Summary
Latent replication-competent HIV-1 persists in individuals on long-term antiretroviral therapy (ART). We developed the Full-Length Individual Proviral Sequencing (FLIPS) assay to determine the distribution of latent replication-competent HIV-1 within memory CD4+ T cell subsets in six individuals on long-term ART. FLIPS is an efficient, high-throughput assay that amplifies and sequences near full-length (~9 kb) HIV-1 proviral genomes and determines potential replication-competency through genetic characterization. FLIPS provides a genome-scale perspective which addresses the limitations of other methods that also genetically characterize the latent reservoir. Using FLIPS, we identified 5% proviruses as intact and potentially replication-competent. Intact proviruses were unequally distributed between T cell subsets, with effector memory cells containing the largest proportion of genetically intact HIV-1 proviruses. We identified multiple identical intact proviruses suggesting a role for cellular proliferation in the maintenance of the latent HIV-1 reservoir.
eTOC Blurb
Latent, replication-competent HIV-1 proviruses pose a significant barrier to HIV-1 cure. Hiener et al. present the assay: Full-Length Individual Proviral Sequencing (FLIPS), to reveal the distribution of genetically intact and potentially replication-competent HIV-1 proviruses in different T-cell subsets isolated from individuals on long-term antiretroviral therapy.

Introduction
Antiretroviral therapy (ART) successfully suppresses HIV-1 replication, reduces viral load, and increases the life expectancy of infected individuals (Palella et al., 1998, Palmer et al., 2008). Despite this, ART is not curative as HIV-1 remains latent in resting memory CD4+ T cells not targeted by ART or the immune system (Finzi et al., 1997). Bruner et al. (2016) recently demonstrated that 93–98% of latent proviruses in HIV-infected individuals on ART are defective and replication-incompetent. Common mechanisms that contribute to defective proviruses include mutations from an error-prone HIV-1 reverse transcriptase (Abram et al., 2010), template switching during reverse transcription (Ho et al., 2013) and/or APOBEC-induced hypermutation (Harris et al., 2003, Lecossier et al., 2003). Despite the high prevalence of defective proviruses, it is clear that replication-competent proviruses persist in individuals on long-term ART as viral load rapidly rebounds if therapy is interrupted (Chun et al., 2010, Davey et al., 1999). Determining the source of latent replication-competent HIV-1 is vital to identifying cellular targets for future curative strategies.
Genetic characterization of the latent HIV-1 reservoir is an important tool for understanding persistent HIV-1 during long-term ART. Single-proviral (Josefsson et al., 2013a) and single-genome (Palmer et al., 2005) sequencing (SPS/SGS) are methods that genetically characterize sub-genomic regions of the HIV-1 genome. SPS/SGS have provided insight into the distribution, dynamics, and persistence of the latent HIV-1 reservoir (Josefsson et al., 2013b, Evering et al., 2012, Chomont et al., 2009, von Stockenstrom et al., 2015), yet these methods are limited because they target sub-genomic regions of the HIV-1 genome, and therefore cannot capture the complete diversity and replication-competency of the HIV-1 proviruses. Furthermore, the use of SPS/SGS has identified large expansions of identical HIV-1 sequences, suggesting that cellular proliferation contributes to the persistence of HIV-1 during therapy, but it remains unknown if these HIV-1 sequences are identical or even intact throughout the entire HIV-1 genome (Laskey et al., 2016). Full-length HIV-1 proviral sequencing methods, which sequence ~9 kb of the HIV-1 genome, overcome the limitations of SPS. Previously available full-length HIV-1 proviral sequencing methods have provided insight into the prevalence and development of defective proviruses (Bruner et al., 2016, Ho et al., 2013). These assays require multiple internal sequencing primers that carry the risk of erroneously identifying defective proviruses and make resolving the entire proviral sequence technically challenging. Additionally, it is possible these methods may not capture the entire population of proviruses present in an individual as, due to the number and complexity of primers used, these methods may be influenced by primer mismatches. In response to these limitations, we and others have developed Next Generation Sequencing (NGS) based assays to sequence near full-length HIV-1 proviruses (Lee et al., 2017, Imamichi et al., 2016). Here, we present the Full-Length Individual Proviral Sequencing (FLIPS) assay: a high-throughput assay utilizing NGS to sequence single, full-length HIV-1 proviruses and predict their potential replication-competency by comparative genomics. We apply FLIPS to determine the distribution of intact and potentially replication-competent proviruses within memory CD4+ T cell subsets isolated from six individuals on long-term ART and demonstrate the advantages of FLIPS over existing sequencing methods.
Results
Full-Length Individual Proviral Sequencing (FLIPS)
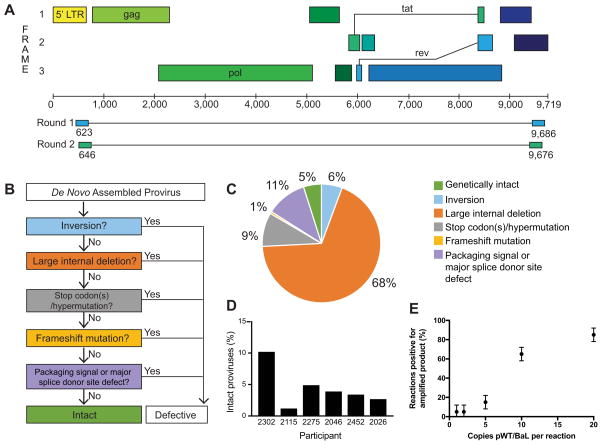
In conducting the FLIPS assay, NGS is used to sequence single (intact and defective) HIV-1 proviruses. Two rounds of nested PCR run at limiting dilution using primers that target the highly conserved 5′ and 3′ U5 LTR regions are used to amplify ~9 kb of the HIV-1 genome (Figure 1A). High-throughput NGS of amplified proviruses is undertaken on the Illumina MiSeq platform, and individual proviral contigs are generated by de novo assembly. To identify defective proviruses, a stringent process of elimination is used (Figure 1B), whereby proviruses containing inversion sequences, large internal deletions (>100 bp), hypermutation/deleterious stop codons, frameshifts, or mutations in the packaging signal or major splice donor (MSD) site are defined as defective and replication-incompetent. Proviruses lacking these defects are considered genetically intact and potentially replication-competent. In optimizing the FLIPS assay, we first determined the extent to which FLIPS amplifies the HIV-1 DNA present in a sample. To do this we amplified the HIV-1 plasmid pWT/BaL (obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 HXB3/BaL Infectious Molecular Clone (pWT/BaL) from Dr. Bryan R. Cullen) at 20-1 copies per reaction and recorded the number of reactions with amplified product (Figure 1E). The number of positive reactions decreased with the number of templates per reaction, with FLIPS amplifying 5% of the reactions when pWT/BaL was diluted to 1 and 2 copies/reaction. In calculating the Poisson Distribution for this experiment, the expected number of amplified templates/reaction closely matched the actual number of copies/reaction, supporting that at dilutions of 1 and 2 copies/well the amplified products originate from a single template.
