Abstract
Objective(s):
Various studies have been conducted to reduce the metastatic behavior of cancerous cells. In this regard, ectopic expression of anti-metastatic microRNAs by miR-mimic and miR-restoration-based therapies could bring new insights to the field. In the present study, the consequences of co-transfecting breast cancer cell lines with miR-193b and miR-31 were investigated via invasion and migration assays.
Materials and Methods:
Double stranded oligonucleotide of mature miR-193b-3p and miR-31-5p were cloned into pcDNA 6.2gw/EmGFP plasmid. The resulting plasmids were used for transfection. Real time-PCR was performed to assess the expression of miR-193b and miR-31 as well as Ras homolog gene family member A (RhoA) and urokinase-type plasminogen activator (uPA) as miR targets. Scratch, Transwell migration and Matrigel invasion assays were carried out to assess the extent of migration and invasion of cell lines.
Results:
The most significant increase in expression of miRs belonged to the single transfection of mimic-miRs in MDA-MB231. Although the co-transfection was not as successful as single transfection in miR expression, it was significantly more effective in inhibition of the cells invasive potential.
Conclusion:
Although the miR-restoration therapy based on co-transfection of two miRs could be less effective in expression of each miRNA, the resulting decrease in metastatic behavior of the cells is more significant due to collective effect of co-transfection to decrease target gene expression. Our results revealed that employing this sort of combinatorial strategies could lead to more efficient reduction in metastatic behavior. It seems that using this strategy would bring about more successful therapeutic outcomes.
Keywords: Breast cancer, Metastasis, miR-31, miR-193b, RhoA, uPA
Introduction
According to the American Cancer Society estimations for 2017, about 252,710 new cases of invasive breast cancer will be diagnosed in women (rank first among other malignancies) and about 40,610 women (rank second among other malignancies) will die from breast cancer. Since metastasis accounts for more than 90% of breast cancer related deaths (1), investigating the molecular mechanisms underlying breast cancer metastasis and epithelial-mesenchymal transition (EMT) seems to be vitally important. In this regard, precisely designed therapeutic approaches would circumvent the insufficiencies of conventional methods exploited against metastasis (2).
As previously described, RNA interference is a compelling method to suppress the gene expression (3-9). MicroRNAs are highly conserved small non-coding RNAs, involved in post-translational gene expression control. Their function is exerted through interaction with 3’ untranslated regions (3’UTR) of target genes. Dysregulation in cell proliferation signaling pathways such as Wnt (10), transforming growth factor beta (TGF-b) (11-13), MAPK and P53 are known to be the driving forces behind metastatic condition. Since key proteins of these signaling pathways are the target of microRNA, the therapeutic approaches based on microRNAs have attained a lot of attention in the field of metastasis investigations (14-16). Recent reports have indicated that the members of metastasis-associated miRNAs family (metastamiR) bear pro- and anti-metastasis effects. Due to their unique nature, they have opened a new direction in the field of metastasis miR-therapy (17). The use of metastamiRs in miRNA restoration-based therapies could be considered as the main approach of microRNA research in this field. Depending on the nature of the employed metastamiRs, the miR-mimic (for anti-metastatic miRNAs (tumor suppressor miRs)) or the antagomiRs approach (for pro-metastasis miRNAs (oncomiRs)) could be utilized (7). In this regard, miR-mimics are double-stranded RNA molecules with a strand identical to the intracellular mature miRNA and a complementary strand. They are usually transferred as bio-conjugates or using a carrier moiety (18). On the other hands, the stable presence of miRNAs in the circulation has profound implications for cancer diagnosis (19). In this regard, several reports have detected elevated amounts of miRNA in patients with breast cancer (20). These miRNAs are listed as the prognostic, diagnostic and therapeutic miRNA-based biomarkers of the breast cancer. These miRNAs are reported to have differential (up/down) expression profiles in the breast cancer cells compared to the normal cells. It was demonstrated that the plasma levels of miR-31 were 5-fold higher in 67 breast cancer patients compared to those in healthy individuals (21). However, in contrast to this report miR-31 was shown to be extremely down expressed in solid breast tumors (22, 23). Different expression profiles of miR-193b were also observed in different cancers including melanoma tissues (24), lung cancer (25), glioma (26), and breast cancer (27, 28). MiR-193b has been identified as an inhibitor of multiple steps of the invasion-metastasis cascade in breast cancer. Also, miR-193b and miR-31 are shown to be metastamiRs of breast cancer with anti-metastatic properties (29). These tumor suppressor miRs could be used to improve the diagnosis and treatment of breast cancer. Therefore, miR-mimic approach could be applied for breast cancer treatment using these metastamiRs (18).
