged from 0.4 nM to 400 μM). Each point represents the mean±SEM of three independent determinations.
Figure 12.
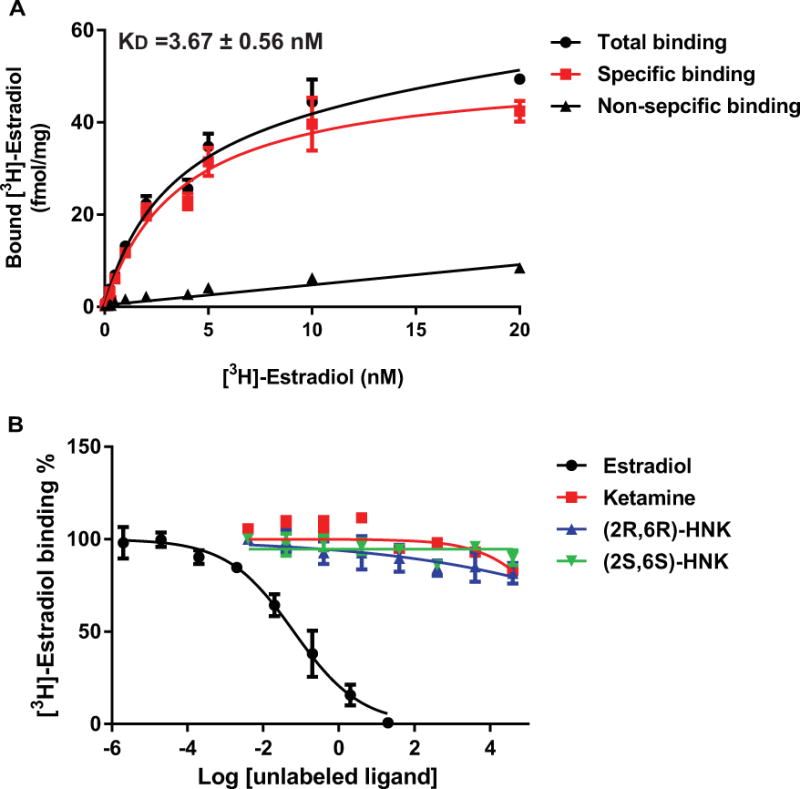
(A) Saturation curves for the binding of increasing concentrations of [3H]-estradiol to ERα (KD=3.67 ± 0.56 nM). (B) [3H]-estradiol competition binding curves. Data are expressed as percentages of specific binding of [3H]-estradiol vs. log of the competitor concentration (non-radioactive E2 concentrations ranged from 0.0002 nM to 20 μM, concentrations of ketamine and 4nM to 40mM for the two metabolites. Each point represents mean±SEM of three independent determinations.
Figure 13.
Immunofluorescence staining of U251-MG cells showing ERα nuclear translocation after the cells were treated with E2 (0.1 nM), or with 400 nM ketamine or its active metabolites.
4. Discussion
Ketamine, a novel and highly promising drug for the treatment of MDD, has a very rapid onset of action [13]. In particular, MDD patients and experimental animal models for MDD both display rapid antidepressant response within hours after ketamine administration [20–22]. Ketamine is known to be an NMDA receptor antagonist. However, a recent study suggested that the antidepressant effect of the two major ketamine active metabolites (2S,6S;2R,6R)-hydroxynorketamine (HNK) depends on the activation of AMPA receptors [15]. In that study, (2S,6S;2R,6R)-HNK were essential for ketamine’s antidepressant effects, but (2R,6R)-HNK lacked some ketamine-related side effects [15]. In addition, several preclinical studies have reported sex-related differences in the antidepressant effect of ketamine. In particular, females displayed greater sensitivity to ketamine than males in rodent models of depression [16–18]. However, the mechanism(s) underlying this sex-related enhanced sensitivity to ketamine in female rodent models has remained unclear.