Figure 1. Full-Length Individual Proviral Sequencing (FLIPS) assay.
(A) Primer binding sites in HIV-1 5′ and 3′ LTR regions used by FLIPS to amplify full-length (defective and intact) HIV-1 proviruses via nested PCR. (B) Process used to identify genetically intact and defective de novo assembled proviruses. (C) Percentage of defective and intact proviruses within the total pool of sequenced proviruses. (D) Percentage of intact proviruses sequenced for each participant. (E) Percentage of PCR reactions positive for amplified product with increasing copies of pWT/BaL, data represented as mean ± SD (n=2).
The classification of a sequenced provirus as intact relies on the absence of deletions, insertions, and/or mutations. It was next important to determine the error rate of the FLIPS assay as polymerase errors could artificially introduce defects within the final proviral contig. We amplified the intact pWT/BaL plasmid at limiting dilution and sequenced 10 amplicons which we compared to the available pWT/BaL reference sequence. None of the sequences contained deletions or insertions; however, one sequence contained a single nucleotide change (C|T) at position 1519 (relative to HXB2). This allowed determination of the error rate for the FLIPS assay as 1 change in every 89,330 nucleotides sequenced (or 1.12×10−5 nt−1) (confidence interval 1 in 16,000 to 1 in 3,530,000).
Latent HIV-1 reservoirs of individuals on long-term ART contain few genetically intact proviruses
Replication-competent proviruses persist in a latent state in individuals on suppressive ART, and viral load rapidly rebounds upon the cessation of therapy (Chun et al., 2010, Davey et al., 1999). Previous studies have identified latent replication-competent HIV-1 in a number of CD4+ T cell subsets including naïve (TN), central (TCM), transitional (TTM), and effector (TEM) memory (Chomont et al., 2009, Soriano-Sarabia et al., 2014). To determine if the distribution of potentially replication-competent HIV-1 proviruses was different between cell subsets, we applied the FLIPS assay to genetically characterize the latent HIV-1 reservoir in different CD4+ T cell subsets isolated from the peripheral blood of participants on long-term suppressive ART. We obtained TN, TCM, TTM, and TEM CD4+ T cell subsets from six participants who were on suppressive ART for 3–17 years (3 initiated ART during acute/early infection (2115, 2275, 2302), and 3 initiated ART during chronic infection (2026, 2046, 2452)) (Table 1, Figure S2).
Table 1.
Participant characteristics
| Participant (SCOPE ID) | Sex | Age | Viral loada (copies/mL) | CD4+ T cell counta (cells per μL) | Time of infection before initiation of therapy (months) | ART durationa (years) | Therapeutic regimenb |
|---|---|---|---|---|---|---|---|
| 2302 | male | 333 | <40 | 696 | 3.4 | 4.6 | FPV, RTV, TDF/FTC |
| 2115 | male | 51 | <40 | 601 | 0.6 | 17.3 | FTC/TDF, NVP |
| 2275 | male | 333 | <40 | 1842 | 0.8 | 15.3 | FTC/TDF, NVP |
| 2452 | male | 333 | <40 | 604 | 24.4 | 3.2 | MVC, RTG, ETR |
| 2026 | male | 59 | <40 | 476 | 117 | 17.7 | TDF, ABC/3TC, RTV, DRV |
| 2046 | male | 50 | <40 | 1099 | 70 | 16.3 | ECV, EFV/TDF/FTC |
At time of sampling
FPV, fosamprenavir; RTV, ritonavir; TDF, tenofovir disoproxil fumarate; FTC, emtricitabine; NVP, nevirapine; MVC, maraviroc; RTG, raltegravir; ETR, Etravirine; ABC, abacavir; 3TC, lamivudine; DRV, darunavir; EFV, efavirenz; ECV, entecavir.
Applying FLIPS, we obtained a total of 531 sequences from the six participants. HIV-1 can also exist in the form of 2-LTR circles within infected cells, albeit at very low levels. However, no 2-LTR circles were identified within total CD4+ T cells from four of the participants with cells available for 2-LTR quantification (Table S1), indicating that the sequences we obtained were most likely derived from proviral DNA (Vandergeeten et al., 2014). Of the 531 sequences, 26 (5%) were characterized as genetically intact and potentially replication-competent (Figure 1C). The total frequency of intact proviruses ranged from 1.3% to 10.3% in the individual participants (Figure 1D). Defective proviruses were identified following our stringent process of elimination (Figure 1B): 6% of proviruses were rendered defective due to an inversion sequence, 68% due to a large internal deletion, 9% by the presence of deleterious stop codons, and 1% by frameshift mutations. We identified 11% of proviruses that were intact in the coding region, but contained deletions in the packaging signal and/or mutations in the MSD site rendering them defective (Bruner et al., 2016, Ho et al., 2013) (Figure 1C). The proportion of intact proviruses in participants who initiated therapy during acute/early infection (6%) and chronic infection (3%) was similar to those obtained by Bruner et al. (2016) (acute 7% and chronic 2%). Additionally, the proportions of the different defective proviruses were similar across the two studies (Table S2).
