Abstract
Structured RNAs and RNA-protein complexes (RNPs) fold through complex pathways that are replete with misfolded traps, and many RNAs and RNPs undergo extensive conformational changes during their functional cycles. These folding steps and conformational transitions are frequently promoted by RNA chaperone proteins, notably by superfamily 2 (SF2) RNA helicase proteins. The two largest families of SF2 helicases, DEAD-box and DEAH-box proteins, share evolutionarily conserved helicase cores but unwind RNA helices through distinct mechanisms. Recent work has advanced our understanding of how their distinct mechanisms enable DEAD-box proteins to disrupt RNA base pairs on the surfaces of structured RNAs and RNPs, while some DEAH-box proteins are adept at disrupting base pairs in the interior of RNPs. Proteins from these families use these mechanisms to chaperone folding and promote rearrangements of structured RNAs and RNPs, including the spliceosome, and may use related mechanisms to maintain cellular mRNAs in unfolded or partially unfolded conformations.
Introduction
Structured RNAs carry out a wide range of regulatory and catalytic functions in all domains of life. RNA folding is a hierarchical process in which rapid formation of local secondary structure, i.e. short helices, allows for formation of tertiary contacts between elements that are widely separated in sequence. With only four standard bases and the potential for non-canonical pairs beyond the standard Watson-Crick pairings, structured RNAs are plagued by a tendency to form non-native secondary structure elements (1, 2). Local non-native structures can be further stabilized by tertiary structure, resulting in misfolded structures that require large-scale remodeling by RNA chaperone proteins to form native conformations at biologically relevant rates (3). Even messenger RNAs (mRNAs), which must adopt single-stranded conformations to be actively translated by processing ribosomes, contain many sequences capable of forming local structures that must be at least transiently unfolded to permit movement of the ribosome in translation (4, 5).
Additionally, many processes in RNA metabolism rely on base pairing between multiple RNAs or formation of RNPs to mark the correct positions for processing steps. Nowhere is this more evident than in eukaryotic pre-mRNA splicing. In each cycle of splicing, small nuclear RNAs (snRNAs) and protein splicing factors must assemble to form a functional spliceosome RNP and then must be extensively remodeled through the splicing process. These assembly steps and rearrangements are accelerated by a set of DEAD-box and DEAH-box proteins (6, 7). There is a clear division of labor between the two helicase families, with DEAD-box proteins accelerating early assembly steps and DEAH-box proteins accelerating RNP rearrangements in the downstream catalytic steps. This striking division raises the possibility that different unwinding mechanisms are required for forming the active spliceosome versus carrying out the splicing reaction (7).
DEAD-box Proteins: Remodeling, One Duplex at a Time
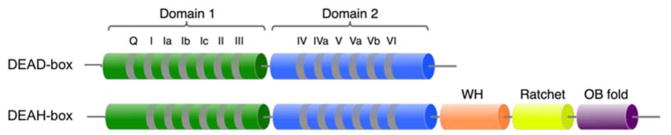
DEAD-box proteins, named for the amino acid sequence of a highly conserved motif, form the largest family of SF2 helicases, with 37 family members in humans and 26 in S. cerevisiae (8, 9). These enzymes function primarily as ATP-driven, non-processive helicases, binding and unwinding short, exposed RNA duplexes before releasing the RNA and repeating the process on another duplex segment (10). All DEAD-box proteins share a highly conserved helicase core that consists of two RecA-like domains (abbreviated D1 and D2) tethered by a short, flexible linker (Fig. 1). The core contains 13 conserved sequence motifs, many of which are implicated in specific steps of substrate binding and RNA duplex unwinding (11). In most DEAD-box proteins, the core is flanked by additional N- and/or C-terminal extensions, which contribute to the functional diversity of this protein family. Many of these extensions direct individual DEAD-box proteins to their functional targets by interacting with protein or RNA components of the targets, and some extensions modulate the activity of the helicase core (12, 13). The core itself binds short RNA duplexes without significant sequence specificity. It also binds ATP, with high specificity relative to other nucleoside triphosphates, to power cycles of RNA duplex unwinding (Table 1) (10, 14).
Figure 1.
