Abstract
Aim
This study was conducted to study the coagulase gene-based genetic diversity of Staphylococcus aureus, isolated from different samples of cattle using restriction fragment length polymorphism (RFLP) and their sequence-based phylogenetic analysis.
Materials and Methods
A total of 192 different samples from mastitic milk, nasal cavity, and pus from skin wounds of cattle from Military Dairy Farm, Jammu, India, were screened for the presence of S. aureus. The presumptive isolates were confirmed by nuc gene-based polymerase chain reaction (PCR). The confirmed S. aureus isolates were subjected to coagulase (coa) gene PCR. Different coa genotypes observed were subjected to RFLP using restriction enzymes Hae111 and Alu1, to obtain the different restriction patterns. One isolate from each restriction pattern was sequenced. These sequences were aligned for maximum homology using the Bioedit softwareandsimilarity in the sequences was inferred with the help of sequence identity matrix.
Results
Of 192 different samples,39 (20.31%) isolates of S. aureus were confirmed by targeting nuc gene using PCR. Of 39 S. aureus isolates, 25 (64.10%) isolates carried coa gene. Four different genotypes of coa gene, i.e., 514 bp, 595 bp, 757 bp, and 802 bp were obtained. Two coa genotypes, 595 bp (15 isolates) and 802 bp (4 isolates), were observed in mastitic milk. 514 bp (2 isolates) and 757 bp (4 isolates) coa genotypes were observed from nasal cavity and pus from skin wounds, respectively. On RFLP using both restriction enzymes, four different restriction patterns P1, P2, P3, and P4 were observed. On sequencing, four different sequences having unique restriction patterns were obtained. The most identical sequences with the value of 0.810 were found between isolate S. aureus 514 (nasal cavity) and S. aureus 595 (mastitic milk), and thus, they are most closely related. While as the most distant sequences with the value of 0.483 were found between S. aureus 514 and S. aureus 802 isolates.
Conclusion
The study, being localized to only one farm, yielded different RFLP patterns as observed from different sampling sites, which indicates that different S. aureus coagulase typeshave a site-specific predilection. Two coa patterns were observed in mastitic milk indicating multiple origins of infection, with 595 bp coa genotype being predominant in mastitic milk. The coa genotypes and their restriction patterns observed in the present study are novel, not published earlier. 514 and 595 coa variants of S. aureus are genetically most related.
Keywords: coagulase, restriction fragment length polymorphism, sequence-based phylogenetic analysis, Staphylococcus aureus
Introduction
Staphylococcus aureus is considered as a significant pathogen of animals and humans. It is regarded as the most common and prominent causative agent in all forms of mastitis. It is the major prevalent organism causing subclinical mastitis in India [1]. Economic losses to dairy industry due to S. aureus result from subclinical mastitis accompanied by a reduction in the quality and quantity of milk production [2]. S. aureus is also associated with pyogenic infections. Staphylococcus species are identified by their biochemical profile, colonial appearance, and hemolytic pattern. Biochemically, they are catalase positive and oxidase negative and utilize maltose. S. aureus produces different types of virulence factors such as capsules and adhesins, enzymes such as coagulase, catalase, hyaluronidase, and staphylokinase, and toxins such as α toxin, β toxin, δ toxin, leukocidin, enterotoxin, exfoliative toxin, and toxic shock syndrome toxin, which cause several disease conditions in humans and animals.
Coagulase is a major virulent factor, which is an enzyme secreted by S. aureus causing the clotting of plasma in the host. It causes conversion of fibrinogen to fibrin and fibrin production may shield Staphylococcus from phagocytosis. Species are classified into two types, coagulase-positive (CoPS) and coagulase-negative S. aureus. Clinical microbiologists consider coagulase production by S. aureus as important criteria for identification of S. aureus. S. aureus is characterized as coagulase-positive staphylococci and known as the main pathogen of mastitis infections in dairy animals [3]. Although the coagulase tube test is the standard phenotypic routine test used to identify S. aureus in biological samples, several groups have implemented the molecular analysis of the coagulase gene as an accurate defined test. The coa gene encoding coagulase protein is highly polymorphic because of the variable sequences (81 bp tandem repeats) at its 3’ end, which allows differentiation of S. aureus species [4]. This coa gene polymorphism is utilized as an epidemiological marker and typing is performed with primers homologous to a conserved region within the coa gene [5]. Since the number of repetitive sequences varies within the coa gene, the resulting polymerase chain reaction (PCR) products of individual strains can be of different lengths.