Genetically intact proviruses are unequally distributed in CD4+ T cell subsets
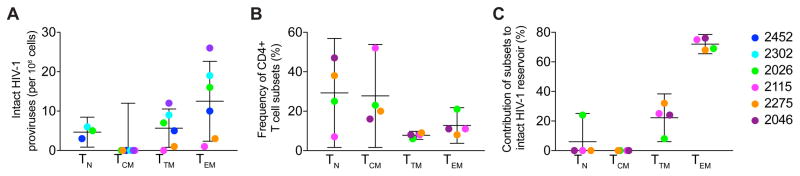
The FLIPS assay was employed to assess the frequency of genetically intact HIV-1 genomes within different CD4+ T cell subsets in a two-step process that firstly measures the number of cells positive for HIV-1, and secondly measures the proportion of these positive cells that have genetically intact HIV-1 genomes. We found strong evidence for a difference in the number of HIV-1 positive cells across the cell subsets (P = 0.003, χ2= 13.70, df = 3, Figure S1A). We observed TN to contain the least HIV-1 positive cells, followed by TCM, TEM, and finally TTM. We also found strong evidence that the proportion of intact proviruses was different across the cell subsets (P < 0.001, χ2 = 20.02, df = 3, Figure S1B). We observed the order from lowest to highest intact proportion to be TCM<TTM<TN<TEM. As the frequency of intact HIV-1 proviruses is a product of the proportion of cells that are HIV-1 positive, and the proportion that are intact, these results provide strong evidence that the frequency of intact HIV-1 proviruses differed across the cell subsets. We observe the order of this frequency from lowest to highest to be TCM<TN<TTM<TEM (Figure 2A). In all participants, TEM contained the highest number of intact proviruses (range 1–26, median 13 per 106 cells). We did not identify any intact proviruses in the TCM subset in any of the six participants despite obtaining 125 sequences from seven million cells (Table 2), however we calculated the upper bound of the confidence interval for TCM to be 12 intact proviruses in 106 cells (Figure 2A). Moreover, the proportion of each cell subset to the total pool of CD4+ T cells can differ between individuals (Figure 2B) (Josefsson et al., 2013b). When the contribution of each cell type to the overall reservoir was calculated for four participants, we found that the highest number of intact HIV-1 genomes were located in TEM cells (range 68–100% of contribution, median 76%, Figure 2C).
Figure 2. Comparison of intact proviruses between cell subsets.
(A) Number of intact proviruses per million cells in each cell subset. (B) Proportion of each cell subset to the total pool of CD4+ T cells (C) Contribution of each CD4+ T cell subset to the overall intact reservoir. Data represented as mean ± CI for all graphs. Confidence intervals are calculated from one sample t-tests, except for TCM in panel A, whose confidence interval upper bound is calculated from the product of the mean proportion of HIV+ cells in the TCM subset, and the upper bound of the confidence interval for the proportion of proviruses that are intact for the TCM subset. See also Figure S1.
Table 2.
Number of cells and sequences obtained for each subset and participant
| Participant | Naïve | Central Memory | Transitional Memory | Effector Memory | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Defective | Intact | # Cells analyzed | Defective | Intact | # Cells analyzed | Defective | Intact | # Cells analyzed | Defective | Intact | # Cells analyzed | |
| 2302 | 15 | 4 | 3,287,933 | 35 | 0 | 704,122 | 31 | 3 | 820,287 | 32 | 6 | 728,025 |
| 2115 | 0 | 0 | 0 | 28 | 0 | 2,294,141 | 29 | 0 | 859,259 | 22 | 1 | 1,888,889 |
| 2275 | 0 | 0 | 0 | 3 | 0 | 251,852 | 21 | 0 | 1,057,688 | 33 | 3 | 1,330,640 |
| 2046 | 0 | 0 | 6,465 | 24 | 0 | 190,647 | 1 | 0 | 4,048 | 23 | 2 | 153,236 |
| 2452 | 35 | 2 | 641,975 | 34 | 0 | 3,440,763 | 37 | 0 | 225,734 | 33 | 3 | 459,055 |
| 2026 | 26 | 0 | 152,727 | 1 | 0 | 47,685 | 10 | 1 | 133,448 | 32 | 1 | 51,046 |
| Total | 76 | 6 | 4,089,100 | 125 | 0 | 6,929,211 | 129 | 4 | 3,100,464 | 175 | 16 | 4,610,891 |
Detection of identical intact HIV-1 proviruses in participants on long-term ART
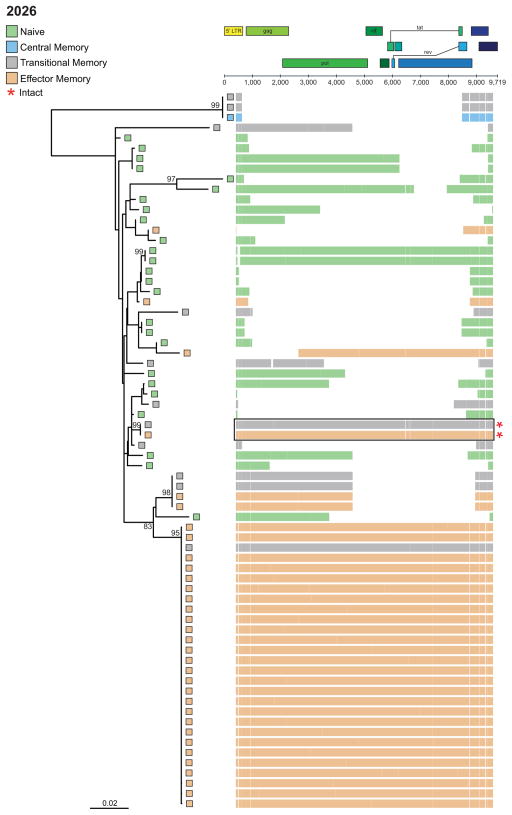
The stability and longevity of the HIV-1 reservoir is thought to be maintained via proliferation of cells containing HIV-1 proviruses (Maldarelli et al., 2014, Wagner et al., 2014, Cohn et al., 2015, Lee et al., 2017), with antigen-driven and/or homeostatic proliferation of CD4+ T cells containing an integrated HIV-1 provirus the likely mechanism (von Stockenstrom et al., 2015, Chomont et al., 2009). Phylogenetic trees of proviral contigs were prepared for each participant (Figure 3 and Data S1A–E). Genetically identical proviruses are indicative of cellular proliferation, as the error rate of the HIV-1 reverse transcriptase limits the transcription of identical proviruses by de novo replication. In all participants, we observed evidence of proliferation via the presence of identical (≥ 2) proviral sequences. The majority (92%) of identical HIV-1 sequence expansions included defective proviruses. Interestingly, in participant 2026, 32% of sequences were derived from an identical HIV-1 sequence expansion located in TEM/TTM cells, which was intact in all coding regions but had a deletion in the packaging loop and MSD site rendering it defective and likely replication-incompetent (Ho et al., 2013) (Figure 3). In three participants (2026, 2046, and 2275; Figure 3, Data S1A, and C respectively) we observed expansions of genetically identical intact proviruses in both TTM and TEM cells. The detection of identical intact proviruses supports previous findings (Simonetti et al., 2016, Lorenzi et al., 2016, Bui et al., 2017), and provides further evidence that the pool of latent replication-competent proviruses is maintained, at least in part, by cellular proliferation of memory CD4+ T cells.