Structural arrangement of conserved domains in DEAD-box and DEAH-box helicases. The protein families share two conserved, RecA-like domains with numbered sequence motifs that are highly conserved within each family. Many DEAD-box proteins have additional N- and C-terminal extensions that are required for their specific functions but are not conserved between different DEAD-box proteins and are not shown here. In contrast, DEAH-box proteins share conserved C-terminal domains consisting of winged helix (WH), ratchet-like, and oligosaccharide binding fold (OB fold) domains.
Table 1.
General Properties of DEAD-box and DEAH-box Helicases
| Family | Unwinding Substrate | NTP Preference | Tail Preference | Translocation | Polarity |
|---|---|---|---|---|---|
| DEAD-box | RNA | ATP | none | none | none |
| DEAH-box | RNA or DNA | Any NTP | 3′ | Yes | 3′ – 5′ |
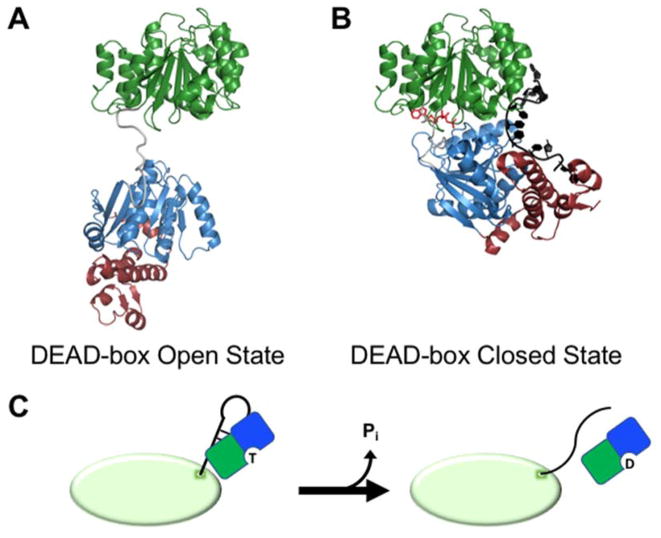
Work with the S. cerevisiae DEAD-box protein Mss116, which is required for efficient splicing of mitochondrial, self-splicing group I and group II introns, has suggested a mechanism by which the two core domains interact to unwind dsRNA. Attached by a flexible linker, the two domains remain ordered but spatially separated prior to substrate binding (Fig. 2A) (15, 16). ATP initially binds to D1, a property that seems to be universal among DEAD-box proteins, and dsRNA binds to D2 of Mss116 (15). In some DEAD-box proteins, initial RNA binding involves interactions with both domains, but upon formation of the ternary complex (enzyme, ATP, and dsRNA), unwinding proceeds similarly (17). A core closure step, which brings the two domains together and creates a functional ATPase active site, is driven by formation of inter-domain contacts between conserved amino acid motifs (18). Core closure significantly distorts the bound RNA, resulting in one strand becoming kinked and retained in a tight complex with the enzyme, while the other strand is released from its base pairs and from the complex (19–25). Mss116 and its homologs are unique among DEAD-box helicases in that they possess a C-terminal extension of D2 that forces a second kink in the retained strand, giving a ‘crimped’ conformation of this strand that may increase unwinding activity (Fig. 2B) (20). Recycling of the enzyme requires hydrolysis of the bound ATP and release of inorganic phosphate (Pi) to allow efficient release of the tightly bound RNA strand (26–29).
Figure 2.
Remodeling of exposed RNA duplexes by DEAD-box proteins. (A) Structural model for the open conformation of a DEAD-box protein (S. cerevisiae Mss116) prior to substrate binding (16). Conserved domains 1 and 2 are colored as in Figure 1. Mss116 and its homologs have an additional C-terminal extension (magenta) that is not present in all DEAD-box proteins. (B) Crystal structure representing the closed state of a DEAD-box protein (Mss116) following strand displacement (PDB: 3I5X) (20). The bound ssRNA strand is shown in black and the bound AMP-PNP molecule is red. (C) Model for unwinding of an exposed RNA duplex from a larger RNA or RNP by a DEAD-box helicase (shown as green and blue D1 and D2 respectively). The ATP-bound DEAD-box protein interacts with an exposed RNA helix on the surface of a structured RNA or RNP (green oval). Protein binding results in unwinding of the helix, allowing the partner strands to form new inter- or intramolecular interactions. ATP hydrolysis and release of Pi then allow the DEAD-box protein to release from the liberated ssRNA.
