Abstract
Although many new assays for HIV have been developed, several labs still use simple and reliable radioactivity-based reverse transcriptase (RT) nucleotide incorporation assays for detection and quantification. We describe here a new assay for detection and quantitation of HIV RT activity that is based on a high affinity DNA aptamer to RT. The aptamer is sequestered on 96-well plates where it can bind to RT and other constituents can be removed by extensive washing. Since the aptamer mimics a primer-template, upon radiolabeled nucleotide addition, bound RT molecules can extend the aptamer and the radioactive signal can be detected by standard methods. In addition to being procedurally simple, the assay demonstrated high sensitivity (detection limits for RT and virions were ≤6,400 molecules (~4 × 10−8 units) and ~100–300 virions, respectively) and was essentially linear over a range of at least 104. Both wild type and drug-resistant forms of HIV-1 RT were detectable as was HIV-2 RT, although there were some modest differences in sensitivity.
Keywords: SELEX, aptamer, reverse transcriptase, HIV quantification
1. INTRODUCTION
Detection and quantitation of HIV has been a longstanding issue for scientists and clinicians since the discovery of the virus in 1984. In addition to assays that detect antibodies to HIV (Haleyur Giri Setty and Hewlett, 2014), several assay methods that directly detect virial components have been developed over time including among others: PCR-based assays that detect HIV RNA (Barletta, Edelman, and Constantine, 2004; Fiscus et al., 2006; Haleyur Giri Setty and Hewlett, 2014; Sun et al., 1998), ELISA assays that detect p24 capsid protein (Fiscus et al., 2006; Haleyur Giri Setty and Hewlett, 2014; Teeparuksapun et al., 2010), and several assays that detect HIV reverse transcriptase (RT).
Reverse transcriptase-based assays include basic poly(rA)-oligo(dT) assays (Rasheed, 1996), enhanced poly(rA)-oligo(dT) assays that can be carried out over several hours due to the high stability of RT (Lee et al., 1987), Scintillation Proximity Assays (SPA) using poly(rA)-oligo(dT) (Van Schoubroeck et al., 2013), assays using fluorescent dye-based detection of RT products (e.g. EnzChek Reverse Transcriptase Assay Kit (Thermo Fisher Scientific)), several assays that combine RT synthesis with PCR enhancement (Brorson et al., 2002; Frezza et al., 2014; Heneine et al., 1995; Hoffmann et al., 2011; Lovatt et al., 1999; Marino-Merlo et al., 2017; Pizzato et al., 2009; Pyra, Boni, and Schupbach, 1994; Sears and Khan, 2003; Vermeire et al., 2012), and assays that combine synthesis on a template with an immune-detection step (e.g. Roche Reverse Transcriptase Assay, Colorimetric; and Cavidi commercial RT assays (Braun et al., 2003; Greengrass et al., 2005; Malmsten et al., 2003; Malmsten et al., 2005; Seyoum et al., 2006; Sivapalasingam et al., 2005)).
Simple poly(rA)-oligo(dT) assays have been reported to be able to detect as few as ~40,000 virion equivalents of RT (Sears, Repaske, and Khan, 1999), while lower detection is possible in assays carried out for several hours (Lee et al., 1987). PCR-based RT assay can detect as low as ~1–10 virions in principle (Fiscus et al., 2006; Haleyur Giri Setty and Hewlett, 2014). The most sensitive non-PCR enhanced RT assay is the Cavidi ExaVir assay. Detection in the most recent version of the assay approaches PCR-enhanced assays and is ~200 virions per ml according to the manufacturer. Several other assays are currently under development with the goal of making detection of the virus in the field and lab easier and more sensitive (for a review see (Haleyur Giri Setty and Hewlett, 2014)).
In this report we describe a new aptamer-based RT assay for use in a laboratory setting with radiolabeled nucleotides. The assay is linear over >4 orders of magnitude, is highly sensitive (detection limit <6,400 RT molecules (~4 × 10−8 units)), and easy to perform. It takes advantage a primer-template mimicking DNA aptamer uncovered using a modified SELEX (Selective Evolution of Ligands by Exponential Enrichment) approach we refer to as “Primer-Template SELEX” (DeStefano and Cristofaro, 2006; DeStefano and Nair, 2008; Nair et al., 2012). Modifications of the recovered aptamers allowed production of a loop-back aptamer that binds extremely tightly to HIV RT. Recently the aptamer, which resembles the HIV polypurine tract (PPT), was used to produce the first crystal structure of HIV RT with nucleic acid in the absence of cross-linking (Miller et al., 2016). A version of this aptamer, when coupled to 96-well plates, can be used to “pull-down” HIV RT. Upon addition of nucleotides, the bound RT extends the recessed 3′ terminus of the aptamer producing a signal that can be detected by gel electrophoresis, binding to DEAE membranes, or scintillation counting.
