ABSTRACT
The wood-rotting mushroom Stereum hirsutum is a known producer of a large number of namesake hirsutenoids, many with important bioactivities. Hirsutenoids form a structurally diverse and distinct class of sesquiterpenoids. No genes involved in hirsutenoid biosynthesis have yet been identified or their enzymes characterized. Here, we describe the cloning and functional characterization of a hirsutene synthase as an unexpected fusion protein of a sesquiterpene synthase (STS) with a C-terminal 3-hydroxy-3-methylglutaryl-coenzyme A (3-hydroxy-3-methylglutaryl-CoA) synthase (HMGS) domain. Both the full-length fusion protein and truncated STS domain are highly product-specific 1,11-cyclizing STS enzymes with kinetic properties typical of STSs. Complementation studies in Saccharomyces cerevisiae confirmed that the HMGS domain is also functional in vivo. Phylogenetic analysis shows that the hirsutene synthase domain does not form a clade with other previously characterized sesquiterpene synthases from Basidiomycota. Comparative gene structure analysis of this hirsutene synthase with characterized fungal enzymes reveals a significantly higher intron density, suggesting that this enzyme may be acquired by horizontal gene transfer. In contrast, the HMGS domain is clearly related to other fungal homologs. This STS-HMGS fusion protein is part of a biosynthetic gene cluster that includes P450s and oxidases that are expressed and could be cloned from cDNA. Finally, this unusual fusion of a terpene synthase to an HMGS domain, which is not generally recognized as a key regulatory enzyme of the mevalonate isoprenoid precursor pathway, led to the identification of additional HMGS duplications in many fungal genomes, including the localization of HMGSs in other predicted sesquiterpenoid biosynthetic gene clusters.
IMPORTANCE Hirsutenoids represent a structurally diverse class of bioactive sesquiterpenoids isolated from fungi. Identification of their biosynthetic pathways will provide access to this chemodiversity for the discovery and synthesis of molecules with new bioactivities. The identification and successful cloning of the previously elusive hirsutene synthase from the S. hirsutum provide important insights and strategies for biosynthetic gene discovery in Basidiomycota. The finding of a terpene synthase-HMGS fusion, the discovery of other sesquiterpenoid biosynthetic gene clusters with dedicated HMGS genes, and HMGS gene duplications in fungal genomes give new importance to the role of HMGS as a key regulatory enzyme in isoprenoid and sterol biosynthesis that should be exploited for metabolic engineering.
KEYWORDS: sesquiterpene synthase (STS), Basidiomycota, mevalonate pathway, hirsutene, 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), horizontal gene transfer
INTRODUCTION
Basidiomycota (also known as mushrooms) produce a plethora of bioactive secondary metabolites. Unlike the Ascomycota, this group of fungi has developed a large repertoire of sesquiterpenoids as their major class of natural products (1). These compounds are generated by the cyclization of the linear 15-carbon precursor farnesyl-pyrophosphate (FPP) catalyzed by sesquiterpene synthases (STSs). Upon enzymatic pyrophosphate cleavage, a reactive carbocation is formed and cyclized into an initial cyclic carbocation that undergoes further reactions until it is finally quenched by proton abstraction or water. FPP itself is synthesized by the condensation of three 5-carbon isoprenyl pyrophosphate units that are derived from acetyl-coenzyme A (acetyl-CoA) through the mevalonate (MVA) pathway in fungi (Fig. 1). The tremendous structural diversity of sesquiterpenoids is achieved by the types of cyclization reactions catalyzed by different STSs and by subsequent modifications of the cyclic products by additional tailoring enzymes. In fungi, genes encoding tailoring enzymes are typically colocated with an STS gene as part of a biosynthetic gene cluster that may also include genes encoding other isoprenoid precursor pathway enzymes (Fig. 1).
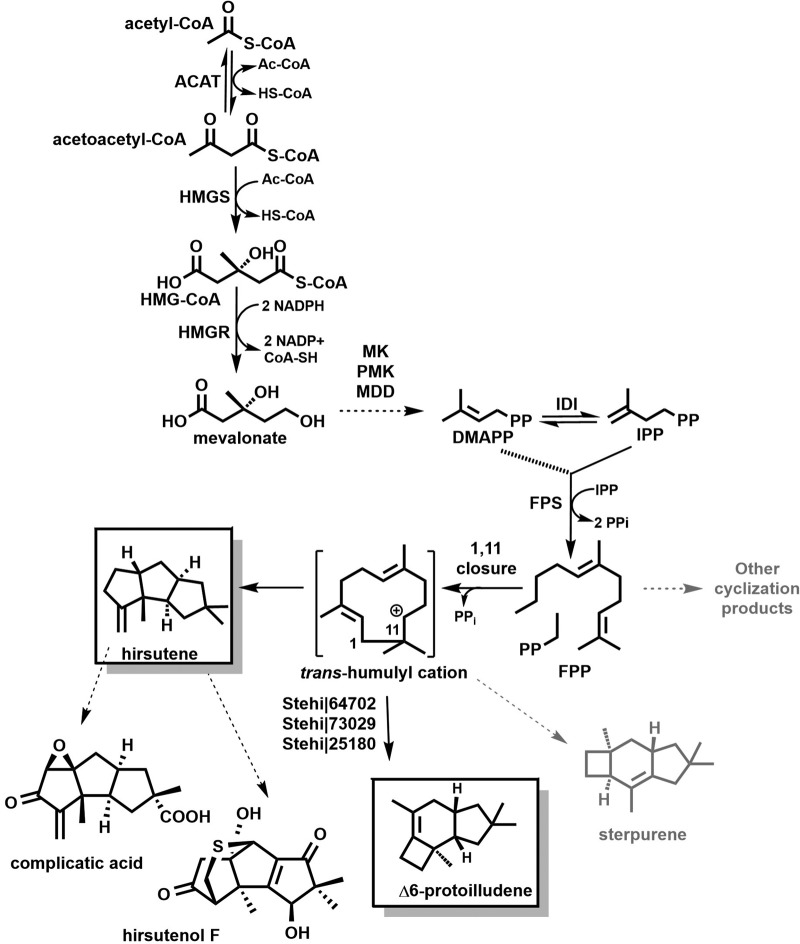
FIG 1.
Overview of pathways to trans-humulyl cation-derived sesquiterpenoids in S. hirsutum. The mevalonate (MVA) pathway converts acetyl-CoA from the central metabolism to the universal 5-carbon isoprenoid precursors isopentenyl-pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). Condensation of DMAPP and IPP (dotted line) followed by a second condensation reaction (solid line) with IPP yields the 15-carbon sesquiterpenoid precursor farnesyl pyrophosphate (FPP). 1,11-Cyclization of FPP by sesquiterpene synthases (STSs) yields a trans-humulyl cation intermediate that is further cyclized into different sesquiterpene scaffolds depending on the STS. Three Δ6-protoilludene synthases have been characterized previously (19), while STSs responsible for hirsutene and sterpurene synthesis remain to be identified. Major volatile sesquiterpenes, hirsutene and Δ6-protoilludene, produced by S. hirsutum, are boxed. Additional biosynthetic steps lead to bioactive hirsutenoids, such as the two examples shown (Fig. S1 shows possible cyclization pathways to hirsutene). ACAT, acetoacetyl-CoA transferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDD, diphosphomevalonate decarboxylase; IDI, IPP isomerase; FPS, FPP synthase.
Many sesquiterpenoid compounds are relevant for the development of new pharmaceuticals, such as antiviral, anticancer, and antimicrobial drugs (2–5). Because of their structural complexity, chemical synthesis is not economically viable, and production requires instead either the isolation of compounds from the native production host or, more commonly, the development of genetically engineered production systems (6–16). The vast majority of Basidiomycota are not genetically tractable, and consequently, the identification of biosynthetic genes of interest and expression in alternative recombinant systems, such as yeast, is necessary to characterize and access this rich portfolio of bioactive compounds from this phylum of fungi.
We have previously functionally characterized several STSs from the model fungus Coprinus cinereus and the anticancer illudin producer Omphalotus olearius. We found that STS sequences appear to clade by the initial 1,10-, 1,11- or 1,6-cyclic carbocation formed upon pyrophosphate cleavage from FPP (references 17–19 and reviewed in reference 20) (Fig. 2A). We then used this information to develop a predictive framework to guide the identification of sesquiterpene biosynthesis pathways as part of a larger effort to produce pharmaceutically relevant compounds from Basidiomycota in genetically tractable heterologous systems. We validated this framework by predicting the activities of 18 STSs identified in the genome of the white rot fungus Stereum hirsutum (19), which is known to synthesize a variety of bioactive sesquiterpenoids, including the namesake hirsutenoids (21–23) (Fig. 1). We successfully predicted, cloned, and confirmed the functions of three 1,11-cyclizing protoilludene synthases, one 1,6-cyclizing STS, and one 1,10-cyclizing germacrenyl-derived STS, verifying the results from our bioinformatics analyses (19). However, although S. hirsutum cultures produced diverse sesquiterpenoids, including Δ6-protoilludene and hirsutene as major compounds, we were unable to obtain an STS responsible for the 1,11-cyclization of FPP to hirsutene (19, 24).
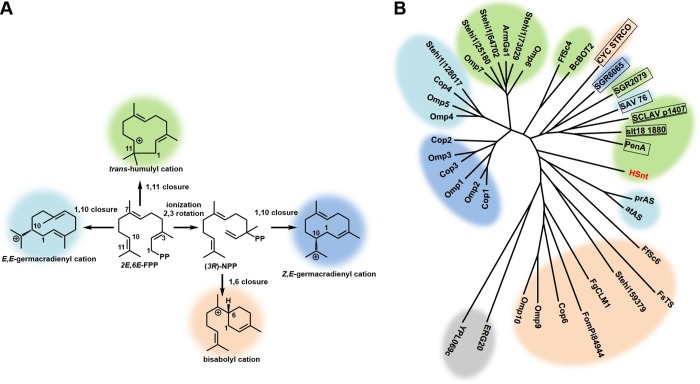
FIG 2.
