ABSTRACT
Microbial degradation of 2-hydroxypyridine usually results in the formation of a blue pigment (nicotine blue). In contrast, the Burkholderia sp. strain MAK1 bacterium utilizes 2-hydroxypyridine without the accumulation of nicotine blue. This scarcely investigated degradation pathway presumably employs 2-hydroxypyridine 5-monooxygenase, an elusive enzyme that has been hypothesized but has yet to be identified or characterized. The isolation of the mutant strain Burkholderia sp. MAK1 ΔP5 that is unable to utilize 2-hydroxypyridine has led to the identification of a gene cluster (designated hpd) which is responsible for the degradation of 2-hydroxypyridine. The activity of 2-hydroxypyridine 5-monooxygenase has been assigned to a soluble diiron monooxygenase (SDIMO) encoded by a five-gene cluster (hpdA, hpdB, hpdC, hpdD, and hpdE). A 4.5-kb DNA fragment containing all five genes has been successfully expressed in Burkholderia sp. MAK1 ΔP5 cells. We have proved that the recombinant HpdABCDE protein catalyzes the enzymatic turnover of 2-hydroxypyridine to 2,5-dihydroxypyridine. Moreover, we have confirmed that emerging 2,5-dihydroxypyridine is a substrate for HpdF, an enzyme similar to 2,5-dihydroxypyridine 5,6-dioxygenases that are involved in the catabolic pathways of nicotine and nicotinic acid. The proteins and genes identified in this study have allowed the identification of a novel degradation pathway of 2-hydroxypyridine. Our results provide a better understanding of the biodegradation of pyridine derivatives in nature. Also, the discovered 2-hydroxypyridine 5-monooxygenase may be an attractive catalyst for the regioselective synthesis of various N-heterocyclic compounds.
IMPORTANCE The degradation pathway of 2-hydroxypyridine without the accumulation of a blue pigment is relatively unexplored, as, to our knowledge, no genetic data related to this process have ever been presented. In this paper, we describe genes and enzymes involved in this little-studied catabolic pathway. This work provides new insights into the metabolism of 2-hydroxypyridine in nature. A broad-range substrate specificity of 2-hydroxypyridine 5-monooxygenase, a key enzyme in the degradation, makes this biocatalyst attractive for the regioselective hydroxylation of pyridine derivatives.
KEYWORDS: 2-hydroxypyridine; 2-hydroxypyridine 5-monooxygenase; Burkholderia sp. MAK1; 2,5-dihydroxypyridine; 2,5-dihydroxypyridine 5,6-dioxygenase
INTRODUCTION
Pyridine and its derivatives are among the most abundant N-heterocyclic compounds in nature (1), including alkaloids, such as nicotine and mimosine, and many derivatives of nicotinic and picolinic acids (2). Pyridine and its derivatives are also produced in large quantities by the chemical industry as part of energy generation (e.g., from coal or shale oil production), pharmaceutical manufacture (e.g., isoniazid, cetylpyridinium chloride, and coramine), and agricultural activities (the herbicides diquat, paraquat, picloram, and fluridone) (3). The wastewater generated during the aforementioned processes contains pyridine and pyridinecarboxylic acids, as well as amino-, chloro-, alkyl-, and hydroxypyridines (4). The heterocyclic nature of pyridine derivatives increases their solubility; therefore, they are easily transported through soil and contaminate groundwater (5). Due to their prevalence, toxicity, and mutagenic potential, pyridines are considered hazardous pollutants (6).