Figure 3. Representative phylogenetic tree of HIV-1 sequences from a participant on long-term ART.
Participant 2026. Horizontal lines adjacent to tips represent the individual contig sequence aligned to HXB2. Red asterisks denote intact sequences; black box indicates identical intact sequences. See also Data S1A–E.
FLIPS provides advantages over existing assays which genetically characterize the latent reservoir
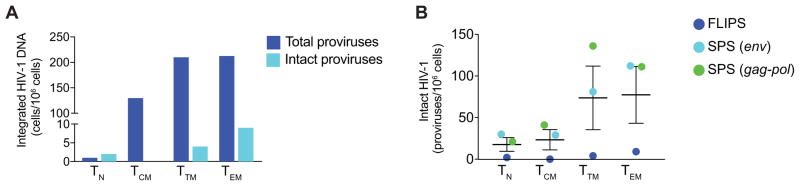
We compared the number of intact proviruses identified using FLIPS to integrated DNA measurements that were available for four participants and were obtained by the method described previously (Vandergeeten et al., 2014). As expected, we found no evidence of a correlation between the number of intact HIV-1 genomes and the number of integrated proviruses per million cells (P = 0.86). We did however find evidence that the number of intact HIV-1 proviruses per million cells measured with the FLIPS assay is lower than the frequency of cells harboring integrated HIV-1 DNA (P < 0.01, Figure 4A).
Figure 4. Comparison of FLIPS to other assays.
(A) Frequency of cells containing either intact or integrated HIV-1 DNA. (B) Number of intact proviruses identified using the FLIPS assay compared to simulated SPS of gag-pol and env regions. Data represented as mean ± CI for all graphs.
To determine if SPS of sub-genomic regions and the FLIPS assay identified equivalent numbers of intact HIV-1 genomes, we compared the number of intact proviruses identified in each cell subset using the FLIPS assay to those that may have been identified if we had employed SPS. To ensure we were working with the same population of cells as the FLIPS assay, we simulated SPS of env (V1–V3; HXB2 positions 6430–7374) and gag-pol (p6 through nucleotides 1-900 of the gene encoding reverse transcriptase; HXB2 positions 1827–3528) (von Stockenstrom et al., 2015), rather than conducting the SPS assay on a different population of cells from each participant. We chose the env and gag-pol regions, as these gene regions have been analyzed in a number of studies (Barton et al., 2016, Josefsson et al., 2012, Josefsson et al., 2011, Josefsson et al., 2013a, Josefsson et al., 2013b, von Stockenstrom et al., 2015). From our full-length contigs, we identified env and/or gag-pol sequences with complete sequencing primer sites (primers E20, E30, E115, E125 (env) and 1849, 3500, 1870, 3410 (gag-pol)) (Josefsson et al., 2013b, von Stockenstrom et al., 2015). This method of simulation may have overestimated the number of env and/or gag-pol sequences obtained as it did not take into account primer mismatches that may have affected PCR amplification prior to sequencing (Kearney et al., 2008). Nonetheless, we identified intact and defective proviruses from our populations of env and gag-pol sequences using the same stringent screening method as the FLIPS assay. When comparing the number of intact proviruses identified using FLIPS to those identified using SPS for env and gag-pol, we found that the number of intact proviruses identified by simulated SPS for these sub-genomic regions increased on average over patients and cell subsets by a factor of 13.2 (P < 0.001, t = 10.9, df = 14) and 17.7 (P < 0.001, t = 10.7, df = 13) respectively (Figure 4B). We identified a number of sequences derived from TCM that were intact in the env and gag-pol regions but were defective when we examined the entire proviral genome using FLIPS. Additionally, as the regions chosen for SPS did not cover the packaging signal and MSD site in the 5′ non-coding region, we were unable to identify sequences that were defective due to deletions or mutations in this region.
Discussion
We have developed a high-throughput and efficient method for genetically characterizing latent proviruses that overcomes the limitations of previous full-length sequencing assays and SPS. Our NGS-based FLIPS approach is able to sequence ~9 kb of the proviral genome without the use of multiple internal Sanger sequencing primers, some of which may not match all viruses within the population. Therefore, unlike the full-length sequencing assays which employ a Sanger approach (Ho et al., 2013, Bruner et al., 2016), FLIPS does not carry the risk of erroneously identifying deletions within variant virus sequences that do not match internal sequencing primers. By eliminating the need to use multiple internal sequencing primers, FLIPS is also able to resolve the entire sequence of an intact or defective provirus, a feat that is technically challenging and has not always been achieved using a Sanger-based approach (Bruner et al., 2016, Ho et al., 2013). It is important to note that although FLIPS uses primers targeted to highly conserved regions of the HIV-1 LTR, it too may be affected by primer mismatches in this region. Clearly, FLIPS has simplified the method for obtaining full-length HIV-1 proviral sequences and improved the quality of the sequences obtained and this may explain the larger number of sequences obtained per participant in this study (average 88 sequences/participant, range 50–144) compared to previous studies which employed the Sanger approach (average 27 sequences/participant (Ho et al., 2013) and average 16 sequences/participant (Bruner et al., 2016)). It is important to note that the proportion of intact proviruses and the different types of defective proviruses obtained in this study were similar to those obtained by Bruner et al. (2016), indicating the reproducibility of obtaining the genetic sequence of HIV-1 proviruses from participants on long-term ART. Recent studies also utilized NGS for sequencing near full-length HIV-1 proviruses (Lee et al., 2017, Imamichi et al., 2016), however the advantages of this approach over SPS and existing sanger-based assays were not presented or discussed.