2. MATERIALS AND METHODS
2.1. Materials
Detergents Tween-20, NP-40, and Triton X-100 were from Thermo Fisher Scientific, US Biologics, and Acros Organics, respectively. dNTPs were from Roche Applied Sciences. Radiolabeled α-P32 dGTP and DEAE Filtermats (90 × 120 mm, product # 1450-522) were from PerkinElmer. Streptavidin coated plates (8 well strips on 96 well plates) were from Pierce™ (~10 pm/well binding capacity, catalog #: 15125). HIV p24 quantification kit was from Zeptometrix. EnzChek Reverse Transcriptase Assay Kit was from Thermo Fisher Scientific. Cell lines (293T and HeLa TZM-bl) and plasmid pNL4-3 (Adachi et al., 1986) were from the NIH AIDS Reagent Program. Oligonucleotides were from Integrated DNA Technologies. The enzyme clone for HIV-1 RT (wild type HXB2 clone, purified as described (Hou et al., 2004)), and AZT-resistant (AZTr) (D67N, K70R, T215F, and K219Q (Arion et al., 1998)) and K65R mutated enzyme were provided by Dr. Michael Parniak (University of Pittsburgh). HIV type E/A and K103N enzymes were provided by Dr. Stefan Sarafianos (University of Missouri). The enzyme clones for HIV-2 RT and HIV-1 RT M184V (purified as described (Achuthan, Singh, and DeStefano, 2017)) were provided by Dr. Stephen Hughes (HIV Dynamics and Replication Program, NIH). Prototype foamy virus RT was from Dr. Edward Arnold (Rutgers University). Klenow (exonuclease minus) and Taq polymerases were from New England Biolabs. Human DNA polymerase α was from Chimerx. All other reagents were obtained from Thermo Fisher Scientific, Inc., Sigma-Aldrich Co., or VWR. Graphs were produced and analyzed using SigmaPlot.
2.2. Aptamer assay protocol
The assay consisted of 6 steps including Aptamer binding, RT binding, RT washing, Aptamer labeling, Aptamer removal, and Aptamer detection. (1) Aptamer binding (Note: this step can be carried out overnight or plates with attached aptamers can be stored for several weeks (see Supplemental Data for a detailed protocol)): After washing plates 3 times with 200 μl of Binding buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 mg/ml Bovine Serum Albumin (BSA), 1 mM EDTA (pH=8), and 0.1% Tween-20), aptamer binding was conducted by incubating 20 pm of 5′-biotinylated or 5′ PC-Biotin biotinylated 33NT2,4methyl+20C (see Fig. 1A) DNA aptamer in each assay well in a volume of 25 μl of Binding buffer. For all steps using the PC-Biotin aptamer, the plate was covered with foil to avoid light exposure. Each well was capable of binding ~10 pm of aptamer in a 100 μl volume with both the floor and walls of the well coated with streptavidin. Therefore, 20 pm was in excess to achieve maximal binding. Aptamer binding was carried out for 2 hours at room temperature and mild shaking at 500 rpm. The wells were then washed four times with 200 μl of Binding buffer. (2) RT binding: RT was added to each well in 25 μl of Binding buffer and incubated at room temperature for 2 hours shaking as above. (3) RT washing: Wells were washed five times with 200 μl of Binding buffer followed by one 200 μl wash with the buffer used in the labeling step (50 mM Tris-HCl, pH=8, 80 mM KCl, 6 mM MgCl2, 1 mM DTT). (4) Aptamer labeling: 2.5 μCi of α-P32 dGTP (3000 Ci/mm, 10 μCi/ul) (H3 dGTP can be used for scintillation detection (see below)) per 25 μl well and unlabeled dGTP (final dGTP concentration in reactions was 0.2 μM) was added to the above labeling buffer. After adding the labeling mix to each well, the wells were covered with a microplate adhesive film (VWR), and placed in a small orbital shaker inside a 37°C incubator. Incubation at 350 rpm was continued for the times indicated with typical assays incubated overnight for 16 hours. After labeling, the radioactive material was carefully removed from wells with a multichannel pipette. Wells were rinsed five times with 200 μl of 0.5 M NaPO4 (pH=7), then incubated in 200 μl of 0.5 M NaPO4 (pH=7) for 30 min at room temperature, shaking at 500 rpm. This was followed by two more NaPO4 rinses and three rinses with 200 μl of water. (5) Aptamer removal: Aptamers were released from the plates by two different methods. For the 5′-biotinylated 33NT2,4methyl+20C aptamer, 25 μl of 90°C formamide buffer (90% formamide, 10 mM EDTA (pH=8), 0.025% bromophenol blue and xylene cyanol) was added to each well and the plates were heated in a microwave for 4 min (plates did not melt but this step should be carefully monitored and may vary with different microwaves). The plates were then incubated at room temperature for 30 min, shaking at 500 rpm. The formamide buffer was then removed from each well and used directly for loading onto PAGE gels (see below). For PC-Biotin 5′-biotinylated 33NT2,4methyl+20C aptamer, 25 μl of water was added to each well. The plate was then exposed to UV light for 10 min using a handheld UV light (UVP Model UVM-57, 302 nm) that was mounted directly on top of the open wells. The water was then dried in the well using a Speedvac vacuum concentrator. Ten μl of water was then used to resuspend material in each well. This was applied directly to DEAE Filtermats. Alternatively, 10 μl from the 25 μl of water in the well can be used directly with a small loss in sensitivity. After air drying, the Filtermats were placed in a container with 0.5 M NaPO4 (pH=7) and incubated with shaking for 30 min at room temperature before decanting the liquid. This was followed by a second 5 min NaPO4 wash and a water rinse. Filters were dried using a heat lamp, wrapped in plastic wrap, and exposed to phosphorimager screens. (6) Aptamer detection: Imaging and quantitation were carried our using a phosphorimager (Fuji FLA 7000 or 5100). Note the PC-Biotin aptamers can also be quantified by scintillation counting with some loss of sensitivity. In this case either P32 or H3 dGTP can be used.