Phylogenetic analysis of sesquiterpene synthases. (A) Color-coded initial cyclization intermediates catalyzed by STSs. (B) Characterized fungal STSs form clades in an unrooted neighbor-joining phylogram based on their catalyzed initial cyclization reaction. Representative examples of characterized bacterial STSs from actinomycetes (boxed) catalyzing different initial cyclization reactions form a separate clade. STSs are colored according to their initial cyclization intermediates using the same color code used in panel A. Unlike previously characterized fungal 1,11-cyclizing STSs, which clade together based on phylum, hirsutene synthase (HSnt) is an outlier and forms its own node between clades formed by 1,10-cyclyzing Ascomycota STSs and bacterial STSs, including the 1,11-cyclizing cucumane synthases (isohirsut-1-ene synthase [SCLAV_p1407] and isohirsut-4-ene synthase [slt18_1880], underlined), pentalenene synthase (PenA), and (+)-caryolan-1-olsynthase (SGR2079). Saccharomyces cerevisiae farnesyl pyrophosphate (ERG20, GenBank accession no. CAA89462.1) and geranylgeranyl pyrophosphate synthase (YPL069c, GenBank accession no. AAT92871.1) were included as an outgroup (colored in gray). The reference protein sequences are as follows: for Basidiomycota STSs, Coprinus cinereus (Cop1 to -4 and Cop6, accession numbers XP_001832573, XP_001836556, XP_01832925, XP_01836356, and XP_01832549, respectively [17]), Omphalotus olearius (Omp1 to 10* [18]), Fomitopsis pinicola (FomPi84944* [18]), Stereum hirsutum (Stehi1∣159379, Stehi1∣128017, Stehi1∣25180, Stehi1∣64702, and Stehi1∣73029* [19]), and Armillaria gallica (ArmGa1, UniProt accession no. P0DL13 [90]); for Ascomycota STSs, Fusarium sporotrichioides (FsTS, UniProt accession no. P13513), Fusarium fujikuroi (Ffsc4, GenBank accession no. HF563560.1) and Ffsc6 (GenBank accession no. HF563561.1), Fusarium graminearum (FgCLM1, GenBank accession no. GU123140) [91–93], Aspergillus terreus (atAS, UniProt accession no. Q9UR08 [94]), Penicillium roqueforti (prAS, GenBank accession no. W6Q4Q9 [95]), and Botrytis cinerea (BcBOT2, GenBank accession no. AAQ16575.1 [96]); for actinomycete STSs, Streptomyces clavuligerus (SCLAV_p1407, UniProt accession no. D5SLU6), Streptomyces lactacystinaeus (slt18_1880, UniProt accession no. A0A097ZQA4 [33, 34]), Streptomyces exfoliatus (PenA, UniProt accession no. Q55012 [97]), Streptomyces griseus (SRG2079, UniProt accession no. B1W019.1 [32]), Streptomyces coelicolor (CYC_STRCO, UniProt accession no. Q9K499.1 [35]), Streptomyces griseus (SGR6065, GenBank accession no. BAG22894.1 [36]), and Streptomyces avermitilis (SAV_76, RefSeq accession no. WP_010981512.1 [37]). For items listed above with an asterisk (*), see the supplemental material for sequences of manually predicted genes and of cloned spliced genes.
Here, we report the successful cloning and functional identification of this elusive hirsutene synthase (HS) as an unexpected fusion protein with a 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) (here referred to as HS-HMGS). We show that both enzyme domains are functional and that the HS domain is a highly product-specific STS. This represents an unusual and surprising example of a natural fusion enzyme catalyzing two nonconsecutive steps in isoprenoid biosynthesis. Sequence analysis and gene structure analysis also provide evidence that the STS domain of the HS-HMGS may have been acquired via horizontal gene transfer. Genome analysis further shows that the duplication of HMGS genes is widespread in fungi, offering new insight into and strategies for isoprenoid pathway engineering.
RESULTS
Identification and cloning of an STS-HMGS fusion gene.
Previously, we predicted six 1,11-cyclizing STSs in the genome of S. hirsutum (25) (Joint Genome Initiative [JGI] Fungal Genome Database [26]) and functionally characterized three of these STSs (annotated as Stehi1∣64702, Stehi1∣73029, and Stehi1∣25180 in the genome) as 1,11-cyclizing protoilludene synthases (19). The three remaining STSs did not cluster tightly with the characterized protoilludene synthases and seem to be located on a more distant branch (19). The genes encoding two of these putative STSs (annotated as Stehi1∣52743 and Stehi1∣122776) appeared each to be part of a biosynthetic gene cluster (Fig. 3 and Tables S1 and S3; also, see below), like the genes encoding the three previously characterized protoilludene synthases (19). In contrast, none of the other 12 STS genes (belonging to 1,6- or 1,10-cyclizing clades) identified in S. hirsutum appeared to be part of a biosynthetic gene cluster. We therefore reasoned that either the Stehi1∣52743 or the Stehi1∣122776 gene product must encode the STS that makes the hirsutene scaffold of the bioactive hirsutenoids isolated from S. hirsutum (21–23) (Fig. 1).
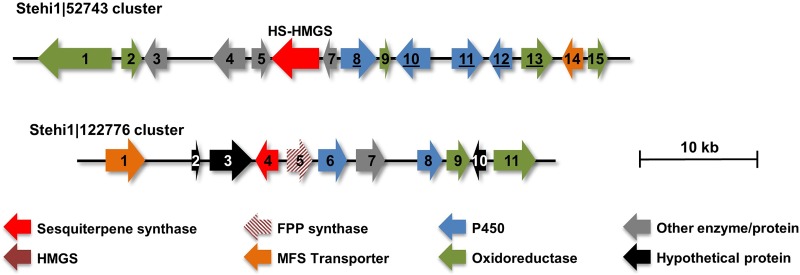
FIG 3.
Biosynthetic gene cluster predictions. Predicted Stehi1∣52743 and Stehi1∣122776 are located in putative gene clusters that were manually annotated. Note that Stehi1∣52743 was initially predicted to encode a standalone STS that is followed downstream by a HMGS gene. The predicted cluster genes are numbered and color-coded according to putative protein functions based on their closest homolog. See Tables S1 and S2 for detailed sequence information and function predictions. Underlined genes indicate that coding sequences were successfully amplified from cDNA. Red arrow indicates predicted STSs, including the HS-HMGS fusion protein identified in this work.
Multiple attempts to clone full-length spliced STS genes from cDNA based on JGI's Stehi1∣52743 and Stehi1∣122776 gene predictions and based on manual predictions with different fungal gene models in Augustus (27) were unsuccessful (see Table 1 for primers used). Failure to amplify spliced genes from cDNA suggested that the coding regions were either not correctly predicted or the genes are not transcribed under laboratory cultivation conditions. The latter was ruled out for the HS, as hirsutene is a major product that is readily detected in the culture headspace of S. hirsutum (19). To confirm the expression of genes, we then designed internal primers corresponding to the highly conserved Mg2+-coordinating active-site D(D/E)XX(D/E) and (N/D)DXX(S/T)XXX(E/D) motifs of STSs (28). A spliced intragenic amplicon of 405 bp could only be obtained for Stehi1∣52743 and was largely consistent with the predicted gene models, except for two additional small introns (Fig. 4). From these results, we hypothesized that Stehi1∣52743 likely encodes the HS, while Stehi1∣122776 appears not to be expressed under laboratory cultivation conditions.
TABLE 1.
Primers used in this study
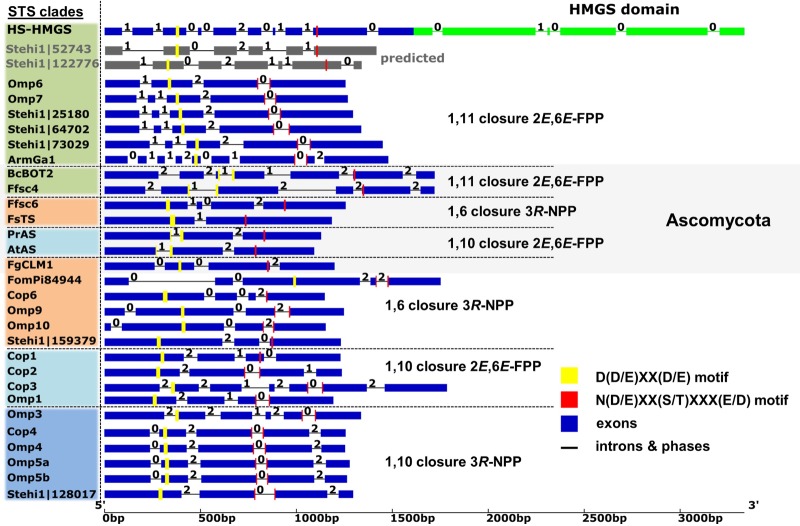
FIG 4.
Comparison of gene structures of fungal sesquiterpene synthases. Intron/exon organization and intron phases are shown for functionally characterized fungal sesquiterpene synthases (STSs) organized by the different clades and phyla (Basidiomycota and Ascomycota [gray box]) depicted in Fig. 2B. STSs are grouped based on catalyzed initial cyclization reaction (1,11-[and 1,10 closure of {2E,6E}-FPP, and 1,10- and 1,6-closure of 3R-NPP]) and color-coded as in Fig. 2A.The HMGS subunit (bright green) of HS-HMGS begins in exon 10. Note the high gene structure and phase conservation within each Basidiomycota (Stehi1, Omp, Cop, FomPi, and ArmGa) and Ascomycota (Ff, Fs, Fg, Bc, and At) cyclization groups. Conserved D(D/E)XX(D/E) motif (yellow) and (N/D)DXX(S/T)XXX(E/D) motif (red) residues are highlighted. Note that the (N/D)DXX(S/T)XXX(E/D) motif is split by an intron between the (N/D)D residues in 18 of 21 Basidiomycota STS genes. The references and accession numbers for fungal STS sequences are listed in Fig. 2.
Basidiomycota have intron-rich genomes (29), and we previously found that computational gene prediction is frequently unreliable and tends to miss small introns in Basidiomycota genes (19); this also appeared to be the case for Stehi1∣52743. To identify potentially erroneously predicted splicing patterns for Stehi1∣52743, we aligned the gene structures of all functionally characterized fungal STSs with the gene structure of Stehi1∣52743, generated by Augustus (27), and, for comparison, also with the Stehi1∣122776 gene structure (Fig. 4). Intron-exon pattern and phase conservation were mostly maintained between STSs belonging to the same cyclization clade (Fig. 2B). In contrast to Ascomycota, most Basidiomycota STS sequences contain an intron between the bases coding for the N and D residues of the conserved (N/D)Dxx(S/T)xxx(E/D) STS active-site motif (28). This intron is absent in both the Stehi1∣52743 and Stehi1∣122776 gene predictions and interestingly also in the (3R)-NPP-cyclizing STSs Stehi1∣159379 and Cop1.