FLIPS overcomes the limitations of SPS by sequencing near full-length proviral sequences as opposed to sub-genomic regions. We have shown that the number of proviruses identified by SPS as intact in sub-genomic regions is 13–17 fold higher than the number identified by FLIPS. Therefore, FLIPS provides a more robust estimate of the number of intact proviruses in a cell population by providing a genome-scale perspective of defective changes. Additionally, nucleotide homology between full-length proviral sequences provides a better prediction of expansions of identical intact sequences than the sequencing of sub-genomic regions.
The FLIPS assay provides a stringent approach to predicting the replication-competency of HIV-1 proviruses without requiring these proviruses to be reactivated and spread in vitro or in vivo. The majority of proviruses were characterized as defective due to the presence of large internal deletions, or deleterious stop codons in multiple coding regions. However, a number of proviruses were defined as defective due to a unique deletion in the packaging signal and MSD site. We believe in vitro studies similar to those performed by Ho et al. (2013) are required to determine the true replication-competency of some sequenced proviruses. Additionally, as we did not determine the integration sites of the sequenced proviruses, it is possible that some proviruses defined as genetically intact may be replication-incompetent due to integration into a centromere (Lewinski et al., 2005).
Genetic characterization of HIV-1 proviruses provides information about the stability, distribution, and dynamics of the latent reservoir. Using FLIPS, we identified a number of identical intact proviruses, which would not have been possible using traditional SPS methods. The detection of identical intact proviruses suggests a role for cellular proliferation in the maintenance and stability of the latent replication-competent reservoir. Unlike Hosmane et al. (2017), Bui et al. (2017) and Lorenzi et al. (2016), who used the definitive viral outgrowth assay to identify identical replication-competent proviruses in participants on ART, we did not employ in vitro studies to confirm the replication-competency of intact proviruses identified by FLIPS. Therefore, we cannot be certain that the identical intact proviruses we identified are truly replication-competent. Additionally, as integration site analysis of identical proviruses was not performed on these participants, we cannot exclude focal infection prior to ART initiation as the source of these identical proviruses. However, the presence of identical defective proviruses in every participant suggests that proliferation of resting memory CD4+ T cells is a common occurrence in participants on long-term ART.
Using the FLIPS assay we observed the distribution of genetically intact HIV-1 proviruses amongst naïve and memory CD4+ T cell populations. The number of intact proviruses was statistically different across the cell subsets and we observed TEM to have the highest number of intact proviruses. We did not directly sequence any intact proviruses from the TCM subset, however using a binomial test we calculated a conservative estimate of the number of intact proviruses in TCM to be at most 12 per 106 cells. The presence of intact HIV-1 proviruses in TCM is supported by previous studies that have identified TCM as a reservoir of replication-competent HIV-1 (Soriano-Sarabia et al., 2014, Chomont et al., 2009). Additionally, as TCM cells differentiate into TEM cells after T cell receptor activation and in response to homeostatic cytokines (Sallusto et al., 2004, Geginat et al., 2003, Sallusto et al., 1999), this may explain why TEM harbors more intact proviruses than TCM. Although we observe no intact provirus in the TCM subset, intact virus may exist at a lower level than the sensitivity of this sample size; therefore, sequencing of HIV-1 from a larger number of TCM cells may be required to detect intact proviruses.
Future studies into HIV-1 latency and eradication will benefit from utilization of the FLIPS assay. For example, further genetic characterization of latent proviruses could be crucial to determining particular coding or non-coding regions which make the virus more responsive to latency reversing agents (LRA). Similarly, sequencing of the virus before and after administration of ART could elucidate if there are any sequence characteristics that could make the virus more likely to become latent. FLIPS could assist in the identification of sites targeted by CRISPR-Cas technology. Lastly, by adapting the FLIPS assay to sequence plasma HIV-1 RNA, the particular subsets that contribute to viral rebound after administration of a LRA or ART interruption could be identified.
Experimental Procedures
Participant cohort
Leukapheresis or peripheral blood samples were obtained from six HIV-1 subtype B positive individuals on long-term suppressive ART (3.2–17.7 years). Participant characteristics, including age and sex are available in Table 1. Participants 2026, 2046 and 2452 initiated therapy during chronic infection and participants 2115, 2302 and 2275 initiated therapy during acute/early infection. Participants were enrolled in the SCOPE (participants 2026participants 2046, 2115 and 2275) or OPTIONS (participants 2452 and 2302) cohorts that are both ongoing longitudinal observational cohorts based at the University of California San Francisco. Viral loads were monitored every four months and all participants maintained viral suppression (<40–75 copies/mL) for at least three years prior to this study.
Written informed consent was obtained from all participants. The study was approved by the institutional review boards at the University of California San Francisco and the Western Sydney Local Health District which includes the Westmead Institute for Medical Research.
Isolation of CD4+ T cell subsets
Naïve and memory CD4+ T cell subsets were isolated by fluorescence-activated cell sorting. For OPTIONS cohort participants (2452 and 2302), cell subsets were isolated from peripheral blood as previously described (von Stockenstrom et al., 2015). For SCOPE cohort participants (2026, 2046, 2115, and 2275), cell subsets were isolated from leukapheresis samples as described below. Both methods sorted memory CD4+ T cell subsets on expression of CD45RO followed by CD27+/CCR7+ (TCM), CD27-/CCR7- (TEM), and CD27+/CCR7- (TTM) to obtain the relevant memory subpopulations.