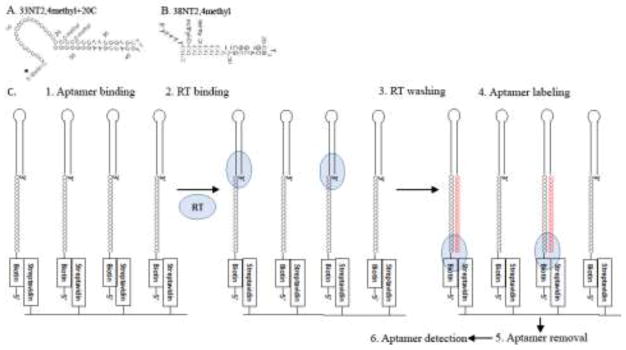
Figure 1.
Aptamer-based Reverse Transcriptase (RT) Detection Assay. (A) Aptamer 33NT2,4methyl+20C used in the assay (O-methyl denotes 2′-O-methyl groups). *The version shown has a biotin group at the 5′ end (used in assays with gel electrophoresis detection) while a second version had a Photo-Cleavable biotin (PC-Biotin) (used in assay with DEAE filter membrane detection or liquid scintillation) at the 5′ end. Shown in panel B is the original aptamer from which 33NT2,4methyl+20C was derived. (C) 96-well plate Aptamer-based Reverse Transcriptase (RT) Detection Assay. The 6 steps of the Aptamer-based RT Detection Assay are illustrated. The key to the assay is the very high affinity and stable binding of HIV RT to the designed aptamer. This allows RT to stay bound through several washing steps that eliminate proteins that might cause assay background or loss of linearity. Also, no secondary detection system (e.g. a secondary antibody in an ELISA) or amplification of the signal (e.g. PCR, or enzyme linked antibody) is required because RT can extend the aptamer that it is bound to with radiolabeled dGTP. The Aptamer removal step was with formamide gel loading buffer for the biotin linkage and UV light exposure for the PC-Biotin linkage. For details of each step refer to Materials and Methods.
2.3. Gel electrophoresis of labeled aptamers
Denaturing polyacrylamide-urea PAGE gels (15% w/v), were prepared and run as described (Sambrook and Russell, 2001). Typically the gels were run until the bromophenol blue dye was about 15 cm down the gel. Gels were dried, exposed to a phosphorimager screen, and processed as described above.
2.4. Virus preparation and quantification
HIV-1 virus was prepared by transfection of 293T cells with plasmid pNL4-3 as described (Rawson, Clouser, and Mansky, 2016). DNase digested viral supernatants were aliquoted and stored at −80°C, or further purified through a 1 ml 20% sucrose cushion by ultracentrifugation with an SW 55 Ti rotor as described (Kutner, Zhang, and Reiser, 2009). Purified virus was resuspended in PBS and aliquoted as described above. The titer of non-sucrose purified virus was determined using an endpoint titration assay on 96-well plates with HeLa TZM-bl (no dextran) (https://www.hiv.lanl.gov/content/nab-reference-strains/html/Protocol-for-Neutralizing-Antibody-Screening-Assay-for-HIV-1-in-TZMbl-cells_Apr2017.pdf). The concentration of HIV p24 protein was determined using an assay kit from Zeptometrix and the manufacturer’s protocol.
3. RESULTS
3.1. Aptamer and RT detection assay protocol
The aptamer used in the RT detection assay is shown in Fig. 1A (referred to as 33NT2,4methyl+20C). This aptamer was derived from the 38NT2,4methyl aptamer shown in Fig. 1B. A schematic diagram of the assay protocol is shown in Fig. 1C and details are provided under Materials and Methods (a more detailed protocol is provided in the Supplemental Material). 38NT2,4methyl was derived from 38NT SELEX (DeStefano and Nair, 2008) by inserting methylated nucleotides at the -2 and -4 positions (relative to the 3′ primer terminus), as previous work showed that rNTPs or 2′-O-methyl nucleotides in these positions enhanced RT binding to nucleic acids (Olimpo and DeStefano, 2010). 38NT2,4methyl binds to HIV RT with a Kd in the low pM range (Miller et al., 2016). Two different versions of 33NT2,4methyl+20C were used in the assay, one had a biotin group attached to the 5′ end while the second had biotin attached to a photocleavable linker group at the 5′ end (PC-Biotin). Note that the 96 well plates used in the assays recommend 100 μl of material in incubation steps as this completely covers the streptavidin coating. The choice to use 25 μl was mainly to decrease the amount of radioactivity and aptamer used in the assays. Assays using 100 μl of material and the same concentration of aptamer and radioactivity yielded similar results (data not shown).
