ABSTRACT
Several members of the Rhodobacterales (Alphaproteobacteria) produce a conserved horizontal gene transfer vector, called the gene transfer agent (GTA), that appears to have evolved from a bacteriophage. The model system used to study GTA biology is the Rhodobacter capsulatus GTA (RcGTA), a small, tailed bacteriophage-like particle produced by a subset of the cells in a culture. The response regulator CtrA is conserved in the Alphaproteobacteria and is an essential regulator of RcGTA production: it controls the production and maturation of the RcGTA particle and RcGTA release from cells. CtrA also controls the natural transformation-like system required for cells to receive RcGTA-donated DNA. Here, we report that dysregulation of the CckA-ChpT-CtrA phosphorelay either by the loss of the PAS domain protein DivL or by substitution of the autophosphorylation residue of the hybrid histidine kinase CckA decreased CtrA phosphorylation and greatly increased RcGTA protein production in R. capsulatus. We show that the loss of the ClpXP protease or the three C-terminal residues of CtrA results in increased CtrA levels in R. capsulatus and identify ClpX(P) to be essential for the maturation of RcGTA particles. Furthermore, we show that CtrA phosphorylation is important for head spike production. Our results provide novel insight into the regulation of CtrA and GTAs in the Rhodobacterales.
IMPORTANCE Members of the Rhodobacterales are abundant in ocean and freshwater environments. The conserved GTA produced by many Rhodobacterales may have an important role in horizontal gene transfer (HGT) in aquatic environments and provide a significant contribution to their adaptation. GTA production is controlled by bacterial regulatory systems, including the conserved CckA-ChpT-CtrA phosphorelay; however, several questions about GTA regulation remain. Our identification that a short DivL homologue and ClpXP regulate CtrA in R. capsulatus extends the model of CtrA regulation from Caulobacter crescentus to a member of the Rhodobacterales. We found that the magnitude of RcGTA production greatly depends on DivL and CckA kinase activity, adding yet another layer of regulatory complexity to RcGTA. RcGTA is known to undergo CckA-dependent maturation, and we extend the understanding of this process by showing that the ClpX chaperone is required for formation of tailed, DNA-containing particles.
KEYWORDS: RcGTA, GTA, HGT, DivL, ClpX, ClpXP, CckA, CtrA, lateral gene transfer, horizontal gene transfer
INTRODUCTION
Horizontal gene transfer (HGT; also called lateral gene transfer) is the transfer of genetic material independently of the production of progeny. Several mechanisms of HGT have been discovered in prokaryotes, and the transfer of traits such as antibiotic resistance has been well documented (1). Gene transfer agents (GTAs) are vectors of HGT that are produced by several phylogenetically distinct prokaryotes, including members of both the Bacteria and Archaea. GTA particles resemble tailed bacteriophages (phages, prokaryotic viruses) of the Siphoviridae, in that they contain protein heads and tails but typically package essentially random segments of the bacterial genome (2, 3). A conserved GTA element is produced by many Rhodobacterales (members of the Alphaproteobacteria), including the purple nonsulfur bacterium Rhodobacter capsulatus and the abundant marine bacteria Roseovarius nubinhibens and Ruegeria pomeroyi (see reference 2 and references therein). Environmental studies have shown a great diversity of Rhodobacterales encoding GTAs in multiple marine environments, and several isolates were confirmed to produce GTA proteins (4–6). While the factors stimulating GTA production in natural environments have not been studied, it is known that nutrient depletion and a high cell concentration stimulate the production in pure cultures of the R. capsulatus GTA (RcGTA) model system (7, 8).
RcGTA is morphologically similar to a tailed phage and is produced by a small subset of the cell population (9, 10). While RcGTA packages ∼4 kb of essentially random bacterial DNA (10), it was recently reported that the DNA packaged by the Dinoroseobacter shibae GTA is not random and may employ a headful-packaging mechanism (11), as has also been proposed for RcGTA.
The genes encoding RcGTA are found at several locations in the bacterial chromosome, with most genes being found in the ∼15-kb RcGTA head and tail gene cluster (2). In laboratory culture, RcGTA particles attach and deliver DNA to other cells, and the incorporation of RcGTA-donated genes into the recipient cell's genome requires a natural transformation-like system (12).
HGT by RcGTA is controlled by several cellular regulators, including a Lux-like quorum sensing system, a (putative) phosphorelay involving the response regulator CtrA, the LexA repressor, and several genes whose functions are not clear (reviewed in reference 13). The regulation of GTAs in members of the Rhodobacterales by CtrA and quorum sensing may be common, as the GTA produced by D. shibae is also regulated by a LuxI homologue and CtrA (11, 14). However, disruption of a LuxI homologue in R. capsulatus decreased GTA production (7), whereas in D. shibae such a disruption increased production (11).
The CtrA response regulator has a central role in regulating HGT and is required both for RcGTA production and for the incorporation of RcGTA-donated DNA by inducing a natural transformation-like system (15, 16). In Alphaproteobacteria, such as the well-studied organism Caulobacter crescentus, CtrA is the terminus of a phosphorelay beginning with the hybrid histidine kinase CckA and including the phosphotransferase ChpT (17). While not biochemically verified, CckA-ChpT-CtrA appears to form a phosphorelay in R. capsulatus, as indicated by genetic experiments.
In C. crescentus, CckA alternates between kinase and phosphatase activity to control the phosphorylation of CtrA, and phosphorylated CtrA (CtrA∼P) induces a set of genes (18). In R. capsulatus, transcription of the ∼15-kb RcGTA gene cluster encoding the capsid and other structural proteins requires CtrA but not CckA (16, 19, 20). The nonphosphorylated CtrA protein appears to be involved in inducing the RcGTA structural genes, as a CtrA(D51A) mutation predicted to prevent phosphorylation increased intracellular RcGTA capsid protein levels (20). Additionally, CtrA phosphorylation is needed for correct assembly of RcGTA and progression to cell lysis (8, 16, 20, 21).
Although the CckA-ChpT-CtrA phosphorelay appears to be conserved throughout the Alphaproteobacteria (22), it serves different roles in different species: CtrA is essential for cell division and viability in members of the Caulobacterales and Rhizobiales (e.g., see references 23 and 24), but it is not essential for members of the Rhodobacterales and Rhodospirillales (e.g., see references 16 and 25). In C. crescentus, CtrA-mediated activation of gene expression is also controlled by the protein SciP (26). A SciP homologue is encoded in R. capsulatus, but a sciP knockout had no effect on RcGTA production (20).
Several regulators of the C. crescentus CckA-ChpT-CtrA phosphorelay have been discovered. DivL, a PAS domain-containing, kinase-deficient sensor kinase homologue, is required for the correct cellular localization of CckA and promotes CckA kinase activity, resulting in increased CtrA phosphorylation. DivL is itself regulated by the response regulator DivK, which interacts with the nonfunctional histidine kinase domain present in DivL (27–32). The C. crescentus CtrA protein is additionally regulated by proteolytic degradation, mediated by the protease ClpXP. The protease recognizes C-terminal residues of the CtrA protein and ensures its rapid removal under certain conditions (33, 34). R. capsulatus and other rhodobacters lack DivK, encode a short DivL homologue missing the nonfunctional histidine kinase domain found in other DivLs (22, 27), and contain ClpXP homologues. Whether DivL and ClpXP have similar roles in the Rhodobacterales has, to our knowledge, not been reported previously.
Here, we report that the R. capsulatus DivL and ClpX(P) homologues regulate CtrA similarly to the way in which they regulate CtrA in C. crescentus, indicating a conserved function in the Rhodobacterales and Caulobacterales. Both disruption of the R. capsulatus divL gene and site-directed mutation of the predicted CckA autophosphorylation (histidine) residue decreased the activity of CtrA∼P-dependent promoters, consistent with a reduced CckA kinase activity. Unexpectedly, both mutants greatly overproduced RcGTA proteins, indicating that a correct balance of CckA kinase and phosphatase activities is important for RcGTA production. Loss of the chaperone and protease component ClpX resulted in increased levels of CtrA, consistent with ClpXP-dependent proteolytic regulation of CtrA. ClpX was required for the assembly of RcGTA particles, and DNA-free, tail-less intermediates incapable of HGT were released in its absence.
RESULTS
The R. capsulatus DivL homologue is a regulator of RcGTA production.