For SCOPE cohort participants, naïve and memory CD4+ T cell subsets were isolated as follows (Figure S2). Peripheral blood mononuclear cells (PBMCs) were obtained from leukapheresis samples by ficoll hypaque density gradient centrifugation followed by isolation of total CD4+ T cells by magnetic negative selection, according to the manufacturer’s protocol (Stem Cell Technologies). Cells were initially sorted using the following antibodies: CD3-Alexa700 (clone UCHT1, BD #557943), CD4-APC (clone RPA-T4, BD #555349), CD14-V500 (clone M5E2, BD #561391), LIVE/DEAD Aqua marker (Invitrogen #L34957), CD45RA-BV650 (clone HI100, Biolegend #304135), and CD45RO-PerCPeFluor710 (clone UCHL1, eBioscience #40-0457-42). CD45RO+/CD45RA− cells were classified as memory cells and further sorted into TCM, TTM, and TEM subsets based on their expression of CD27 and CCR7 using the antibodies CD27-APCeFluor780 (cloneO323, eBioscience #47-0279-42) and CCR7-PECF564 (clone 150503, BD #562381) with a purity for each T cell subset of 95–100%, 67–92%, and 85–98%, respectively. CD45RO−/CD45RA+ cells were further sorted into TN based on expression of CD27, CCR7, CD127, and CD95 using the additional antibodies CD127-PE (cloneHIL-7R-M21, BD #557938) and CD95-PECy7 (clone DX2, BD #561633), with a purity of 100%.
The number of cells obtained ranged from 5 × 104 to 12 × 106 for both sorting strategies. Cell pellets were stored at −80°C. Cells were lysed in 1 00 μL lysis buffer (10 mM Tris-HCl (Invitrogen), 0.5% Nonidet P-40, 0.5% Tween-20 (Sigma) and 0.3 mg/ml proteinase K (Ambion)) per 106 cells by incubating at 55°C for one hour followed by incubation at 85°C for 15 min to obtain genomic DNA for PCR amplification.
2-LTR circle & Integrated DNA quantification
2-LTR circles from total CD4+ T cells and integrated DNA from naïve and memory T cell subsets were quantified as previously described (Vandergeeten et al., 2014).
Full-length Individual Proviral Sequencing (FLIPS) assay
Nested PCR
To amplify single full-length HIV-1 proviruses, lysed CD4+ T cells were serially diluted 1:3, 1:9, 1:27, and 1:81 and 20 reactions at each dilution were performed with two rounds of nested PCR using primers specific to the HIV-1 5′ and 3′ U5 LTR region. This PCR amplification has been modified from previously reported assays (Bruner et al., 2016, Ho et al., 2013, Li et al., 2007). Briefly, for the first round PCR, 2 μL of diluted DNA was amplified in a 40 μL reaction containing 1 μM primers (BLOuterF: 5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′, HXB2 position 623–649 and BLOuterR: 5′-TGAGGGATCTCTAGTTACCAGAGTC-3′, HXB2 position 9662–9686), 1X High Fidelity Buffer (600 mM Tris-SO4 (pH 8.9), 180 mM (NH4)2SO4), Invitrogen), 2 mM MgSO4 (Invitrogen), 0.2 mM dNTPs (Promega), and 0.025 U/μL Platinum Taq High Fidelity (Invitrogen). Each PCR plate contained 85 PCR reactions: 80 sample reactions, 4 negative control reactions, and a positive control reaction containing pWT/BaL (obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 HXB3/BaL Infectious Molecular Clone (pWT/BaL), Catalogue number 11414, from Dr. Bryan R. Cullen). PCR conditions for the first round were 94°C for 2 min; then 94°C for 30 s, 64°C for 30 s, 68°C for 10 min for 3 cycles; 94°C for 30 s, 61°C for 30 s, 68°C for 10 min for 3 cycles; 94°C for 30 s, 58°C for 30 s, 68°C for 10 min for 3 cycles; 94°C f or 30 s, 55°C for 30 s, 68°C for 10 min for 21 cycles; and then 68°C for 10 min. The first roun d PCR reaction was diluted 1:3 in Tris-HCl (5 mM, pH 8), and 2 μL of the diluted reaction was transferred to the 30 μL second round reaction (primers 275F: 5′-ACAGGGACCTGAAAGCGAAAG-3′, HXB2 positions 646–666 and 280R: 5′-CTAGTTACCAGAGTCACACAACAGACG-3′, HXB2 positions 9650–9676). Second round PCR conditions were identical to the first round except that an additional 10 cycles at 94°C for 30 s, 55°C for 30 s, and 68°C for 10 min were added. Wells positive for amplified HIV-1 proviruses were identified by diluting the second round PCR reaction 1:3 with Tris-HCl (5 mM, pH 8) followed by visualization on a 1% agarose gel. According to Poisson distribution, the dilution at which 30% of PCR reactions are positive has an 80% probability of containing a single amplified provirus. Therefore, the dilution where approximately 30% of the amplicons were positive was selected, and additional PCRs were completed until 30–40 single proviruses were amplified per T cell subset.
DNA purification and quantification
Proviral DNA from each single amplified HIV-1 provirus was purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions with minor modifications. These modifications included heating the elution buffer (Buffer EB) to 60°C and incubating the buffer on the column for 3 min prior to the final spin. Each HIV-1 provirus was then quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) and diluted in water to a final concentration of 0.2 ng DNA/μL as input for Nextera XT library preparation.
Nextera XT library preparation
Amplified proviruses were prepared for NGS using the Nextera XT DNA Library Preparation Kit (Illumina) with indexing of 96-samples per run. All tagmentation, PCR amplification, and clean-up steps were performed according to the manufacturer’s instructions except that reagent and input DNA volumes were halved and libraries were normalized manually. To do this, the concentration of each individual provirus Nextera XT library was determined using the Kapa Illumina Universal NGS qPCR kit (KAPA Biosystems) and then combined in equimolar amounts to a concentration of 4 nM. The final pooled 96-sample library was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) with the average fragment lengths determined by bioanalyzer (Agilent). The pooled library was denatured by combining equal volumes (5 μl each) with 0.2 N NaOH before neutralization with 5 μL of 200 mM Tris-HCl. The final library was then diluted to 12.5 pM with chilled Hybridization Buffer (HT1) immediately prior to sequencing. Pooled libraries underwent 2 × 150 nt paired-end sequencing on the Illumina MiSeq platform (300 cycle v2 kit), which was performed by the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia). This yielded approximately 20 million paired-end reads per run or 200,000 reads per individual provirus library for analysis following de-multiplexing.