3.2. Assay optimization
Tween-20 at 0.1% was used as the detergent in the assay as it yielded the best results compared to NP-40 and Triton X-100 which have also been used for HIV lysis. The RT binding step was also performed for different times and no further gains in activity were evident beyond a 2 hour incubation. The RT wash step was salt sensitive with a clear reduction in activity observed when 500 mM vs. 150 mM NaCl was used in the binding buffer in this step (data not shown).
The concentration of non-radioactive dGTP in the assay Labeling step was also varied. Radiolabeled α-32P-dGTP was held constant at 2.5 μCi (3000 Ci/mmol, 10 μCi/μl, equivalent to ~0.033 μM in the 25 μl assay). Unlabeled dGTP was added to bring the final concentration to 0.05, 0.1, 0.2, 0.5, or 1 μM dGTP and assays were conducted for 4 hours. The highest signal was yielded with 0.1 μM dGTP and the signal was slightly reduced at 0.05 μM or 0.2 μM, while larger losses were observed at 0.5 μM or 1 μM (Fig. S1). The 0.2 μM concentration was chosen for standard assays as it yielded both high sensitivity and greater linearity.
A time course showed a steady increase in signal over times up to at least 20 hours for the Labeling step (Fig. S2). Signal increased ~4 fold between 2 hours and 20 hours. Overnight 16 hour assays were used for most experiments, however, shorter assay times would be adequate for most application where very high sensitivity is not required.
To test the activity of the HIV RT preparation used in these assays, the enzyme was evaluated using a commercially available, standardized RT assay (EnzChek Reverse Transcriptase Assay Kit) (Fig. S3). The indicated detection limit for the assay is 0.02 units. The activity of HXB2 wild type RT is approximately 3.6 units per pmole enzyme, or about 1.7 × 1011 RT molecules per unit of enzyme (Le Grice, Cameron, and Benkovic, 1995). The detection limit for our enzyme in the EnzChek assay was ~0.75 × 1010 molecules of RT (~0.04 units). Therefore, our enzyme appears to be a little less active than the company’s reference enzyme, but the difference is quite small (only about 2-fold). Based on these results the HIV RT used in these experiments would be comparable to other wild type HIV-1 RTs.
3.3. Assay sensitivity and linearity with RT and HIV virions
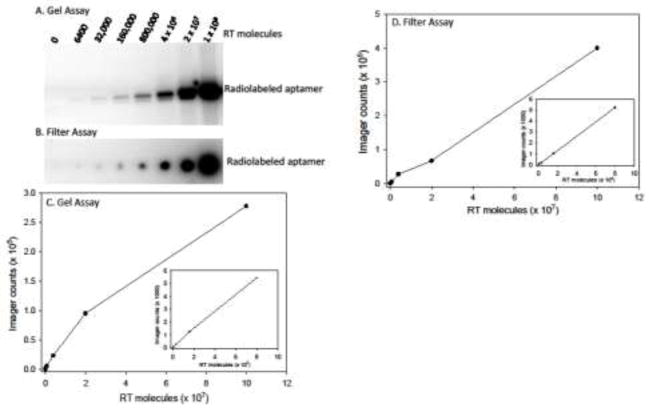
To test the detection limits of the assay with purified HIV RT, RT was diluted in binding buffer over a range from 6,400–1 × 108 molecules of RT. After the RT binding and RT washing steps, labeling buffer was added and incubations were continued at 37°C for 16 hours. The released aptamers were processed and quantified as described above. In general, detection of 33NT2,4methyl+20C on PAGE gels was more sensitive and showed a lower background than detection of PC-Biotin-33NT2,4methyl+20C on DEAE filters (Figs. 2A and 2B, respectively). Although detection was modestly better using PAGE, the filter assay is less time consuming and easier to perform. As a general laboratory assay, we would recommend using the filter assay unless very high sensitivity is desired. PC-Biotin aptamers can also be quantified by liquid scintillation using either P32 or H3 dGTP. This approach is also desirable when high sensitivity is not necessary.
Figure 2.
Sensitivity and range of HIV-1 RT detection in the Aptamer-based RT Detection Assay. Examples of a Gel-based (A) and Filter-based (B) detection assays are shown. The results for the Gel-based (C) and Filter-based (D) assays were plotted. The inserted graphs show an expanded view of the lower RT molecule data points. Imager counts are in arbitrary units that are dependent on the exposure time and the area selected for evaluation and cannot be directly compared between the gel and filter assays. See Materials and Methods and Fig. 1 for experimental details.
Assays with both filter and PAGE Aptamer detection steps were generally near linear over a range from 6,400 to 1 × 108 RT molecules (Fig 2C and 2D). Linearity was in general limited by the concentration of dGTP included in the reactions as assays with less than 0.2 μM dGTP plateaued at lower RT concentrations while those with higher concentrations extended linearity at the expense of signal sensitivity (data not shown). Overall the assay was highly quantitative over ~4 orders of magnitude.