In the present study, we evaluated the possibility of inhibiting the invasive behavior of metastatic cells by restoring miR-193b-3p and miR-31-5p expression levels. In this regard, MDA-MB231 cell line was used as metastatic breast cancer cells; MCF-10A cell line was used as non-tumorigenic cell; and MCF-7 cells were used as tumorigenic but non metastatic cells. Our results indicated that by restoring the expression of these miRs and consequently downregulation of their target genes like urokinase-type plasminogen activator (uPA) and Ras homolog gene family member A (RhoA), the metastatic behavior of tumor cells could be inhibited.
Materials and methods
Cell culture
MDA-MB231 was grown in Dulbecco’s modified Eagle’s Minimal Essential Medium (DMEM- high glucose; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin solution, in a humidified atmosphere of 5% CO2 in air. The medium for MCF-10A cells is DMEM/F12 supplemented with 5% donor horse serum, 20 ng/ml epidermal growth factor (EGF), 10 μg/ml insulin and 0.5 μg/ml hydrocortisone. MCF-7 cell line was propagated in DMEM/F12, 10% FBS and 1% penicillin/streptomycin.
Recombinant vector generation and target gene search
The mature miR-31-5p and miR-193b-3p sequences were obtained from miRBase database (http://www.mirbase.org). These oligomers were designed based on BLOCK-iT ™ Pol II miR RNAi Expression Vector Kits (Invitrogen). Then, the digested sequences were ligated into pcDNA 6.2gw/ EmGFP vector. To identify potential target genes of miR-193b and miR-31, the TargetScan (http://www.targetscan.org), miRanda (http://www.microrna.org), miRDB (http://www.mirdb.org/miRDB/) and PicTar (http://pictar.mdc-berlin.de) databases were used.
Optimization of transfection
Our study was performed in 3 different 24-well plates for three cell lines (MCF-10A, MCF7, and MDA-MB231). Each plate was devoted to 5 study groups (in triplicate): untreated group, miR-negative, miR-193b-3p-mimic, miR-31-5p-mimic, co-transfection of miR-193b-3p-mimic and miR-31-5p-mimic. According to the manufacturer’s recommendations, 2 µl of Lipofectamine 2000 (Invitrogen) was used with 1 µg of pcDNA6.2 for single transfections and 2 µl of Lipofectamine 2000 was used with 500 ng of each pcDNA6.2 (1 µg total pcDNA6.2) for co-transfection. Efficiency of the transfection was determined by green fluorescent protein (GFP) and flowcytometry 72 hr post-transfection. Blasticidin (2.5 µg/ml concentration, Invitrogen, Carlsbad, CA, USA) was used for antibiotic selection. Transfected cells were isolated from un-transfected cells using this antibiotic.
microRNA isolation and Real Time PCR
Using High pure miRNA isolation kit (Roche), miRNAs were extracted 48 hr after infection from all samples according to manufacturer’s instruction. Then, cDNA was synthesized by Universal cDNA Synthesis kit (Exiqon). Real time PCR for mature miR-31-5p (NO:204236), miR-193b-3p (NO:204226) and miR-191 (NO:204306) (as endogenous control) was performed with miRCURY LNA Universal RT microRNA PCR kit. The employed primers for each miR were LNA microRNA from Exiqon Company. Rotorgene 3000 series PCR machine (Corbett Research, San Francisco, USA) was used to evaluate gene expression levels. All reactions were performed in triplicate and the average values were used for relative quantification. We used the Pfaffl method to determine the relative quantity of expression. Briefly, we normalized the cycle of the threshold (Ct) values of the target gene to the endogenous control and compared it with a calibrator (same miR as the control without any treatment).
RNA extraction and Real time PCR
Total RNA was extracted (48 hr after infection) from three cell lines by RNeasy mini kit (Qiagen, Hilden, Germany). cDNA was randomly primed from total RNA using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Thermo Fisher Scientific, Waltham, MA). Real time PCR assays were performed in triplicate by FastSrart SYBR Green MasterMix (Roche) on a Corbett Rotor-gene real-time PCR using primers listed in Table 1. Relative expression levels were determined using
Table 1.