This cycle produces a nonconventional helicase mechanism for DEAD-box proteins that does not involve translocation. Instead, unwinding is limited to short RNA duplexes and achieved in a single cycle of ATP-dependent conformational changes (28, 30). Instead of advancing along the RNA duplex, DEAD-box proteins rely on stochastic separation of the base pairs adjacent to the unwound section. Consequently, the unwinding process becomes less efficient as duplex length increases and re-annealing becomes more favorable upon incomplete unwinding by the DEAD-box protein. Though the basic mechanism is similar to unwinding, some DEAD-box proteins function primarily as nucleotide dependent RNA clamps and RNP assembly platforms. In these cases, associated protein factors trap the helicase core in the RNA-bound state by regulating steps in the ATPase cycle. A notable example is eIF4A-III, which forms the core of the exon junction complex (EJC) by remaining stably bound to mRNA following splicing. The MAGOH-Y14 dimer, also made up of EJC components, interacts with eIF4A-III and stabilizes its ADP-Pi-bound state, which binds tightly to RNA and enables the ‘clamping’ activity within the EJC (9, 31, 32).
In vitro experiments using the Tetrahymena ribozyme, a well-studied group I intron derivative, have shown how the local unwinding activity of a DEAD-box helicase can lead to remodeling of a complex, structured RNA (Fig. 2C). In vitro, the Tetrahymena ribozyme predominantly folds into a stable, misfolded state that is structurally similar to the native state but requires significant remodeling of secondary and tertiary structure to refold into an active conformation (33, 34). CYT-19, an Mss116 homolog from Neurospora crassa, was shown to accelerate this transition significantly in the presence of ATP, suggesting that it acts as a general RNA chaperone and raising the questions of how a helicase can accelerate folding transitions that include loss of tertiary contacts, and how a general chaperone is able to favor the formation of native structure over misfolded structure (35, 36). Single molecule FRET experiments showed that instead of prying open tertiary contacts, CYT-19 relies on a mechanism of “helix capture,” in which the enzyme binds to transiently exposed helical structures, preventing them from re-forming tertiary contacts with the larger RNA structure and weakening any other secondary or tertiary contacts that form cooperatively (37). CYT-19 can then unwind the captured helix, further disrupting local structure and exposing more helices for binding. This result was bolstered by the finding that ribozyme mutations that decrease tertiary stability and lead to less compact structures also increase stimulation of CYT-19’s ATPase activity (38). The dependence of CYT-19 activity on exposed duplex structures suggests that DEAD-box proteins act preferentially on nonnative structures that lack the extensive compaction and tertiary contacts found in natively folded RNAs.
For larger RNAs, some remodeling steps likely require multiple cycles of helix capture, unwinding, and ATP hydrolysis, which can be accelerated by tethering the DEAD-box helicase to the RNA substrate. Both CYT-19 and Mss116 contain arginine-rich C-tails that bind introns non-specifically and are thought to tether the helicase cores to perform multiple rounds of unwinding on helices within an RNA structure (39, 40). Recently it was shown that decreasing the number of arginines in the CYT-19 C-tail led to decreased unwinding activity on an exposed Tetrahymena ribozyme duplex, and increasing the number of arginines increased the unwinding activity (41). These changes tracked directly with changes in functional RNA affinity. Although the basic C-tail is not ubiquitous among DEAD-box proteins, a similar mechanism of tethering the helicase core to structured RNAs or RNPs could be mediated by other unstructured domains, by assembly of the DEAD-box protein into a larger protein complex, or by specific interactions between an ancillary domain and a substrate RNA or RNP. A well-studied example of this last mechanism is the bacterial protein DbpA/YxiN, in which a folded ancillary domain localizes the protein by interacting specifically with an RNA helix within the large ribosomal subunit during ribosome biogenesis (42, 43). Recent results have suggested that the human DEAD-box protein DDX43 may follow a similar pattern, with an N-terminal K-homology nucleic acid-binding domain interacting with flanking RNA and imparting a preference for 5′-tailed dsRNA substrates (44).