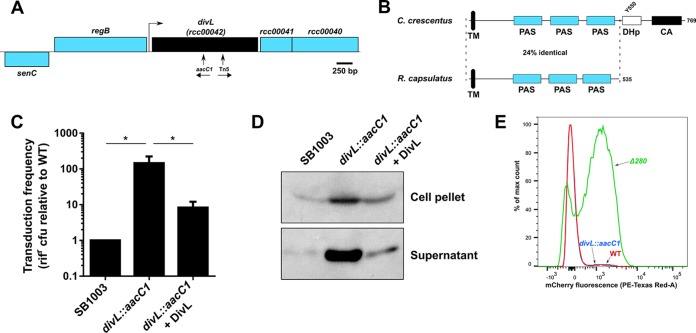
To improve our understanding of the regulation of HGT in the Rhodobacterales, we performed a transposon mutagenesis screen for new regulators of RcGTA production in the purple nonsulfur bacterium R. capsulatus, the model GTA producer. As a reporter, we utilized the plasmid pXCA-ghsA containing the CckA-dependent ghsAB head spike promoter fused to the Escherichia coli lacZ coding region (35). A transposon insertion into the open reading frame (ORF) rcc00042 (Fig. 1A) markedly reduced reporter activity. This ORF is annotated to encode a PAS/PAC sensor domain protein and is the BLASTP reciprocal best hit of the C. crescentus protein DivL (CcDivL; the prefixes “Cc” and “Rc” are used here as needed to differentiate between C. crescentus and R. capsulatus protein homologues, respectively), which promotes CckA autophosphorylation and localization in C. crescentus (27, 30, 32). The encoded Rcc00042 protein is 24% identical to the N-terminal 535 residues of CcDivL but does not contain the repurposed, nonfunctional histidine kinase domains dimerization histidine phosphotransfer (DHp) and catalytic ATP binding (CA) (Fig. 1B) present in CcDivL that interact with the DivK response regulator (a protein absent from rhodobacters [22]). Because of these similarities and differences, rcc00042 (hereinafter named divL) was chosen for further study, and a targeted disruption of divL (divL::aacC1; Fig. 1A) was constructed to verify the transposon screen and used for subsequent experiments.
FIG 1.
DivL regulates RcGTA production. (A) R. capsulatus divL (rcc00042) is located 3′ of regB (encoding the sensor histidine kinase of the redox-responding two-component system RegB/RegA). The insertion site of the transposon (Tn5) and the targeted disruption (aacC1) of divL are indicated, and the transcriptional directions of aacC1 and aph (in the transposed fragment) are indicated by arrows. A predicted promoter (BPROM) between regB and divL is indicated by a bent arrow. (B) Diagram of C. crescentus DivL and the short R. capsulatus DivL. Predicted domains, the Tyr550 residue, and the amino acid sequence identity between the two DivL proteins over the conserved length are indicated. TM, transmembrane domain. The domains were predicted using the EMBL SMART database. Panels A and B are drawn approximately to scale. (C) Gene transfer frequencies of culture supernatant from RcGTA-producing WT strain SB1003, the SB1003 divL::aacC1 mutant, and the mutant complemented with divL in trans (pABW627). Bars represent the means, error bars represent the standard deviations for three biological replicates, and significance was determined by the t test (*, P < 0.05). rifr, rifampin resistant. (D) RcGTA capsid production by SB1003, SB1003 divL::aacC1, and the mutant complemented with divL in trans (pABW627). An immunoblot of the culture supernatant (extracellular) and cell pellet (intracellular) fractions with proportionally equal loadings was probed with anti-RcGTA capsid serum. (E) Subpopulation expression of the RcGTA head-tail gene cluster. The distribution of cells producing the mCherry reporter from the RcGTA promoter in SBpG (WT) and the derived divL::aacC1 and Δ280 mutants (conferring the RcGTA overproduction phenotype) is shown. PE, phycoerythrin.
RcGTA maturation, lytic release, and head spike formation as well as gene transduction require the CckA protein (16, 20, 21, 35). Genetic experiments indicate that this is due to the role of CckA in forming CtrA∼P in R. capsulatus; for example, a phosphomimetic CtrA protein restored capsid release from a ΔcckA mutant (20). Based on studies of C. crescentus (27, 28, 30), we predicted that a loss of RcDivL would lead to reduced CtrA∼P formation, impaired activity of CtrA∼P-dependent promoters, and, therefore, reduced RcGTA transduction, due to the importance CtrA∼P in RcGTA maturation and release (13). To our surprise, the SB1003 divL mutant showed a greatly increased RcGTA transduction frequency (145-fold) relative to the parental strain (Fig. 1C). Similarly, greatly increased levels of RcGTA capsid protein were observed in immunoblots of the divL mutant compared to those of the parental strain, indicating that increased numbers of RcGTA particles were produced and released (Fig. 1D).
RcGTA is produced by a small subset (0.15 to 3%) of the cells in laboratory culture (9, 10). Using a fluorescent reporter for RcGTA production, we did not detect an increase in the subset of cells producing RcGTA for the divL mutant compared to the parental strain (Fig. 1E). We therefore attribute the increase in RcGTA production to result from an increased number of RcGTA particles per RcGTA-producing cell.
divL is required for maximal CtrA∼P formation, and divL expression is regulated by CtrA.
The observation that a divL mutation increased RcGTA production appeared to be at odds with either the prediction that RcDivL promotes CtrA∼P formation, as does the C. crescentus homologue (36), or the model that CtrA∼P formation is required for RcGTA transduction (13). Because RcDivL lacks the nonfunctional histidine kinase domains DHp and CA present in CcDivL, it appeared to be possible that it could have a different function than CcDivL, and we therefore tested whether RcDivL was involved in CtrA∼P formation.
Both expression of ghsAB and endolysin-holin-mediated cell lysis of R. capsulatus require an intact CckA-ChpT-CtrA phosphorelay (21, 35). We confirmed the assumption that CtrA∼P formation is required for ghsAB transcription by measuring expression in cells encoding phosphomimetic D51E and nonphosphorylatable D51A versions of CtrA (see Fig. S1A in the supplemental material). Similar experiments indicated that CtrA∼P was required for detection of cell lysis of an overproducer mutant (Fig. S1B), confirming the capsid release results obtained in the wild-type (WT) strain SB1003 background by Mercer et al. (20). Measurements of ghsAB promoter activity and cell lysis therefore appear to be valid reporters for CtrA∼P formation in R. capsulatus.
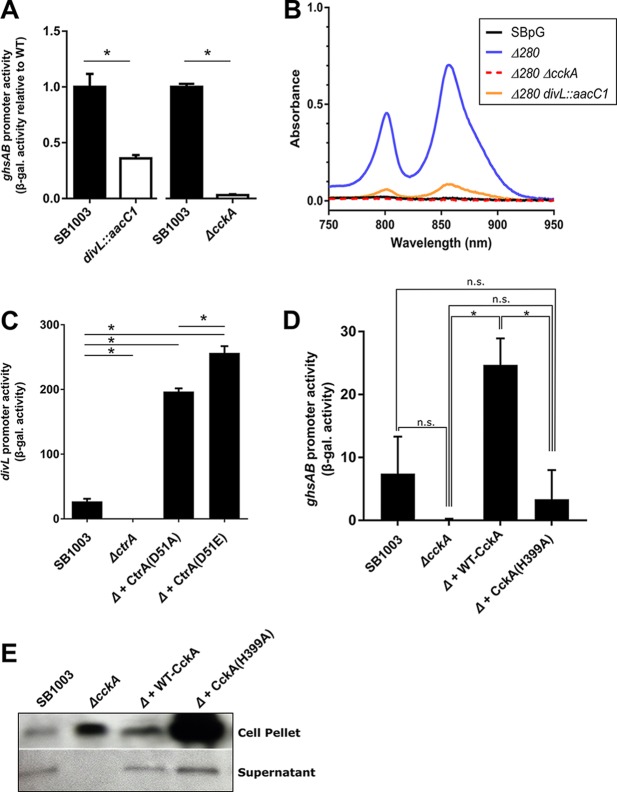
Consistent with the findings obtained with the transposon mutant, introduction of a targeted divL mutation into WT strain SB1003 decreased the ghsAB promoter activity to 36% of that in the strain without the mutation (Fig. 2A), indicating reduced CtrA∼P formation. Similarly, introduction of the divL mutation into the RcGTA overproducer SBpG Δ280 (see Materials and Methods) greatly inhibited cell lysis (Fig. 2B). However, the ΔcckA mutation had a much more dramatic effect (3.1% promoter activity and no detectable cell lysis; Fig. 2A and B) than the divL mutation. These intermediate levels of ghsAB expression and cell lysis in the divL mutant indicate that the levels of CtrA∼P in the divL mutant are reduced compared to those in WT cells and that DivL promotes but is not essential for CtrA∼P formation.
FIG 2.