De Novo assembly of unique proviruses
Following amplification and sequencing, we de novo assembled individual proviruses from the paired-end reads using a custom designed workflow in CLC Genomics Workbench 9 (Qiagen) (Figure S3). This workflow involved the following specific steps. (1) Quality Control: Removal of Illumina adapter sequences and ambiguous nucleotides, trimming of 5′ and 3′ terminal nucleotides, and a stringent quality limit of 0.001 that corresponds to a QC phred score of 30. (2) Merge overlapping reads: Paired forward and reverse reads with overlapping regions were merged to form single extended reads. (3) De Novo Assembly: 10,000 non-overlapping paired reads were subsampled to obtain an expected coverage of ~200X and were then de novo assembled using the native CLC Genomics assembler with a word size of 30 nt and a maximum bubble size of 65 nt. (4) Map reads to de novo assembled contig: The full read set was then re-mapped to the de novo assembled contig before extraction of a final majority consensus sequence. The final de novo assembled contig length was compared to the size of the corresponding band on the original gel. The minimum acceptable average coverage was 1,000 reads. The final consensus underwent further quality control measures before being confirmed as a unique HIV-1 proviral sequence as described below.
The majority of the proviral contigs were assembled with the above workflow. In some circumstances, multiple contigs with a similar coverage were assembled. These contigs were then aligned to a full-length HIV-1 (9 kb) reference from the same participant using the Align Contigs tool in the CLC Genome finishing module, which allowed for manual assembly of a final single consensus. As before, all reads were re-mapped to this single consensus and then accepted as a final HIV-1 proviral sequence if the read coverage was even throughout the assembly and absent of single nucleotide polymorphisms (SNPs) with a frequency of >40%.
Further quality control measures
(1) BLAST Search: Non-HIV contigs from non-specific amplification were identified through BLAST search. The amount of non-HIV sequences identified ranged from 1–14% in the participants and were mainly evident when the infection frequency of a cell subset was low. These contigs were not included in further analyses. (2) Identification of mixtures: To further ensure that each contig represented a single provirus, we screened all contigs and identified those assembled from multiple amplified HIV-1 proviral templates (mixtures) on the basis of read coverage and variant calling. Reads were mapped to a full-length HIV-1 (9 kb) reference from the same participant. Mixtures were subsequently identified by uneven read coverage (e.g., when proviral contigs of different lengths were co-amplified) or by the presence of SNPs with a frequency of >40%. The percentage of amplified proviruses that were identified as mixtures ranged from 3–15% per participant. Mixed populations were not included in subsequent analyses.
Assembled proviral contigs were imported into Molecular Evolutionary Genetics Analysis (MEGA) 6 and aligned to the HIV-1 reference genome HXB2 using MAFFT version 7 and manual editing where appropriate. To identify cases of inter-participant or positive control pWT/BaL contamination, an alignment and neighbor-joining tree were prepared containing all participants and the pWT/BaL sequence (MEGA 6). No contamination was observed as each participant’s sequences formed a unique monophyletic group distinct from the control sequence.
Identification of intact proviruses
To identify genetically intact proviruses, we developed a workflow that identifies HIV-1 proviral contigs with inversions, large internal deletions, deleterious stop codons and hypermutation, frameshift mutations, and defects in the packaging signal/MSD (Figure 1B).
Inversions
Inversions, which are regions of reverse complementarity, were identified manually in the alignment stage since they included regions that did not align to the reference sequence HXB2, however following reverse complementation were able to be aligned. All contigs containing inversions were subsequently omitted from the final alignment of full-length sequences for each T cell subset as homologous alignments could not be prepared from these sequences in any orientation.
Large internal deletions
Contigs containing large internal deletions (>100 nucleotides) were identified at the alignment stage and defined as defective.
Deleterious stop codons and hypermutation
All contigs of length >8900 nucleotides were assumed to be nearly full-length and were checked for the presence of deleterious stop codons and hypermutation. Deleterious stop codons were identified using the Gene Cutter tool (Los Alamos HIV Database), which divides the contigs into the open reading frames (gag to nef) and translates them to amino acids. Any contigs containing a stop codon in any gene, excluding nef (Foster and Garcia, 2007), were defined as defective. Additionally, contigs containing nucleotide deletions or insertions causing frameshift mutations were identified as defective. APOBEC-induced G-A hypermutation was identified in the remaining full-length proviruses using the Los Alamos HIV Database Hypermut tool (http://www.hiv.lanl.gov) and a consensus of the corresponding participant’s full-length provirus as a reference.
Defects in cis-acting elements
We defined contigs with defects in the cis-acting elements of the non-coding 5′ LTR, including the packaging signal and MSD site, as defective. These sites are essential to HIV-1 replication, as proviruses with a deletion in the four stem loops (SL1, SL2, SL3 and SL4) of the packaging signal, or a point mutation in the MSD (sequence GT) have previously been identified as replication-incompetent (Ho et al., 2013).
Construction of phylogenetic trees
Maximum likelihood phylogenetic trees were estimated for each participant using MEGA6. The best fit model of nucleotide substitution was determined for each tree using the MEGA6 model finder tool. Models used in this study included generalized time-reversible, and Tamura and Nei, incorporating gamma-distributed (gamma category 4) and/or invariant sites where appropriate. Our heuristic tree search strategy used the nearest neighbor interchanges branch-swapping algorithm. Branch support was inferred using 1000 bootstrap replicates. Annotated tree images were constructed using the ggtree tool (Yu et al., 2016) and including bootstrap values >75.
Statistical Analyses
Statistical analysis was performed in R: A language and environment for statistical computing version 3.3.2. As per the Poisson distribution, the average number of HIV-1 positive cells per PCR plate was estimated as npcr = wlog(p) where w is the total number of wells per plate and p is the estimated true proportion of negative wells (number of negative wells adjusted for sequenced positive wells that were not HIV-1, a mixture, or were unable to be assembled). The proportion of cells positive for HIV-1 was then calculated as npcr/nt where nt is the estimated number of cells used per plate. This proportion is used to estimate the frequency of cells HIV-1 positive per 106 cells. A linear mixed effects model was used to compare the estimated frequency of HIV-1 positive cells per 106 cells across memory subsets and adjust for potential correlation of multiple observations within each patient (library nlme, function lme). The fixed effect of memory subset was assessed with a likelihood ratio test on maximum likelihoods. The frequency of HIV-1 positive cells was log transformed to ensure normality of residuals and the power variance function varPower() was used to ensure heteroscedasticity. A linear fixed-effect model with a fixed effect for each subject, was also analyzed and provided similar conclusions (function lme).