The predicted Stehi1∣52743 gene structure exhibited a splicing pattern surrounding the conserved DDxxD/E motif similar to those of the other 1,11-cyclizing Basidiomycota STSs, but its splicing pattern diverged significantly upstream and downstream from this motif. When inspecting the genomic region surrounding Stehi1∣52743, we noticed that the immediately downstream predicted open reading frame (ORF) Stehi1∣53380 was located on the same strand and separated by only 198 bp from the predicted Stehi1∣52743 ORF. We therefore considered that the 3′-coding region of Stehi1∣52743 may not be correctly predicted, and we hypothesized that the Stehi1∣52743 gene may form an unpredicted fusion with its downstream gene Stehi1∣53380. This gene is predicted to encode an HMG-CoA synthase (HMGS) which catalyzes the second, and first committed reaction, in the MVA isoprenoid precursor pathway (Fig. 1). To test this hypothesis, we designed primers (Table 1, primers 3 and 4) matching the 5′ and 3′ regions of the two predicted genes for the amplification of spliced gene products from cDNA. Indeed, a discrete 2,526-bp amplicon (Data Set S1) was obtained that combined Stehi1∣52743 and Stehi1∣53380 into a single ORF encoding an unprecedented STS HMG-CoA synthase fusion protein (referred to here as HS-HMGS).
The gene structure (Fig. 4) surrounding the fusion junction differs substantially from the predicted splicing pattern, explaining previous unsuccessful amplification attempts. Interestingly, the predicted HMGS start codon in Stehi1∣53380 is maintained as M365 in the fusion protein, enabling the identification of a distinct terpene synthase (bp 1 to 1092, amino acids [aa] 1 to 364) and a predicted HMGS domain (bp 1093 to 2526, aa 365 to 842).
Given that the putative HS-HMGS appears to be quite different in sequence and gene structure from other previously characterized fungal STSs, we performed an extended sequence alignment and phylogenetic analysis of the N-terminal STS domain (HSnt) with fungal (20) and bacterial characterized STSs. From roughly 40 sesquiterpene synthases identified in bacteria (30), we selected 1,11-cyclizing enzymes and representative examples of 1,6- and 1,10-cyclizing enzymes that have been cloned and functionally characterized from different Streptomyces strains. These enzymes included 1,11-cyclyzing pentalenene synthase (PenA) (31), (+)caryolan-1-ol synthase (SGR2079) (32), two cucumane synthases (isohirsut-1-ene synthase [SCLAV_p1407] and isohirsut-4-ene synthase [slt18_1880]) which form triquinane sesquiterpenes similar in structure to hirsutene (33, 34), 1,6-cyclizing epi-isozizaene synthase (CYC_STRCO) (35), and 1,10-cyclizing epicubenol (SGR6065) (36) and avermitol (SAV_76) synthases (37) (Fig. 2B and S2 for an alignment of 1,11-cyclizing STSs). Our extended analysis confirmed that HSnt is an outlier that shows no close relationship with other Basidiomycota STSs; instead, it appears to be more closely related to Ascomycota 1,10-cyclizing aristolochene synthases atAS and prAS, 1,6-cyclizing acorenol synthase Ffsc6, and bacterial STSs.
STS domain is a highly specific hirsutene synthase.
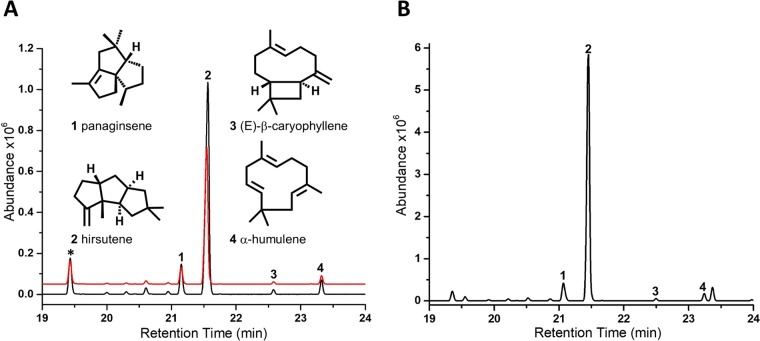
To confirm that the fusion gene in fact encodes a functional hirsutene synthase, both the cloned full-length fusion protein (HS-HMGS, aa 1 to 842) and a truncated version containing only the N-terminal STS domain (HSnt, aa 1 to 364) were expressed in Escherichia coli, and sesquiterpene production in the volatile culture headspace was analyzed by gas chromatography-mass spectrometry (GC-MS), as described previously (38). The full-length and the truncated versions (Fig. 5A) produced in E. coli had nearly identical levels and relative quantities of the trans-humulyl-derived sesquiterpenes hirsutene (85.3%) as the major product and panaginsene (7.5%), (E)-β-caryophyllene (0.6%), and α-humulene (3.1%) as minor products, in ratios similar to those previously detected in the culture headspace of S. hirsutum (19). Several minor peaks (∼3.5% of the total sesquiterpene products) were also detected that could not be unequivocally identified (see Fig. S2 for complete annotation of peaks and corresponding mass spectra). These results confirm that the amplified gene encodes a functional hirsutene synthase and that its activity does not appear to be influenced by the C-terminally fused HMGS domain.
FIG 5.
Analysis of volatile sesquiterpene production. (A) GC-MS product profiles of E. coli overexpressing the HS-HMGS fusion protein (black) or hirsutene synthase domain HSnt (red). The major products of both proteins are hirsutene (2), followed by detectable quantities of the 1,11 cyclization products of (2E,6E)-FPP panaginsene (1), (E)-β-caryophyllene (3), and α-humulene (4) (structures are shown) and several very minor peaks with characteristic sesquiterpene m/z 204 that could not be identified (see Fig. S2 for complete annotation of profiles and for mass spectra). (B) Purified HS-HMGS incubated with 2 μM (2E,6E)-FPP produced the same major peaks as in E. coli. Indole (*) is a natural breakdown product of tryptophan metabolism in E. coli and was used as an internal standard.
To compare the sesquiterpene synthase activity of this fusion protein with those of previously characterized enzymes from S. hirsutum (19), recombinant His-tagged HS-HMGS was purified from E. coli cultures and characterized in vitro. The purified enzyme was assayed with (2E,6E)-FPP as the substrate to determine its product profile and kinetic parameters, as described previously (19). The purified HS-HMGS catalyzed the cyclization of FPP into the same products and in the same ratios as observed with recombinant E. coli cultures (Fig. 5B). The kinetic parameters of the HS-HMGS fusion protein with FPP (Km = 6.3 × 10−6 ± 3.3 × 10−6 M and kcat = 4.0 × 10−3 ± 0.5 × 10−3 · s−1 [kcat/Km = 630 ± 140 M−1 · s−1]) were comparable to those of previously characterized STSs from S. hirsutum and other Basidiomycota (see references 17–19, and reviewed in reference 20), further confirming that the HMGS domain does not affect terpene synthase activity.
HMG-CoA synthase domain is functional in yeast.
Sequence analysis of the predicted C-terminal HMGS domain suggests it to be a functional enzyme. The critical residues required for activity are conserved (39–41), and its amino acid sequence is closely related to other Basidiomycota HMGSs (39, 42–44), including three other homologs encoded in the genome of S. hirsutum (Fig. S4). To confirm that the HMGS domain is functional, we performed a conditional complementation test in S. cerevisiae with a wild-type HS-HMGS gene and an inactivated version of the gene. HMGS catalysis is dependent on a triad of conserved residues (C497-H644-E463, HS-HMGS numbering) (40). We therefore created a C497A HS-HMGS mutation (HS-HMGS*C497A) known to inactivate HMGS (39, 41).
An S. cerevisiae HMGSΔ haploid (mevalonate auxotroph) was transformed with either the wild-type HS-HMGS or mutated HS-HMGS*C497A gene placed under the control of a galactose-inducible promoter for expression. The same plasmids were transformed into a heterozygous diploid S. cerevisiae HMGS/HMGSΔ strain as a positive control. As expected, under noninducing conditions, the haploid transformants failed to grow with either of the two plasmids. Haploid growth was only rescued upon the induction of wild-type HS-HMGS expression on galactose-containing plates, confirming that the HMGS domain is functional in yeast. The heterozygous diploid control transformants grew as expected under both growth conditions (Fig. 6).
FIG 6.
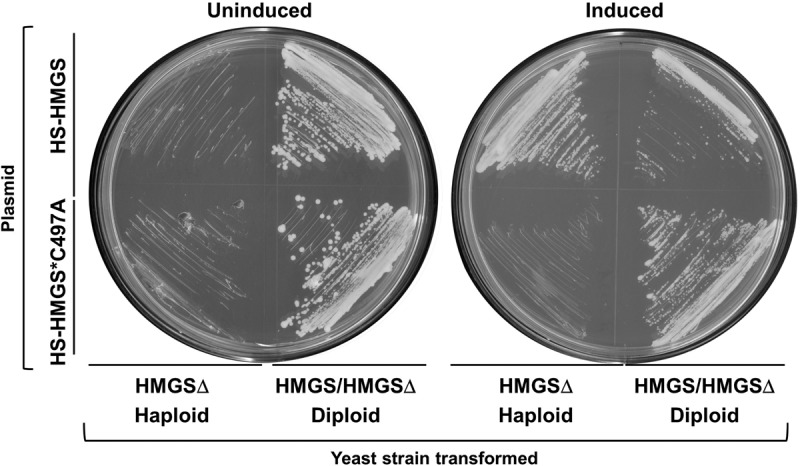
Functional complementation of S. cerevisiae HMGSΔ haploid with wild-type HS-HMGS and mutated HS-HMGS*C497A. Haploid HMGSΔ S. cerevisiae and a positive-control heterozygous diploid (HMGS/HMGSΔ) parent strain were transformed with galactose-inducible HS-HMGS or mutated HS-HMGS*C497A and streaked on selective plates with (induced) and without (uninduced) galactose. Heterozygous diploid positive-control transformants grew under all conditions, while only haploids that expressed the wild-type HS-HMGS (under induced conditions) grew.
We then wondered about the biological relevance of linking the expression of an STS with that of an HMGS and speculated that it may boost isoprenoid precursor supply for hirsutene biosynthesis. HMG-CoA reductase (HMGR) and not HMGS, however, is generally considered the key rate-limiting and regulated step in the mevalonate pathway of yeasts (Fig. 1). HMGR has therefore been the target of metabolic engineering efforts to increase terpenoid production in yeast (10, 45–47). Yet, the genomic identification in S. hirsutum of two additional predicted HMGS genes (Stehi1∣166728 and Stehi1∣119554) that each are located in protoilludene STS biosynthetic gene clusters (19) seems to suggest that HMGS may play a role in increasing mevalonate precursor supply for sesquiterpenoid secondary metabolite production. A fourth predicted HMGS gene (Stehi1∣168705) is not part of a biosynthetic gene cluster and likely encodes the housekeeping HMGS for general isoprenoid precursor biosynthesis. To test this hypothesis, we transformed wild-type HS-HMGS or mutated HS-HMGS*C497A into S. cerevisiae CEN.PK2 and quantified hirsutene production after the induction of heterologous gene expression with galactose for 48 h. However, hirsutene production levels (ranging from 54 to 62 μg/ml culture for four biological replicates of HS-HMGS or mutated HS-HMGS*C497A; Fig. S5) were not significantly different between strains expressing the fully functional and HMGS domain-inactivated fusion proteins. This suggests that at least in the yeast S. cerevisiae, the expression of an additional HMGS gene copy does not increase flux through the mevalonate pathway and, consequently, does not increase sesquiterpene production. Similar results were also obtained for expression studies in Pichia pastoris (reassigned to Komagataella phaffii) as another commonly engineered yeast system (data not shown).