Roles of DivL and CckA autophosphorylation in CtrA phosphorylation and RcGTA production. (A) Effect of the divL::aacC1 mutation on the CtrA∼P-dependent ghsAB promoter. The β-galactosidase (β-gal.) activities of SB1003, SB1003 divL::aacC1, and SB1003 ΔcckA containing the ghsAB promoter-lacZ reporter plasmid pXCA-ghsA are shown. (B) Amount of membrane-bound LH2 pigment in the culture supernatant of SBpG (WT), the RcGTA overproducer SBpG Δ280, and the SBpG Δ280-derived divL::aacC1 and ΔcckA double mutants. Representative curves are shown. (C) Effect of CtrA on divL promoter activity. The β-galactosidase activity from the chromosomally integrated divL promoter-lacZ reporter (pABW910) in SB1003, SB1003 ΔctrA, or the ΔctrA mutant (Δ) with nonphosphorylatable CtrA(D51A) or phosphomimetic CtrA(D51E) encoded on pD51A or pD51E, respectively, is shown. (D) Effect of the CckA(H399A) mutation on the CtrA∼P-dependent ghsAB promoter. The β-galactosidase activity from the chromosomally integrated ghsAB promoter-lacZ reporter (pABW848) in SB1003, SB1003 containing a ΔcckA mutation, and plasmid-encoded WT CckA (pRCckA) or ΔcckA and the predicted autophosphorylation-deficient CckA(H399A) (pCW104) is shown. (E) Effect of the CckA(H399A) mutation on RcGTA capsid production. The results for SB1003 or SB1003 containing a ΔcckA mutation and plasmid-encoded WT CckA (pRCckA) or ΔcckA and predicted autophosphorylation-deficient CckA(H399A) (pCW104) are shown. The immunoblot with proportionally equal loadings of the culture supernatant (extracellular) and cell pellet (intracellular) fractions was probed with anti-RcGTA capsid serum. Bars indicate means, and error bars indicate standard deviations for three biological replicates. *, a statistically significant difference, as follows: by a t test for the ghsAB promoter (P < 0.05) (A), by one-way ANOVA (F3,8 = 1,298; P < 0.0001) followed by Tukey's test (alpha = 0.05) (only selected significant results are indicated) (C), and by one-way ANOVA (F3,8 = 19.55; P = 0.005) followed by Tukey's test (alpha = 0.05) (D). n.s., not significant.
The expression of divL in Rhodobacter species was predicted to be regulated by CtrA (22), and a stimulatory role of RcCtrA on divL (then designated rcc00042) was observed in a microarray experiment (19). Using a divL promoter-lacZ reporter, we confirmed that CtrA is required for maximal divL expression (Fig. 2C). Unexpectedly, CtrA phosphorylation did not appear to be involved in the regulation of divL, as both the nonphosphorylatable CtrA(D51A) and the phosphomimetic CtrA(D51E), encoded in trans, restored reporter activity in the ΔctrA mutant. Therefore, divL expression requires CtrA in R. capsulatus, but CtrA phosphorylation does not appear to be a major contributor to divL regulation, at least under the conditions tested.
Impairment of CckA autophosphorylation greatly increases RcGTA protein production.
The role of CckA and CtrA in RcGTA production is not fully understood: overexpression of cckA decreases RcGTA production by an unknown mechanism (37). Although a nonphosphorylatable CtrA(D51A) mutant had increased intracellular levels of capsid protein (20), no apparent change in total (intracellular plus extracellular) capsid protein was observed for the cckA deletion mutant (20, 21, 37).
DivL was reported to stimulate CckA kinase activity in C. crescentus (36). To test the hypothesis that increased RcGTA production in the divL mutant was caused by a decrease in the ratio of CckA kinase to phosphatase activity, we mutated the predicted RcCckA autophosphorylation residue H399A. The analogous mutation was reported to block CcCckA kinase activity, but not phosphatase activity, in vitro (18). The ghsAB reporter activity in cells with the H399A mutation was reduced to 14% of that in cells with trans-encoded WT CckA, indicating a large impairment in CtrA∼P formation (Fig. 2D). This activity was higher than (although not statistically different from) the activity in ΔcckA cells (completely lacking the CckA protein); compared to the activity in WT cells, the decrease for cells producing CckA(H399A) was the same as that for cells producing the nonphosphorylatable CtrA(D51A) protein (both were decreased by 45%; Fig. 2D and S1A), indicating that little or no CtrA∼P was formed in cells producing CckA(H399A).
Similar to the findings for the divL mutant, cells producing RcCckA(H399A) as the sole CckA protein had greatly elevated levels of RcGTA capsid protein production (Fig. 2E). The great majority of the capsid produced was found in the cell fraction, consistent with weak lysis, attributed to very low levels of CtrA∼P.
Therefore, two R. capsulatus mutations [divL and CckA(H399A)] that are predicted to reduce the kinase activity of CckA and result in decreased CtrA∼P formation greatly increased the levels of the RcGTA capsid protein (and likely other proteins encoded in the ∼15-kb RcGTA cluster), similar to the phenotype of mutants with a CtrA(D51A) mutation (20). Our data therefore support a model in which a short DivL homologue regulates CckA activity and in which CckA and the relative levels of CtrA and CtrA∼P regulate RcGTA protein production (see Fig. 5).
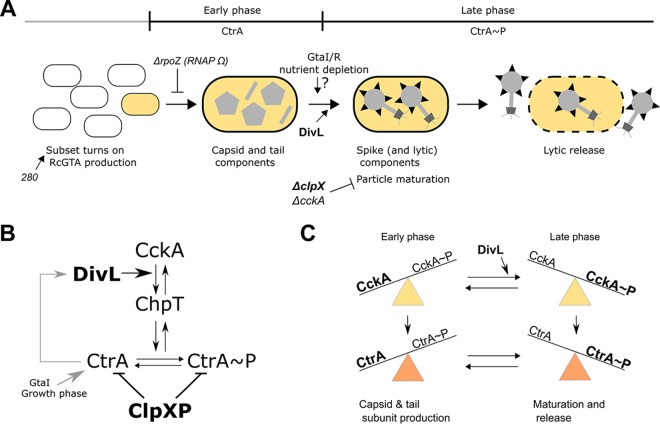
FIG 5.
Proposed model of RcGTA production and its regulation. (A) RcGTA production is turned on in a subset of the cell population by a poorly understood mechanism. In the proposed model (see the text for details), nonphosphorylated CtrA is present early on during RcGTA production and stimulates the production of RcGTA capsid and likely tail proteins. CtrA phosphorylation initiates the second phase of RcGTA production with the production of head spikes, the maturation of RcGTA particles, and, finally, the lytic release of functional RcGTA particles. DivL stimulates the transition into the late phase. The 280 allele affects the subset of cells producing RcGTA, and appreciable RcGTA production requires the rpoZ-encoded RNA polymerase omega subunit (RNAP Ω) (8). RcGTA production is stimulated by nutrient depletion (8) and is controlled by GtaI/R (7, 85), possibly by influencing protein levels or the phosphorylation status of CtrA. A loss of cckA (21) or clpX interferes with maturation. (B) Proposed regulatory model for the CckA-ChpT-CtrA phosphorelay in R. capsulatus, based on genetic evidence (20). CckA autophosphorylation initiates a phosphorylation cascade through the phosphotransferase ChpT to CtrA. DivL stimulates CckA kinase activity, resulting in the increased phosphorylation of CtrA. CtrA is further regulated transcriptionally by growth phase and an acyl-homoserine lactone produced by GtaI (44) and at the protein level by ClpXP-mediated proteolysis. Expression of divL is regulated by CtrA. (C) DivL appears to act as a stimulatory switch on CckA, promoting the transition from the early to the late phase of RcGTA production by increasing the relative ratio of CckA kinase to phosphatase activity. This in turn increases CtrA phosphorylation.
The ClpXP protease regulates the CtrA protein in R. capsulatus.
In C. crescentus, the protease ClpXP recognizes C-terminal residues of CtrA and degrades CtrA at specific points in the cell cycle (33, 38). Although the CtrA proteins of Alphaproteobacteria are highly conserved along most of their length (Fig. S2A), the C-terminal sequence is diverse, making it unclear whether proteolytic regulation occurs in species other than C. crescentus. We therefore set out to test whether ClpXP regulates the amount of CtrA in R. capsulatus and what the effects of a loss of ClpXP might have on RcGTA production.
The ClpXP protease is composed of the subunits ClpX and ClpP (39). In R. capsulatus, ClpP and ClpX are encoded adjacent to each other in an apparent operon (Fig. 3A and S2B), similar to their arrangement in the gammaproteobacterium E. coli (40, 41). Inspection of the genomes of several model Alphaproteobacteria (Fig. S2B) indicated that this organization is common. In contrast, C. crescentus has a different arrangement (Fig. S2B), with CicA (42) being encoded by a sequence between clpP and clpX and transcribed in the opposite direction (43). C. crescentus clpX is transcribed independently of clpP from its own set of promoters (43), whereas in E. coli, clpX appears to be expressed predominantly from the clpP promoter, with a weak promoter being present between clpP and clpX (41). Similar to the situation in E. coli, an intercistronic promoter-like sequence is present in R. capsulatus (Fig. 3A). No Rho-independent transcriptional terminator was predicted between clpP and clpX in R. capsulatus (using the tool ARNold [data not shown]), unlike in C. crescentus (43).