In the present study, we set out to determine whether estrogen might play a role in regulation of the expression of the glutamate receptors that are among the therapeutic targets for ketamine in the treatment of MDD. We chose to perform these studies using U251-MG cells, cells derived from an astrocytoma, which resulted in data comparable with results generated using primary human astrocytes (Figure 3 and 6). Use of this cell line also made it possible for us to subsequently pursue functional genomic studies, which would have been impractical using primary astrocytes due to their high cost and the lengthy time required for differentiation. Our results indicated that E2 could induce the expression of three of the four AMPA subunits. We also found that ketamine and the active ketamine metabolites (2R,6R)-HNK and (2S,6S)-HNK behaved in a similar fashion. Specifically, all three compounds could induce the mRNA expression of CYP2B6 and CYP2A6 in cell lines obtained from both brain and liver (Figure 6). (2S,6S)-HNK and (2R,6R)-HNK are the major HNK metabolites found in the plasma of both humans and mice; and both of these metabolites are being considered as possible next generation rapid acting antidepressant agents [15, 32]. It should also be pointed out that female subjects who received ketamine infusion therapy appeared to have higher concentrations of these active ketamine metabolites than did male subjects [19], which might have occurred, in part, because females had higher expression and activity of CYP2A6 and CYP2B6 [26–28] and because the formation of (2S,6S;2R,6R)-HNK results from biotransformation catalyzed primarily by these two CYP enzymes [23], enzymes that are known to be estrogen inducible [26]. These observations led us to ask whether ketamine and the two ketamine metabolites might—like E2—be ER ligands. Our radioligand binding and SPR studies showed that all three compounds bound to ERα—although the site of binding remains unclear. Further studies such as protein-ligand docking will be needed to determine the position and orientation of their binding to ERα.
The present study used a variety of cell lines to perform functional genomic studies. However, each of those cell lines has limitations. Therefore, our observations will have to be replicated by future studies conducted with clinical samples and/or with cell lines derived from patients, i.e, iPS cells. However, the present results represent a potentially important step in the process of obtaining functional insight into mechanism of action for ketamine and its active metabolites, and the present study is the first to demonstrate the binding of ketamine and its active metabolites to ERα. Ketamine and the two major ketamine metabolites that we studied were all able to regulate the expression of ESR1, the gene that encodes ERα (Figure 4A), as well as the genes encoding three of the four AMPA receptor subunits and CYP2A6 and CYP2B6 in a concentration dependent fashion. Strikingly, estrogens exerted additive effects on ERα expression during exposure of cells to ketamine and the two ketamine active metabolites (2R,6R)-HNK and (2S,6S)-HNK. The present study also demonstrated that non-radioactive ketamine displaced [3H]-ketamine binding to ERα with affinities similar to those for (2R,6R)-HNK and (2S,6S)-HNK (Figure 11). Therefore, ketamine and the ketamine metabolites (2R,6R)-HNK and (2S,6S)-HNK appear to be novel ERα ligands, although the site to which they bind remains unclear. These results also suggested the existence of a positive feedback loop by which estrogen can augment the effects of ketamine and its (2R,6R)-HNK and (2S,6S)-HNK metabolites on the transcription of ERα (Figure 4A). We also found that ERα intracellular localization was altered when cells were exposed to ER ligands, in this case E2, or ketamine and its two active metabolites (Figure 13). Specifically, when cells were treated with E2, or ketamine or its metabolites, ERα trafficked from the cytoplasm to the nucleus where it acts as a ligand activated transcription factor. Both CYP2A6 and CYP2B6 can be induced by E2, in part, due to functional estrogen response elements (EREs) in their promoter regions, which could interact with ERα and result in transcription regulation, consistent with previous reports [26–28]. The present study demonstrated that knockdown of ERα resulted in downregulation of the expression of AMPA receptors, and the induction of AMPA expression by E2 or ketamine treatment was lost (Figure 8). Future studies are warranted to determine molecular mechanisms underlying the additive effects on AMPA expression of ketamine plus E2 treatment.