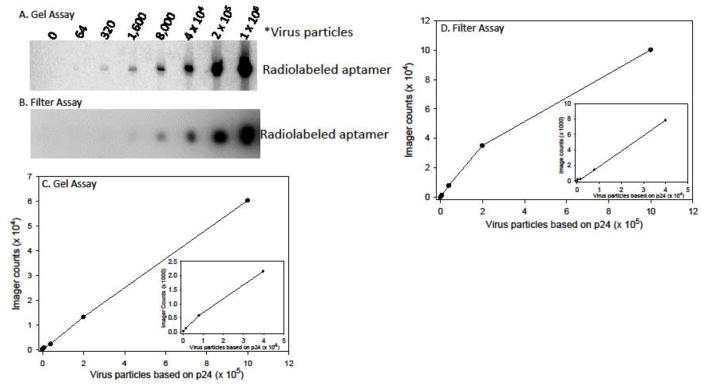
Reverse transcriptase was also detectable from purified virus (Fig. 3). For these assays, the amount of virions was determined from p24 quantitation of sucrose purified virus. An estimate of 104 virus particles per picogram of p24 was used. Three separate preparations of virus were tested. A PAGE and filter assay with one preparation, along with quantifications, is shown in Fig. 3. For this preparation the lowest amount of virus (~64 particles) was barely detectable by PAGE analysis while 320 virus particles was detectable in the filter assay. For the other two virus preparations, the lowest level of detection was at the 320 virion level on PAGE. Assays were essentially linear up to the highest level of virus tested (1 × 106 particles). A direct comparison with purified RT indicated that 10–20 active RT molecules were detectable in each virus particles while typical virions presumably contain 50–100 molecules (Julias et al., 2001; Ma and Khan, 2009). This suggested that detection of virus is 5–10 fold less sensitive compared to pure RT. Several attempts were made to improve the sensitivity of the assay for virions. These included: (1) increasing the amount and changing the type of detergent used in the assays (see above), (2) preincubating the virus in higher concentrations of detergent (0.5, 1, or 2%) before performing the RT Binding step, and (3) including RNase in the binding step to degrade viral RNA that may have been bound to RT. None of these steps improved sensitivity.
Figure 3.
Sensitivity and range of virus particle detection in the Aptamer-based RT Detection Assay. Examples of a Gel-based (A) and Filter-based (B) detection assays are shown. The results for the Gel-based (C) and Filter-based (D) assays were plotted. The inserted graphs show an expanded view of the lower virus particle data points. Imager counts are in arbitrary units that are dependent on the exposure time and the area selected for evaluation and cannot be directly compared between the gel and filter assays. Virus particle numbers were calculated for sucrose purified virus preparations by measuring the level of p24 capsid protein and using an estimate of 104 particles per pg of p24. See Materials and Methods and Fig. 1 for experimental details.
Virus was also detectable from infected cell media. In these experiments, 293T cells were transfected with pNL4-3 plasmid and virus was recovered by slow speed centrifugation to remove cell debris. Media from non-transfected cells produced a background equivalent to the no enzyme controls in both PAGE and filter assays indicating that no cellular polymerases that could extend the aptamer were bound to the plate (Fig. 4). In the assay shown detection was evident at the lowest concentration of virus used which was equivalent to 0.016 TCID50 (corresponding to ~0.0015 μl of viral supernatant for this preparation). Note that for HIV-1, there are typically several thousand virus particles for each TCID50 unit and values are highly dependent on the assay procedure used for quantitation (O’Doherty, Swiggard, and Malim, 2000; Thomas, Ott, and Gorelick, 2007). For a comparison, a standard assay with purified HIV RT over a range from ~5,860 to 3.75 × 105 molecules is also shown.
Figure 4.

Detection of virus in culture media in the Aptamer-based RT Detection Assay. (A) HIV-1 virus was prepared from plasmid pNL4-3 using 293T cells as described in Materials and Methods. Cell-free media was assayed for infectious virus using a limit-dilution assay on HeLa TZM-bl cells in order to calculate TCID50 values. One μl of cell media from non-transfected 293T cells was used for the “0” virus control. Note that for HIV-1, there are typically several thousand virus particles for each TCID50 unit and values are highly dependent on the assay procedure used for quantitation. (B) An assay with purified HIV RT is shown for comparison. See Materials and Methods and Fig. 1 for experimental details.
3.4. Assay detection of drug-resistant type B HIV-1 RTs, type A/E HIV-1 RT, HIV-2 RT, and other RTs and DNA polymerases
Several other RTs were tested in the filter-based assay. These included drug-resistant mutants of type B HIV-1 RT: AZTr, K65R, M184V, and K103N; wild type RTs from HIV-1 type A/E and HIV-2; and RTs from Moloney murine leukemia virus (MuLV), avian myeloblastosis virus (AMV), and Prototype human foamy virus (PFV). The drug-resistant RTs were identical to the type B enzyme used for all assays described above except for the noted amino acid changes. All the HIV enzymes were detected by the assay (Table S1) and detection was generally in the same range as the standard wild type HIV-1. There were some notable differences in levels, especially with AZTr and HIV-2 RTs showing decreased responses. Since equal amounts of each RT by weight was used in the assays, the modestly different levels of detection may stem from some differences in activity between the preparations. In contrast, MuLV and PFV RT were detected with much lower sensitivity, while AMV RT, like Klenow, Taq and human DNA polymerase α were not detected under the conditions used.