Sequences used in this study: A) miR oligo strands, B) forward and reverse primers
| A | ||
| hsa-miR-31-5p | 5’-AGGCAAGAUGCUGGCAUAGCU-3’ | |
| hsa-miR-193b-3p | 5’-AACUGGCCCUCAAAGUCCCGCU-3’ | |
| hsa-miR-191-5p | 5’- CAACGGAAUCCCAAAAGCAGCUG-3’ | |
| B | ||
| primers | sequence 5’-3’ | amplicon size |
| uPA - forward | CCAAAGAAGGAGGACTACATCG | 152 bp |
| uPA - reverse | TTCAGCAAGGCAATGTCG | |
| RhoA - forward | CGAGGTGGATGGAAAGCAGG | 202 bp |
| RhoA - reverse | TGATGGGCACGTTGGGACAG | |
| β-actin – forward | TCCCTGGAGAAGAGCTAC | 131 bp |
| β-actin – reverse | GTAGTTTCGTGGATGCCACA | |
comparative quantification characteristic of the Rotorgene software. All mRNA quantification data were normalized to β-actin and the comparative Ct (Pfaffl) method was used to determine the expression fold change.
Scratch test
All three cell lines were cultured on 6-well plates in duplicate. Cells were given time to reach 70% confluency in complete medium. To perform scratch test, a vertical line along a diameter of a well was ticked by a tip. Isolated cells were gently washed several times with phosphate buffered saline (PBS) to remove cell debris and incubated for 24 hr. Photography was performed at time zero and 24 hr after incubation. Cells migrated into wound surface and the average distance of migrating cells was determined under an inverted microscopy at designated time points.
Migration assay
To perform the migration assays, 2×104 MDA-MB231 cells that were serum starved for 24 hr, were plated into the upper chamber of the non-coated membrane (24-well insert; pore size, 8 μm) (Millipore, Billerica, MA, USA). This chamber was placed in the 24-well plate with FBS-free and FBS-containing medium, and the cells were allowed to migrate towards the lower chamber. After incubation for 24 hr at 37 °C in a 5% CO2 humidified incubator, cells in the upper chambers were removed by wiping with a cotton swab, and the cells migrated to the lower surface of filter were fixed in 4% formaldehyde for 30 min and stained with 0.5% crystal violet for 10 min. Cell migration was scored by counting 10 random fields per filter under a light microscope by ImageJ software.
Invasion assay
The cell invasion was analyzed with Matrigel-coated Transwell cell culture chambers (8-μm pore size). Briefly, cells (2.5×104 cells/well) were serum starved for 24 hr and plated in the upper insert. Two kinds of upper chamber were used: with (M+) and without Matrigel (M-) (Invitrogen). These chambers were placed in the 24-well plate that contained 10% serum (FBS) medium. The cells were incubated for 24 hr to migrate toward the lower chamber. Cells on the upper side of the filters were mechanically removed by scrubbing with a cotton swab, after which the membrane was fixed with 4% formaldehyde and stained with 0.5% crystal violet. Ultimately, invasive cells were counted at ×200 magnification from 10 accidental pictures of each filter by ImageJ software.
Statistical analysis
Two-tailed Student’s t-test was utilized for statistical analysis in our study. An asterisk means significant that shows P<0.05. All graphs and statistical analyses were performed using Prism 6 statistical software (GraphPad Software, Inc.), unless otherwise stated. The results are expressed as mean±standard deviation (SD) in three experimental repeats.
Results
miR-vector generation and their targets
The DNA sequencing results indicated that miR-mimic recombinant vectors were successfully built by miR strands hybridization and their cloning into pcDNA 6.2gw / EmGFP plasmid. According to all of the employed databases, the potential targets for miR-31-5p (by breast cancer correlation) were determined to be PRKCE, RDX, GNA13, MPRIP, ITGA5, Fzd3, MMP16 and RhoA. In the case of miR-193b-3p, the potential targets were determined to be YWHAZ, ESR1, AKR1C2, SHMT2, SMAD3 and uPA (Figure 1).
Figure 1.