Molecular typing of S. aureus can be done based on different typing techniques. In the past decade, numerous molecular techniques have been developed and used for the identification and comparison of S. aureus isolates in epidemiological studies. The conventional typing procedures have several drawbacks; for example, phage typing as well as genotyping methods using pulsed-field gel electrophoresis are labor-intensive and time-consuming and can be performed only in specialized laboratories. Identification based on PCR-restriction fragment length polymorphism (RFLP) of the coa gene has been considered a more simple and accurate typing method for epidemiological investigations of bovine mastitis [6]. PCR-RFLP is a rapid, reproducible, simple, and efficient method for typing S. aureus isolated from a clinical specimen of human and animal sources. This typing technique helps in establishing genetic relations (lineages) between isolates from different sources [7]. PCR products (amplicons) are digested with the restriction enzymes AluI [8] or HaeIII. DNA fragments are separated by gel electrophoresis which can be compared by their RFLP. PCR products of different strains showing different RFLP patterns can be sequenced for both the strands of coa gene. The sequences can be aligned, and evolutionary lineages (phylogeny) based on the pair-wise distance between strains can be inferred.
This study aimed to study the coagulase gene-based genetic diversity of Staphylococcus aureus, isolated from different samples of cattle using restriction fragment length polymorphism (RFLP) and their sequence-based phylogenetic analysis.
Materials and Methods
Ethical approval
Ethical approval is not needed to pursue this type of the study. However samples were collected as per standard sample collection procedure without harming the animals.
Samples
A total of 192 samples were collected aseptically from cattle of different age groups from Military dairy farm, Belicharana, Jammu, in a study period from October 2015 to May 2016. Of total 192 samples, 98 were mastitic milk samples (from clinical as well as subclinical mastitis), 44 pus samples from open wounds on skin, and 50 nasal samples. The samples were immediately taken to laboratory and stored at 4°C until further processing.
Cultural identification of S. aureus
Mastitic milk samples were enriched in peptone water (PW) enrichment broth (HiMedia Pvt. Ltd., India). A loop full of milk sample was homogenized with sterile enrichment broth PW and incubated for 24 h at 37°C. The selective medium used for the isolation of S. aureus from mastitic milk was Baird Parker Agar (BPA) (HiMedia Pvt. Ltd., India). A loop full of inoculum from enrichment broth was streaked on BPA and incubated for 48 h at 37°C. The characteristic appearance of jet black colonies surrounded by a white halo was considered to be presumptive S. aureus. However, pus and nasal swab samples were streaked directly on selective medium Mannitol Salt Agar (MSA). The appearance of golden-yellow colonies was presumed to be S. aureus.
Phenotypic detection of S. aureus
Phenotypic identification was done by demonstration of the typical cellular morphology in Gram’s stained smears, and Gram-positive cocci that occurred in clusters under the microscope were subjected to preliminary biochemical tests.
Catalase test
A loopful of the young culture of bacterial isolates was mixed with a drop of 3% hydrogen peroxide over a clean glass slide. The production of gas bubbles or any effervescence within a few seconds was considered as catalase positive.
Oxidase test
A loopful of test bacterial culture was placed on oxidase disc. The appearance of dark purple color within few seconds was noted as oxidase positive.
Deoxyribonuclease (DNase) activity
For DNase test, appropriate quantity of DNase Test Agar with toluidine blue (HiMedia Pvt. Ltd., India) was dissolved in the required volume of distilled water and sterilized by autoclaving at 121°C for 15 min and poured in sterile Petri dishes. This medium was used for detecting the DNase activity of the bacteria after inoculating the plate with test organism. DNase produced by organism polymerizes DNA present in the media. This results in formation of a clear zone around the microbial growth which is visualized by flooding the plate with HCl.
Tube coagulase test
Tube coagulase test was performed by placing 0.5 ml of rabbit plasma (HiMedia Pvt. Ltd., India) placed in small (7 mm) test tube. Two drops of an overnight broth culture of the S. aureus were added. The tube was rotated gently to mix the contents and then incubated at 37°C. The positive test was confirmed by clotting of plasma within 2-4 h.
Extraction of bacterial DNA
Suspensions of isolated colonies of phenotypically identified S. aureus, isolated from all the three sampling sites, subculturedon nutrient agar (NA) slant were prepared in 1.5 ml microcentrifuge tubes in 250 µl of sterile double-distilled water by gentle mixing. The samples were boiled for 10 min, cooled on ice for 10 min, and centrifuged at 10,000× g in a table-top microcentrifuge (Biofuge Stratos, Heraeus) for 5 min. 2 µL of the supernatant was used as the template for PCR.