HS-HMGS is in a biosynthetic gene cluster.
S. hirsutum is known to produce structurally diverse hirsutenoid secondary metabolites that are derived from the hirsutene scaffold (21–23) (Fig. 1). Biosynthesis genes tend to be clustered in fungi, and it was therefore expected that the HS would be clustered with scaffold-tailoring enzymes, such as P450 monooxygenases, oxidoreductases, and small-molecule transporters previously annotated in other STS biosynthetic gene clusters from Basidiomycota (17–19). Manual reprediction of the genomic region spanning ±50 kb on either side of the HS-HMGS identified 15 ORFs that include four cytochrome P450s, five oxidoreductases, and a major facilitator superfamily (MFS) transporter (Fig. 3 and Table S1).
Hypothesizing that the four P450s (ORF8 and ORF10 to ORF12) and a predicted FAD-containing oxidoreductase (ORF13) are the most likely enzymes catalyzing the initial oxidative modification of the hirsutene scaffold, we set out to clone these genes for expression in yeast. S. cerevisiae is generally considered a suitable host for the expression of microsomal cytochrome P450 monooxygenases, and we have previously successfully functionally expressed two P450s clustered with an α-cuprenene synthase from the ink cap mushroom Coprinus cinereus (17). Full-length spliced genes could be amplified from cDNA (Table S1), confirming that the biosynthesis genes are expressed in S. hirsutum. The genes were cloned into galactose-inducible expression vectors (Table 2). Different P450 combinations with and without oxidoreductase were then transformed into S. cerevisiae expressing HS-HMGS. The culture medium and headspace were analyzed by liquid chromatography (LC) and GC-MS for the production of terpenoid compounds. However, despite exhaustive attempts, including the coexpression of a cognate cytochrome P450 NADPH reductase (ShCPR1, amplified from cDNA S. hirsutum; see the supplemental material), none of the recombinant strains produced detectable levels of modified hirsutene compounds. Similar coexpression attempts in P. pastoris did not yield any new compounds, suggesting that these yeasts may not provide a suitable environment for the functional expression of these enzymes.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14 gyrA96 recA1 relA1 endA1 thi hsdR17 | New England BioLabs |
| E. coli C2566 (T7 Express) | fhuA2 lacZ::T7 gene 1 [lon] ompT gal sulA11 R(mcr-73::miniTn10-Tets)2 [dcm] R(zgb-210::Tn10-Tets) endA1 Δ(mcrC-mrr)114::IS10 | New England BioLabs |
| E. coli Rosetta (DE3)pLysS | F− ompT hsdSB (rB− mB−) gal dcm (DE3) pLysSRARE (Camr) | EMD Millipore |
| S. cerevisiae CEN.PK2 | MATa/MATα ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 MAL2-8C/MAL2-8C SUC2/SUC2 | Euroscarf accession no. 30000D |
| S. cerevisiae YML126c het/dip | MATa/MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0/MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Yml126C (ERG13 HMGS)::KanMX/Yml126c | Dharmacon |
| S. cerevisiae YML126cΔ haploid | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Yml126C (ERG13Δ)::KanMX | This study |
| S. hirsutum FP-91666 SS1 | Wild-type strain | 19 |
| Plasmids | ||
| pUCBB-eGFP | Constitutive lacP′ promoter with eGFP reporter, BioBrick restriction sites, Ampr | 38 |
| pUCBB-HS | HS-HMGS cloned into BamHI and NotI sites of pUCBB, replacing eGFP | This study |
| pUCBB-eGFP-ctH6 | pUCBB-eGFP with C-terminal His×6 purification tag to allow cloning in-frame with XhoI site | 38 |
| pUCBB-HSnt-ctH6 | HSnt cloned into BamHI/XhoI sites of pUCBB-ctH6 | This study |
| pUCBB-NtH6-eGFP | pUCBB-eGFP with N-terminal His×6 purification tag to allow cloning in-frame with NdeI restriction site | 38 |
| pUCBB-NtH6-HS | HS-HMGS cloned into pUCBB-NtH6 using NdeI/NotI restriction sites | This study |
| pET28a | T7 promoter, Nt-His tag with thrombin cleavage site when gene is inserted with NdeI restriction enzyme | EMD Millipore |
| pET28a-HS | HS-HMGS cloned into pET28a using NdeI/NotI restriction sites | This study |
| pESC-HIS | Yeast 2u plasmid, galactose-inducible GAL1-10 promoter | Stratagene |
| pESC-HIS-HS-HMGS | HS-HMGS inserted between EcoRI and SpeI restriction sites | This study |
| pESC-HIS-P450#8 | P450#8 inserted between NotI and PacI restriction sites | This study |
| pESC-HIS-P450#10 | P450#10 inserted between NotI and PacI restriction sites | This study |
| pESC-HIS-P450#11 | P450#11 inserted between NotI and PacI restriction sites | This study |
| pESC-URA-P450#12 | P450#12 inserted between NotI and PacI restriction sites | This study |
| pESC-URA-FAD#13 | FAD#13 inserted between NotI and PacI restriction sites | This study |
| pESC-TRP-HS-HMGS | HS-HMGS between EcoR1 and SpeI restriction sites | This study |
| pESC-LEU-ShCPR1 | CPR1 from S. hirsutum (see Data Set S1 for sequence) cloned between the BamH1 and HindIII restriction sites | This study |
eGFP, enhanced green fluorescent protein; Tets, tetracycline sensitivity; Camr, chloramphenicol resistance; Ampr, ampicillin resistance.
HMGS is duplicated in fungal genomes.
Genomic analysis of S. hirsutum identified four putative HMGS genes, including three in putative STS gene clusters, whereas the fourth appears to be the housekeeping enzyme. In contrast, only a single HMGR was identified in the S. hirsutum genome (Table S3). Although HMGR has been established as the rate-limiting enzyme of the mevalonate pathway (10, 45–47), duplication and clustering of HMGSs with terpenoid biosynthetic pathways in S. hirsutum suggested that HMGS may play an equal role in controlling isoprenoid precursor supply in fungi. We therefore performed homology searches of Basidiomycota and Ascomycota genomes in JGI's Fungal Genome Database (Tables S4 and S5) with the five mevalonate pathway enzymes from S. cerevisiae (Fig. 1) to check whether HMGS and any of the other mevalonate pathway genes are duplicated in fungi. We found that both HMGS and HMGR are widely duplicated in fungal genomes. Roughly 20% of the sequenced Basidiomycota genomes have multiple HMGS and/or HMGR gene copies, and about 6% of the genomes contain multiple copies of both genes. Similarly, between 25 and 30% of sequenced Ascomycota genomes have multiple HMGR and/or HMGS copies, and about 16% of the genomes have duplicated both genes, although these numbers are somewhat biased because of the relatively large fraction of sequenced Aspergillus and Penicillium genomes that tend to have multiple HMGS and HMGR genes. Few fungal genomes contain several copies of any of the downstream mevalonate pathway enzymes mevalonate kinase (MK), phosphomevalonate kinase (PMK), and diphosphomevalonate decarboxylase (MDD), suggesting that in fungi, both HMGS and HMGR are key enzymes in the regulation of isoprenoid precursor flux. Further analysis of fungal genomes (Tables S4 and S5) with one HMGR gene and three or more HMGS copies comparable to S. hirsutum led to the identification of several additional examples of putative STS biosynthetic gene clusters with an HMGS gene, including two examples of putative STS-HMGS fusion proteins (Table S6). Our genomic analysis therefore supports the duplication of HMGS for sesquiterpenoid secondary metabolite biosynthesis in fungi.
DISCUSSION
The hirsutane scaffold was first identified in 1947 with the isolation of the antibiotic hirsutic acid C from S. hirsutum extract (2). Bioactive hirsutene-derived compounds have since then been isolated from diverse marine and terrestrial fungi (reviewed in references 2, 48, and 49). Many of these compounds feature intriguing scaffold modifications (22, 23, 50), suggesting that their biosynthesis pathways involve new catalytic activities that would be of interest for the recombinant production of bioactive hirsutenoids and other modified sesquiterpenoids. Our goal was therefore to identify in S. hirsutum a hirsutene-specific STS and potentially clustered biosynthesis genes responsible for hirsutenoid production. Here, we used our previously proven bioinformatic approach (19) to predict and subsequently functionally identify this enzyme (HS-HMGS) as a new type of fusion protein between an STS (HS) and an HMG-CoA synthase (HMGS) domain. Further, sequence analysis and comparison with previously characterized fungal and representative bacterial STSs (Fig. 2B) suggested that the HS domain seems to be similarly evolutionarily related to bacterial 1,11-cyclizing STSs and to three STSs from Ascomycota. The HMGS domain, on the other hand, was closely related to other fungal HMGS sequences.