DEAH-Box Proteins as Molecular Winches
DEAH-box proteins share many sequence and structural similarities with DEAD-box proteins, but they have adopted a very different mechanism of duplex unwinding (7). The DEAH-box core consists of two RecA-like domains, which contain the same set of conserved motifs as DEAD-box proteins but have notable differences in their sequences (Fig. 1). These core domains are flanked by N- and C-terminal extensions (8, 45). The N-terminal extensions show little conservation, and, as with enzymes from other families in SF2, often seem to be involved in recruiting the helicase to the correct complex or subcellular location (46). Unlike DEAD-box proteins, which contain a wide variety of non-conserved C-terminal extensions, DEAH-box proteins share a highly-conserved C-terminus made up of three domains: a winged helix (WH), a ratchet-like domain, and an oligosaccharide binding (OB) fold (45). Crystal structure show that these C-terminal domains interact strongly with the helicase core, yielding an enzyme with less flexibility in the core than DEAD-box proteins (47–49).
Whereas DEAD-box proteins use simple cycles of RNA duplex binding, unwinding, and release, DEAH-box proteins function as translocating helicases, advancing in the 3′–>5′ direction to disrupt nucleic acid structures (25, 50, 51). Instead of binding directly to structured RNA elements, DEAH-box helicases require 3′ single-stranded regions for unwinding activity (50, 52, 53). Unlike DEAD-box proteins, which are highly specific for dsRNA, some members of the DEAH-box family can act on both DNA and RNA, leading to unwinding of helices and, for some DEAH-box proteins, four-stranded G-quadruplex structures (54, 55). DEAH-box proteins also lack specificity for ATP, binding and hydrolyzing all four NTPs to power cycles of directional movement (Table 1) (8, 52, 56). Although many questions remain, recent structural and biochemical data have provided new insights into how these enzymes load onto a nucleic acid strand, couple ATP hydrolysis with translocation, and remodel RNP structures.
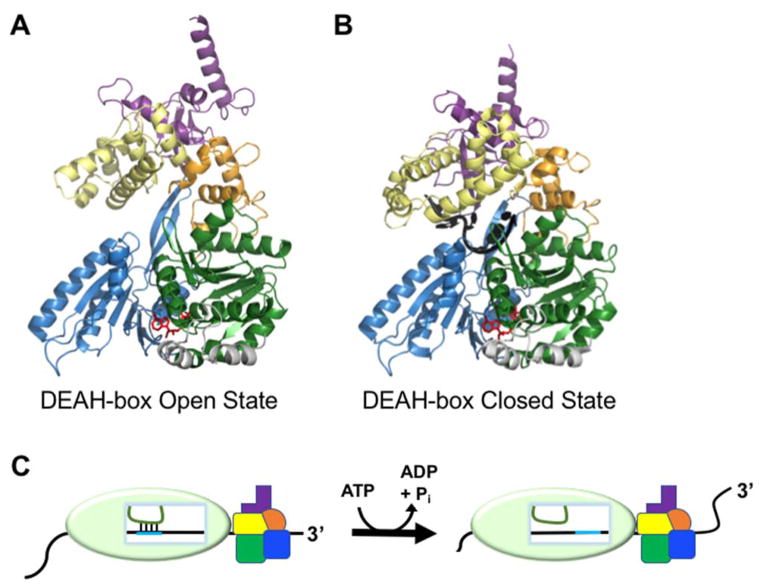
Recent structures of Prp43 from C. thermophilum have provided insight into how DEAH-box proteins load onto ssRNA adjacent to their target structures to initiate unwinding (57). Among other roles, Prp43 is required in the final step of pre-mRNA splicing, where it disrupts base pairing between the U2 snRNP and the intron branch point, releasing the U2, U5, and U6 snRNPs from the lariat intron (58–62). Like other DEAH-box proteins, Prp43 lacks sequence specificity for RNA substrates, a property that was rationalized from the crystal structure of the enzyme bound to ADP-BeF3 and U7 RNA, as the majority of contacts are formed with the sugar-phosphate backbone of the RNA, not the uracil bases (57, 63). A similar structure without RNA showed that large conformational changes in the C-terminal domains, particularly the ratchet-like and OB-fold domains, lead to an opening of the RNA-binding tunnel between D2 and the C-terminal domains (Fig. 3A,B). When this opening step was blocked by a disulfide bridge, the mutant Prp43 was decreased substantially in RNA unwinding ability, suggesting that opening is required for efficient substrate loading (57).