Molecular detection of nuc gene of S. aureus isolates
The S. aureus isolates were confirmed by PCR using species-specific thermonuclease (nuc) gene (270 bp) primers nuc-F: GCGATTGATGGTGATACGGTT and nuc-R: AGCCAAGCCTTGACGAACTAAAGC [9]. The PCR amplification for nuc gene was performed in 25 µl reaction volume in 0.2 ml thin-walled PCR tubes using Mastercycler Gradient (Eppendorf, Hamburg, Germany). The reaction consisted of 2.0 µl template DNA, 5 µl of 5X buffer, 0.5 µl of 10 mM dNTP mix, 2.5 µl of 25 mM Mgcl2, 2 U of Taq DNA polymerase (Promega, USA), 0.5 µM concentration of each primer, and sterile nuclease-free water (Promega). The PCR conditions in Mastercycler Gradient Thermal cycler (Eppendorf, Germany) consisted of initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 1 min. This was followed by final extension at 72°C for 7 min.
Electrophoresis and documentation
Agarose gel (1% w/v) was made by heating the appropriate amount of agarose (Sigma-Aldrich, St. Louis, USA) with 30 ml 1X Tris-acetate EDTA (TAE) buffer in a 500 ml Erlenmeyer flask. The flask was cooled to 60°C, and ethidium bromide added to the final concentration of 0.5 µg/ml. The warm agarose solution was poured into a plastic holder with suitable comb (with 0.5-mm or 1-mm wells) and allowed to completely set at room temperature for 30 min. DNA samples were mixed with 6X loading dye and loaded in separate wells on the submerged gel. Standard molecular weight marker, 100 bp (Promega, USA), was also loaded in one well. The voltage 1-5 V/cm was applied across the gel unit tracking dye (Bromophenol blue) migrated to appropriate distance. The gel was removed and DNA bands were visualised under ultraviolet illumination and photographed with Gel Documentation System (BioDocAnalyze, Biometra, Germany). The molecular sizes of the DNA bands were analyzed in relation to molecular weight marker.
Molecular detection of coagulase (coa) gene
The primers used for amplification of coa gene of S. aureus are coa F: ATAGAGATGCTGGTACAGG and coa R: GCTTCCGATTGTTCGATGC [10]. The PCR amplification was performed in 25 µl in 0.2 ml thin-walled PCR tubes (Eppendorf, Germany). The PCR mixture contained a final concentration of 2.5 µl of 5X coloured buffer, 2.5 µl of 25 mM MgCl2, 0.25 µl 25 µM concentration of each primer (Table-1), 0.5 mM concentrations of each 2’-deoxynucleoside 5’-triphosphate, and 2 U of Taq DNA polymerase (Promega, USA). The amplification cycles in Mastercycler Gradient Thermal cycler (Eppendorf, Germany) consisted of 94°C for 45 s, followed by 30 cycles of 94°C for 20 s, 57°C for 15 s, and 70°C for 15 s and final extension at 72°C for 2 min.
Table-1.
S. aureus isolates obtained from different samples using nuc gene PCR.
| Samples | Source | Number of samples | S. aureus isolates (nuc positive) (%) |
|---|---|---|---|
| Mastitic milk | Cattle | 98 | 22 (22.4) |
| Nasal swabs | Cattle | 50 | 8 (16) |
| Pus from wounds | Cattle | 44 | 9 (20.4) |
| Total | Cattle | 192 | 39 (20.31) |
S. aureus=Staphylococcus aureus, PCR=Polymerase chain reaction
RFLP of coagene product of S. aureus
Variation in restriction patterns of coa variable S. aureus isolates was determined by PCR-RFLP using HaeIII and AluI restriction endonucleases (Promega, U.S.A). For digestion with AluI restriction endonuclease, 10 µl of coa PCR product, 18 µl of nuclease free water, 2 µl of 10X Tango buffer, and 2 unit of AluI enzyme were added, mixed gently in a 0.2 ml PCR tube, and incubated at 37°C for 2 h in a water bath. Similarly, for digestion with HaeIII, 2 µl of 10*Buffer, 2 units of HaeIII enzyme, 10 µl of coa PCR product, and 18 µl of nuclease-free water were added and then mixed gently in a 0.2 ml PCR tube and incubated in the same fashion as during AluI digestion.After 2 h of incubation, the digested products were run in 2.75% agarose gel prepared in 1X TAE buffer containing 0.5 µg/ml of EtBr. The digested products were electrophoresed for 1 h at 100 V and then visualized and documented under Gel Doc System (BioDocAnalyze, Biometra, Germany).
Results
Culture findings
Of total 98 mastitic milk samples, 84 samples (85.7%) revealed jet-black colonies surrounded by white halo indicating lecithinase activity on BPA medium, characteristic of S. aureus, as shown in Figure-1. Of 94 pus and nasal samples, 78 samples (82.9%) showed characteristic golden-yellow colonies of S. aureus (Figure-2). One pure well-isolated colony from all the BPA and MSA plates presumed to be S. aureus was subcultured on NA slant, to obtain 162 presumptive S. aureus cultures.