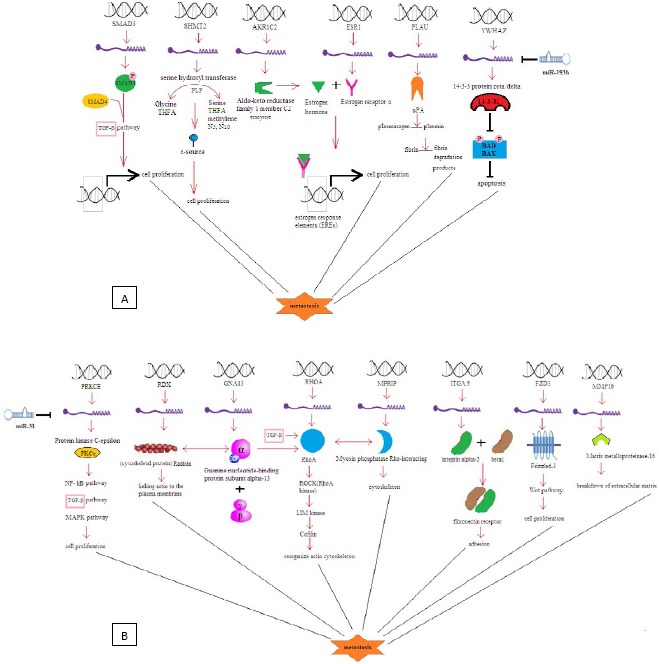
Signaling pathways involved by A) miR-193b-3p and B) miR-31-5p in breast cancer
Transfection
Lipofectamine 2000 was used for transfection, and the quality of cell lines was analyzed by fluorescence microscopy and the flowcytometry. For quantification, these data were analyzed by Flowjo software, which indicated the average transfection rate of 57% (Figure 2). Transfection rate was increased up to 90% by Blasticidin selection.
Figure 2.
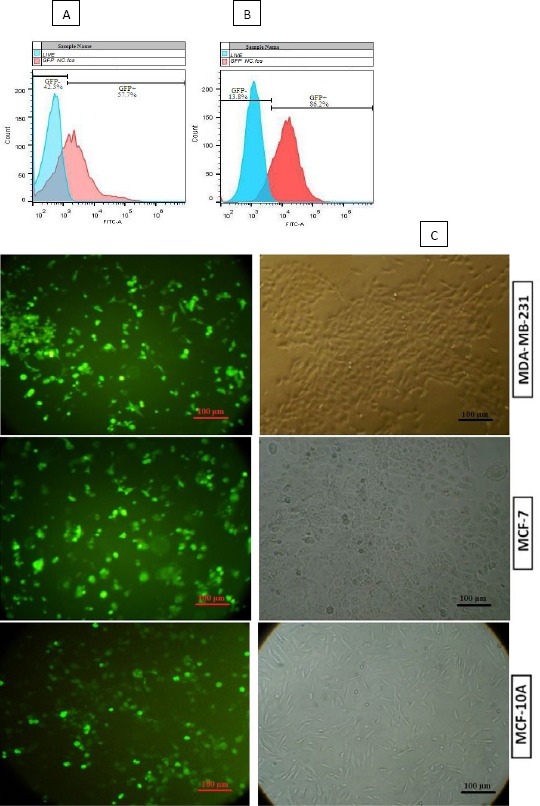
In quantification, Flowjo software indicated the average rate of three cell line transfection before (A) and after Blasticidin selection (B). C) All cell lines were analyzed for green fluorescent protein (GFP) expression by fluorescence microscopy, after Blasticidin selection
Real Time PCR for miRs
The highest increase in miR expression belonged to the single transfection of ectopic miR expression. Increase in the relative expression for miR-193b-3p and miR-31-5p was respectively 65.3 and 75.7 fold change in MDA-MB231 cell line (Figure 3A).
Figure 3.