The gene structure of HS is distinct from those of characterized fungal STSs and was therefore initially not correctly predicted using Augustus (27) and sequence alignments. The HS gene region has a significantly larger number of introns than typically found in fungal STS genes (9 introns versus 4 introns, on average). In addition, it does not contain an intron that typically splits the conserved catalytic ND motif in Basidiomycota STS genes (Fig. 4). High intron density in fungal genes has been correlated with horizontal gene transfer from bacteria, which was recently shown for the acquisition by S. hirsutum of an α-amylase gene (Stehi1∣78757) from an actinomycete several hundred million years ago (29). Inspection of the GC content (which is typically >60% in actinomycetes) of the acquired amylase gene, however, shows that it is the same as for S. hirsutum, suggesting that the codon usage of the bacterial gene must have undergone adaptation since its acquisition. The GC content of the HS domain (and its HMGS domain) is also as expected for S. hirsutum, eliminating recent gene transfer from GC-rich bacteria. Acquisition of biosynthesis genes from bacteria, however, is not uncommon in fungi; for example, the biotin pathway of S. cerevisiae has been acquired by horizontal gene transfer from multiple bacterial species (51). Horizontal gene transfer between fungal phyla is also common; e.g., we recently provided evidence for the acquisition of a Basidiomycota STS (protoilludene synthase) gene cluster by an endophytic ascomycete (52). Phylogenetic analysis of characterized fungal STSs and representative characterized bacterial STSs (which are mostly from actinomycetes) (Fig. 2) showed that HS appears to be similarly related to the bacterial STSs and to a small cluster of 1,6- and 1,10-cyclizing Ascomycota STSs (prAS, atAS, and FfSc6) (13) (12). Intriguingly, the bacterial STS clade includes several 1,11-cyclizing enzymes, including the recently discovered cucumene synthases (slt18 1880 and SCLAV p1407) that produce 1- and 4-cucumene (aka isohirsut-1-ene and isohirsut-4-ene, respectively) (33, 34, 53). The cucumane scaffold differs from the hirsutane scaffold only in the position of one methyl side group (2) as the result of a divergent cyclization path following the initial cyclization of FPP into the 1,11 trans-humulyl cation (20). Like the hirsutenoids, cucumanes exhibit antimicrobial or cytotoxic activity (2, 48) and may play a similar defense role in their fungal and bacterial producers. However, without additional functionally characterized HS homologs, we can therefore at this point only speculate about its evolutionary origin.
Unlike the HS domain, the C-terminal HMGS domain is most closely related to other Basidiomycota HMGSs (Fig. S4). Likewise, predicted cytochrome P450s and other putative biosynthesis proteins located in the putative HS-HMGS biosynthesis gene cluster (Fig. 3 and Table S1) are closely related to other Basidiomycota sequences, leaving room for future studies into the origination and evolution of fungal biosynthetic gene clusters (reviewed in reference 54) where a key scaffold-generating enzyme has been acquired from a bacterial source. Fusion of the STS and HMGS genes may have been a serendipitous event without direct apparent functional advantage other than clustering of the two proteins. The two enzymes are five pathway steps removed from each other (Fig. 1), ruling out potential substrate channeling between them to increase pathway flux. Translational fusion of enzymes has been widely used in isoprenoid pathway metabolic engineering to increase terpenoid production, but fusions were made between a terpene synthase/cyclase and the enzyme producing its prenyl diphosphate substrate (see, e.g., references 55–59).
Homology searches of fungal genomes for genes encoding mevalonate pathway enzymes (Tables S4 and S5) revealed that about 20 to 30% of the searched genomes contain multiple copies of HMGR and/or HMGS genes (Tables S4 and S5). In addition, we identified additional sesquiterpenoid biosynthetic gene clusters that include a putative HMGS gene, including gene clusters that contain an STS-HMGS fusion gene (Table S6). Our analysis therefore suggests that both enzymes play a key role in controlling isoprenoid precursor flux in fungi. Until now, however, only HMGR was generally considered the rate-limiting enzyme of the mevalonate pathway (46) and was consequently targeted for increasing isoprenoid biosynthesis in yeast (10, 45–47).
Increasingly, research in fungi and plants shows that HMGS, like HMGR, is highly regulated at multiple levels and controls sterol and terpenoid production (43, 60). Yet, unexpectedly, hirsutene production was not increased in recombinant S. cerevisiae strains that expressed the HS-HMGS fusion protein compared to that in strains that expressed only the HS domain, suggesting that mevalonate pathway regulation could be different in these two yeast strains from that in other fungi, plants, and animals. Comparisons of HMGR and HMGS sequences across phyla (see Table S3 for sequences) point to substitutions of conserved amino acids important for regulating mevalonate pathway flux in different organisms. The HMGRs from S. cerevisiae and P. pastoris (Komagataella phaffii/K. pastoris) lack a conserved serine residue (S872, human HMGR numbering) that is present in HMGRs from other organisms, including in fungi and other yeasts. This residue is phosphorylated in response to a low cellular energy state and decreases HMGR activity in most organisms; HMGRs from S. cerevisiae and P. pastoris enzymes, however, are not controlled by this mechanism (46). In HMGSs, the substitution of a conserved His residue to N (H188 in Brassica juncea HMGS) has been shown to result in a loss of substrate inhibition by acetoacetyl-CoA (61). This substitution is found in the human mitochondrial HMGS isoform, which is part of the ketogenesis pathway. Interestingly, HMGSs from Basidiomycota, unlike enzymes from other fungi and plants (Table S3), also have the H188N substitution, suggesting a lack of acetoacetyl-CoA substrate inhibition. Together, these differences in the regulation of the two key enzymes in the mevalonate pathway between S. hirsutum and the two yeast expression hosts likely explain why the expression of an extra copy of HMGS has no effect on hirsutene production in these yeasts. From a metabolic engineering perspective, HMGS and HMGR should both be considered potential targets for increasing mevalonate pathway flux; the combination of an HMGS and an HMGR from Basidiomycota may prove to be a successful strategy toward this goal.
Attempts to functionally express predicted hirsutene scaffold modifying P450 and oxidoreductase enzymes in recombinant yeast were unsuccessful. Numerous examples have shown that filamentous fungi compartmentalize secondary metabolite biosynthesis into subcellular structures (62–66). A well-known example are the toxisomes made by Fusarium graminearum that serve as hubs for trichothecene sesquiterpenoid mycotoxin production by colocalizing P450s, terpene synthase, and HMGR to specialized membrane structures (63, 67). It is therefore likely that heterologous reconstitution of hirsutenoid biosynthesis will require a more sophisticated cellular environment than can be offered by yeasts which have no significant native secondary metabolism.
In summary, we describe the functional characterization of a previously unknown STS in a Basidiomycota, thereby completing the identification of STSs producing all major sesquiterpenes detected in S. hirsutum. The characterized HS-HMGS is part of a large putative hirsutenoid biosynthetic gene cluster, offering access to a new group of sesquiterpenoids and opening the door for the heterologous production of these pharmaceutically relevant compounds. Further, this work offers insights into the generation of new biosynthesis pathways in Basidiomycota via the acquisition of a bacterial gene by horizontal gene transfer, and it provides new avenues for strain engineering to increase terpenoid and sterol production. Finally, our study urges the development of new fungal chassis organisms, including Basidiomycota, with genetic tractability and engineerability similar to those of current yeast and some fungal systems to access the chemodiversity of Basidiomycota. Functional characterization of enzymes involved in the biosynthesis of diterpenoid pleuromutilins by the basidiomycete Clitopilus was very recently accomplished using Aspergillus oryzae as a heterologous expression system (68), thereby providing support for the development and testing of different fungal chassis systems.
MATERIALS AND METHODS
S. hirsutum cultivation and cDNA isolation.
Fungal strain growth conditions, sesquiterpene detection, mRNA extractions, and cDNA preparations were carried out as described previously (19). Briefly, Stereum hirsutum FP-91666 SS-1 was grown at 22°C in rich medium in the dark for 14 days in 100-ml liquid cultures (500-ml flask). S. hirsutum mycelial tissue was dried and frozen at −80°C. Tissue aliquots were ground in liquid N2 by sterile mortar and pestle for mRNA extraction using TRIzol reagent (Life Technologies, Grand Island, NY, USA). cDNA was then prepared using the Superscript III first-strand synthesis system (Life Technologies) with oligo(dT)20 primers for reverse transcription-PCR (RT-PCR), followed by RNase H treatment.
Gene predictions and cloning of HS-HMGS.
Initial STS gene annotation, alignment, and gene structure prediction were performed as described previously (19). Briefly, gene predictions were made in Augustus (69) with different fungal gene models using the genomic region 10 to 15 kb flanking the putative hirsutene synthase ORFs Stehi1∣122776 and Stehi1∣52743, including downstream ORF Stehi1∣53380, which was predicted to be an HMGS. STS gene predictions were manually aligned with functionally characterized fungal STSs using MEGA7 (70). Primers for the amplification of Stehi1∣52743 and Stehi1∣122776 from cDNA were designed based on these gene predictions (Table S1). The complete ORF comprising Stehi1∣52743 to Stehi1∣53380 was amplified from cDNA with primers 5 and 6 (Table 1) using touchdown PCR (annealing temperature, 68 to 53°C) and the Phusion high-fidelity PCR kit (New England BioLabs, Ipswich, MA). The resulting 2,526-bp PCR product was gel purified, cloned using the Zero Blunt TOPO system (Life Technologies, Grand Island, NY), and confirmed by Sanger sequencing.
E. coli strains and cultivation.
E. coli JM109 cells were grown in LB medium at 37°C and supplemented with the appropriate antibiotic(s) of ampicillin (100 μg · ml−1), chloramphenicol (50 μg · ml−1), zeocin (50 μg · ml−1), or kanamycin (100 μg · ml−1), which were used for cloning and plasmid maintenance. Constitutive expression of HS-HMGS and HSnt from our in-house pUCBB plasmid (see below) was performed in E. coli C2566 cells grown at 30°C in 50 ml LB supplemented with the appropriate antibiotics in 250-ml shake flasks (Table 2). For protein purification, HS-HMGS was expressed from a pET28a plasmid in E. coli Rosetta cells (EMD Millipore, Billerica, MA) (see below).
Construction of E. coli expression plasmids.
The HS-HMGS ORF was PCR amplified from the TOPO vector using primers 7 and 8 (Table 1), adding BamHI and NotI sites for subcloning into a similarly digested pUCBB plasmid (38) for constitutive expression in E. coli. Truncated HSnt (lacking the HMGS domain) was created by PCR using primers 7 and 9, replacing the predicted HMGS start codon methionine residue (M365) with an in-frame XhoI site, and subsequently ligated into the similarly digested pUCBB-eGFP-ctHis6 plasmid (38) to create pUCBB-HSnt-ctHis6 (Table 2). Primers 10 and 11 were used to generate HS-HMGS with flanking NdeI/NotI sites for insertion after the NtHis6 tag of the pUCBB-ntH6-eGFP plasmid (38) and the pET28a vector (EMD Millipore) to generate ntH6-HS-HMGS. All ligations and restriction digestions were performed in accordance with the conditions recommended by NEB.
GC-MS analysis of sesquiterpenoids.
Volatile sesquiterpenes of E. coli cultures constitutively expressing the full-length fusion HS-HMGS or HSnt from pUCBB were analyzed and identified as described previously (19). Briefly, volatile sesquiterpenes were extracted from E. coli culture flask headspace by inserting a 100 μm polydimethylsiloxane (PDMS) solid-phase microextraction (SPME) fiber (Supelco, Bellefonte, PA) through a tin foil seal for 10 min. Volatiles from overnight in vitro assays with purified HS-HMGS (quantity) and 2 μM (2E,6E)-FPP as the substrate were similarly extracted by PDMS SPME for 10 min from GC-MS vials. The extracted terpenoid products were separated and analyzed by GC-MS on an HP 7890A GC coupled to an anion-trap mass spectrometer HP MSD triple axis detector (Agilent Technologies, Santa Clara, CA), using an HP-5MS capillary column (30 m by 0.25 mm by 1.0 μm) with an injection port temperature of 250°C and helium as a carrier gas. The oven temperature started at 60°C and was increased at 6°C · min−1 to a final temperature of 250°C. Terpenoid compounds were identified by comparing mass spectra and retention indices to the MassFinder software (version 4) terpene library (71).