Figure-1.

Typical jet-black colonies of Staphylococcus aureus surrounded by white halo showing lecithinase activity on Baird Parker Agar.
Figure-2.
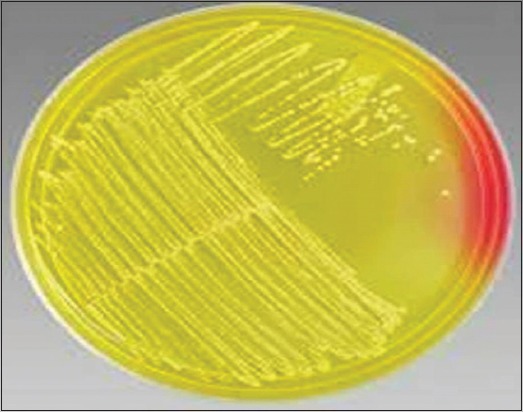
Typical golden-yellow colonies of Staphylococcus aureus on Mannitol Salt Agar.
Biochemical characterization
Subsequently, the colonies from NA slant were subjected to gram staining for morphological characterization and bacteria were typically Gram-positive cocci occurring in bunched, grape-like irregular clusters, which are characteristic of Staphylococcus spp. These Staphylococcus cultures were characterized biochemically for catalase and oxidase activity. Of 162 presumptive cultures, 114 (70.37%) cultures revealed positive catalase and negative oxidase activity (Figure-3a and b). Of 114 cultures, 75 (65.7%) cultures were positive for DNase test and showed typical depolymerization of DNA around the bacterial colonies on DNase agar as shown in Figure-4. Among the 114 cultures, only 22 (28.07%) cultures showed a positive result for tube coagulase test observed by the formation of the clot as shown in Figure-5.
Figure-3.
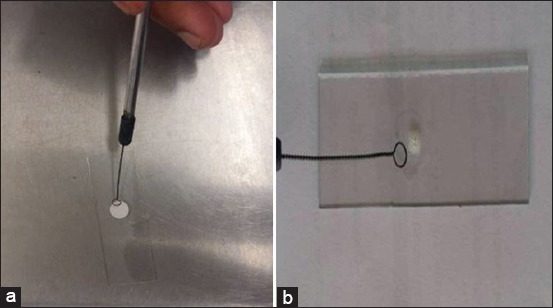
(a) Negative oxidase test. (b) Positive catalase test.
Figure-4.
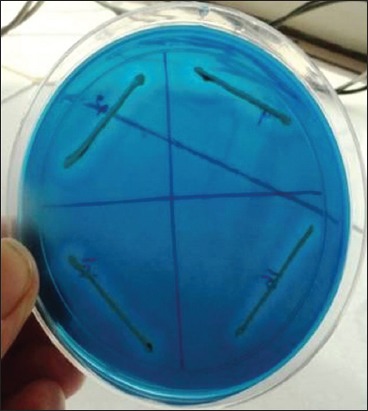
Staphylococcus aureus showing DNase activity on DNase agar.
Figure-5.
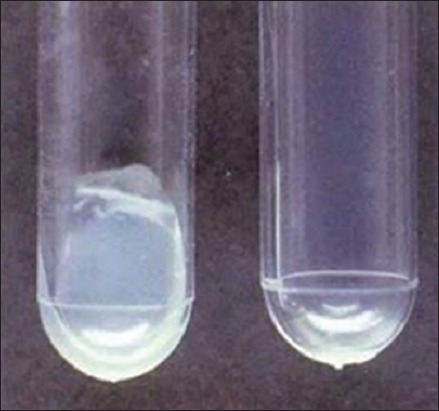
Coagulase positive (Clot), Coagulase negative (No Clot).
Molecular confirmation of S. aureus
To validate our identification of S. aureus, all 114 cultures were subjected to genotypic characterization by PCR using species-specific nuc gene amplification. 39 (34.2%) cultures which were morphologically and biochemically identified as S. aureus revealed the presence of nuc gene by the amplification of 270 bp product of species-specific nuc gene (Figure-6 and Table-1).
Figure-6.
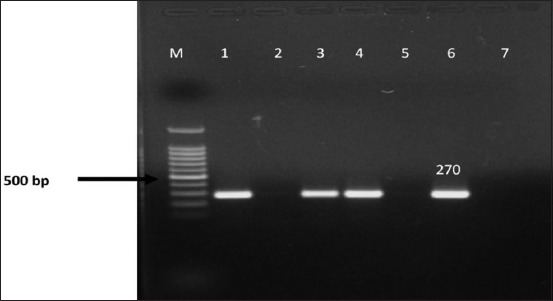
Polymerase chain reaction amplification of species-specific nuc gene of presumptive Staphylococcus aureus isolates. Lane M - 100 bp DNA ladder. Lane 1,3,4 - positive isolates. Lane 2 - negative isolate. Lane 5 - negative control. Lane 6 - positive control.