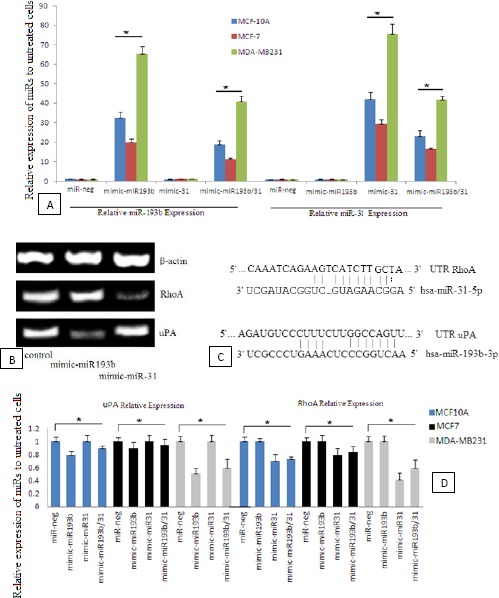
A) Relative miR-193b (left) and miR-31 (right) expression compared to the untreated cells. The results normalized by miR-191 for Pfaffl method. Data represent means±SD of three separate tests (*P<0.05). B) qRT-PCR of urokinase-type plasminogen activator (uPA) and Ras homolog gene family member A (RhoA) expression in single miR transfection of MDA-MB231 cell line. C) miR binding site and seed sequence of miR-193b and miR-31. D) Relative expression of the genes compared to the untreated cells. The results normalized by β-actin as the housekeeping gene for Pfaffl method (*P<0.05)
Quantitative real-time PCR (qRT-PCR)
qRT-PCR showed less uPA and RhoA expression in the case of single miR transfection (Figure 3B). It indicates that most uPA down-regulation occurred after miR-193b transfection, while RhoA down-regulation occurred after miR-31 transfection (Figure 3C). Relative expression of the genes (compared to β-actin as the housekeeping gene) was obtained according to Pfaffl relevance. The highest observed decrease for uPA gene expression in MDA-MB231 cells was 49%, while 58% reduction was the highest observed expression decrease for RhoA (Figure 3D).
Scratch test
The cell migration and motility power without any miR-treatment into the scratch was analyzed by Image J analysis software. The distance between two edges of the scratch was calculated by Image J software (Figure 4). For each cell line, the distance between two edges at time zero was presupposed as 100% (30) (Figure 4D, E, F). This assumption was performed by software computation. After 24 hr, the distances between two edges were calculated to be 96%, 42% and 6% (compared to the zero time distance or 100%) in MCF-10A, MCF-7 and MDA-MB231, respectively (Figure 4G, H, I). In other word, our analyses showed 4%, 58% and 94% of cell migration into wound surface in MCF-10A, MCF-7 and MDA-MB231, respectively. Our results suggested that the MDA-MB231 cell line has higher motility. Therefore, this cell line was selected for miR treatments to reduce its invasion and migration properties.
Figure 4.
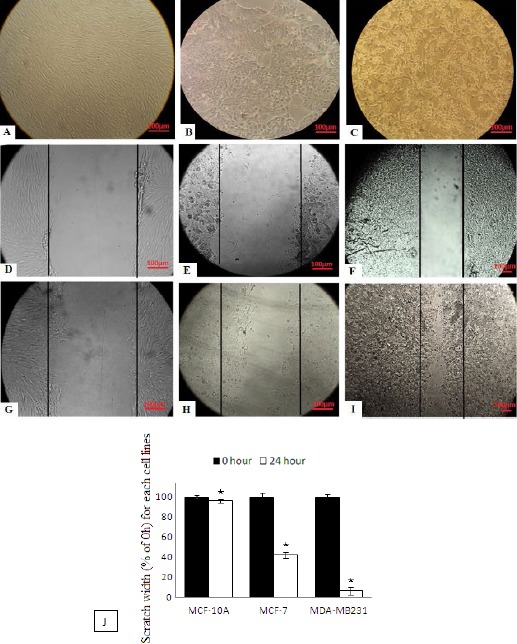
Scratch test: A) MCF-10A, B) MCF-7, C) MDA-MB231 without any miR-treatment are before and D) MCF-10A, E) MCF-7, F) MDA-MB231 are after performing a scratch - a vertical line along a diameter of a well was ticked by a tip. This distance between two edges at zero time (D, E and F) are presupposed as a 100%. G) MCF-10A, H) MCF-7 and I) MDA-MB231 are 24 hr after performing the scratch. J) The scratch chart (*P<0.05)
Invasion and migration
After transfection, the data from Transwell migration and Matrigel invasion assays indicated the reduction in both migration and invasion ability of cells compared to the control group (Figure 5). Intriguingly, the co-transfection led to an extreme reduction in both migration and invasion (93%).
Figure 5.