Figure 3.
Remodeling of internal RNA duplexes by DEAH-box proteins. (A) Crystal structure of Prp43 with bound ADP-BeFx showing the open conformation, which facilitates RNA loading (PDB: 5LTK)(57). Domains are colored as in Fig. 1. (B) Crystal structure of the closed conformation of Prp43. The structure includes bound ADP-BeFx and U7 RNA and reflects the conformation that follows RNA loading (PDB: 5LTA)(57). (C) Model for DEAH-box helicase-catalyzed disruption of an RNA duplex within a larger RNP complex by molecular winching (68). Translocation of the protein against the RNP surface results in movement of the RNA (black) relative to the interior of the RNAP. Because its base-pairing partner (green) is held in place relative to the RNP, the movement of the black RNA results in disruption of the base pairs between the two RNAs.
Structural and biochemical experiments with Prp43 have also suggested a model for ATP-dependent translocation. Two extended amino acid hairpin structures serve as 3′ and 5′ bookends to the bases of a bound RNA stack, and their movement relative to the RNA strand during cycles of NTP hydrolysis is suggested to drive 3′->5′ translocation (51). The 3′ hairpin includes an extended motif Ib on domain 1, which is highly conserved among DEAH-box helicases and related DExH helicases, but varies significantly from those of other SF2 families (51, 64). The 5′ hairpin is a D2 structure that does not correspond to previously identified SF2 core motifs but is present in other DEAH/RHA proteins, the viral NS3 helicase, and as a shorter version in the related ski2-like helicases (64–67). These bookends trap the bound nucleic acid bases between them, and they move in cycles to incorporate a new base on the 5′ side and exclude one on the 3′ side, resulting in directional translocation of the helicase. This cycle is driven by changing interactions between conserved motifs as NTPs are bound, hydrolyzed, and released (64). The directional translocation is suggested to confer RNA unwinding activity by displacing the complementary strand(55).
In pre-mRNA splicing, several DEAH-box proteins are required to promote a series of base-pairing changes between spliceosomal snRNAs and the mRNA substrate within a larger RNP during the splicing process. While the basic biochemical properties of DEAH-box proteins outlined above would suggest that they would be capable of unwinding these helices, the burial of the helices inside the spliceosome has led to the question of how the helicase proteins gain access to their substrates. Recent biochemical experiments with the yeast DEAH-box proteins Prp16 and Prp22 have provided an exciting new insight, suggesting a model in which DEAH-box proteins can unwind helices without traversing through them (Fig 3C) (68). Following the branching step of splicing, Prp16 is required for repositioning the RNA substrate to enable recognition of the 3′ splice site, and it also plays a role in branch site selection (69). Prp22 serves similar functions downstream in the process, regulating selection of the 3′ splice site and releasing mRNA from the spliceosome following exon ligation (70). The key observation was that as long as Prp16 and Prp22 could load onto a downstream segment of ssRNA, their activity was not abrogated by DNA nucleotide substitutions between the loading site and the remodeled regions, even though the proteins are unable to translocate through these DNA stretches. These seemingly paradoxical results were reconciled with a model in which the DEAH-box proteins load onto a 3′ extension and begin translocating. However, when they are prevented from moving relative to the spliceosome by contact with the protein surface, the result of continued translocation is to pull on the RNA strand, disrupting base pairs in a process termed winching (68). This novel model explains how DEAH-box proteins can promote unwinding of RNA helices that are buried inside the spliceosome without the helicases burrowing into the RNP. It remains to be determined whether this mechanism is shared by other DEAH-box proteins involved in splicing, such as Prp2 and Prp43, and whether it extends beyond the spliceosomal DEAH-box proteins.