Given the interaction of ketamine and its metabolites with ERα, one question is whether there may be any potential concerns relating to estrogen-related cancers. It is known that endogenous estrogen levels are directly associated with risk for developing breast cancer [33]. The use of estrogens in the treatment of infertility has been associated with an increased risk of breast cancer but only if the women achieved a 10+ week pregnancy [34], which is a potential reason, to avoid pregnancies in women receiving ketamine. In the placebo-controlled Women’s Health Initiative Study of unopposed conjugated equine estrogens in menopausal women without a uterus, there was a decrease in the incidence of breast cancer [35]. The main issue with unopposed estrogens appears to be the risk for endometrial cancer [36], which might indicate that monitoring postmenopausal women treated with ketamine for endometrial abnormalities would be prudent.
In conclusion, two thirds of MDD patients are women, which might be due, at least in part, to biological and hormonal factors unique to women. Estrogens can cross the blood brain barrier and interact with cells in the central nervous system. ERα is a ligand-activated transcription factor, and reduced ERα mRNA expression was previously reported in the basomedial nucleus of the amygdala in patients with MDD [37]. The present study is the first to identify ketamine and its metabolites (2R,6R)-HNK and (2S,6S)-HNK as possible novel ERα ligands. Our results also showed that estrogens and ketamine can act in an additive fashion to induce the expression of three of the four AMPA receptor subunits, and it has been reported that these receptors may play a major role in the antidepressant effects of ketamine. Finally, silencing of ERα blocked induction of the expression of genes encoding CYP2A6, CYP2B6 and three of the four AMPA receptor subunits. This series of observations highlight(s) the possible role that ERα may play in the transcriptional effects of ketamine and the two major ketamine active metabolites, thus contributing to ketamine therapeutic response in major depressive disorder (MDD)—a disease for which two thirds of patients are women.
Acknowledgments
The ketamine metabolites: (2R,6R)-hydroxynorketamine (lot number: NCGC00378227-18) and (2S,6S)-hydroxynorketamine (lot number: NCGC00373033-12) were synthesized and characterized in Dr Craig Thomas’s laboratory at the National Center for Advancing Translational Sciences. We thank Dr. Thomas and his team for these compounds. This work was supported in part by National Institutes of Health grants U19 GM61388 (the Pharmacogenomics Research Network –PGRN), P50 CA11620, RO1 CA196648, RO1 GM28157, RO1 MH108348, U54 GM114838 and RO1 CA138461.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
Drs. Richard Weinshilboum and Liewei Wang are co-founders and stockholders in OneOme, a pharmacogenomics decision support company.
References
- 1.Murrough JW, Abdallah CG, Mathew SJ. Targeting glutamate signalling in depression: progress and prospects. Nat Rev Drug Discov. 2017 doi: 10.1038/nrd.2017.16. advance online publication. [DOI] [PubMed] [Google Scholar]
- 2.Kuehner C. Why is depression more common among women than among men? The Lancet Psychiatry. 2017;4:146–58. doi: 10.1016/S2215-0366(16)30263-2. [DOI] [PubMed] [Google Scholar]
- 3.Lépine J-P, Briley M. The increasing burden of depression. Neuropsychiatric Disease and Treatment. 2011;7:3–7. doi: 10.2147/NDT.S19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez AD, Murray CCJL. The global burden of disease, 1990-2020. Nat Med. 1998;4:1241–3. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- 5.Gillies GE, McArthur S. Estrogen Actions in the Brain and the Basis for Differential Action in Men and Women: A Case for Sex-Specific Medicines. Pharmacological Reviews. 2010;62:155–98. doi: 10.1124/pr.109.002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade TJ, Cairney J, Pevalin DJ. Emergence of Gender Differences in Depression During Adolescence: National Panel Results From Three Countries. Journal of the American Academy of Child & Adolescent Psychiatry. 2002;41:190–8. doi: 10.1097/00004583-200202000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Hankin BL, Abramson LY, Moffitt TE, Silva PA, McGee R, Angell KE. Development of depression from preadolescence to young adulthood: emerging gender differences in a 10-year longitudinal study. J Abnorm Psychol. 1998;107:128–40. doi: 10.1037//0021-843x.107.1.128. [DOI] [PubMed] [Google Scholar]