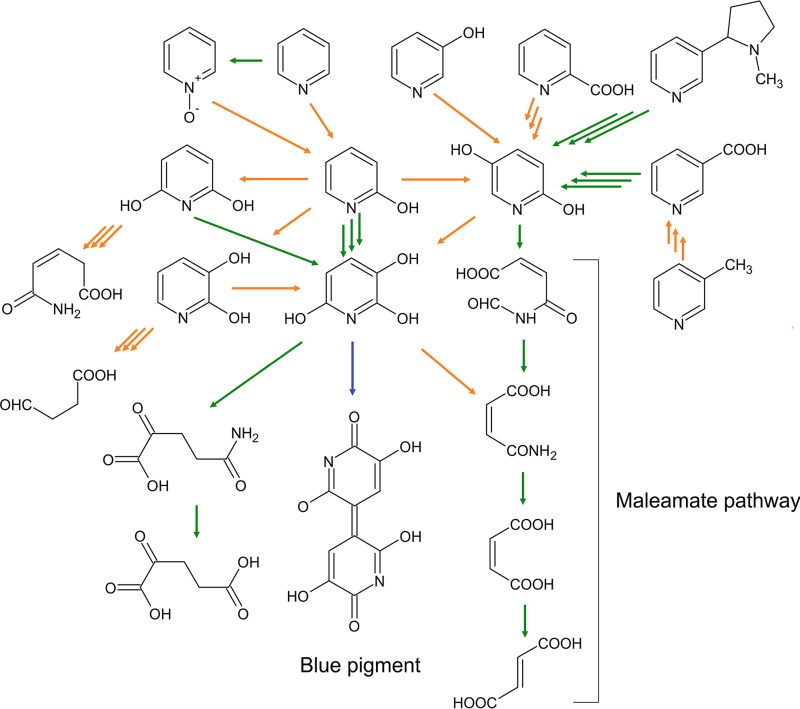
The microbial degradation of pyridine compounds usually involves the formation of hydroxylated intermediates (Fig. 1). Nevertheless, for decades, it was assumed that the biodegradation of pyridine did not include initial hydroxylation (1). However, more recent results oppose this theory. The data obtained by gas chromatography-mass spectrometry (GC-MS) methods suggest that Arthrobacter sp. strain KM-4 consumes pyridine via the formation of 2-hydroxypyridine (2HP) and 2,3-dihidroxypyridine (23DHP) (7). Likewise, MS analysis has shown the formation of dihydroxypyridines as metabolites of the pyridine degradation pathway in Rhodococcus sp. strain Chr9 (8), although no enzymes involved in these conversions have been identified thus far. In the case of phenol-degrading bacteria, the conversion of pyridine to pyridine-N-oxide (PNO) is catalyzed by the phenol monooxygenase (9). In Nocardia spp., the degradation of PNO begins with an enzymatic transformation of PNO to 2HP before undergoing further catabolism (10). Various forms of hydroxypyridines (2HP, PNO, 3-hydroxypyridine, and 4-hydroxypyridine) have been detected in urine samples of animals dosed with pyridine, suggesting that hydroxylation of pyridine is ubiquitous in nature (11). The most common hydroxylated intermediate in the degradation of pyridine derivatives appears to be 2,5-dihydroxypyridine (25DHP) (2). In the nicotinic acid degradation pathway, the formation of 25DHP from 6-hydroxynicotinic acid is catalyzed by the monooxygenase NicC (12). Two studies describe the formation of 25DHP from 6-hydroxy-3-succinoylpyridine catalyzed by monooxygenases HspB (13) and VppD (14). The occurrence of 25DHP has also been detected during the catabolism of picolinic acid (1) or 3-hydroxypyridine (15). However, the enzymes responsible for these biotransformations have not been identified so far. The best-studied 25DHP degradation pathway involves ring opening by 25DHP 5,6-dioxygenase, followed by the three-step catalytic cascade (maleamate pathway) leading to the formation of fumarate (16).
FIG 1.
Proposed degradation pathways of pyridine derivatives in bacteria through the formation of hydroxypyridines. Green arrows represent pathways or reactions for which appropriate genes and/or enzymes have been identified. Orange arrows show proposed pathways or reactions. The blue arrow represents spontaneous autooxidation. Triple arrows indicate more than one reaction.