Conclusions
We have discussed two distinct mechanisms by which RNA helicases can act as chaperones for RNAs and RNPs by disrupting RNA base pairs to permit remodeling steps. DEAD-box proteins rely on binding to exposed RNA duplexes and local, non-processive strand displacement to promote stepwise loss of compaction and ultimately allow RNAs a chance to refold. Though this activity is ubiquitous in all cells, the division of labor between DEAD-box and DEAH-box proteins in eukaryotic splicing suggests that this mechanism is not sufficient to remodel RNA structures occluded within the center of a larger RNP complex. The discovery that a subset of DEAH-box proteins use translocation to winch an RNA strand out of a larger structure, disrupting upstream base-pairing without translocating through the complex, suggests a simple solution to this problem.
It remains unclear whether these two mechanisms are universal for enzymes in their respective families. Although several well-characterized DEAD-box proteins have shown non-processive unwinding activities, partner proteins can dramatically alter their properties. Such appears to be the case for human eIF4A, which has been reported to undergo stepwise, processive duplex unwinding in the presence of eIF4G and eIF4B accessory proteins (71). It is also possible that some DEAD-box and/or DEAH-box proteins have evolved specifically for their individual functions in ways that endow them with properties quite distinct from the conventional properties of their families. Analysis of RNA and RNP remodeling reactions of a broader set of helicases from each family will ultimately shed light on the universality of these mechanisms.
Beyond the pathways discussed here, recent high-throughput experiments have shown that for mRNAs, duplex and G-quadruplex structures are actively unwound by robust, ATP-dependent RNA chaperone activities (72, 73). We still know relatively little about which RNA helicases are responsible, but DEAD-box and DEAH-box helicases are strong candidates for these activities. Indeed, the local duplex unwinding and winching mechanisms we have described for these helicases are likely to play important roles in these and other steps of RNA metabolism.
Acknowledgments
We thank Anna Mallam for providing the structural model of the open state of the DEAD-box protein core.
Funding Information
Research in the Russell laboratory is supported by grants from the National Institutes of Health (R01 GM070456 and P01 GM066275) and the Welch Foundation (F-1563).
Abbreviations
- dsRNA
double-stranded RNA
- ssRNA
single-stranded RNA
- FRET
Förster resonance energy transfer
- lncRNA
long non-coding RNA
- Pi
inorganic phosphate
- snRNP
small nuclear ribonucleoprotein complex
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270(36):20871–4. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Karpel RL, Swistel DG, Miller NS, Geroch ME, Lu C, Fresco JR. Acceleration of RNA renaturation by nucleic acid unwinding proteins. Brookhaven Symp Biol. 1975;(26):165–74. [PubMed] [Google Scholar]
- 3.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3(4):495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 5.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12(9):641–55. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Jarmoskaite I, Russell R. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem. 2014;83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28(2):253–63. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Mohr G, Del Campo M, Turner KG, Gilman B, Wolf RZ, Lambowitz AM. High-throughput genetic identification of functionally important regions of the yeast DEAD-box protein Mss116p. J Mol Biol. 2011;413(5):952–72. doi: 10.1016/j.jmb.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390(12):1237–50. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 14.Mallam AL, Sidote DJ, Lambowitz AM. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 2014;3:e04630. doi: 10.7554/eLife.04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490(7418):121–5. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, et al. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A. 2011;108(30):12254–9. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samatanga B, Klostermeier D. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 2014;42(16):10644–54. doi: 10.1093/nar/gku747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105(2):548–53. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35(5):598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313(5795):1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 22.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126(4):713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Collins R, Karlberg T, Lehtio L, Schutz P, van den Berg S, Dahlgren LG, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J Biol Chem. 2009;284(16):10296–300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16(3):247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 25.Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biol. 2012;9(6):751–71. doi: 10.4161/rna.20090. [DOI] [PubMed] [Google Scholar]
- 26.Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, et al. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol. 2011;409(3):399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aregger R, Klostermeier D. The DEAD box helicase YxiN maintains a closed conformation during ATP hydrolysis. Biochemistry. 2009;48(45):10679–81. doi: 10.1021/bi901278p. [DOI] [PubMed] [Google Scholar]