Molecular detection of coagulase gene (coa)
Twenty-five (64.10%) of 39 S. aureus isolates were positive for coa gene and four different amplicons of coa gene were detected (Table-2 and Figure-7). 22 isolates among them were positive for tube coagulase test.
Table-2.
coa gene PCR amplification showing polymorphism ranging from 500 to 800 bp.
| Samples | Source | S. aureus isolates | coa positive S. aureus (%) | Coa gene amplicon (bp) |
|---|---|---|---|---|
| Mastitic milk | Cattle | 22 | 19 (86.3) | 595 (15 isolates) 802 (4 isolates) |
| Nasal swabs | Cattle | 8 | 2 (25) | 514 (2 isolates) |
| Pus | Cattle | 9 | 4 (44.4) | 757 (4 isolates) |
| Total | Cattle | 39 | 25 (64.1) | 514, 595, 757, 802 |
PCR=Polymerase chain reaction, S. aureus=Staphylococcus aureus
Figure-7.
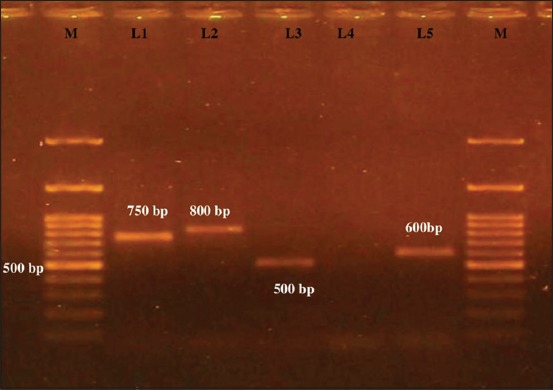
Polymerase chain reaction amplification of coagulase gene (coa) of Staphylococcus aureus isolates. M - 100 bp DNA ladder. L1 - 750 bp (approximately). L2 - 800 bp (approximately). L3 - 500 bp (approximately). L4 - negative control. L5 - 600 bp (approximately).
Detection of coa gene variants by RFLP
On HaeIII and AluI digestion of coa gene PCR product, four different RFLP patterns of coa gene variants were obtained (Figures-8 and 9, Table-3). Thus, four different RFLP patterns of coa gene variants were obtained in the present study. One strain each from different RFLP pattern was randomly selected and sent for sequencing (Xcelris Labs Ltd.). The sequences are yet to be submitted to NCBI, so accession number of the sequences are not generated yet. These sequences were aligned for maximum homology using Bioedit software (Figure-10). The similarity in the sequences was inferred with the help of sequence identity matrix as shown in Table-4 and a dendrogram was obtained (Figure-11). The most identical sequences with the value of 0.810 were found between isolate S. aureus 514, obtained from nasal cavity, and isolate S. aureus 595, obtained from mastitic milk. Thus, they are the most closely related, while as the isolates S. aureus 514 and S. aureus 802 are comparatively distant because of their least similarity with the value of 0.483 (Table-4).
Figure-8.
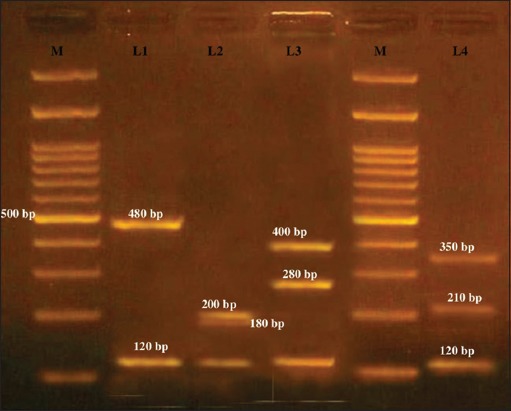
Hae111 restriction enzyme digestion of coa variant polymerase chain reaction products. M - 100 bp ladder. L1 - Hae111 digestion of approximately 600 bp coa gene variant into approximately 480 and 120 bp (P2). L2 - Hae111 digestion of approximately 500 bp coa gene variant into approximately 200,180, and 120 bp (P1). L3 - Hae111 digestion of approximately 800 bp coa gene variant into approximately 400, 280, and 120 bp (P4). L4 - Hae111 digestion of approximately 750 bp coa gene variant into approximately 350, 210, and 120 bp (P3).
Figure-9.