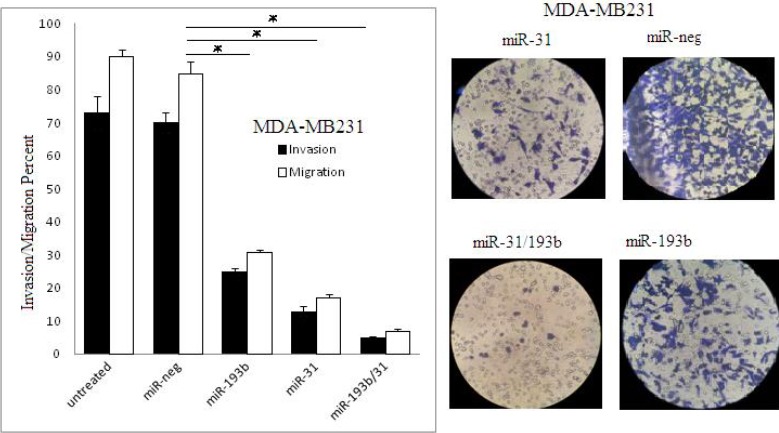
Migration and invasion assay in MDA-MB231 cell line 48 hr after transfection by miR-neg, miR-31, miR-193b, and miR-31/193b (*P<0.05)
Discussion
Currently, miR restoration therapy has garnered a lot of attention in cancer and metastasis inhibition (18). Our findings showed that the miR-193b-3p and miR-31-5p molecules are involved in cross-interacting with breast cancer signaling pathways and they are down-regulated during the metastatic stage of the disease. These properties make them to be amenable targets for miR restoration approach (22, 25-27, 31-33). MiR-31 is a pleiotropic anti-metastatic miRNA whose expression significantly decreases in metastatic breast cancer cells. This miR has multiple roles in metastasis cascades, anoikis, invasion and colonization. Its functions are exerted through disturbing the structural order of actin cytoskeleton, extracellular matrix or plasma membrane. RHOA, RDX, ITGA5, MMP16, Fzd3 and MRIP genes are the targets of miR-31 for its anti-metastatic functions (5, 22). Amongst, RhoA is a small GTPase protein of Rho family, which reorganizes the actin cytoskeleton of the cell by recruiting some mediator proteins. TGF-β1 is a tumor suppressive growth factor (3, 34), which directly activates RhoA in epithelial cells. Myosin phosphatase Rho-interacting protein (MRIP) is another enzyme that has been shown to interact with RhoA (35). Given the large network of cross-interacting targets of miR-31, we chose RhoA to assess the outcomes of miR-31-restoration based therapy on its expression rates. On the other hands, miR-193b has been implicated as a tumor suppressor in breast cancer (29). Several hundreds of genes have been identified by miRanda and PicTar servers to be down-regulated by miR-193b-3p. The sequence analyses of these genes that are related to metastasis and cell signaling have been confirmed to contain miR binding sites. Amongst, estrogen receptor (ERα) and uPA were the genes involved in breast cancer pathology, which are targeted by miR-193b-3p strand (27, 28). Among the employed three cell lines, MCF-7 is the only ER+ cell line. Therefore, analyzing its expression in all three cell lines does not seem to be appropriate for anti-metastatic evaluations. However, uPA as a urokinase-type plasminogen activator and as a serine protease is involved in degradation of the extracellular matrix and possibly tumor cell migration and proliferation, especially in breast cancer invasion and metastasis (36). It has been shown that uPA can play a pivotal role as a trigger of Epithelial-Mesenchymal Transition (EMT). Since the employed cell lines were all uPA+, we chose uPA as the target gene to assess the anti-metastatic functions of miR-193b-3p.
Relative expression levels of RhoA and uPA indicated that miR-31 and miR-193b can directly target 3’UTR of their mRNAs (23, 37). Intriguingly, we indicated that higher levels of miRs expression caused reduced expression levels in their targets. These results were in line with previous studies and confirmed the targets of miRs. It is interesting that miR-31 was more successful than the miR-193b to decrease the invasion rate, which indicates the higher potency of miR-31 as a tumor suppressor miR (22, 31, 32). This miR can control metastatic signaling pathways by deactivating various intermediate molecules, which could be considered as “triggers” for initiation of metastatic cascades (22). Although co-transfection results were associated with less effective miR expression, more effective inhibition of invasion behavior was observed using this approach.
