Abstract
Inosine is a base located at wobble position 34 of the tRNA anticodon stem–loop, enabling the recognition of more than one codon in the translation process. A heterodimer consists of ADAT3 and ADAT2 and is involved in the adenosine-to-inosine conversion in tRNA. Here, we report the second novel ADAT3 mutation in a patient with microcephaly, intellectual disability, and hyperactivity. These findings constitute a second mutation and expand the clinical spectrum of extremely rare ADAT3 mutations.
Adenosine (A)-to-inosine (I) RNA editing is a post-transcriptional RNA process capable of generating RNA and protein diversity1. Inosine at wobble position 34 of tRNA anticodons can translate codons ending in uracil, cytosine, or adenine2. The modification, which creates an I from an A at position 34 (wobble position) of tRNA, is catalyzed by the heterodimeric enzyme, adenosine deaminase, tRNA-specific 3 (ADAT3)/ADAT22.
Alazami et al. described a homozygous ADAT3 mutation (c.382 G > A, p.Val128Met) in 24 affected individuals with autosomal-recessive mental retardation 36 (MRT36; MIM*615286) from eight consanguineous Arab families3. Very recently, El-Hattab et al. reported an additional 15 patients with an identical homozygous ADAT3 mutation in 15 affected individuals from 11 Arab families. In the previous reports, strabismus, microcephaly, failure to thrive, and abnormal brain structure were frequently seen in such patients.
We encountered a 6-year-old female presenting with intellectual disability, mild cognitive impairment, attention deficit, hyperactivity disorder, neurodevelopmental delay, speech delay, and microcephaly. The patient’s face was asymmetric, and her nasal bridge was depressed. She was born to healthy Iranian consanguineous parents (Fig. 1a). The proband visited our genetic center seeking a genetic testing service. Considering the clinical findings, the targeted sequencing of 12 genes associated with microcephaly (SLC25A19, STIL, ASPM, CEP135, MCPH1, CDK5RAP2, CENPJ, CEP152, WDR62, ZNF335, ADAT3, and EFTUD2) was provided. After obtaining informed consent, genomic DNA of peripheral blood leukocytes was extracted and used for the genome partitioning. Targeted capture was performed using the GeneRead DNAseq Custom Panel V2 (QIAGEN, Hilden, Germany), and the libraries were sequenced to mean >80–100 × coverage on a HiSeq2000 sequencing platform (Illumina, San Diego, CA, USA). For read mapping and variant analysis, sample sequences were aligned to the human reference genome (GRCh37/hg19) using Burrows-Wheeler Aligner4. To identify variants relevant to the disease, the obtained data were manipulated using picard and processed with the Genome Analysis Toolkit (GATK refv1.2905)5.
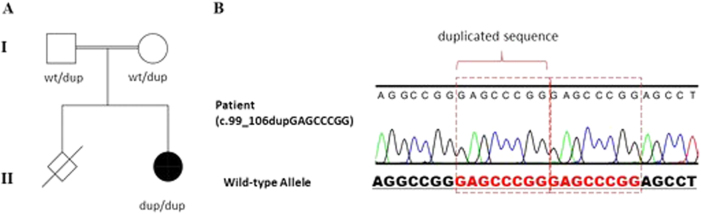
Fig. 1. Segregation status of the mutation and Sanger confirmation of c.99_106dupGAGCCCGG mutation in the proband.
a Pedigree information and segregation status of the ADAT3 8-bp duplication. b Chromatogram of the c.99_106dupGAGCCCGG, p.(Glu36Glyfs*44) mutation
Through our targeted sequencing, we identified a homozygous 8-bp duplication in ADAT3 (c.99_106dupGAGCCCGG, p.(Glu36Glyfs*44); Fig. 1b). This variant was not previously registered in the 1000 genomes database (http://browser.1000genomes.org/index.html), ExAC browser (http://exac.broadinstitute.org/), or EVS (http://evs.gs.washington.edu/EVS/). Since this gene has one coding exon, the frameshift mutation might produce a truncated protein. We confirmed both parents as heterozygous carriers (Fig. 1b), agreeing with the autosomal-recessive mode of inheritance.
The proband we present here shared many clinical features with the patients reported by El-Hattab and Alazami3, including hyperactivity, developmental delay, microcephaly, depressed nasal bridge, and asymmetric face, which were commonly seen in the current patient. In contrast, our patient showed speech delay, while El-Hattab reported speech incapability (no words) in patients with the c.382 G > A mutation. Most patients with the c.382 G > A mutation in ADAT3 have been characterized with moderate to severe cognitive impairment3,6, while the present patient was a sufferer from mild intellectual disability. Moreover, previous reports on ADAT3 mutation noted strabismus as an accompanying sign of cognitive impairment in patients with the c.382 G > A mutation3,6; however, this patient did not show strabismus. Other clinical findings were consistent with previous reports3,6 (Table 1); therefore, the difference in clinical features might be due to the different mutational effects of respective mutations.
Table 1.
Clinical features of the patient compared to previous report of ADAT3-related cognitive impairment
| This report | Previous report |
|---|---|
| Cognition | |
| Intellectual disability | Intellectual disability |
| Mild to moderate cognitive impairment | Moderate to severe cognitive impairment |
| Attention deficit hyperactivity disorder (ADHD) | Aggressive/hyperactivity |
| Development | |
| Neurodevelopmental delay | Developmental delay |
| Speech delay | No speech ability |
| Face–skull | |
| Microcephaly | Microcephaly |
| Asymmetric face | Elongated face with prominent nose |
| Depressed nasal bridge | Depressed nasal bridge |
| No strabismus | Strabismus |
In conclusion, we report a novel and second ADAT3 mutation in a patient with intellectual disability and propose that ADAT3 sequencing should be considered for intellectual disability in the Middle East.
Acknowledgements
We would like to thank the patient and her family members for participating in this study.
HGV Database
The relevant data from this Data Report are hosted at the Human Genome Variation Database at 10.6084/m9.figshare.hgv.1942.
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tang W, Fei Y, Page M. Biological significance of RNA editing in cells. Mol. Biotechnol. 2012;52:91–100. doi: 10.1007/s12033-012-9498-7. [DOI] [PubMed] [Google Scholar]
- 2.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 3.Alazami AM, et al. Mutation in ADAT3, encoding adenosine deaminase acting on transfer RNA, causes intellectual disability and strabismus. J. Med. Genet. 2013;50:425–430. doi: 10.1136/jmedgenet-2012-101378. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Hattab AW, et al. ADAT3-related intellectual disability: further delineation of the phenotype. Am. J. Med. Genet. A. 2016;170a:1142–1147. doi: 10.1002/ajmg.a.37578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The relevant data from this Data Report are hosted at the Human Genome Variation Database at 10.6084/m9.figshare.hgv.1942.