- 8.Thapar A, Collishaw S, Pine DS, Thapar AK. Depression in adolescence. Lancet. 2012;379:1056–67. doi: 10.1016/S0140-6736(11)60871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler RC, McGonagle KA, Swartz M, Blazer DG, Nelson CB. Sex and depression in the National Comorbidity Survey I: Lifetime prevalence, chronicity and recurrence. Journal of Affective Disorders. 1993;29:85–96. doi: 10.1016/0165-0327(93)90026-g. [DOI] [PubMed] [Google Scholar]
- 10.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol Psychiatry. 2015;20:1057–68. doi: 10.1038/mp.2015.91. [DOI] [PubMed] [Google Scholar]
- 11.Newport D Jeffrey, Carpenter Linda L, McDonald William M, Potash James B, Tohen Mauricio, Nemeroff Charles B, et al. Ketamine and Other NMDA Antagonists: Early Clinical Trials and Possible Mechanisms in Depression. American Journal of Psychiatry. 2015;172:950–66. doi: 10.1176/appi.ajp.2015.15040465. [DOI] [PubMed] [Google Scholar]
- 12.Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, et al. Antidepressant effects of ketamine in depressed patients. Biological Psychiatry. 2000;47:351–4. doi: 10.1016/s0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 13.Zarate CA, Jr, Machado-Vieira R. Ketamine: translating mechanistic discoveries into the next generation of glutamate modulators for mood disorders. Mol Psychiatry. 2017;22:324–7. doi: 10.1038/mp.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheuing L, Chiu C-T, Liao H-M, Chuang DM. Antidepressant mechanism of ketamine: perspective from preclinical studies. Frontiers in Neuroscience. 2015;9 doi: 10.3389/fnins.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanos P, Moaddel R, Morris PJ, Georgiou P, Fischell J, Elmer GI, et al. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature. 2016;533:481–6. doi: 10.1038/nature17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franceschelli A, Sens J, Herchick S, Thelen C, Pitychoutis PM. Sex differences in the rapid and the sustained antidepressant-like effects of ketamine in stress-naïve and “depressed” mice exposed to chronic mild stress. Neuroscience. 2015;290:49–60. doi: 10.1016/j.neuroscience.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Carrier N, Kabbaj M. Sex differences in the antidepressant-like effects of ketamine. Neuropharmacology. 2013;70:27–34. doi: 10.1016/j.neuropharm.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar A, Kabbaj M. Sex Differences in Effects of Ketamine on Behavior, Spine Density, and Synaptic Proteins in Socially Isolated Rats. Biological Psychiatry. 2016;80:448–56. doi: 10.1016/j.biopsych.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarate CA, Brutsche N, Laje G, Luckenbaugh DA, Vattem Venkata SL, Ramamoorthy A, et al. Relationship of Ketamine’s Plasma Metabolites with Response, Diagnosis, and Side Effects in Major Depression. Biological Psychiatry. 2012;72:331–8. doi: 10.1016/j.biopsych.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapidus KAB, Levitch CF, Perez AM, Brallier JW, Parides MK, Soleimani L, et al. A Randomized Controlled Trial of Intranasal Ketamine in Major Depressive Disorder. Biological Psychiatry. 2014;76:970–6. doi: 10.1016/j.biopsych.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarate CA, Jr, Brutsche NE, Ibrahim L, Franco-Chaves J, Diazgranados N, Cravchik A, et al. Replication of Ketamine’s Antidepressant Efficacy in Bipolar Depression: A Randomized Controlled Add-On Trial. Biological Psychiatry. 2012;71:939–46. doi: 10.1016/j.biopsych.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarate CA, Jr, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, et al. A randomized trial of an n-methyl-d-aspartate antagonist in treatment-resistant major depression. Archives of General Psychiatry. 