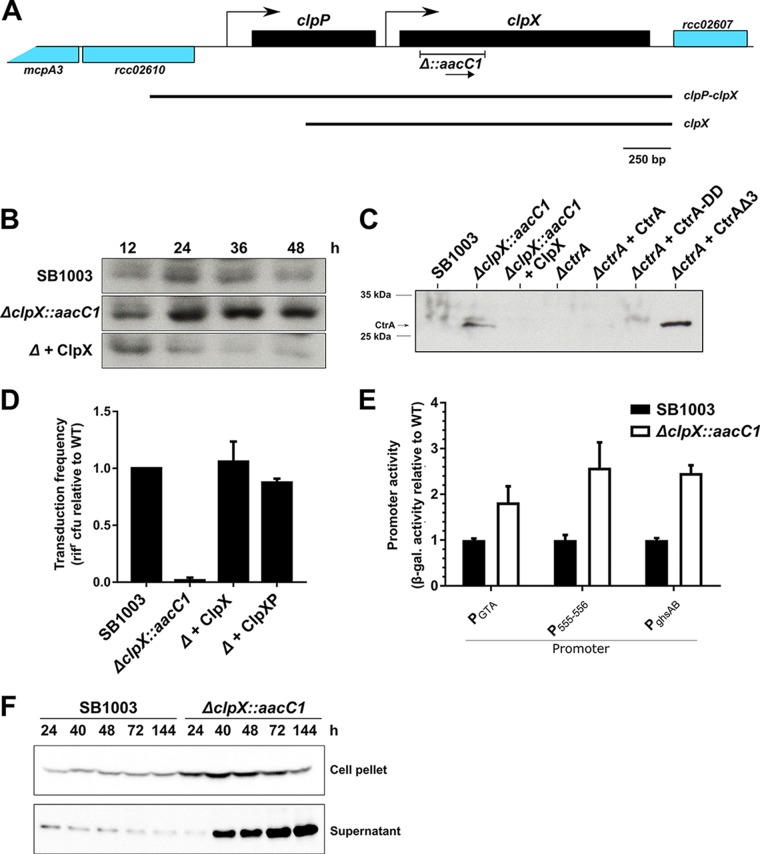
FIG 3.
ClpX(P) regulates CtrA levels and is required for RcGTA-mediated gene transfer. (A) Genetic context of clpX and clpP in R. capsulatus (see Fig. S2B in the supplemental material for the genetic context in other bacteria). Predicted promoters (bent arrows), the fragment replaced by aacC1 to create the ΔclpX::aacC1 mutant, and fragments included in the complementation plasmids pABW710 and pLK718 are indicated. The transcriptional direction of aacC1 is indicated by an arrow. The figure is drawn approximately to scale. (B and C) Immunoblots of CtrA levels in cells. The membranes were probed using C. crescentus CtrA antiserum. (B) A time course experiment of CtrA in WT strain SB1003, the SB1003 ΔclpX::aacC1 mutant, and the mutant containing clpX in trans on pABW710. (C) CtrA levels in SB1003 and the derived ΔclpX::aacC1 and ΔctrA mutants encoding ClpX (pABW710), WT CtrA (pLK754), CtrA-DD (pLK755), or CtrAΔ3 (pLK756) in trans, as indicated. (D) RcGTA-mediated gene transfer frequency of the culture supernatant of SB1003, the SB1003 ΔclpX::aacC1 mutant (Δ), and the mutant containing clpX (pABW710) or clpP-clpX (pLK718) in trans. (E) Activities of three RcGTA promoters in SB1003 and SB1003 ΔclpX::aacC1 mutant cells. The β-galactosidase activity of cells containing the RcGTA head-tail cluster reporter p601-g65 (PGTA), the endolysin-holin reporter pXCA-555 (P555-556), and the ghsAB head spike reporter pXCA-ghsA (PghsAB) is shown. (F) Production and release of cleaved RcGTA capsid protein by SB1003 and SB1003 ΔclpX::aacC1. An immunoblot of the supernatant (extracellular) and cell pellet (intracellular) fractions is shown. The membranes were probed using RcGTA capsid antiserum. Cells were cultured for an equivalent number of cell divisions (C to E) or harvested after the indicated times (in hours) of incubation after inoculation (B and F). Bars represent means, and error bars represent the standard deviations for two biological replicates (D and E).
Because ClpP is a member of a collection of proteases including ClpAP (39), we opted to delete and replace a segment of the R. capsulatus clpX gene with aacC1 (Fig. 3A), to maximize the likelihood that the mutation would be specific only for the ClpXP protease. ΔclpX::aacC1 mutants (here referred to as ΔclpX mutants) were readily obtained but showed a doubling time (3.9 h) slower than that of the parental strain (2.2 h).
ctrA expression is increased at later growth stages in R. capsulatus cultures (44); however, immunoblots (Fig. 3B) of WT cells (probed using CcCtrA antiserum) revealed that RcCtrA (26.7 kDa) levels were relatively constant over the entire growth phase, indicating that CtrA levels are maintained at a low level by a posttranscriptional mechanism in R. capsulatus. The RcCtrA band was greatly increased in the SB1003 ΔclpX mutant at later time points (24, 36, and 48 h postinoculation) but not at an early time point (12 h) compared to its level in strain SB1003, indicating that ClpXP is required for correct CtrA protein levels in R. capsulatus. Because ClpXP recognition of CcCtrA requires the last three C-terminal amino acids of the protein, we constructed plasmids that harbored RcCtrA either that lacked the three C-terminal amino acids, Val-Gly-Ala (CtrAΔ3), or in which the two last amino acids were converted to Asp (CtrA-DD), alterations that stabilized CcCtrA against degradation by ClpXP (33). Both CtrA alleles restored RcGTA transduction to a ΔctrA mutant, indicating that they encoded proteins that were functional (Fig. S2C). A band that migrated slightly slower than the WT RcGTA protein and that was of increased intensity was observed for the CtrA-DD mutant (Fig. 3C). In contrast, the amount of CtrA in cells producing CtrAΔ3 was clearly increased compared to that in cells producing WT CtrA, indicating that the C-terminal sequence of RcCtrA is required for the correct regulation of CtrA levels in R. capsulatus (Fig. 3C).
Therefore, the R. capsulatus CtrA protein appears to be proteolytically regulated by ClpXP and ClpXP maintains CtrA at relatively low levels in the cell population throughout the growth phase.
ClpX is required for RcGTA-mediated horizontal gene transfer.
We anticipated that the elevated levels of RcCtrA in the ΔclpX mutant would increase the production of RcGTA, because RcCtrA is required for expression of RcGTA genes (13, 16). To our surprise, this mutation nearly abolished RcGTA-mediated gene transfer (1.7% of the WT level, restored by providing clpX or clpP-clpX in trans; Fig. 3D). Further analyses indicated that the ΔclpX mutation had a novel effect on RcGTA production: the promoters for the RcGTA gene cluster, the lytic endolysin-holin system, and the ghsAB head spike genes were highly active in the ΔclpX mutant (Fig. 3E), indicating formation of CtrA∼P, and the mutant produced and released greatly elevated levels of the capsid protein (Fig. 3F). In contrast, all capsid-producing but gene transfer-deficient mutants previously discovered (the ΔcckA, ΔchpT, and Δ1866 mutants) do not release RcGTA proteins to the culture supernatant and, where investigated, do not express the lysis or head spike genes (20, 21, 45).
Maturation of RcGTA heads requires ClpX.
The results presented above indicated that ClpX could be required for the maturation of RcGTA to functional particles. RcGTA morphologically resembles a small, tailed double-stranded DNA (dsDNA) phage and appears to have separate head and tail assembly pathways, as in many phages (35). The head structure (the capsid) of tailed dsDNA phages (Caudovirales) matures through several intermediate (pro)head stages, which often involve cleavage of the capsid (46). Similarly, the capsid protein of RcGTA is proteolytically cleaved by the prohead protease to form a smaller peptide (16; our unpublished observations).
Because the ΔclpX mutant produced capsid protein of the same size as the cleaved form (Fig. 3F), we suspected that a prohead II-like or later maturation structure was formed, since proteolytic cleavage occurs concomitantly with the transition from prohead I to prohead II in phage HK97 (47).
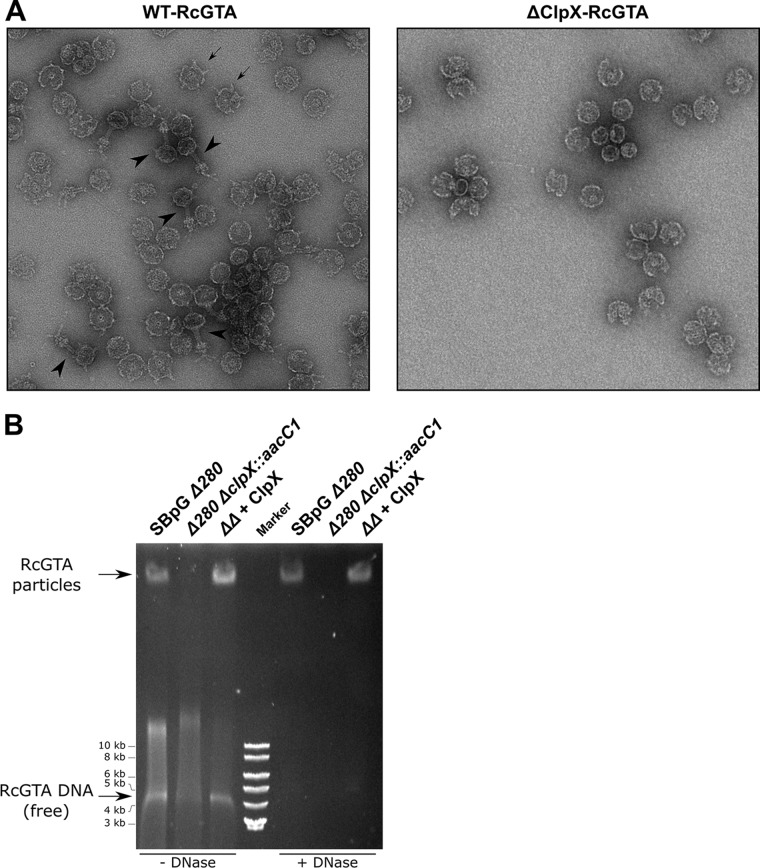
We purified His-tagged RcGTA particles (35) from a ΔclpX mutant (ΔClpX-RcGTA) and compared them to the particles from the parental strain (WT-RcGTA) using transmission electron microscopy. Numerous tailed particles with head spikes and spike-containing heads were observed for the WT-RcGTA (Fig. 4A). In contrast, all ΔClpX-RcGTA particles lacked tails, and no head spikes were observed (Fig. 4A).