The binding affinity (Kd) of each tested enzyme to the parent aptamer (Fig. 1B) was also tested in nitrocellulose filter binding assays (Table S1). All the HIV enzymes bound tightly to the aptamer (Kd < ~200 pM) although enzymes with N-terminal histidine tags bound even more strongly (Kd < ~15 pM). The strength of binding did not always directly correlate with the activity of the enzyme in the Aptamer-based RT assay. For example, HIV-2 RT had ~5-fold lower activity in the assay than HIV-1 wild type RT even though it bound modestly more tightly to the aptamer. Both MuLV and PFV RT showed very low activity despite binding with pM affinity to the aptamer. The differences may be due to the complex nature of the assay. Binding is only one factor in the level of detection as the assays were performed for several hours and enzymes may have different stabilities. There may also be differences in the ability of a given enzyme to extend the aptamer which has methylated nucleotides in the template strand and a long run of poly dC. As expected, enzymes that bound with relatively low affinity showed no activity in the assay.
4. DISCUSSION
In this report an aptamer-based assay for detection and quantitation of HIV RT is described. The assay demonstrated excellent limits of detection (<6,400 RT molecules and ~100–300 virus particles) and was highly quantitative over a large range (>104). In this unique assay, the aptamer is used both to sequester RT, and provide a substrate for amplifying the radioactive signal through RT incorporation. Unlike most ELISA assays, no secondary binding step is required for signal generation and amplification. The use of radioactivity augments detection and the range of quantitation over most colorimetric assays. Depending on the sensitivity required, the assay can be completed in 1–2 days.
The enhanced sensitivity and linearity of this assay over standard poly(rA)-oligo(dT) assays probably stems from RT being “purified” away from potentially inhibitory contaminants in the RT wash step. This step also serves to remove other protein components that may incorporate nucleotides on the substrate and this reduces the background to nearly undetectable levels. Still the detection limit is not as great as virion RT assays that are coupled to PCR (see Introduction). To some extent, this probably results from low enzyme turnover and limited substrate availability on the plate assay. Aptamers are sequestered on the floor and walls of the well so the vast majority of liquid in the well has no substrate in it. Previous results showed that the aptamer loses its affinity for RT when extension occurs to reduce the length of the 5′ overhang (DeStefano and Nair, 2008). Therefore RT would be expected to release from the aptamer after extension. However, binding to another aptamer is then governed by diffusion within a solution that has concentrated aptamer only on the periphery. This issue is supported by the relatively small 4-fold increase in signal over about 18 hours (from 2–20 hours) in the assay (Fig. S2). Since HIV RT is known to be highly stable for several hours (Lee et al., 1987), the smaller than expected increase is unlikely to result from a loss of activity and probably results from a combination of absorption of the protein to the well and inefficient turnover.
The apparent 5–10 fold reduction of sensitivity (based on p24 levels) for the assay with virus particles may have resulted from inefficient release of RT from virions or lower than expected RT activity in the preparations. Reports have indicated that p24 levels as well as RNA levels correlate quite well with RT activity levels in various quantitation assays (Vermeire et al., 2012), while correlation was not as strong in other reports where different virus strains were used (Marozsan et al., 2004). Sucrose purified virus preparations used here may have lost some RT due to damage of the virions during purification. It is also possible that not all the virions underwent complete proteolytic processing or RT dimer formation. It would be interesting to make the virus in other cell types to see how this effects sensitivity. RT was also detectable in unpurified virus, but the possible presence of p24 from lysed cells makes it difficult to correlate RT levels to the p24 levels in these preparations. RT detection in unpurified virus corresponding to much less than 1 TCID50 unit is consistent with the low infectious ratio of HIV preparations and the detection of RT activity in virus particles that do not lead to an infection in standard titer assays (O’Doherty et al., 2000; Thomas et al., 2007). The ratio of infectious to total virus particles may also have been relatively low in these assays as steps that improve infectious virus detections were not employed in the endpoint dilution assays used for TCID50 quantifications. Assays were done in the absence of dextran or polybrene, and no spinoculture procedures were used. These processes are known to improve detection of infectious HIV (Castro et al., 1988; O’Doherty et al., 2000; Thomas et al., 2007).
The ability of this assay to easily detect drug-resistant mutants and even HIV-2 RT was not surprising. The aptamer used here mimics a primer-template which is the natural substrate for RT. Previous experiments showed that strong binding of the aptamer to RT resulted mostly from the PPT mimicking run of G residues at the 3′ end (DeStefano and Nair, 2008). Since drug-resistant RTs have the same basic substrate binding site as wild type RT and still use the same PPT and HIV-2 uses a nearly identical PPT compared to HIV-1, it is not surprising that these RTs would also bind tightly to the aptamer.
5. CONCLUSIONS
In conclusion, the aptamer-based RT assay demonstrated here is part of a growing list of aptamer-based pathogen detection platforms for viruses (for a review see (Wandtke, Wozniak, and Kopinski, 2015)). Although the current version uses radioactivity as the output signal, this platform could potentially be converted to other outputs (e.g. colorimetric or ELISA assays using the aptamer in place of antibody, or incorporation of fluorescent nucleotides), although this would affect sensitivity and range which are two current strengths of the assay. Among the strengths of this assay are the simple procedures required to complete the protocol, and broad detection of viral RTs. Complexity is modestly greater than a simple poly(rA)-oligo(dT) assay while linearity, sensitivity, and quantitation range are far greater.