While quite a number of different bacteria (Arthrobacter, Rhodococcus, Achromobacter, and Nocardia spp.) have been found to degrade 2HP (1, 15, 17), neither enzymes nor genes implicated in this process have been extensively studied to date. The most researched degradation pathways of 2HP involve the formation of 2,3,6-trihydroxypyridine (THP). The THP intermediate autooxidizes spontaneously to form a blue pigment, 4,5,4′,5′-tetrahydroxy-3,3′-diazadiphenoquinone-(2,2′) (18, 19). It has been proposed that THP originates through the oxidation of 2,5-dihydroxypyridine (25DHP), 2,6-dihydroxypyridine (26DHP), or 23DHP. However, the enzymes involved in these conversions are mostly unknown, and only the 26DHP 3-hydroxylase from Arthrobacter nicotinovorans (20) and HpyB monooxygenase from Arthrobacter sp. strain PY22 (21) have been identified to date. Most recently, it has been demonstrated that Rhodococcus sp. strain PY11 encodes a four-component dioxygenase, HpoBCDF, that transforms 2HP to 3,6-dihydroxy-1,2,3,6-tetrahydropyridin-2-one, which is further oxidized by HpoE to THP (22). Also, 2HP degradation without the formation of a blue pigment was observed. In three Achromobacter species, the degradation of 2HP resulted in an accumulation of maleamate, formic acid, ammonium, maleate, and fumarate, indicating the involvement of maleamate degradation pathway (15). The cell extracts of these bacteria also demonstrate 25DHP 5,6-dioxygenase activity, suggesting 25DHP to be a putative intermediate. However, the first step of 2HP degradation in this pathway remains enigmatic, since although the involvement of a putative 2HP 5-monooxygenase has been implied, neither the enzyme itself nor its gene have been identified/characterized thus far.
In our previous work (23), we demonstrated that Burkholderia sp. strain MAK1 cells pregrown in the presence of 2HP are able to hydroxylate various pyridine and pyrazine derivatives regioselectively. In this study, we present data on the genes and enzymes involved in the degradation of 2HP in Burkholderia sp. MAK1, a strain that uses 2HP as the sole source of carbon and energy without the formation of a blue pigment. The identified gene cluster hpd contains genes required for 2HP assimilation. The first steps of 2HP degradation in Burkholderia sp. MAK1 have been elucidated both by analysis of reaction products and identification of the enzymes responsible for these conversions.
RESULTS AND DISCUSSION
Identification of the gene cluster responsible for the catabolism of 2HP in Burkholderia sp. MAK1.
The ability of the 2HP-induced Burkholderia sp. MAK1 cells to hydroxylate pyridines at the C-5 position suggested that 2HP was degraded via the formation of 25DHP. Later, we noticed that 2HP-induced Burkholderia sp. MAK1 cells turned deep blue when grown on agar plates with indole, whereas the color of uninduced cells remained unchanged. It is well known that the enzymatic synthesis of indigo dye from indole involves various types of mono- or dioxygenases (24). Therefore, we assumed that the degradation of 2HP by Burkholderia sp. MAK1 begins with the formation of 25DHP, catalyzed by a 2HP-inducible monooxygenase, which is also able to hydroxylate indole. Following this hypothesis, a mutant of Burkholderia sp. MAK1 in which the appropriate gene was disrupted would neither turn blue in the presence of indole nor be able to grow with 2HP as the sole carbon and energy source.