The single transfections were performed using 1 µg of the miR containing plasmids, while the co-transfection was performed using 0.5 µg of each plasmid (1 µg of plasmids in total). Therefore, the miR containing liposomes would be loaded with 50% of the each plasmid observed in single transfection method. It was expected that the ectopic expression of each miR in co-transfection approach to be equal to 50% of the expression level in single transfection approach. However, higher levels of expression were observed, which could be rooted in the synergistic effect of these miRs on each other. The lower expression level of the target proteins (RhoA and uPA) confirmed the synergistic effect of the miRs in the case of co-transfection. It should be noted that, although the decrease in individual expression levels of RhoA and uPA was more significant in the single transfection mode, the collected amount of expression decrease for both RhoA and uPA was more significant in co-transfection mode. The higher decrease in the metastasis behavior of the cell lines is the direct consequence of higher collective decrease in RhoA and uPA expression. Therefore, co-miR-transfection in miR-mimic and miR-restoration-based therapy could be suggested as an efficient anti-metastatic strategy for cancer therapy.
Conclusion
It should be pointed out that miR-31 and miR-193b as powerful members of metastamiR family could inhibit a lot of key components of metastasis behavior. These complicated pathways are common in numerous molecules and have crossed with each other; this property makes these miRs to be deemed as a pleiotropic remedy in cancer therapy. These miRs could be of clinical significance for miR-therapy of cancers, owing to their pleiotropic anti-metastatic capabilities.
Acknowledgement
The present study has been supported by a grant from Tehran University of Medical Sciences (Tehran, Iran) and the results described in this paper were part of a student thesis. grant number:93-03-87-2672.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashemi ZS, Forouzandeh Moghadam M, Soleimani M. Comparison of TGFbR2 down-regulation in expanded HSCs on MBA/DBM scaffolds coated by UCB stromal cells. In Vitro Cell Dev Biol Anim. 2015;51:495–506. doi: 10.1007/s11626-014-9854-y. [DOI] [PubMed] [Google Scholar]
- 4.Choghaei E, Khamisipour G, Falahati M, Naeimi B, Mossahebi-Mohammadi M, Tahmasebi R, et al. Knockdown of microRNA-29a Changes the Expression of Heat Shock Proteins in Breast Carcinoma MCF-7 Cells. Oncol Res. 2016;23:69–78. doi: 10.3727/096504015X14478843952906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farokhimanesh S, Forouzandeh Moghadam M, Ebrahimi M, Ghamnak A, Hashemi ZS. Synthetic canonical miR-31, a pleiotropically acting anti-metastasis miR-based therapeutic in invasive breast cancer. Res Pharm Sci. 2012;7:S495. [Google Scholar]
- 6.Farokhimanesh S, Forouzandeh Moghadam M, Ebrahimi M, Hashemi ZS, Ghamnak A. Stemness inhibiting mir-200 family target sequence for translational repression of therapeutic gene in cancer stem cell rich MDA-MB231 cell line;POSTER PRESENTATION. Cell J. 2012 [Google Scholar]
- 7.Hashemi ZS, Khalili S, Forouzandeh Moghadam M, Sadroddiny E. Lung cancer and miRNAs: a possible remedy for anti-metastatic, therapeutic and diagnostic applications. Expert Rev Respir Med. 2017;11:147–157. doi: 10.1080/17476348.2017.1279403. [DOI] [PubMed] [Google Scholar]
- 8.Kouhkan F, Mohamadi Mosahebi M, Mohamadi Sh, Khamisipour Gh, Soufizomorrod M, Hashemi ZS, et al. Evaluation of microRNA-16 functions in breast cancer;POSTER PRESENTATION (Iran) Cell J. 2012 [Google Scholar]
- 9.Langroudi L, Forouzandeh M, Soleimani M, Atashi A, Golestaneh AF. Induction of differentiation by down-regulation of Nanog and Rex-1 in cord blood derived unrestricted somatic stem cells. Mol Biol Rep. 2013;40:4429–4437. doi: 10.1007/s11033-013-2533-3. [DOI] [PubMed] [Google Scholar]
- 10.Mohammadpour H, Khalili S, Hashemi ZS. Kremen is beyond a subsidiary co-receptor of Wnt signaling: an in silico validation. Turk J Biol. 2015;39:501–510. [Google Scholar]