- 28.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105(51):20209–14. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, Putnam AA, Jankowsky E. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry. 2014;53(2):423–33. doi: 10.1021/bi401540q. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105(51):20203–8. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–9. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15(1):67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363(2):531–44. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell D, 3rd, Jarmoskaite I, Seval N, Seifert S, Russell R. The long-range P3 helix of the Tetrahymena ribozyme is disrupted during folding between the native and misfolded conformations. J Mol Biol. 2013;425(15):2670–86. doi: 10.1016/j.jmb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449(7165):1014–8. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103(45):16698–703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan C, Potratz JP, Cannon B, Simpson ZB, Ziehr JL, Tijerina P, et al. DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture. PLoS Biol. 2014;12(10):e1001981. doi: 10.1371/journal.pbio.1001981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarmoskaite I, Bhaskaran H, Seifert S, Russell R. DEAD-box protein CYT-19 is activated by exposed helices in a group I intron RNA. Proc Natl Acad Sci U S A. 2014;111(29):E2928–36. doi: 10.1073/pnas.1404307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46(11):3013–22. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, et al. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375(5):1344–64. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busa VF, Rector MJ, Russell R. The DEAD-box Protein CYT-19 Uses Arginine Residues in Its C-tail to Tether RNA Substrates. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem. 2005;280(42):35499–505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 43.Rudolph MG, Klostermeier D. When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol Chem. 2015;396(8):849–65. doi: 10.1515/hsz-2014-0277. [DOI] [PubMed] [Google Scholar]
- 44.Talwar T, Vidhyasagar V, Qing J, Guo M, Kariem A, Lu Y, et al. The DEAD-box protein DDX43 (HAGE) is a dual RNA-DNA helicase and has a K-homology domain required for full nucleic acid unwinding activity. J Biol Chem. 2017;292(25):10429–43. doi: 10.1074/jbc.M117.774950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozgur S, Buchwald G, Falk S, Chakrabarti S, Prabu JR, Conti E. The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 2015;282(5):850–63. doi: 10.1111/febs.13198. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4(10):1216–29. [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11(3):180–6. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29(13):2194–204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami K, Nakano K, Shimizu T, Ohto U. The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family. Acta Crystallogr F Struct Biol Commun. 2017;73(Pt 6):347–55. doi: 10.1107/S2053230X17007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30(6):743–54. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y, Staley JP, Andersen GR, Nielsen KH. Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA. RNA. 2017;23(7):1110–24. doi: 10.1261/rna.060954.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44(28):9795–803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 53.Smaldino PJ, Routh ED, Kim JH, Giri B, Creacy SD, Hantgan RR, et al. Mutational Dissection of Telomeric DNA Binding Requirements of G4 Resolvase 1 Shows that G4-Structure and Certain 3′-Tail Sequences Are Sufficient for Tight and Complete Binding. PLoS One. 2015;10(7):e0132668. doi: 10.1371/journal.pone.0132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, et al. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem. 2005;280(46):38117–20. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 55.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45(20):6510–21. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tauchert MJ, Fourmann JB, Luhrmann R, Ficner R. Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. Elife. 2017:6. doi: 10.7554/eLife.21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36(4):583–92. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fourmann JB, Dybkov O, Agafonov DE, Tauchert MJ, Urlaub H, Ficner R, et al. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. Elife. 2016:5. doi: 10.7554/eLife.15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL, et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013;27(4):413–28. doi: 10.1101/gad.207779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39(3):385–95. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, et al. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25(21):9269–82. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37(7):263–73. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prabu JR, Muller M, Thomae AW, Schussler S, Bonneau F, Becker PB, et al. Structure of the RNA Helicase MLE Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling. Mol Cell. 2015;60(3):487–99. doi: 10.1016/j.molcel.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107(2):521–8. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, et al. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol. 2011;405(5):1139–53. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107(27):12139–44. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164(5):985–98. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohrt T, Odenwalder P, Dannenberg J, Prior M, Warkocki Z, Schmitzova J, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA. 2013;19(7):902–15. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349(6309):487–93. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Garcia C, Frieda KL, Feoktistova K, Fraser CS, Block SM RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science. 2015;348(6242):1486–8. doi: 10.1126/science.aaa5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485):701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016;353(6306) doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]