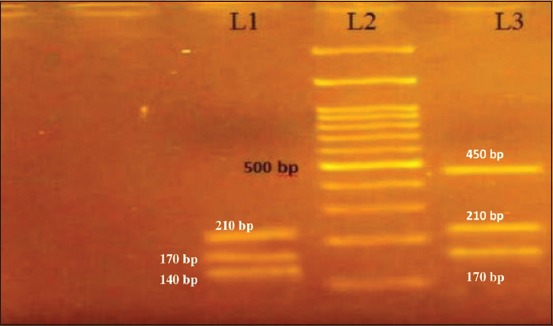
Alu1 restriction enzyme digestion of 750 and 600 bp coa gene polymerase chain reaction products. L1 -Alu1 digestion of approximately 600 bp coa gene variant into approximately 210, 170, and 140 bp (P2). L2 - 100 bp ladder. L3-Alu1 digestion of approximately 750 bp coa gene variant into approximately 450, 210, and 170 bp (P3).
Table-3.
Restriction digestion pattern of coa variable isolates using restriction enzymes HaeIII and AluI.
| Coa amplicon (approx. bp) | Number of isolates | Source | HaeIII digestion (bp approximately) | AluI digestion (bp approximately) | Pattern |
|---|---|---|---|---|---|
| 500 | 2 | Nasal swab | 200, 180, 120 | P1 | |
| 600 | 15 | Mastitic milk | 480, 120 | 210, 170, 140 | P2 |
| 750 | 4 | Pus | 350, 210, 120 | 450, 210, 170 | P3 |
| 800 | 4 | Mastitic milk | 400, 280, 120 | 450, 260 | P4 |
Figure-10.

Sequence alignment using Bioedit software, to find maximum homology between the sequences of isolates, one each from four different restriction fragment length polymorphism patterns
Table-4.
Sequence identity matrix showing genetic homology among four coa variable isolates.
| Seq‑> | S. aureus 757 | S. aureus 595 | S. aureus 514 | S. aureus 802 |
|---|---|---|---|---|
| S. aureus 757 | ID | 0.659 | 0.577 | 0.782 |
| S. aureus 595 | 0.659 | ID | 0.810 | 0.539 |
| S. aureus 514 | 0.577 | 0.810 | ID | 0.483 |
| S. aureus 802 | 0.782 | 0.539 | 0.483 | ID |
S. aureus=Staphylococcus aureus
Figure-11.
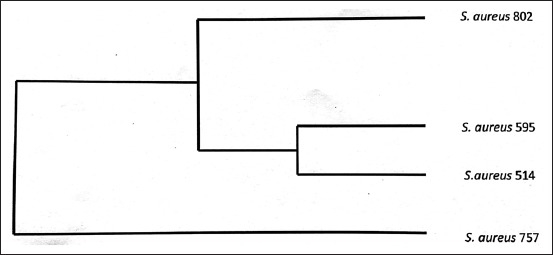
Dendrogram showing similarity among four coa variables having unique restriction patterns.
Discussion
In our study, 39 samples comprising of 22 mastitic milk, 9 pus samples from skin wounds, and 8 nasal swab samples were found positive for S. aureus, which indicate their occurrence of 22.4%, 20.4%, and 16%, respectively in Military dairy farm, Belicharana, Jammu. The previous study showed that S. aureus accounts for 23% of etiological agents associated with clinical mastitis in 30 milk recording dairy herds [11]. Another report showed a prevalence of 33.9% of S. aureus in pus samples from cattle [12]. Similarly, 5.06% prevalence of S. aureus was reportedin the nasal cavity of cattle [13] in Iran. In another report in Norway, a prevalence of 13.9% of S. aureus in the nasal cavity of cattle was reported [14].
For molecular identification of S. aureus isolates, molecular targeting of species-specific nuc gene of S. aureus coding for the extracellular thermostable nuclease protein (TNase) of S. aureus was demonstrated by Louie et al., and the same was utilized in the present study. PCR utilizing synthetic oligonucleotide primers of 21 and 24 bases has been used to amplify a segment of the nuc gene that is specific for S. aureus [15]. In our study, of 114 phenotypically identified Staphylococci, only 39 (34.2%) S. aureus isolates could be confirmed by nuc gene PCR. A study carried out on 68 Staphylococcus strains obtained from mastitic milk samples revealed 53 (78%) as positive for nuc gene [16]. Low detection of S. aureus inthe study may be attributed to non-utilization of other genotypic species-specific molecular procedures such as 23S rRNA-specific PCR, utilization of the crude method of DNA extraction, and lack of quantification that might increase the chances of false-negative results [17].