- 11.Hashemi ZS, Forouzandeh Moghadam M, Soleimani M, Hafizi M, Amirizadeh N. TGF-b downregulation by RNAi technique in ex vivo-expanded HSCs on 3D DBM scaffold. Teh Univ Med Sci. 2012:70. [Google Scholar]
- 12.Khalili S, Rasaee M, Bamdad T. 3D structure of DKK1 indicates its involvement in both canonical and non-canonical Wnt pathways. Mol Biol (Mosk) 2017;51:155–166. doi: 10.7868/S0026898417010098. [DOI] [PubMed] [Google Scholar]
- 13.Hashemi ZS, Forouzandeh Moghadam M, Soleimani M. Comparison of the ex vivo expansion of UCB-derived CD34+in 3D DBM/MBA scaffolds with USSC as a feeder layer. Iran J Basic Med Sci. 2013;16:1075–1087. [PMC free article] [PubMed] [Google Scholar]
- 14.Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 15.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Banaei-Esfahani A, Moazzeni H, Nosar PN, Amin S, Arefian E, Soleimani M, et al. MicroRNAs that target RGS5. Iran J Basic Med Sci. 2015;18:108–114. [PMC free article] [PubMed] [Google Scholar]
- 17.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Z, Ye Y, Jiao D, Qiao J, Cui S, Liu Z. miR-155 and miR-31 are differentially expressed in breast cancer patients and are correlated with the estrogen receptor and progesterone receptor status. Oncol Lett. 2012;4:1027–1032. doi: 10.3892/ol.2012.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valastyan S, Weinberg RA. miR-31: a crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010;9:2124–2129. doi: 10.4161/cc.9.11.11843. [DOI] [PubMed] [Google Scholar]
- 23.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, Li S, Liu J, Ni B. MicroRNA-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin (Shanghai) 2012;44:424–340. doi: 10.1093/abbs/gms018. [DOI] [PubMed] [Google Scholar]
- 26.Zhong Q, Wang T, Lu P, Zhang R, Zou J, Yuan S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J Neurosci Res. 2014;92:619–626. doi: 10.1002/jnr.23339. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Yan P, Shao Z. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 28.Leivonen S, Mäkelä R, Östling P, Kohonen P, Haapa-Paananen S, Kleivi K, et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 29.Gruppen F. MicroRNAs in breast cancer. 2011 [Google Scholar]
- 30.Tang F, Zhang R, He Y, Zou M, Guo L, Xi T. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One. 2012;7:e35435. doi: 10.1371/journal.pone.0035435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasheed SAK, Teo CR, Beillard EJ, Voorhoeve PM, Zhou W, Ghosh S, et al. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Mol Cancer. 2015;14:67. doi: 10.1186/s12943-015-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Körner C, Keklikoglou I, Bender C, Wörner A, Münstermann E, Wiemann S. MicroRNA-31 Sensitizes Human Breast Cells to Apoptosis by Direct Targeting of Protein Kinase C ϵ(PKCϵ) J Biol Chem. 2013;288:8750–8761. doi: 10.1074/jbc.M112.414128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J, Ji Z, Jiang K, Wang Z, Meng G. miR-193b modulates resistance to doxorubicin in human breast cancer cells by downregulating MCL-1. Biomed Res Int. 2015;2015 doi: 10.1155/2015/373574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashemi ZS, Forouzandeh Moghadam M, Soleimani M. Comparison of TGFbR2 down-regulation in expanded HSCs on MBA/DBM scaffolds coated by UCB stromal cells. In Vitro Cell Dev Biol Anim. 2015;51:495–506. doi: 10.1007/s11626-014-9854-y. [DOI] [PubMed] [Google Scholar]
- 35.Surks HK, Richards CT, Mendelsohn ME. Myosin phosphatase-Rho interacting protein. A new member of the myosin phosphatase complex that directly binds RhoA. J Biol Chem. 2003;278:51484–51493. doi: 10.1074/jbc.M305622200. [DOI] [PubMed] [Google Scholar]
- 36.Tang L, Han X. The urokinase plasminogen activator system in breast cancer invasion and metastasis. Biomed Pharmacother. 2013;67:179–182. doi: 10.1016/j.biopha.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Nanbu R, Menoud P-A, Nagamine Y. Multiple instability-regulating sites in the 3'untranslated region of the urokinase-type plasminogen activator mRNA. Mol Cell Biol. 1994;14:4920–4928. doi: 10.1128/mcb.14.7.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]