FIG 4.
RcGTA assembly and DNA packaging defects in the absence of ClpX. (A) Transmission electron microscopy of chimeric RcGTA purified from the ClpX-positive RcGTA overproducer DE442 (WT-RcGTA) or the DE442 ΔclpX::aacC1 mutant (ΔClpX-RcGTA). Examples of tailed particles with head spikes (arrowheads) and head-only spiked particles (arrows) are indicated. The 6His-tagged capsid protein was carried on pRhoG5CTH. (B) Native agarose gels stained for DNA. Culture supernatants from SBpG Δ280, the SBpG Δ280 ΔclpX double mutant (ΔΔ), or the double mutant complemented with ClpX in trans (pABW710) were tested. DNase treatment is indicated. Arrows indicate the migration of the DNase-resistant RcGTA particle (top) and the DNase-sensitive ∼4-kb free DNA band derived from RcGTA (bottom).
To test whether a DNA-containing intermediate was formed, we performed native gel analysis of the capsid-containing culture supernatant. No DNase-resistant band or enrichment of the ∼4-kb RcGTA DNA band was observed for the ΔclpX mutant (Fig. 4B). Therefore, ClpX is required for maturation of the RcGTA prohead to a DNA-containing head structure, and this appears to be independent of its role in CtrA regulation, as the CtrAΔ3 protein supported the production of transduction-competent RcGTA (Fig. S2C).
In summary, we have identified DivL and ClpX(P) to be new regulators of the R. capsulatus CckA-ChpT-CtrA pathway and RcGTA production: DivL was required for maximal phosphorylation of CtrA, and an impairment in CtrA phosphorylation due to a loss of DivL or mutation of the CckA autophosphorylation residue greatly increased RcGTA capsid protein production. The ClpXP protease was found to control the levels of the CtrA protein, and ClpX was required for maturation of RcGTA particles.
DISCUSSION
R. capsulatus and several other Rhodobacterales transfer genetic material by particles called GTAs (2, 3). Allele exchange mediated by RcGTA has properties of both transduction and transformation, and both processes (RcGTA production and uptake/recombination of RcGTA-donated DNA) are controlled by cellular regulators (reviewed in reference 13). In this study, we further extend the repertoire of known regulators of RcGTA production to include DivL and ClpX(P). Our results indicate that they both regulate CtrA, a response regulator that has a central role in controlling GTA production in R. capsulatus (13, 16) and D. shibae (14). We speculate that these regulators could control GTAs produced by other Rhodobacterales species.
Members of the Rhodobacterales are not alone in regulating processes that allow for HGT (48). For example, regulation of natural competence genes in Streptococcus pneumoniae involves ClpP (ClpX was not investigated) (49), the two-component system ComD-ComE that senses a secreted peptide, and the alternative sigma factor ComX (50–53). Despite the increasing knowledge of RcGTA regulation, the transcription factor(s) acting directly on the RcGTA promoters is still a mystery: there is no evidence that CtrA binds to or near any RcGTA promoters (unpublished results of gel mobility shift experiments), and a rigorous attempt to identify a potential sigma factor(s) involved in regulating RcGTA using gene knockouts failed to show altered RcGTA production (54).
The CtrA protein is conserved in the Alphaproteobacteria, and our results show that the regulation of CtrA by DivL and ClpXP is also conserved, at least between the Caulobacterales (36) and the Rhodobacterales. Several members of the Alphaproteobacteria lack both DivK and the nonfunctional histidine kinase domain present in CcDivL. Our data show that at least one of these short DivL homologues promotes CtrA∼P formation similarly to the full-length CcDivL, and so as reported by Childers et al., the DivL histidine kinase domain appears to be repurposed as an input domain for sensing the DivK phosphorylation status (27). Our data are consistent with a regulatory model where DivL enhances CckA kinase activity and, thereby, CtrA∼P formation in R. capsulatus (Fig. 5). In this model, the dysregulation or loss of CckA kinase activity due to either the divL or the CckA(H399A) mutation greatly increased the production of RcGTA proteins, and for the divL mutant, this also resulted in a large increase in extracellular RcGTA proteins and gene transfer. It therefore appears that the reduced but substantial levels of CtrA∼P (inducing 36% ghsAB promoter activity) in the divL mutant are sufficient to allow the maturation and release of RcGTA particles, whereas the very low CtrA∼P levels in the CckA(H399) mutant are not. A similar, but smaller, increase in capsid production occurred in cells producing the nonphosphorylatable CtrA variant CtrA(D51A) (20). We therefore suggest that RcGTA production is divided into at least two phases and that the switch between the phases is controlled by the kinase activity of CckA (Fig. 5A and C): in the early phase of RcGTA production, CckA has very low or no kinase activity and CtrA is present but not phosphorylated. In this phase, cells produce large amounts of RcGTA head and tail components encoded in the ∼15-kb gene cluster. In the late phase, CckA is active as a kinase, CtrA becomes phosphorylated, and additional RcGTA genes, including the ghsAB head spike and lytic genes, are expressed, resulting in complete assembly and eventually the release of RcGTA particles from cells by controlled lysis. This is somewhat analogous to the temporal control of gene expression found in bacteriophages, such as bacteriophage lambda, where DNA replication occurs early, followed by virion production and, later, cell lysis (55, 56). In the RcGTA system, it appears that the timing of this transition was disrupted by the divL or CckA(H399A) mutation, and a postponed (divL) or abolished [CckA(H399A)] entry into the late phase results in cells that produce increased amounts of early (e.g., capsid) proteins and, in the case of divL, subsequent maturation and lysis. Consistent with this model, we did not observe an increase in the number of RcGTA-producing cells for the divL mutant. We note that bacteriophages with mutations that delay the onset of cell lysis can have a drastically increased number of phages produced per infected cell compared to the parental phage (57, 58), and the RcGTA system may similarly have evolved for an optimal timing of release rather than maximum particle production. We suggest that in R. capsulatus, ClpXP-mediated degradation of CtrA decreases or resets the DivL-modulated kinase activity of CckA, because divL expression requires CtrA (Fig. 2C). This may be of particular importance to the non-RcGTA-producing and nonlysing cells (the majority of cells) in cultures. It is unclear why a ΔcckA mutant does not drastically increase the production of RcGTA proteins, but we speculate that such increases relate to the ratio of CckA kinase/phosphatase activity, as opposed to the complete absence of the CckA protein.
The maturation defect caused by the absence of ClpX may be related to the failure of ΔcckA cells to produce transduction-competent RcGTA particles (21). In C. crescentus and Sinorhizobium meliloti, CckA regulates ClpXP-mediated degradation of CtrA, mediated by phosphorylation of the protein CpdR (59–61). Although members of the Rhodobacter genus lack homologues of CpdR (22), it is conceivable that another protein has a similar function.
E. coli ClpX has been reported to have biological roles both in the association with ClpP and as an independent chaperone (39). The mechanistic role of ClpX in RcGTA head maturation remains elusive, but we note that our observations (the absence of DNA packaging and spikes) would be consistent with a lack of capsid expansion, as the attachment of phage-decorating proteins (analogous to the RcGTA head spikes), such as phage T4 hoc and soc proteins and the phage T5 pb10 protein, requires the head expansion that occurs concomitantly with DNA packaging (62–64). Little is known about how RcGTA DNA packaging is initiated, and ClpX may be involved in the DNA packaging process. It is noteworthy that in C. crescentus, ClpXP cleaves the DnaX clamp loader protein to generate the short γ-like form (65). Alternatively, several phages require cellular proteases (66–68), such as in the degradation of the phage lambda O protein (required for phage DNA replication) by ClpXP (69, 70). A ClpP-independent role for ClpX in phage lambda and phage Mu production has also been reported (71, 72). Clearly, further research is required to unravel the mechanistic role(s) of ClpX in RcGTA production.