In our study, among all 39 isolates of S. aureus screened genotypically for coa gene, 25 strains (64.10%) amplified coa gene and are referred to as CoPS. All the 114 phenotypically identified Staphylococci cultures were subjected to tube coagulase test, which yielded positive coagulase test for 22 cultures. Three strains which were detected phenotypically by tube coagulase test as coagulase-negative were genotypically identified as coagulase positive. This emphasizes the use of molecular methods in the accurate detection of coa gene in S. aureus. A similar study carried out in 2015 found that 10 isolates, classified as coagulase-negative by tube coagulase test, were found to be positive with coa gene PCR [18].
PCR-RFLP is a rapid, reproducible, simple, and efficient method for typing S. aureus isolated from a clinical specimen. The variable region of coa gene is comprised of 81 bp tandem short sequence repeats. In our study, all the coa-positive isolates showed a single amplicon but of approximately four different sizes, 500 bp, 600 bp, 750 bp, and 800 bp. S. aureus isolates from mastitic milk source possessed coa amplicon of approximately 600 bp (15 isolates) and 800 bp (4 isolates). This finding is in agreement with a study carried out in 2007, which reported the presence of many variants of coagulase genotypes from regions of their study, but they indicated that only a few genotypes were predominant [19]. In a study carried out in 2015, all PCR products for coa gene were detected at 750 bp [18]. In our study, polymorphism of 4 coa gene products by RFLP using HaeIII and AluI restriction endonucleases was studied, and four RFLP patterns p1, p2, p3, and p4 were obtained. HaeIII showed better discrimination between the species than AluI. This is substantiated by a study carried out in 2014, which also concluded that HaeIII discriminatory power is better than AluI for typing S. aureus isolates [20]. One representative sample from each pattern was sequenced to obtain actual size of four coa types to be 514 bp, 595 bp, 757 bp, and 802 bp. Sequences of all the four patterns had unique restriction patterns. This was substantiated by unique restriction profile shown by these isolates on PCR-RFLP. Therefore, the study records the isolation of S. aureus with coagulase gene sequences having unique restriction patterns, not reported in earlier studies.
The sequences obtained were aligned for maximum homology between the sequences using Bioedit software [21], and similarity in the sequences was inferred with the help of sequence identity matrix. When the sequences from each of the distinct RFLP pattern were analyzed, it was found that the S. aureus isolate with a coa size of 514 bp, isolated from nasal cavity, had a highest similarity (81%) with the S. aureus isolate having coa size of 595 bp, isolated from mastitic milk. These results suggest that the possibility of S. aureus originating from nasal infections in the present study has evolved from mastitic milk strains and vice versa. When the genetic similarity of the other isolates was compared, it was found that a similarity of 48%, 53%, 57%, 65%, and 78% existed between the isolates S. aureus 802and S. aureus 514 (milk and nasal), S. aureus 595and S. aureus 802(both milk), S. aureus 757and S. aureus 514(pus from skin wound and nasal), S. aureus 602and S. aureus 757(milk and pus from skin wound),and S. aureus 800and S. aureus 757(milk and pus from skin wound),respectively. The genetic similarity among two representatives of S. aureus isolates from mastitic milk showed similarity only around 50% indicating evolutionary diversity in the strains causing mastitis in bovines.
Conclusion
Thus, it is concluded from the present study that both nuc gene and 23S rRNA gene-specific PCR should be used simultaneously for the molecular identification of S. aureus as these molecular tests when used individually may not cover all the S. aureus isolates from the samples. Further,the present study was localized to only one farm and different RFLP patterns of coagulase gene were observed from different sampling sites, indicating that S. aureus coagulase types have site-specific predilection. Two coa patterns were observed in mastitic milk, indicating multiple origins of infection, with 595 bp coa genotype being predominant. 514 and 595 coa variants of S. aureus are genetically most related.
Authors’ Contributions
AT and MAB designed the study. Laboratory work was done by FJ and assisted by AT and MAB. TAS helped in making understand the molecular part. GAB and MM prepared the manuscript and analyzed the data. All authors read and approved the final manuscript.