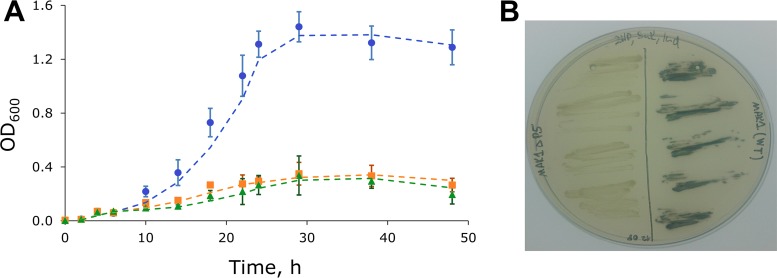
To generate random mutants, the competent cells of Burkholderia sp. MAK1 were transformed with the plasposon pTnMod-OKm′ (25) by electroporation and then plated on EFA medium[10.0 g/liter K2HPO4, 4.0 g/liter KH2PO4, 0.5 g/liter yeast extract, 1.0 g/liter (NH4)2SO4, 0.2 g/liter MgSO4·7H2O (pH 7.0)] containing 2HP (as an inducer), succinate (Suc; as a carbon source), kanamycin (Km), and indole (Ind). The first screening step involved the identification of the white colony-forming mutants among the blue-forming ones. Then, such mutants were transferred onto another 2HP-Suc-Km-Ind agar plate to ensure that the cells were kanamycin resistant and remained white. To test their inability to consume 2HP, the mutant cells were transferred onto agar plates containing 2HP as the sole source of carbon and energy. In this manner, we were able to select a proper mutant, designated Burkholderia sp. MAK1 ΔP5, with no capacity to synthesize indigo dye (in the presence of indole and 2HP) or to degrade 2HP (Fig. 2).
FIG 2.
Growth kinetics and indigo dye production of wild-type Burkholderia sp. MAK1 and MAK1 ΔP5 mutant. (A) Blue circles, growth of wild-type bacterium in EFA medium containing 2HP (1.0 g/liter); orange squares, MAK1 ΔP5 mutant in EFA medium containing 2HP (1.0 g/liter); green triangles, wild-type bacterium in EFA medium without the carbon source. Data points are averages of the results from three experiments, and error bars show the standard deviation. Dashed lines represent trendlines using moving average data approximation (period = 2). (B) Burkholderia sp. MAK1 ΔP5 bacterium (on the left) and wild-type Burkholderia sp. MAK1 (on the right) grown on EFA medium supplemented with 2HP, succinate, and indole.
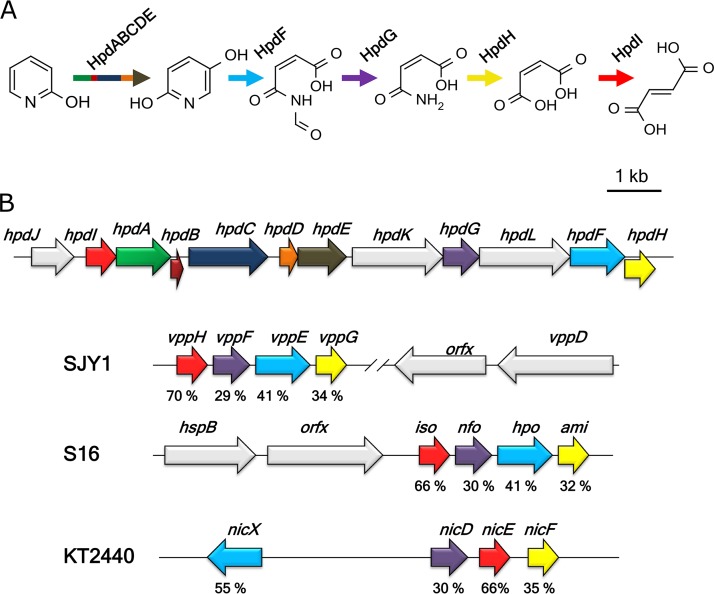
To retrieve the transposon insertion site, the chromosomic DNA of the Burkholderia sp. MAK1 ΔP5 was digested with one of the restriction enzymes (SalI or DraI), both of which had recognition sites flanking a kanamycin resistance cassette, and was self-ligated. Thus, the recombinant circular DNA molecules containing a pMB1 ori sequence were produced, so that they could function as the plasmids in Escherichia coli and could be used for the transformation and screening. The insertion site flanking sequences (plasmids pSal_4 [5,714 bp] and pDra_1 [8,249 bp]) were determined applying a primer walking strategy. Merging these sequences resulted in a continuous DNA fragment. Then, the derived sequence was compared to that obtained from whole-genome sequencing. Notably, the analysis of the whole-genome sequencing data alone did not allow for the identification of 2HP assimilation genes. However, by combining the data from the analysis of kanamycin resistance cassette-flanking regions and whole-genome sequencing, the gene cluster (designated hpd) responsible for the degradation of 2HP in Burkholderia sp. MAK1 (Fig. 3B) was identified.