In conclusion, in the Rhodobacterales, HGT mediated by GTAs is regulated by numerous cellular regulators, and the CckA-ChpT-CtrA phosphorelay has a central role. Here, we have shown that the proteins DivL and ClpX are peculiar regulators of CtrA activity and RcGTA production in the GTA model system of R. capsulatus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The rifampin-resistant RcGTA-producing WT strain SB1003 of R. capsulatus (73) was used to study RcGTA-mediated gene transfer, RcGTA capsid protein production, promoter activities, and cell morphology. Measurement of population-level production by flow cytometry was performed using SBpG, an SB1003-derived strain containing a chromosomally integrated RcGTA head-tail cluster promoter-mCherry reporter (37). For measurement of cell lysis and RcGTA maturation, strain SBpG (37) containing the Δ280 overproducer mutation was used. This mutation greatly increases the percentage of cells that produce (Fig. 1E) and release RcGTA but otherwise appears to behave like WT strains (H. Ding, unpublished data). For affinity purification of RcGTA, the chimeric RcGTA overproduction system DE442/pRhoG5CTH (35) was used. For gene transfer assays, rifampin-sensitive strain B10 (74) was used as the recipient of the rifampin resistance allele. The genome of SB1003 has been sequenced (75).
Cells were cultured in the minimal medium RCV (76) for general growth, promoter activity, subpopulation production, and CtrA level measurements. The lysis-promoting medium RCVm containing a low concentration (0.5 mM) of KPO4 (21) was used for all RcGTA production and release experiments. Cells for affinity purification of RcGTA were cultured in the complex medium YPS (77). Cultures were inoculated to an optical density at 660 nm (OD660) of 0.15 (approximately 7.2 × 107 CFU per milliliter) and incubated photoheterotrophically for 24 h (promoter activity experiments) or 40 h (gene transfer, capsid production, maturation, and lysis experiments) or for equivalent durations based on doubling times for experiments involving the ΔclpX mutant at 30°C in 16.5-ml screw-cap glass tubes, unless otherwise noted. RcGTA recipient cells were cultured chemoheterotrophically in the dark at 30°C. For affinity purification, cells were cultured photoheterotrophically in 200-ml screw-cap glass bottles as described previously (35). Cultures were supplemented with tetracycline HCl (0.5 μg/ml), spectinomycin (10 μg/ml), kanamycin sulfate (10 μg/ml), or gentamicin sulfate (3 μg/ml), as appropriate.
For general cloning work, E. coli strain DH5α (78) lambda pir was used. E. coli S17-1 (79) lambda pir and TEC5 (80) were used for conjugation of plasmids to R. capsulatus. E. coli was cultured in LB medium (78) supplemented with ampicillin (100 μg/ml), tetracycline HCl (10 μg/ml), kanamycin sulfate (50 μg/ml), or gentamicin sulfate (10 μg/ml), as appropriate.
Construction of mutants and plasmids.
The plasmids used in this study are listed in Table 1. The transposon mutagenesis screen was performed using the Tn5-derived plasmid pRL27 (81) in strain DE442 (an RcGTA overproducer strain of uncertain provenance) containing the reporter plasmid pXCA-ghsA (which confers β-galactosidase activity [35]), and cells were spread on RCV agar containing tetracycline, kanamycin, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 20 μg/ml). Kanamycin-resistant colonies with altered β-galactosidase activity were further investigated and screened for the insertion site by sequencing as described by Larsen et al. (81).
TABLE 1.
Plasmids used in this study
| Name | Reference or source | Description |
|---|---|---|
| pRL27 | 81 | Transposon mutagenesis plasmid, Kanr |
| pUC19 | Invitrogen | General cloning vector, Ampr |
| pRK415 | 82 | Broad-host-range vector, Tetr |
| pCM62 | 83 | Broad-host-range vector, Tetr |
| pZJD29A | Unpublished construct (J. Jiang and C. E. Bauer) | Suicide plasmid, R6K origin of replication, Gmr |
| pXCA-601 | 86 | Promoter-lacZ reporter plasmid, replicating, Tetr |
| p601-g65 | 85 | RcGTA head-tail cluster reporter, promoter-lacZ fusion plasmid, Tetr |
| pXCA-555 | 21 | Endolysin-holin system reporter, promoter-lacZ fusion plasmid, Tetr |
| pXCA-ghsA | 35 | ghsAB head spike reporter, promoter-lacZ fusion plasmid, Tetr |
| pABW573 | This work | divL disruption plasmid, divL::aacC1, Ampr Gmr |
| pABW627 | This work | DivL complementation plasmid, Tetr |
| pABW710 | This work | ClpX complementation plasmid, Tetr |
| pLK718 | This work | ClpP-ClpX complementation plasmid, Tetr |
| pUCckA | 21 | ΔcckA mutation donor plasmid, Ampr Kanr |
| pRCckA | 21 | CckA complementation plasmid; Tetr |
| pCW104 | This work | Plasmid encoding CckA(H399A), a predicted autophosphorylation-deficient protein; Tetr |
| pD51A | 20 | Plasmid encoding CtrA(D51A), a nonphosphorylatable protein; Tetr |
| pD51E | 20 | Plasmid encoding CtrA(D51E), a phosphomimetic protein; Tetr |
| pLK754 | This work | WT CtrA complementation plasmid, Tetr |
| pLK755 | This work | Plasmid encoding CtrA with the last two amino acids converted to Asp-Asp (CtrA-DD), Tetr |
| pLK756 | This work | Plasmid encoding CtrA lacking the last three amino acids, Val-Gly-Ala (CtrAΔ3); Tetr |
| pABW848 | This work | ghsAB head spike reporter, promoter-lacZ fusion on suicide plasmid for chromosomal integration, Gmr |
| pABW910 | This work | divL reporter, promoter-lacZ fusion on suicide plasmid for chromosomal integration, Gmr |
| pABW901 | This work | Empty promoter-reporter suicide plasmid, accepts a PstI-BamHI or SphI-BamHI fragment to create a translationally in-frame promoter-lacZ reporter, Gmr |
| pRhoG5CTH | 35 | RcGTA affinity purification plasmid for use with DE442, 6His-tagged RcGTA capsid protein, Kanr Specr |
The targeted divL disruption mutants (divL::aacC1 mutants) were constructed by amplification of divL (rcc00042) by PCR with the primers ATATGAGCTCCTGGCGCGAGGACGAGACCG and ATATTCTAGAGCACTCTAGCGGCCCTGCCC. The fragment was cloned into pUC19 using SacI and XbaI. A SmaI-cut aacC1 amplicon (conferring gentamicin resistance [21]) was cloned into the NaeI site present in divL in the opposite transcriptional orientation to make pABW573. Mutants were created by RcGTA-mediated gene transfer of divL::aacC1 as previously described (80). The complementation plasmid pABW627 was constructed by amplification of divL and ∼500 bp of the upstream (5′) sequence by PCR using primers ATATGAGCTCCTATCGCTACCCCGAGCTGG and ATATTCTAGATCTTGCATGGCCGCACTCTA. The 2.2-kb amplicon was ligated into the broad-host-range vector pRK415 (82) cut with XbaI and SacI.
The insertional partial deletions of clpX (ΔclpX::aacC1) were constructed by amplifying the entire clpX (rcc02608) ORF including ∼500 bp of the upstream (5′) sequence using the primers CCTCGCGATCTAGAACACCATGC and CCACCGAGCTCCAGTGTTTTGC. The 1.8-kb amplicon was cloned into the broad-host-range vector pCM62 (83) using XbaI and SacI to create pABW710 (Fig. 3A). The aacC1 fragment was inserted between the AgeI and NcoI sites of pABW710 in the same transcriptional orientation as clpX, deleting 20% of clpX, and mutants were created by RcGTA-mediated gene transfer of the ΔclpX::aacC1 fragment. To create plasmid pLK718, containing ∼500 bp of the upstream sequence of clpP and the clpP-clpX ORFs (Fig. 3A), the 2.7-kb amplicon obtained using primers ATATTCTAGACGGTGACGAAAGCCTCGGTG and ATATGAGCTCCACCTCCGCATCTTTGGCCC was inserted between the SacI and XbaI sites of pCM62 (83).
The Δ280 ΔcckA mutant was constructed by RcGTA-mediated gene transfer of the kanamycin-marked mutation to SBpG Δ280, as previously described (21). The plasmid pCW104, encoding the autophosphorylation-deficient CckA(H399A) protein, was constructed by site-directed mutagenesis (SDM) of pRCckA (21) using the mutagenic primers CGGGCGGGGTTGCGGCGGATTTCAACAACTTG and CAAGTTGTTGAAATCCGCCGCAACCCCGCCCG. WT CtrA complementation plasmid pLK754 was constructed by amplifying ctrA and ∼450 bp of the 5′ sequence using ATATTCTAGACTTGGCTAAACCCAGCGTTA and ATATGAGCTCCAGCGCATCATAGTCAGCAT as primers, and the sequence was cloned into pCM62 using XbaI and SacI. Plasmid pLK755, encoding CtrA with the last two amino acids converted to Asp-Asp, was constructed by SDM using the mutagenic primers CATGGTCGTGGACGACTGAGTCTCCGCT and AGCGGAGACTCAGTCGTCCACGACCATG. pLK756, encoding CtrA lacking the three last amino acids, was constructed by inverse-PCR FastCloning (84) using the primers GCATGGTCTGAGTCTCCGCTTCTCTCTGG and GAGACTCAGACCATGCGGCGGTCAAG.