HS-HMGS purification and enzyme kinetics.
N-terminally His-tagged HS-HMGS was expressed using the pET28a plasmid and E. coli Rosetta cells (EMD Millipore) in 500 ml LB medium with appropriate antibiotics (2-liter flask) at 30°C and induced with 0.5 mM isopropyl β-d-1-thiogalactopyrranoside at an optical density at 600 nm (OD600) of ∼0.3. Cultures were then grown for 3 h after induction, monitored for hirsutene production by SPME as described above, pelleted by 10 min of centrifugation at 4,000 × g, and immediately frozen at −80°C. The pellets were resuspended in 30 ml STS purification buffer (50 mM Tris [pH 8], 250 mM NaCl, 5 mM imidazole, 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with cOmplete protease inhibitor (Sigma-Aldrich, St. Louis, MO) prior to a 6-min sonication (1 s on, 2 s off), followed by 30 min of centrifugation at 25,000 × g. Soluble lysate was then purified using Talon resin (TaKaRa Clontech, Mountain View, CA) and eluted into STS elution buffer (50 mM Tris [pH 8], 250 mM NaCl, 250 mM imidazole, 10 mM MgCl2, 1 mM PMSF). This sample was then concentrated using Amicon (EMD Millipore) Ultra-15 10,000 nominal molecular weight limit (NMWL) centrifugation filter units and desalted using PD-10 columns (GE Healthcare, Chicago, IL). Protein concentrations were determined using Bradford reagent (Amresco, Solon, OH), according to the manufacturer's instructions. Proteins were analyzed by 15% SDS-PAGE gels using standard methods. The kinetic parameters of the purified desalted HS-HMGS (2.55 μg/ml) were determined using the PiPer pyrophosphate assay (Thermo Fisher Scientific, Waltham, MA), according to the manufacturer's instructions, over a substrate range of 0 to 100 μM FPP in the presence of 0.5 mM MgCl2, as described previously (19).
S. cerevisiae strains and cultivation.
S. cerevisiae CEN.PK2 was used for yeast vector recombination cloning (see below) and comparison of sesquiterpenoid production of strains transformed with pESC-HIS-HS-HMGS or pESC-HS-HMGS*C497A. Complementation experiments with HS-HMGS and HS-HMGS*C497A were performed with the heterozygous ERG13 strain Yml126c (ERG13Δ::KanMX/ERG13) (note that ERG13 encodes HMGS) (72) (Table 2), obtained from the GE Dharmacon (Lafayette, CO) yeast knockout collection. The haploid strain Yml126cΔ (Table 2) used as a control in the complementation experiments was obtained after sporulation, as described below.
Yeast strains were maintained on yeast extract-peptone-dextrose (YPD) medium, and strains transformed with galactose-inducible expression plasmid pESC-HIS (harboring a histidine [HIS] auxotrophic selection marker) were cultivated in synthetic dextrose (SD) medium lacking histidine (1.7 g · liter−1 yeast nitrogen base without amino acids [Sigma-Aldrich], 1.4 g · liter−1 yeast synthetic dropout supplement [Sigma-Aldrich], 120 mg · liter−1 uracil, 120 mg · liter−1 leucine, 50 mg · liter−1 tryptophan, 20 g · liter−1 dextrose). Protein expression was induced in synthetic galactose (SG) medium, which has the same composition as the SD medium, except that dextrose is replaced with galactose. Twenty grams per liter agar was added to make solid medium. Liquid cultures were cultivated at 220 rpm and 30°C.
Construction of yeast expression plasmids pESC-HIS-HMGS and pESC-HIS-HMGS*C497A.
The yeast expression plasmid pESC-HIS-HS-HMGS (Table 2) was made via yeast recombination cloning (YRC) (73). pESC-HIS (HIS auxotrophic marker; Stratagene, La Jolla, CA) was digested with EcoRI and SpeI. HS-HMGS was PCR amplified from the pUCBB-HS-HMGS plasmid with 40 bp of homology to the linearized pESC-HIS vector using primers 12 and 13 (Table 1) to enable insertion between the EcoRI and SpeI sites. Gel-purified PCR product (25 μl) and digested vector (25 μl) were cotransformed into yeast CEN.PK2 (74) using the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/SS carrier DNA-PEG) method (75), plated on SD-HIS selective plates, and incubated for 2 days. Yeast colonies containing pESC-HIS-HS-HMGS plasmids were patched onto selective plates and grown overnight. For pESC-HIS-HS-HMGS plasmid isolation, 3-ml liquid cultures (SD-HIS) of yeast transformants were grown overnight. Cells were pelleted for 5 min at full speed in an Eppendorf 5810 R tabletop centrifuge (Hamburg, Germany), resuspended in 250 μl with 2 mg/ml Zymolase (1.2 M sorbitol, 0.1 M KPO4 [pH 7.5]), and incubated for 2 h at 37°C, followed by DNA isolation using the Wizard Plus SV miniprep DNA purification system (Promega, Madison, WI). To create an inactive HMGS C497A domain mutant, primers 14 and 15 (Table 1) were used to replace C497 with alanine via Q5 site-directed mutagenesis (New England BioLabs, Ipswich, MA) using pESC-HIS-HS-HMGS as the template, creating pESC-HIS-HS-HMGS C497A.
S. cerevisiae ERG13Δ complementation.
Heterozygous/diploid strain YML126c (ERG13Δ::KanMX/ERG13) was cultivated in 5 ml sporulation medium (10 g · liter−1 potassium acetate [KOAc], 1 g · liter−1 yeast extract, 0.5 g · liter−1 dextrose) for 9 days at 30°C. The haploid spores (ERG13Δ::KanMX or ERG13) were enriched for random spore analysis before plating on selective YPD plus 200 μg · ml−1 G418, with replica plating on nonselective (YPD) and ERG13 auxotroph selective plates (YPD plus G418, plus 5 mg · ml−1 mevalonate) (76) to select for ERG13Δ::KanMX colonies. Individual colonies from the mevalonate auxotroph plates were replica plated on G418 and mevalonate plates and on G418-only plates with heterozygous diploid YML126c controls to confirm the isolated mevalonate auxotrophic phenotype. Isolates were then PCR screened for KanMX insertion at the correct genomic location using primers described by Winzeler et al. (72), and for mating type using MATa/MATα locus primers described by Huxley et al. (77), confirming the MATa haploid ERG13(HMGS)Δ auxotroph. The heterozygous/diploid strain YMl126c (ERG13/ERG13Δ::KanMX) as a control and auxotrophic MATa haploid ERG13Δ::KanMX were transformed with either pESC-HIS-HS-HMGS or pESC-HIS-HS-HMGS*C497A. Transformants were replica plated on SD-HIS (uninduced) or SG-HIS (induced) plates to investigate complementation by HS-HMGS and inactivated HS-HMGS*C497A. Growth was monitored for 3 days. A minimum of three isolates of each plasmid transformant were tested for complementation of the ERG13Δ genotype.
Hirsutene production in S. cerevisiae expression HS-HMGS or HS-HMGS*C497A.
Four independent S. cerevisiae CEN.PK2 transformants with either pESC-HS-HMGS or pESC-HS-HMGS*C497A were inoculated into 14-ml conical shake tubes overnight in 3 ml SD medium and grown overnight at 30°C with 220 rpm shaking. One milliliter of overnight culture was used to inoculate 50 ml SD medium in 250-ml unbaffled shake flasks and allowed to grow overnight at 30°C and 220 rpm. Cells were washed 2 times with 10 ml sterile distilled water (dH2O) and resuspended to an OD of 1.0 into 50 ml of SG induction medium. Five milliliters of dodecane overlay [containing 100 μg · ml−1, 95% pure (−)-caryophyllene oxide (Sigma-Aldrich) as an internal control] was added to cultures to absorb volatile sesquiterpene products, as described previously (10, 78). Cultures were grown for 48 h in SG induction medium before collection of the dodecane layer by centrifugation. Five microliters of each dodecane extract was injected with methyl tert-butyl ether (MTBE) and ethyl acetate as the first and second wash buffers, respectively, for GC-MS-flame ionization detector (GC-MS-FID) analysis. The oven temperature started at 60°C and was increased at 10°C · min−1 to a final temperature at 250°C. The FID was kept at 250°C, with a He flow of 30 ml · min−1 and airflow rate of 400 ml · min−1. Hirsutene concentration was quantified by comparison to a (−)-caryophyllene oxide standard curve.
Bioinformatics analysis.
Sequence alignments and phylogenetic analysis were performed in MEGA7 (79) using MUSCLE (80) for protein alignments. Protein alignments (Fig. S3) were exported to Jalview 2.0 (81), with active-site residues manually annotated and colored for similar amino acid properties. Phylogenetic analysis of the STS HS-HMGS domain (residues 1 to 364) with microbial STSs (Fig. 2B) was done using the neighbor-joining method using the Poisson correction method with 500 bootstrap replications (82, 83). The gene structures shown in Fig. 4 were visualized using Gene Structure Display Server (GSDS 2.0) (84). The HMGS domain phylogeny of HS-HMGS (residues 365 to 842) (Fig. S4) was inferred by comparison with 16 HMGS protein sequences using the maximum likelihood method. The HMGS sequences aligned were from Homo sapiens (hHMGS1 and hHMGS2, UniProt accession numbers P54868 and Q01581, respectively [40]), Brassica juncea (bjHMGS, GenBank accession no. AAF69804.1 [85]), Staphylococcus aureus (saHMGS, UniProt accession no. Q9FD87 [86]), Enterococcus faecalis (efHMGS, UniProt accession no. Q9FD71 [87]), Hevea brasiliensis (hbHMGS, UniProt accession no. Q6QLW8 [39]), Phycomyces blakeseeanus (hmgS, GenBank accession no. AJ297414 [88]), S. pombe (hcs, GenBank accession no. AAB17601.1 [89]), Ganoderma lucidum (glHMGS, UniProt accession no. K7PL94 [43]), Phaffia rhodozyma (HMGS, GenBank accession no. E50999.1 [42]), Ustilago maydis (hcs1, RefSeq accession no. 23565277 from EMBL [44]), Saccharomyces cerevisiae (ERG13 and YML126c, UniProt accession no. P54839 [46]), and from S. hirsutum (putative HMGSs Stehi1∣166728, Stehi1∣119554, and Stehi1∣168705 identified by BLAST search of the S. hirsutum FP-91666 SS1 version 1.0 genome sequence [https://genome.jgi.doe.gov] using the S. cerevisiae ERG13 sequence). Duplications of HMGS homologues in fungi were identified by a BLAST search of 307 Basidiomycota and 510 Ascomycota genome sequences in JGI's fungal database using a 10−5 E value cutoff and searching with the S. cerevisiae ERG13 sequence and with the two characterized Basidiomycota HMGS, Ustilago maydis Hcs1 (44) and Ganoderma lucidum glHMGS (43). Manual annotation of the HS-HMGS Stehi1∣52743 and Stehi1∣122776 (Fig. 3) gene cluster was done as previously described for other STS-containing gene clusters from S. hirsutum (19).