2006;63:856–64. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- 23.Desta Z, Moaddel R, Ogburn ET, Xu C, Ramamoorthy A, Venkata SLV, et al. Stereoselective and regiospecific hydroxylation of ketamine and norketamine. Xenobiotica; the fate of foreign compounds in biological systems. 2012;42:1076–87. doi: 10.3109/00498254.2012.685777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Koudsi N, Hoffmann EB, Assadzadeh A, Tyndale RF. Hepatic CYP2A6 levels and nicotine metabolism: Impact of genetic, physiologic, environmental, and epigenetic factors. European journal of clinical pharmacology. 2010;66:239–51. doi: 10.1007/s00228-009-0762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, et al. Hepatic CYP2B6 Expression: Gender and Ethnic Differences and Relationship to CYP2B6 Genotype and CAR (Constitutive Androstane Receptor) Expression. Journal of Pharmacology and Experimental Therapeutics. 2003;307:906–22. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 26.Choi S-Y, Koh KH, Jeong H. Isoform-Specific Regulation of Cytochromes P450 Expression by Estradiol and Progesterone. Drug Metabolism and Disposition. 2013;41:263–9. doi: 10.1124/dmd.112.046276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, et al. Human CYP2A6 Is Induced by Estrogen via Estrogen Receptor. Drug Metabolism and Disposition. 2007;35:1935–41. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- 28.Koh KH, Jurkovic S, Yang K, Choi S-Y, Jung JW, Kim KP, et al. Estradiol induces cytochrome P450 2B6 expression at high concentrations: Implication in estrogen-mediated gene regulation in pregnancy. Biochemical Pharmacology. 2012;84:93–103. doi: 10.1016/j.bcp.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li N, Lee B, Liu R-J, Banasr M, Dwyer JM, Iwata M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science (New York, NY) 2010;329:959–64. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harraz MM, Tyagi R, Cortés P, Snyder SH. Antidepressant action of ketamine via mTOR is mediated by inhibition of nitrergic Rheb degradation. Molecular psychiatry. 2016;21:313–9. doi: 10.1038/mp.2015.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Venkata SLV, Moaddel R, Luckenbaugh DA, Brutsche NE, Ibrahim L, et al. Simultaneous population pharmacokinetic modelling of ketamine and three major metabolites in patients with treatment-resistant bipolar depression. British Journal of Clinical Pharmacology. 2012;74:304–14. doi: 10.1111/j.1365-2125.2012.04198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdallah CG. What’s the Buzz About Hydroxynorketamine? Is It the History, the Story, the Debate, or the Promise? Biological Psychiatry. 2017;81:e61–e3. doi: 10.1016/j.biopsych.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Key TJ, Appleby PN, Reeves GK, Roddam AW, Helzlsouer KJ, Alberg AJ, et al. Circulating sex hormones and breast cancer risk factors in postmenopausal women: reanalysis of 13 studies. Br J Cancer. 2011;105:709–22. doi: 10.1038/bjc.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei C, Deroo LA, Sandler DP, Weinberg CR. Fertility drugs and young-onset breast cancer: results from the Two Sister Study. J Natl Cancer Inst. 2012;104:1021–7. doi: 10.1093/jnci/djs255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manson JE, Chlebowski RT, Stefanick ML, Aragaki AK, Rossouw JE, Prentice RL, et al. Menopausal hormone therapy and health outcomes during the intervention and extended poststopping phases of the Women’s Health Initiative randomized trials. Jama. 2013;310:1353–68. doi: 10.1001/jama.2013.278040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grady D, Gebretsadik T, Kerlikowske K, Ernster V, Petitti D. Hormone replacement therapy and endometrial cancer risk: a meta-analysis. Obstet Gynecol. 1995;85:304–13. doi: 10.1016/0029-7844(94)00383-O. [DOI] [PubMed] [Google Scholar]
- 37.Perlman WR, Webster MJ, Kleinman JE, Weickert CS. Reduced glucocorticoid and estrogen receptor alpha messenger ribonucleic acid levels in the amygdala of patients with major mental illness. Biological Psychiatry. 2004;56:844–52. doi: 10.1016/j.biopsych.2004.09.006. [DOI] [PubMed] [Google Scholar]