Supplementary Material
HIGHLIGHTS.
Aptamer-based assay is highly sensitive and quantitative in comparison to other radioactivity-based HIV detection protocols that are still widely used.
Capable of detecting reverse transcriptase (RT) from HIV-1 and HIV-2 and drug-resistant viruses.
Approach is unique as the aptamer is used both as the vehicle to capture RT and a substrate for RT catalysis.
Acknowledgments
FUNDING
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (AI116389) and the National Institute of General Medical Sciences (GM116645)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achuthan V, Singh K, DeStefano JJ. Physiological Mg2+ Conditions Significantly Alter the Inhibition of HIV-1 and HIV-2 Reverse Transcriptases by Nucleoside and Non-Nucleoside Inhibitors in Vitro. Biochemistry. 2017;56:33–46. doi: 10.1021/acs.biochem.6b00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- Barletta JM, Edelman DC, Constantine NT. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am J Clin Pathol. 2004;122:20–7. doi: 10.1309/529T-2WDN-EB6X-8VUN. [DOI] [PubMed] [Google Scholar]
- Braun J, Plantier JC, Hellot MF, Tuaillon E, Gueudin M, Damond F, Malmsten A, Corrigan GE, Simon F. A new quantitative HIV load assay based on plasma virion reverse transcriptase activity for the different types, groups and subtypes. Aids. 2003;17:331–6. doi: 10.1097/00002030-200302140-00006. [DOI] [PubMed] [Google Scholar]
- Brorson K, Xu Y, Swann PG, Hamilton E, Mustafa M, de Wit C, Norling LA, Stein KE. Evaluation of a quantitative product-enhanced reverse transcriptase assay to monitor retrovirus in mAb cell-culture. Biologicals. 2002;30:15–26. doi: 10.1006/biol.2001.0290. [DOI] [PubMed] [Google Scholar]
- Castro BA, Weiss CD, Wiviott LD, Levy JA. Optimal conditions for recovery of the human immunodeficiency virus from peripheral blood mononuclear cells. J Clin Microbiol. 1988;26:2371–6. doi: 10.1128/jcm.26.11.2371-2376.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano JJ, Cristofaro JV. Selection of primer-template sequences that bind human immunodeficiency virus reverse transcriptase with high affinity. Nucleic Acids Res. 2006;34:130–9. doi: 10.1093/nar/gkj426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano JJ, Nair GR. Novel aptamer inhibitors of human immunodeficiency virus reverse transcriptase. Oligonucleotides. 2008;18:133–44. doi: 10.1089/oli.2008.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Balestrieri E, Marino-Merlo F, Mastino A, Macchi B. A novel, cell-free PCR-based assay for evaluating the inhibitory activity of antiretroviral compounds against HIV reverse transcriptase. J Med Virol. 2014;86:1–7. doi: 10.1002/jmv.23748. [DOI] [PubMed] [Google Scholar]
- Greengrass VL, Turnbull SP, Hocking J, Dunne AL, Tachedjian G, Corrigan GE, Crowe SM. Evaluation of a low cost reverse transcriptase assay for plasma HIV-1 viral load monitoring. Curr HIV Res. 2005;3:183–90. doi: 10.2174/1570162053506955. [DOI] [PubMed] [Google Scholar]
- Haleyur Giri Setty MK, Hewlett IK. Point of Care Technologies for HIV. AIDS Res Treat. 2014;2014:497046. doi: 10.1155/2014/497046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneine W, Yamamoto S, Switzer WM, Spira TJ, Folks TM. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J Infect Dis. 1995;171:1210–6. doi: 10.1093/infdis/171.5.1210. [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Garcia AD, Harrigan PR, Johnston IC, Nakasone T, Garcia-Lerma JG, Heneine W. Measuring enzymatic HIV-1 susceptibility to two reverse transcriptase inhibitors as a rapid and simple approach to HIV-1 drug-resistance testing. PLoS One. 2011;6:e22019. doi: 10.1371/journal.pone.0022019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou EW, Prasad R, Beard WA, Wilson SH. High-level expression and purification of untagged and histidine-tagged HIV-1 reverse transcriptase. Protein Expr Purif. 2004;34:75–86. doi: 10.1016/j.pep.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J Virol. 2001;75:6537–46. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Le Grice SF, Cameron CE, Benkovic SJ. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- Lee MH, Sano K, Morales FE, Imagawa DT. Sensitive reverse transcriptase assay to detect and quantitate human immunodeficiency virus. J Clin Microbiol. 1987;25:1717–21. doi: 10.1128/jcm.25.9.1717-1721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovatt A, Black J, Galbraith D, Doherty I, Moran MW, Shepherd AJ, Griffen A, Bailey A, Wilson N, Smith KT. High throughput detection of retrovirus-associated reverse transcriptase using an improved fluorescent product enhanced reverse transcriptase assay and its comparison to conventional detection methods. J Virol Methods. 1999;82:185–200. doi: 10.1016/s0166-0934(99)00111-1. [DOI] [PubMed] [Google Scholar]