Cloning and expression of P450s and FAD-oxidoreductase.
Predicted ORFs (Table S1) were amplified and cloned from cDNA, first into E. coli cloning vector and then into pESC yeast expression vectors, as described above for HS-HMGS. Genes were cloned into pESC vector backbones with different auxotrophic selection markers (Trp/Ura/Leu/His) to allow for coexpression with HS-HMGS and cytochrome P450 reductases (CPR) cloned into pESC backbones with different auxotrophic selection markers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in whole or part by the National Institutes of Health grant GM080299 (to C.S.-D.). C.M.F. was supported by a doctoral dissertation fellowship from the University of Minnesota.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00036-18.
REFERENCES
- 1.Schmidt-Dannert C. 2015. Biosynthesis of terpenoid natural products in fungi. Adv Biochem Eng Biotechnol 148:19–61. doi: 10.1007/10_2014_283. [DOI] [PubMed] [Google Scholar]
- 2.Abraham WR. 2001. Bioactive sesquiterpenes produced by fungi–are they useful for humans as well? Curr Med Chem 8:583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- 3.Elisashvili V. 2012. Submerged cultivation of medicinal mushrooms: bioprocesses and products (review). Int J Med Mushrooms 14:211–239. doi: 10.1615/IntJMedMushr.v14.i3.10. [DOI] [PubMed] [Google Scholar]
- 4.Evidente A, Kornienko A, Cimmino A, Andolfi A, Lefranc F, Mathieu V, Kiss R. 2014. Fungal metabolites with anticancer activity. Nat Prod Rep 31:617–627. doi: 10.1039/C3NP70078J. [DOI] [PubMed] [Google Scholar]
- 5.Misiek M, Hoffmeister D. 2007. Fungal genetics, genomics, and secondary metabolites in pharmaceutical sciences. Planta Med 73:103–115. doi: 10.1055/s-2007-967104. [DOI] [PubMed] [Google Scholar]
- 6.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. 2010. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boettger D, Hertweck C. 2013. Molecular diversity sculpted by fungal PKS-NRPS hybrids. Chembiochem 14:28–42. doi: 10.1002/cbic.201200624. [DOI] [PubMed] [Google Scholar]
- 8.Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y. 2013. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J Am Chem Soc 135:4616–4619. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin HC, Tsunematsu Y, Dhingra S, Xu W, Fukutomi M, Chooi YH, Cane DE, Calvo AM, Watanabe K, Tang Y. 2014. Generation of complexity in fungal terpene biosynthesis: discovery of a multifunctional cytochrome P450 in the fumagillin pathway. J Am Chem Soc 136:4426–4436. doi: 10.1021/ja500881e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D. Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. 2013. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 11.Tsunematsu Y, Ishiuchi K, Hotta K, Watanabe K. 2013. Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat Prod Rep 30:1139–1149. doi: 10.1039/c3np70037b. [DOI] [PubMed] [Google Scholar]
- 12.Wawrzyn GT, Bloch SE, Schmidt-Dannert C. 2012. Discovery and characterization of terpenoid biosynthetic pathways of fungi. Methods Enzymol 515:83–105. doi: 10.1016/B978-0-12-394290-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 13.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A 110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiemann P, Keller NP. 2014. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol 41:301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 15.Yaegashi J, Oakley BR, Wang CC. 2014. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J Ind Microbiol Biotechnol 41:433–442. doi: 10.1007/s10295-013-1386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spakowicz DJ, Strobel SA. 2015. Biosynthesis of hydrocarbons and volatile organic compounds by fungi: bioengineering potential. Appl Microbiol Biotechnol 99:4943–4951. doi: 10.1007/s00253-015-6641-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agger S, Lopez-Gallego F, Schmidt-Dannert C. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, Schmidt-Dannert C. 2012. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem Biol 19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quin MB, Flynn CM, Wawrzyn GT, Choudhary S, Schmidt-Dannert C. 2013. Mushroom hunting by using bioinformatics: application of a predictive framework facilitates the selective identification of sesquiterpene synthases in Basidiomycota. Chembiochem 14:2480–2491. doi: 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quin MB, Flynn CM, Schmidt-Dannert C. 2014. Traversing the fungal terpenome. Nat Prod Rep 31:1449–1473. doi: 10.1039/C4NP00075G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comer FW, McCapra F, Qureshi IH, Scott AI. 1967. The structure and chemistry of hirsutic acid. Tetrahedron 23:4761–4768. doi: 10.1016/S0040-4020(01)92573-6. [DOI] [PubMed] [Google Scholar]
- 22.Yoo NH, Kim JP, Yun BS, Ryoo IJ, Lee IK, Yoon ES, Koshino H, Yoo ID. 2006. Hirsutenols D, E and F, new sesquiterpenes from the culture broth of Stereum hirsutum. J Antibiot (Tokyo) 59:110–113. doi: 10.1038/ja.2006.16. [DOI] [PubMed] [Google Scholar]
- 23.Yun BS, Lee IK, Cho Y, Cho SM, Yoo ID. 2002. New tricyclic sesquiterpenes from the fermentation broth of Stereum hirsutum. J Nat Prod 65:786–788. doi: 10.1021/np010602b. [DOI] [PubMed] [Google Scholar]
- 24.Feline TC, Mellows G, Jones RB, Phillips L. 1974. Biosynthesis of hirsutic acid C using 13C nuclear magnetic resonance spectroscopy. J Chem Soc Chem Commun 2:63–64. doi: 10.1039/c39740000063. [DOI] [Google Scholar]
- 25.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 26.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao XL, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoff KJ, Stanke M. 2013. WebAUGUSTUS–a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res 41:W123–W128. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rynkiewicz MJ, Cane DE, Christianson DW. 2001. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci U S A 98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Da Lage J-L, Binder M, Hua-Van A, Janecek S, Casane D. 2013. Gene make-up: rapid and massive intron gains after horizontal transfer of a bacterial alpha-amylase gene to Basidiomycetes. BMC Evol Biol 13:40. doi: 10.1186/1471-2148-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickschat JS. 2016. Bacterial terpene cyclases. Nat Prod Rep 33:87–110. doi: 10.1039/C5NP00102A. [DOI] [PubMed] [Google Scholar]
- 31.Lesburg CA, Zhai G, Cane DE, Christianson DW. 1997. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science 277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 32.Nakano C, Horinouchi S, Ohnishi Y. 2011. Characterization of a novel sesquiterpene cyclase involved in (+)-caryolan-1-ol biosynthesis in Streptomyces griseus. J Biol Chem 286:27980–27987. doi: 10.1074/jbc.M111.265652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada Y, Arima S, Nagamitsu T, Johmoto K, Uekusa H, Eguchi T, Shin-ya K, Cane DE, Ikeda H. 2015. Novel terpenes generated by heterologous expression of bacterial terpene synthase genes in an engineered Streptomyces host. J Antibiotics 68:385–394. doi: 10.1038/ja.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Kuzuyama T, Komatsu M, Shin-ya K, Omura S, Cane DE, Ikeda H. 2015. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci U S A 112:857–862. doi: 10.1073/pnas.1422108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin X, Hopson R, Cane DE. 2006. Genome mining in Streptomyces coelicolor: molecular cloning and characterization of a new sesquiterpene synthase. J Am Chem Soc 128:6022–6023. doi: 10.1021/ja061292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano C, Tezuka T, Horinouchi S, Ohnishi Y. 2012. Identification of the SGR6065 gene product as a sesquiterpene cyclase involved in (+)-epicubenol biosynthesis in Streptomyces griseus. J Antibiot (Tokyo) 65:551–558. doi: 10.1038/ja.2012.68. [DOI] [PubMed] [Google Scholar]
- 37.Chou WK, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE. 2010. Genome mining in Streptomyces avermitilis: cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc 132:8850–8851. doi: 10.1021/ja103087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vick JE, Johnson ET, Choudhary S, Bloch SE, Lopez-Gallego F, Srivastava P, Tikh IB, Wawrzyn GT, Schmidt-Dannert C. 2011. Optimized compatible set of BioBrick vectors for metabolic pathway engineering. Appl Microbiol Biotechnol 92:1275–1286. doi: 10.1007/s00253-011-3633-4. [DOI] [PubMed] [Google Scholar]
- 39.Sirinupong N, Suwanmanee P, Doolittle RF, Suvachitanont W. 2005. Molecular cloning of a new cDNA and expression of 3-hydroxy-3-methylglutaryl-CoA synthase gene from Hevea brasiliensis. Planta 221:502–512. doi: 10.1007/s00425-004-1463-7. [DOI] [PubMed] [Google Scholar]
- 40.Shafqat N, Turnbull A, Zschocke J, Oppermann U, Yue WW. 2010. Crystal structures of human HMG-CoA synthase isoforms provide insights into inherited ketogenesis disorders and inhibitor design. J Mol Biol 398:497–506. doi: 10.1016/j.jmb.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 41.Misra I, Narasimhan C, Miziorko HM. 1993. Avian 3-hydroxy-3-methylglutaryl-CoA synthase. Characterization of a recombinant cholesterogenic isozyme and demonstration of the requirement for a sulfhydryl functionality in formation of the acetyl-enzyme reaction intermediate. J Biol Chem 268:12129–12135. [PubMed] [Google Scholar]
- 42.Miao L, Chi S, Tang Y, Su Z, Yin T, Guan G, Li Y. 2011. Astaxanthin biosynthesis is enhanced by high carotenogenic gene expression and decrease of fatty acids and ergosterol in a Phaffia rhodozyma mutant strain. FEMS Yeast Res 11:192–201. doi: 10.1111/j.1567-1364.2010.00705.x. [DOI] [PubMed] [Google Scholar]
- 43.Ren A, Ouyang X, Shi L, Jiang A-L, Mu D-S, Li M-J, Han Q, Zhao M-W. 2013. Molecular characterization and expression analysis of GlHMGS, a gene encoding hydroxymethylglutaryl-CoA synthase from Ganoderma lucidum (Ling-zhi) in ganoderic acid biosynthesis pathway. World J Microbiol Biotechnol 29:523–531. doi: 10.1007/s11274-012-1206-z. [DOI] [PubMed] [Google Scholar]