The promoter-lacZ reporter plasmids p601-g65 (an ∼15-kb RcGTA gene cluster promoter), pXCA-555 (an endolysin-holin system promoter), and pXCA-ghsA (a ghsAB head spike promoter) have been described previously (21, 35, 85). To measure promoter activities in the presence of trans expression plasmids, we constructed several promoter-lacZ reporter suicide plasmids for chromosomal integration: the promoter ghsAB-lacZ reporter plasmid pABW848 was constructed by inverse PCR of pZJD29A using primers ATATctgcagGAGTGGGGAGG (PstI) and ATATtctagaCTTGAACGAATTGTTAGGTG (XbaI), removing PPuc-sacB and lacZα (the lowercase nucleotides indicate the restriction enzyme sequences). The ghsAB promoter lacZ fusion present in pXCA-ghsA (35) was amplified using ATATctgcagCATGCggatccGTCGACTCGCGGGGTG (PstI-SphI-BamHI) and ATATtctagaCTGCCCGGTTATTATTATT (XbaI). The two resulting amplicons were joined using PstI and XbaI, resulting in pABW848. An empty lacZ reporter plasmid was constructed by excising the ghsAB promoter from pABW848 using the two BamHI sites present. This plasmid (pABW901) can accept a promoter on a PstI-BamHI fragment, similar to the reporter plasmid pXCA-601 (86), or a SphI-BamHI fragment, to create a translationally in-frame promoter-lacZ reporter for chromosomal integration. To construct the promoter divL-lacZ reporter plasmid pABW910, the divL promoter region was amplified with the primers ATATatgcatCCGAAGAGCTGGTCGACGAT (NsiI) and ATATagatcttcCATGACTGCAGCCCTCTCGC (BglII) and cloned into the compatible PstI/BamHI sites of pABW901. Plasmids were introduced into R. capsulatus by conjugation.
RcGTA gene transfer frequency.
Transduction frequencies were determined as previously described (21). In short, culture supernatants from rifampin-resistant donor cultures were filtered (0.2-μm pore size), and the filtrate and dilutions thereof were added to the rifampin-sensitive recipient strain B10. After incubation to allow transfer and recombination of the rifampin resistance allele, cells were plated on RCV medium containing 80 μg/ml rifampin and colonies were enumerated after 3 days. Control assays without B10 cells or without added filtered supernatant were included.
Measurement of cell lysis.
Cell lysis was determined by measuring the amount of light-harvesting complex 2 pigments, components of intracellular membranes (87), released to the culture supernatant as previously described (21).
Flow cytometry.
Subpopulation production of RcGTA was determined using SBpG (expressing an mCherry reporter from the RcGTA gene cluster promoter [37]) and derived strains. Samples for flow cytometry were harvested after 24 h of photoheterotrophic growth, and the mCherry reporter protein was recovered in the presence of oxygen (88): cultures were diluted 1:50 in modified phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM KPO4, pH 6.8) containing 50 μg/ml chloramphenicol and incubated aerobically on a rotator for 1 h at 30°C. Samples were counted on a BD Life Sciences LSR-II flow cytometer containing a 561-nm laser and analyzed using FlowJo software essentially as previously described (9) using the strain SB1003 (lacking mCherry) for gating.
Native gel migration.
Cultures were centrifuged (16,000 relative centrifugal force [rcf]), and the supernatant was treated with 100 μg/ml DNase I for 1 h at 30°C. Samples were separated by electrophoresis in agarose gels (0.8%), and the bands were visualized by staining the DNA with ethidium bromide as previously described (21, 35).
Promoter activity.
Promoter activity was determined by measuring the β-galactosidase activity of cells containing promoter-lacZ reporters (see above), and the activity was normalized to the total protein concentration determined by the Lowry assay as previously described (21).
TEM microscopy of affinity-purified RcGTA.
Purification and transmission electron microscopy (TEM) of RcGTA were performed as previously described (35). In short, RcGTA was affinity purified from the culture supernatant of DE442 cells harboring pRhoG5CTH, which encodes a 6His-tagged RcGTA capsid protein, using Ni-nitrilotriacetic acid agarose. Purified particles were adsorbed to carbon-coated copper grids, stained with uranyl formate, and examined using a Tecnai Spirit transmission electron microscope (FEI) operating at an acceleration voltage of 120 kV. Images were acquired using a 4K Eagle charge-coupled device (CCD) camera (FEI).
Immunoblots.
Proteins were separated in 12% SDS-polyacrylamide gels, blotted onto nitrocellulose membranes, probed using either RcGTA major capsid antiserum (catalog number AS08 365; Agrisera AB) or C. crescentus CtrA antiserum (a gift from Lucy Shapiro, Stanford University), and detected with horseradish peroxidase-conjugated secondary antibody as previously described (9). The amount of sample loaded on the gel was normalized using the OD660 of the culture.
Statistical analysis.
The statistical significance of the results was evaluated by a t test or one-way analysis of variance (ANOVA) followed by multiple comparisons using Tukey's test in GraphPad Prism (version 7) software. A P value of 0.05 was set as a cutoff for significance, and significance is indicated in the figures. Significance tests performed with a P value of >0.05 were deemed not significantly different (n.s.), which is indicated in the figures unless otherwise noted.
Bioinformatic tools.
DNA and protein sequences were routinely inspected and analyzed using the Artemis tool (http://www.sanger.ac.uk/science/tools/artemis), the BLAST tool (http://blast.ncbi.nlm.nih.gov), the PFAM database (http://pfam.xfam.org), and the SMART database (http://smart.embl-heidelberg.de). In silico assembly of DNA sequences was performed using Serial Cloner software (http://serialbasics.free.fr/Serial_Cloner). Global amino acid sequence alignment was performed using the EMBOSS NeedleP program (http://www.ebi.ac.uk/Tools/psa/emboss_needle), and multiple-sequence alignment was performed using the EMBOSS Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) (89). Prediction of promoters and Rho-independent terminators was performed using the BPROM (Softberry) and ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold/) tools.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. A. Johnson for help with construction of the clpX mutant, L. Shapiro for the C. crescentus CtrA antiserum, and the University of British Columbia Flow Cytometry Facility for assistance.
This work was supported by Canadian Institutes of Health research operating grant 93779 to J.T.B. and a Natural Sciences and Engineering Research Council of Canada discovery grant (RGPIN 418157-12) to C.K.Y.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00275-18.