Acknowledgments
The authors wish to acknowledge the financial support provided by Dean, FVSc & A.H SKUAST-J and DBT (BT/PR12582/BPA/118/50/2014), New Delhi, the funding agency for projects in the division, for carrying out the research work.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Hedge R, Isloor S, Prabhu K.N, Shome B.R, Rathnamma D, Suryanarayana V.V.S. Incidence of subclinical mastitis and prevalence of major mastitis pathogens in organized farms and unorganized sectors. Indian J. Microbiol. 2013;53:315–320. doi: 10.1007/s12088-012-0336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harmon R.J. Physiology of mastitis and factors affecting somatic cell counts. J. Dairy Sci. 1994;77:2103. doi: 10.3168/jds.S0022-0302(94)77153-8. [DOI] [PubMed] [Google Scholar]
- 3.Windria S, Widianingrum D.C, Salasia S.I.O. Identification of Staphylococcus aureus and coagulase-negative staphylococci isolates from mastitis milk of Etawa crossbred goat. Res. J. Microbiol. 2016;11:11–19. [Google Scholar]
- 4.Goh S.H, Byrne S.K, Zhang J.L, Chow A.W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarzkoph A. Coagulase gene polymorphism in Staphylococcus aureus -A new epidemiological marker. Immun. Infekt. 1995;23:9–14. [PubMed] [Google Scholar]
- 6.Dendani Z.C, Bezille P, Arcangioli M.A. PCR and PCR-RFLP genotyping of Staphyloccocus aureus coagulase gene: Convenience compared to pulse-field gel electrophoresis. Comp. Clin. Pathol. 2016;25:1061–1064. [Google Scholar]
- 7.Karakulska J, Pobucewicz A, Nawrotek P, Muszynska M, Furowicz A.J, Czernomysy-Furowicz D. Molecular typing of Staphylococcus aureus based on PCR-RFLP of coa gene and RAPD analysis. Pol. J. Vet. Sci. 2011;14:285–286. doi: 10.2478/v10181-011-0044-5. [DOI] [PubMed] [Google Scholar]
- 8.Dehkordi A.A, Tajbakhsh E, Tajbakhsh F, Khamesipour F, Shahraki M.M, Momeni H. Molecular typing of Staphylococcus aureus strains from Iranian raw milk and dairy products by coagulase gene polymorphisms. Adv. Stud. Biol. 2015;7:169–177. [Google Scholar]
- 9.Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor A.E. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 2002;40:2786–2790. doi: 10.1128/JCM.40.8.2786-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hookey J.V, Richardson J.F, Cookson B.D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keane O.M, Budd K.E, Flynn J, McCoy F. Pathogen profile of clinical mastitis in Irish milk recording herds reveals a complex etiology. Vet. Rec. 2013;21:12–15. doi: 10.1136/vr.101308. [DOI] [PubMed] [Google Scholar]
- 12.AL-Tuffylie Y.I. Clinical and bacteriological study of subcutaneous abscesses caused by gram-positive bacteria in cow and sheep in Al-Qadissiyia province. AL-Qadisiya. J. Vet. Med. Sci. 2012;11:17–24. [Google Scholar]
- 13.Rahimi H, Saei H.D, Ahmadi M. Nasal carriage of Staphylococcus aureus : Frequency and antibiotic resistance in Healthy Ruminants. Microbiology. 2015;8:e22413. doi: 10.5812/jjm.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mork T, Kvitle B, Jorgensen H.J. Reservoirs of Staphylococcus aureus in meat sheep and dairy cattle. Vet. Microbiol. 2011;155:81–87. doi: 10.1016/j.vetmic.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Khan A.U, Sultan A, Tyagi A, Zahoor S, Akram M, Kaur S, Shahid M, Vaishnavi C.V. Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J. Infect. Dev. Ctries. 2007;1:289–295. [PubMed] [Google Scholar]
- 16.Bekhit M.M.S, Muharram M.M, Alhosiny I.M, Hashim M.E.S.Y. Molecular detection of genes encoding virulence determinants in Staphylococcus aureus strains isolated from bovine mastitis. J. Appl. Sci. Res. 2010;6:121–128. [Google Scholar]
- 17.Straub J.A, Hertel C, Hammes W.P. A 23S r RNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J. Food. Protect. 1999;62:1150–1156. doi: 10.4315/0362-028x-62.10.1150. [DOI] [PubMed] [Google Scholar]
- 18.Hamza D.A, Dorgham S.M, Arafa A. Coagulase gene typing with emphasis on methicillin-resistance Staphylococci: Emergence to public health. Adv. Infect. Dis. 2015;5:196–203. [Google Scholar]
- 19.Aslantas O, Demir C, Turutoglu H, Cantekin Z, Ergun Y, Dogruer G. Coagulase gene polymorphism of Staphylococcus aureus isolated from subclinical bovine mastitis. Turkish J. Vet. Anim. Sci. 2007;31:253–257. [Google Scholar]
- 20.Roodmajani H.K, Sarvari J, Bazargani A, Ghahraman M.R.K, Nazari A, Motamedifar M. Molecular typing of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from Shiraz teaching hospitals by PCR-RFLP of coagulase gene. Iran. J. Microbiol. 2014;6:246–252. [PMC free article] [PubMed] [Google Scholar]
- 21.Khademi F, Ghanbar F, Mellmann A, Najafzadeh M.J, Khaledi A. Phylogenetic relationships among Staphylococcus aureus isolated from clinical samples in Mashhad, Iran. Infect. Public Health. 2016;9:639–644. doi: 10.1016/j.jiph.2016.01.003. [DOI] [PubMed] [Google Scholar]