- 44.Winterberg B, Uhlmann S, Linne U, Lessing F, Marahiel MA, Eichhorn H, Kahmann R, Schirawski J. 2010. Elucidation of the complete ferrichrome A biosynthetic pathway in Ustilago maydis. Mol Microbiol 75:1260–1271. doi: 10.1111/j.1365-2958.2010.07048.x. [DOI] [PubMed] [Google Scholar]
- 45.Goldstein JL, Brown MS. 1990. Regulation of the mevalonate pathway. Nature 343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 46.Burg JS, Espenshade PJ. 2011. Regulation of HMG-CoA reductase in mammals and yeast. Prog Lipid Res 50:403–410. doi: 10.1016/j.plipres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma SM, Garcia DE, Redding-Johanson AM, Friedland GD, Chan R, Batth TS, Haliburton JR, Chivian D, Keasling JD, Petzold CJ, Lee TS, Chhabra SR. 2011. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab Eng 13:588–597. doi: 10.1016/j.ymben.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Kramer R, Abraham W-R. 2011. Volatile sesquiterpenes from fungi: what are they good for? Phytochem Rev 11:15–37. doi: 10.1007/s11101-011-9216-2. [DOI] [Google Scholar]
- 49.Fraga BM. 2013. Natural sesquiterpenoids. Nat Prod Rep 30:1226–1264. doi: 10.1039/c3np70047j. [DOI] [PubMed] [Google Scholar]
- 50.Liermann JC, Schuffler A, Wollinsky B, Birnbacher J, Kolshorn H, Anke T, Opatz T. 2010. Hirsutane-type sesquiterpenes with uncommon modifications from three basidiomycetes. J Org Chem 75:2955–2961. doi: 10.1021/jo100202b. [DOI] [PubMed] [Google Scholar]
- 51.Hall C, Dietrich FS. 2007. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177:2293–2307. doi: 10.1534/genetics.107.074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Sena Filho JG, Quin MB, Spakowicz DJ, Shaw JJ, Kucera K, Dunican B, Strobel SA, Schmidt-Dannert C. 2016. Genome of Diaporthe sp. provides insights into the potential inter-phylum transfer of a fungal sesquiterpenoid biosynthetic pathway. Fungal Biol 120:1050–1063. doi: 10.1016/j.funbio.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow J-Y, Tian B-X, Ramamoorthy G, Hillerich BS, Seidel RD, Almo SC, Jacobson MP, Poulter CD. 2015. Computational-guided discovery and characterization of a sesquiterpene synthase from Streptomyces clavuligerus. Proc Natl Acad Sci U S A 112:5661–5666. doi: 10.1073/pnas.1505127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slot JC. 2017. Fungal gene cluster diversity and evolution. Adv Genet 100:141–178. doi: 10.1016/bs.adgen.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Zhao J, Bao X, Li C, Shen Y, Hou J. 2016. Improving monoterpene geraniol production through geranyl diphosphate synthesis regulation in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 100:4561–4571. doi: 10.1007/s00253-016-7375-1. [DOI] [PubMed] [Google Scholar]
- 56.Sarria S, Wong B, Garcia Martin H, Keasling JD, Peralta-Yahya P. 2014. Microbial synthesis of pinene. ACS Synth Biol 3:466–475. doi: 10.1021/sb4001382. [DOI] [PubMed] [Google Scholar]
- 57.Ignea C, Pontini M, Maffei ME, Makris AM, Kampranis SC. 2014. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth Biol 3:298–306. doi: 10.1021/sb400115e. [DOI] [PubMed] [Google Scholar]
- 58.Zhou YJ, Gao W, Rong Q, Jin G, Chu H, Liu W, Yang W, Zhu Z, Li G, Zhu G, Huang L, Zhao ZK. 2012. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc 134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 59.Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH. 2011. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol 77:1033–1040. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, Nagegowda DA, Rawat R, Bouvier-Nave P, Guo D, Bach TJ, Chye M-L. 2012. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol J 10:31–42. doi: 10.1111/j.1467-7652.2011.00631.x. [DOI] [PubMed] [Google Scholar]
- 61.Nagegowda DA, Bach TJ, Chye ML. 2004. Brassica juncea 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase 1: expression and characterization of recombinant wild-type and mutant enzymes. Biochem J 383:517–527. doi: 10.1042/BJ20040721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He RL, Zhang C, Guo W, Wang LX, Zhang DY, Chen SL. 2013. Construction of a plasmid for heterologous protein expression with a constitutive promoter in Trichoderma reesei. Plasmid 70:425–429. doi: 10.1016/j.plasmid.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Kistler HC, Broz K. 2015. Cellular compartmentalization of secondary metabolism. Front Microbiol 6:68. doi: 10.3389/fmicb.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim FY, Keller NP. 2014. Spatial and temporal control of fungal natural product synthesis. Nat Prod Rep 31:1277–1286. doi: 10.1039/C4NP00083H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roze LV, Chanda A, Linz JE. 2011. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Gen Biol 48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin JF, Ullan RV, Garcia-Estrada C. 2012. Role of peroxisomes in the biosynthesis and secretion of beta-lactams and other secondary metabolites. J Ind Microbiol Biotechnol 39:367–382. doi: 10.1007/s10295-011-1063-z. [DOI] [PubMed] [Google Scholar]
- 67.Menke J, Weber J, Broz K, Kistler HC. 2013. Cellular development associated with induced mycotoxin synthesis in the filamentous fungus Fusarium graminearum. PLoS One 8:e63077. doi: 10.1371/journal.pone.0063077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamane M, Minami A, Liu C, Ozaki T, Takeuchi I, Tsukagoshi T, Tokiwano T, Gomi K, Oikawa H. 2017. Biosynthetic machinery of diterpene pleuromutilin isolated from basidiomycete fungi. Chembiochem 18:2317–2322. doi: 10.1002/cbic.201700434. [DOI] [PubMed] [Google Scholar]
- 69.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Joulain D, Konig WA. 1998. The atlas of spectral data of sesquiterpene hydrocarbons. E. B. Verlag, Hamburg, Germany. [Google Scholar]
- 72.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 73.Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY, Verrips CT, Vindelov J, Pronk JT. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26:706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 75.Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 76.Lum PY, Edwards S, Wright R. 1996. Molecular, functional and evolutionary characterization of the gene encoding HMG-CoA reductase in the fission yeast, Schizosaccharomyces pombe. Yeast 12:1107–1124. doi:. [DOI] [PubMed] [Google Scholar]
- 77.Huxley C, Green ED, Dunham I. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6:236. doi: 10.1016/0168-9525(90)90190-H. [DOI] [PubMed] [Google Scholar]
- 78.Martin VJ, Yoshikuni Y, Keasling JD. 2001. The in vivo synthesis of plant sesquiterpenes by Escherichia coli. Biotechnol Bioeng 75:497–503. doi: 10.1002/bit.10037. [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 83.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 84.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (Oxford, England) 31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pojer F, Ferrer JL, Richard SB, Nagegowda DA, Chye ML, Bach TJ, Noel JP. 2006. Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases. Proc Natl Acad Sci U S A 103:11491–11496. doi: 10.1073/pnas.0604935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campobasso N, Patel M, Wilding IE, Kallender H, Rosenberg M, Gwynn MN. 2004. Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase–crystal structure and mechanism. J Biol Chem 279:44883–44888. doi: 10.1074/jbc.M407882200. [DOI] [PubMed] [Google Scholar]
- 87.Steussy CN, Vartia AA, Burgner JW Jr, Sutherlin A, Rodwell VW, Stauffacher CV. 2005. X-ray crystal structures of HMG-CoA synthase from Enterococcus faecalis and a complex with its second substrate/inhibitor acetoacetyl-CoA. Biochemistry 44:14256–14267. doi: 10.1021/bi051487x. [DOI] [PubMed] [Google Scholar]
- 88.Ruiz-Albert J, Cerda-Olmedo E, Corrochano LM. 2002. Genes for mevalonate biosynthesis in Phycomyces. Mol Genet Genomics 266:768–777. doi: 10.1007/s004380100565. [DOI] [PubMed] [Google Scholar]
- 89.Katayama S, Adachi N, Takao K, Nakagawa T, Matsuda H, Kawamukai M. 1995. Molecular cloning and sequencing of the hcs gene, which encodes 3-hydroxy-3-methylglutaryl coenzyme A synthase of Schizosaccharomyces pombe. Yeast 11:1533–1537. doi: 10.1002/yea.320111509. [DOI] [PubMed] [Google Scholar]
- 90.Engels B, Heinig U, Grothe T, Stadler M, Jennewein S. 2011. Cloning and characterization of an Armillaria gallica cDNA encoding protoilludene synthase, which catalyzes the first committed step in the synthesis of antimicrobial melleolides. J Biol Chem 286:6871–6878. doi: 10.1074/jbc.M110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCormick SP, Alexander NJ, Harris LJ. 2010. CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl Environ Microbiol 76:136–141. doi: 10.1128/AEM.02017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hohn TM, Beremand PD. 1989. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79:131–138. doi: 10.1016/0378-1119(89)90098-X. [DOI] [PubMed] [Google Scholar]
- 93.Brock NL, Huss K, Tudzynski B, Dickschat JS. 2013. Genetic dissection of sesquiterpene biosynthesis by Fusarium fujikuroi. Chembiochem 14:311–315. doi: 10.1002/cbic.201200695. [DOI] [PubMed] [Google Scholar]
- 94.Shishova EY, Costanzo LD, Cane DE, Christianson DW. 2007. X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry 46:1941–1951. doi: 10.1021/bi0622524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Caruthers JM, Kang I, Rynkiewicz MJ, Cane DE, Christianson DW. 2000. Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti. J Biol Chem 275:25533–25539. doi: 10.1074/jbc.M000433200. [DOI] [PubMed] [Google Scholar]
- 96.Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgant G, Collado IG, Cane DE, Viaud M. 2008. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem Biol 3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cane DE, Sohng JK, Lamberson CR, Rudnicki SM, Wu Z, Lloyd MD, Oliver JS, Hubbard BR. 1994. Pentalenene synthase. Purification, molecular cloning, sequencing, and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry 33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.