REFERENCES
- 1.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 2.Lang AS, Westbye AB, Beatty JT. 2017. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu Rev Virol 4:87–104. doi: 10.1146/annurev-virology-101416-041624. [DOI] [PubMed] [Google Scholar]
- 3.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, MacLeod DM, Rivkin RB, Chen F, Buchan A, Lang AS. 2010. High diversity of Rhodobacterales in the subarctic North Atlantic Ocean and gene transfer agent protein expression in isolated strains. Aquatic Microb Ecol 59:283–293. doi: 10.3354/ame01398. [DOI] [Google Scholar]
- 5.Sun FL, Wang YS, Wu ML, Jiang ZY, Sun CC, Cheng H. 2014. Genetic diversity of bacterial communities and gene transfer agents in northern South China Sea. PLoS One 9:e111892. doi: 10.1371/journal.pone.0111892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Wang K, Budinoff C, Buchan A, Lang A, Jiao N, Chen F. 2009. Gene transfer agent (GTA) genes reveal diverse and dynamic Roseobacter and Rhodobacter populations in the Chesapeake Bay. ISME J 3:364–373. doi: 10.1038/ismej.2008.115. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westbye AB, O'Neill Z, Schellenberg-Beaver T, Beatty JT. 2017. The Rhodobacter capsulatus gene transfer agent is induced by nutrient depletion and the RNAP omega subunit. Microbiology 163:1355–1363. doi: 10.1099/mic.0.000519. [DOI] [PubMed] [Google Scholar]
- 9.Fogg PCM, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772. doi: 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes AP, Mercer RG, Watton DE, Buckley CB, Lang AS. 2012. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol Microbiol 85:314–325. doi: 10.1111/j.1365-2958.2012.08113.x. [DOI] [PubMed] [Google Scholar]
- 11.Tomasch J, Wang H, Hall ATK, Patzelt D, Preusse M, Petersen J, Brinkmann H, Bunk B, Bhuju S, Jarek M, Geffers R, Lang AS, Wagner-Dobler I. 2018. Packaging of Dinoroseobacter shibae DNA into gene transfer agent particles is not random. Genome Biol Evol 10:359–369. doi: 10.1093/gbe/evy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brimacombe CA, Ding H, Johnson JA, Beatty JT. 2015. Homologues of genetic transformation DNA import genes are required for Rhodobacter capsulatus gene transfer agent recipient capability regulated by the response regulator CtrA. J Bacteriol 197:2653–2663. doi: 10.1128/JB.00332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westbye AB, Beatty JT, Lang AS. 2017. Guaranteeing a captive audience: coordinated regulation of gene transfer agent (GTA) production and recipient capability by cellular regulators. Curr Opin Microbiol 38:122–129. doi: 10.1016/j.mib.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Ziesche L, Frank O, Michael V, Martin M, Petersen J, Schulz S, Wagner-Dobler I, Tomasch J. 2014. The CtrA phosphorelay integrates differentiation and communication in the marine alphaproteobacterium Dinoroseobacter shibae. BMC Genomics 15:130. doi: 10.1186/1471-2164-15-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brimacombe CA, Ding H, Beatty JT. 2014. Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol Microbiol 92:1260–1278. doi: 10.1111/mmi.12628. [DOI] [PubMed] [Google Scholar]
- 16.Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci U S A 97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. 2009. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol 191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, Beatty JT, Lang AS. 2010. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol 192:2701–2710. doi: 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331:53–62. doi: 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 21.Westbye AB, Leung MM, Florizone SM, Taylor TA, Johnson JA, Fogg PC, Beatty JT. 2013. Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195:5025–5040. doi: 10.1128/JB.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- 24.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 25.Greene SE, Brilli M, Biondi EG, Komeili A. 2012. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol 194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. 2010. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell 39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Childers WS, Xu Q, Mann TH, Mathews II, Blair JA, Deacon AM, Shapiro L. 2014. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol 12:e1001979. doi: 10.1371/journal.pbio.1001979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce DL, O'Donnol DS, Allen RC, Javens JW, Quardokus EM, Brun YV. 2006. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J Bacteriol 188:2473–2482. doi: 10.1128/JB.188.7.2473-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sciochetti SA, Ohta N, Newton A. 2005. The role of polar localization in the function of an essential Caulobacter crescentus tyrosine kinase. Mol Microbiol 56:1467–1480. doi: 10.1111/j.1365-2958.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 30.Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Ohta N, Zhao JL, Newton A. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci U S A 96:13068–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iniesta AA, Hillson NJ, Shapiro L. 2010. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci U S A 107:7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415–424. doi: 10.1016/S0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 34.Joshi KK, Chien P. 2016. Regulated proteolysis in bacteria: Caulobacter. Annu Rev Genet 50:423–445. doi: 10.1146/annurev-genet-120215-035235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westbye AB, Kuchinski K, Yip CK, Beatty JT. 2016. The gene transfer agent RcGTA contains head spikes needed for binding to the Rhodobacter capsulatus polysaccharide cell capsule. J Mol Biol 428:477–491. doi: 10.1016/j.jmb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Tsokos CG, Laub MT. 2012. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol 15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuchinski KS, Brimacombe CA, Westbye AB, Ding H, Beatty JT. 2016. The SOS response master regulator LexA regulates the gene transfer agent of Rhodobacter capsulatus and represses transcription of the signal transduction protein CckA. J Bacteriol 198:1137–1148. doi: 10.1128/JB.00839-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenal U, Fuchs T. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J 17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottesman S, Clark WP, de Crecy-Lagard V, Maurizi MR. 1993. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem 268:22618–22626. [PubMed] [Google Scholar]
- 41.Yoo SJ, Seol JH, Kang MS, Ha DB, Chung CH. 1994. clpX encoding an alternative ATP-binding subunit of protease Ti (Clp) can be expressed independently from clpP in Escherichia coli. Biochem Biophys Res Commun 203:798–804. doi: 10.1006/bbrc.1994.2253. [DOI] [PubMed] [Google Scholar]
- 42.Fuchs T, Wiget P, Osteras M, Jenal U. 2001. Precise amounts of a novel member of a phosphotransferase superfamily are essential for growth and normal morphology in Caulobacter crescentus. Mol Microbiol 39:679–692. doi: 10.1046/j.1365-2958.2001.02238.x. [DOI] [PubMed] [Google Scholar]
- 43.Østerås M, Stotz A, Schmid Nuoffer S, Jenal U. 1999. Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease. J Bacteriol 181:3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung MM, Brimacombe CA, Beatty JT. 2013. Transcriptional regulation of the Rhodobacter capsulatus response regulator CtrA. Microbiology 159:96–106. doi: 10.1099/mic.0.062349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hynes AP, Shakya M, Mercer RG, Grull MP, Bown L, Davidson F, Steffen E, Matchem H, Peach ME, Berger T, Grebe K, Zhaxybayeva O, Lang AS. 2016. Functional and evolutionary characterization of a gene transfer agent's multilocus “genome.” Mol Biol Evol 33:2530–2543. doi: 10.1093/molbev/msw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson JE. 2010. Virus particle maturation: insights into elegantly programmed nanomachines. Curr Opin Struct Biol 20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hendrix RW, Johnson JE. 2012. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol 726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- 48.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 49.Chastanet A, Prudhomme M, Claverys JP, Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J Bacteriol 183:7295–7307. doi: 10.1128/JB.183.24.7295-7307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 51.Johnsborg O, Håvarstein LS. 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev 33:627–642. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- 52.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pestova EV, Havarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 54.Mercer RG, Lang AS. 2014. Identification of a predicted partner-switching system that affects production of the gene transfer agent RcGTA and stationary phase viability in Rhodobacter capsulatus. BMC Microbiol 14:71. doi: 10.1186/1471-2180-14-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casjens SR, Hendrix RW. 2015. Bacteriophage lambda: early pioneer and still relevant. Virology 479-480:310–330. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendrix RW, Casjens S. 2006. Bacteriophage lambda and its genetic neighbourhood. In Calendar R. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY. [Google Scholar]
- 57.Josslin R. 1970. The lysis mechanism of phage T4: mutants affecting lysis. Virology 40:719–726. doi: 10.1016/0042-6822(70)90216-3. [DOI] [PubMed] [Google Scholar]
- 58.Reader RW, Siminovitch L. 1971. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology 43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- 59.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol 183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A 103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. 2009. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol 73:586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aksyuk AA, Rossmann MG. 2011. Bacteriophage assembly. Viruses 3:172–203. doi: 10.3390/v3030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrascosa JL. 1978. Head maturation pathway of bacteriophages T4 and T2. IV. In vitro transformation of T4 head-related particles produced by mutants in gene 17 to capsid-like structures. J Virol 26:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huet A, Duda RL, Hendrix RW, Boulanger P, Conway JF. 2016. Correct assembly of the bacteriophage T5 procapsid requires both the maturation protease and the portal complex. J Mol Biol 428:165–181. doi: 10.1016/j.jmb.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vass RH, Chien P. 2013. Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc Natl Acad Sci U S A 110:18138–18143. doi: 10.1073/pnas.1311302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottesman S. 2003. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 67.Hellen CU, Wimmer E. 1992. The role of proteolytic processing in the morphogenesis of virus particles. Experientia 48:201–215. doi: 10.1007/BF01923512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hershko A, Fry M. 1975. Post-translational cleavage of polypeptide chains: role in assembly. Annu Rev Biochem 44:775–797. doi: 10.1146/annurev.bi.44.070175.004015. [DOI] [PubMed] [Google Scholar]
- 69.Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. 1999. Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J Biol Chem 274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 70.Mott ML, Berger JM. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 71.Levchenko I, Luo L, Baker TA. 1995. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev 9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 72.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. 1995. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J 14:1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yen HC, Marrs B. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol 126:619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weaver PF, Wall JD, Gest H. 1975. Characterization of Rhodopseudomonas capsulata. Arch Microbiol 105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 75.Strnad H, Lapidus A, Paces J, Ulbrich P, Vlcek C, Paces V, Haselkorn R. 2010. Complete genome sequence of the photosynthetic purple nonsulfur bacterium Rhodobacter capsulatus SB 1003. J Bacteriol 192:3545–3546. doi: 10.1128/JB.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beatty JT, Gest H. 1981. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch Microbiol 129:335–340. doi: 10.1007/BF00406457. [DOI] [Google Scholar]
- 77.Wall JD, Weaver PF, Gest H. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol 105:217–224. doi: 10.1007/BF00447140. [DOI] [PubMed] [Google Scholar]
- 78.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 79.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic-engineering—transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 80.Taylor DP, Cohen SN, Clark WG, Marrs BL. 1983. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol 154:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 82.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 83.Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065–2075. doi: 10.1099/00221287-147-8-2065. [DOI] [PubMed] [Google Scholar]
- 84.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. 2011. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol 11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leung MM, Brimacombe CA, Spiegelman GB, Beatty JT. 2012. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol Microbiol 83:759–774. doi: 10.1111/j.1365-2958.2011.07963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adams CW, Forrest ME, Cohen SN, Beatty JT. 1989. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J Bacteriol 171:473–482. doi: 10.1128/jb.171.1.473-482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feick R, van Grondelle R, Rijgersberg CP, Drews G. 1980. Fluorescence emission by wild-type- and mutant-strains of Rhodopseudomonas capsulata. Biochim Biophys Acta 593:241–253. doi: 10.1016/0005-2728(80)90062-6. [DOI] [PubMed] [Google Scholar]
- 88.Zhang C, Xing XH, Lou K. 2005. Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol Lett 249:211–218. doi: 10.1016/j.femsle.2005.05.051. [DOI] [PubMed] [Google Scholar]
- 89.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.