The proportion of sequenced HIV-1 proviruses that were intact was calculated with a mixed logistic model to account for potential correlation of multiple observations within each patient (library lme4, function glmer). A likelihood ratio test was used to assess the fixed effect of memory subset. A fixed effect logistic regression with a fixed effect for each subject was also analyzed and provided similar conclusions (function glm). To calculate the frequency of intact HIV-1 cells per 106 cells, the proportion of HIV-1 positive cells per 106 cells was multiplied by the proportion of HIV-1 intact genomes. The comparison of the FLIPS estimate of intact virus to the SPS estimate was analyzed using a paired t-test on the logarithm of the ratio of estimates within each patient and memory subset. The logarithm of the ratio of estimates was approximately normally distributed.
To compare the distribution of intact HIV proviruses across the memory subsets from the FLIPs assay, and that what would have been detected by existing full-length sequencing assays (Bruner et al., 2016), a multinominal regression was carried out with fixed effects of patient and experiment type, using function “multinom” in package “nnet” (Venables, 2002). Likelihood ratio tests were used to calculate P-values.
Supplementary Material
Highlights.
FLIPS utilizes NGS to sequence and genetically characterize HIV-1 proviruses
FLIPS identifies genetically intact & likely replication-competent HIV-1 proviruses
Identical HIV-1 proviruses suggest maintenance of reservoir by cell replication
Demonstrate advantages of FLIPS over other common HIV-1 sequencing assays
Acknowledgments
This work was supported the Delaney AIDS Research Enterprise (DARE) to Find a Cure (1U19AI096109 and 1UM1AI126611-01), an amfAR Research Consortium on HIV Eradication (ARCHE) Collaborative Research Grant from The Foundation for AIDS Research (amfAR 108074-50-RGRL), Australian Centre for HIV and Hepatitis Virology Research (ACH2), UCSF-GIVI Center for AIDS Research (P30 AI027763), and the Australian National Health and Medical Research Council (AAP1061681). We would like to thank Dr. Joey Lai, Genomics Facility Manager at The Westmead Institute for Medical Research for his training in Nextera XT library preparation and use of his facility; Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia) for conducting MiSeq. We acknowledge with gratitude the participants who donated samples for this study.
Footnotes
Author Contributions
B.H. and J-S.E. designed the assay; B.H. and B.A.H. conducted FLIPS on participant samples and analyzed data; R.H., S.G.D., F.M.H., E.S., J.M.M., S.v.S., L.O., N.C., and R.F. enrolled the participants, collected and/or sorted cell subsets from the participant samples; N.C. and R.F., conducted the 2-LTR and integrated DNA assays; E.L., T.L., E.A.B., D.D., S.v.S., and L.O. prepared participant samples; K.B. and J-S.E. designed analysis and data visualization workflows. T.E.S conducted and interpreted the statistical analysis, and contributed to review and editing of the results and methods. B.H. wrote the original manuscript. S.P. designed the study, supervised the work performed, and edited the manuscript.
Accession Numbers
All sequences for this study have been deposited in the GenBank Nucleotide database with the accession codes KY778264-KY778681 and KY766150-KY766212.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol. 2010;84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K, Hiener B, Winckelmann A, Rasmussen TA, Shao W, Byth K, Lanfear R, Solomon A, Mcmahon J, Harrington S, et al. Broad activation of latent HIV-1 in vivo. Nat Commun. 2016;7:12731. doi: 10.1038/ncomms12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22:1043–9. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 2017;13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, Sheth PM, Kaul R, Ostrowski M, Moir S, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24:2803–8. doi: 10.1097/QAD.0b013e328340a239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, Mcelrath MJ, Lehmann C, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–32. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96:15109–14. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 2012;8:e1002506. doi: 10.1371/journal.ppat.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Foster JL, Garcia JV. Role of Nef in HIV-1 replication and pathogenesis. Adv Pharmacol. 2007;55:389–409. doi: 10.1016/S1054-3589(07)55011-8. [DOI] [PubMed] [Google Scholar]
- Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory and effector memory CD4+ T cells. Pathol Biol (Paris) 2003;51:64–6. doi: 10.1016/s0369-8114(03)00098-1. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–51. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, Siliciano RF. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med. 2017;214:959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’doherty U, Paxinos EE, Fauci AS, Lane HC. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 2016;113:8783–8. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, Eriksson S, Sinclair E, Ho T, Killian M, Epling L, Shao W, Lewis B, Bacchetti P, Loeb L, et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis. 2012;206:28–34. doi: 10.1093/infdis/jis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, et al. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci U S A. 2011;108:11199–204. doi: 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, Kearney M, Shao W, Kottilil S, Sneller M, et al. Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLoS Pathog. 2013a;9:e1003432. doi: 10.1371/journal.ppat.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, Von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A. 2013b;110:E4987–96. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Palmer S, Maldarelli F, Shao W, Polis MA, Mican J, Rock-Kress D, Margolick JB, Coffin JM, Mellors JW. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey SB, Pohlmeyer CW, Bruner KM, Siliciano RF. Evaluating Clonal Expansion of HIV-Infected Cells: Optimization of PCR Strategies to Predict Clonality. PLoS Pathog. 2016;12:e1005689. doi: 10.1371/journal.ppat.1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 2017;127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol. 2005;79:6610–9. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gladden AD, Altfeld M, Kaldor JM, Cooper DA, Kelleher AD, Allen TM. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol. 2007;81:193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi JC, Cohen YZ, Cohn LB, Kreider EF, Barton JP, Learn GH, Oliveira T, Lavine CL, Horwitz JA, Settler A, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci U S A. 2016;113:E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–83. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–60. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–13. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105:3879–84. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A. 2016;113:1883–8. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol. 2014;88:14070–7. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandergeeten C, Fromentin R, Merlini E, Lawani MB, Dafonseca S, Bakeman W, Mcnulty A, Ramgopal M, Michael N, Kim JH, et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol. 2014;88:12385–96. doi: 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S. 4. Springer; New York: 2002. [Google Scholar]
- Von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, et al. Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. J Infect Dis. 2015;212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Mclaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–3. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 2016
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.