- Ma YK, Khan AS. Evaluation of different RT enzyme standards for quantitation of retroviruses using the single-tube fluorescent product-enhanced reverse transcriptase assay. J Virol Methods. 2009;157:133–40. doi: 10.1016/j.jviromet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Malmsten A, Shao XW, Aperia K, Corrigan GE, Sandstrom E, Kallander CF, Leitner T, Gronowitz JS. HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J Med Virol. 2003;71:347–59. doi: 10.1002/jmv.10492. [DOI] [PubMed] [Google Scholar]
- Malmsten A, Shao XW, Sjodahl S, Fredriksson EL, Pettersson I, Leitner T, Kallander CF, Sandstrom E, Gronowitz JS. Improved HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J Med Virol. 2005;76:291–6. doi: 10.1002/jmv.20360. [DOI] [PubMed] [Google Scholar]
- Marino-Merlo F, Frezza C, Papaianni E, Valletta E, Mastino A, Macchi B. Development and evaluation of a simple and effective RT-qPCR inhibitory assay for detection of the efficacy of compounds towards HIV reverse transcriptase. Appl Microbiol Biotechnol. 2017;101:8249–8258. doi: 10.1007/s00253-017-8544-6. [DOI] [PubMed] [Google Scholar]
- Marozsan AJ, Fraundorf E, Abraha A, Baird H, Moore D, Troyer R, Nankja I, Arts EJ. Relationships between infectious titer, capsid protein levels, and reverse transcriptase activities of diverse human immunodeficiency virus type 1 isolates. J Virol. 2004;78:11130–41. doi: 10.1128/JVI.78.20.11130-11141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Tuske S, Das K, DeStefano JJ, Arnold E. Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Protein Sci. 2016;25:46–55. doi: 10.1002/pro.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GR, Dash C, Le Grice SF, DeStefano JJ. Viral reverse transcriptases show selective high affinity binding to DNA-DNA primer-templates that resemble the polypurine tract. PloS One. 2012;7:e41712. doi: 10.1371/journal.pone.0041712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–80. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olimpo JT, DeStefano JJ. Duplex structural differences and not 2′-hydroxyls explain the more stable binding of HIV-reverse transcriptase to RNA-DNA versus DNA-DNA. Nucleic Acids Res. 2010;38:4426–35. doi: 10.1093/nar/gkq169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D, McClure MO. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods. 2009;156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Pyra H, Boni J, Schupbach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc Natl Acad Sci U S A. 1994;91:1544–8. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S. Detection, quantitation, and characterization, of human retroviruses. In: Adolph KW, editor. Viral Genome Methods. CRC Press; 1996. pp. 111–166. [Google Scholar]
- Rawson JMO, Clouser CL, Mansky LM. Rapid determination of HIV-1 mutant frequencies and mutation spectra using an mCherry/EGFP dual- reporter viral vector. In: Prasad VR, Kalpana GV, editors. HIV Protocols. Humana Press; New York: 2016. pp. 71–88. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sears JF, Khan AS. Single-tube fluorescent product-enhanced reverse transcriptase assay with Ampliwax (STF-PERT) for retrovirus quantitation. J Virol Methods. 2003;108:139–42. doi: 10.1016/s0166-0934(02)00287-2. [DOI] [PubMed] [Google Scholar]
- Sears JF, Repaske R, Khan AS. Improved Mg2+-based reverse transcriptase assay for detection of primate retroviruses. J Clin Microbiol. 1999;37:1704–8. doi: 10.1128/jcm.37.6.1704-1708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyoum E, Wolday D, Girma M, Malmsten A, Meselle T, Gronowitz JS, Britton S. Reverse transcriptase activity for quantitation of HIV-1 subtype C in plasma: relation to RNA copy number and CD4 T-cell count. J Med Virol. 2006;78:161–8. doi: 10.1002/jmv.20523. [DOI] [PubMed] [Google Scholar]
- Sivapalasingam S, Essajee S, Nyambi PN, Itri V, Hanna B, Holzman R, Valentine F. Human immunodeficiency virus (HIV) reverse transcriptase activity correlates with HIV RNA load: implications for resource-limited settings. J Clin Microbiol. 2005;43:3793–6. doi: 10.1128/JCM.43.8.3793-3796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Ku J, Jayakar H, Kuo JC, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–9. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeparuksapun K, Hedstrom M, Wong EY, Tang S, Hewlett IK, Mattiasson B. Ultrasensitive detection of HIV-1 p24 antigen using nanofunctionalized surfaces in a capacitive immunosensor. Anal Chem. 2010;82:8406–11. doi: 10.1021/ac102144a. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J Virol. 2007;81:4367–70. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schoubroeck B, Van Loock M, Ivens T, Dehertogh P, Jochmans D, Dams G. Identification of HIV-1 reverse transcriptase inhibitors using a scintillation proximity assay. Methods Mol Biol. 2013;1030:19–24. doi: 10.1007/978-1-62703-484-5_3. [DOI] [PubMed] [Google Scholar]
- Vermeire J, Naessens E, Vanderstraeten H, Landi A, Iannucci V, Van Nuffel A, Taghon T, Pizzato M, Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PloS One. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandtke T, Wozniak J, Kopinski P. Aptamers in diagnostics and treatment of viral infections. Viruses. 2015;7:751–80. doi: 10.3390/v7020751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.