FIG 3.
(A) Proposed 2-hydroxypyridine degradation pathway in Burkholderia sp. MAK1. (B) Organization of the hpd gene cluster. hpdABCDE, 2HP 5-monooxygenase (green [hpdA], dark red [hpdB], dark blue [hpdC], orange [hpdD], and brown [hpdE] arrows); hpdF, 25DHP 5,6-dioxygenase (cyan); hpdG, N-formylmaleamate deformylase (purple); hpdH, maleamate amidase (yellow); hpdI, maleate isomerase (red). hpdL (chaperone), hpdJ (putative regulator), hpdK (permease) and other genes are presented as gray arrows. Homologous genes are colored the same as in well-characterized pyridine derivative-degrading bacteria (Ochrobactrum sp. SJY1, Pseudomonas putida S16, and Pseudomonas putida KT2440). Identities (%) of amino acid sequence between hpd proteins and their homologues are indicated under the corresponding ORFs.
Bioinformatic analysis of the hpd gene cluster.
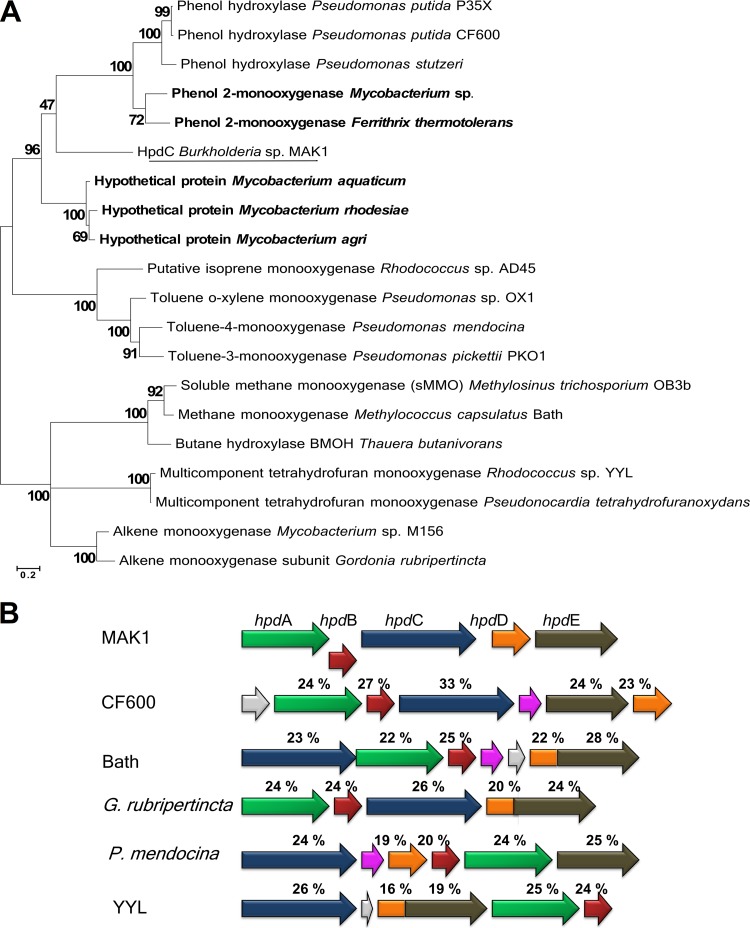
The hpd gene cluster is a 13-kb-long DNA fragment containing 12 open reading frames (Table 1). Four genes show high sequence similarity to those encoding the catabolism of 25DHP (25DHP 5,6-dioxygenase, N-formylmaleamate deformylase, maleamate amidase, and maleate isomerase) in nicotine- and nicotinate-degrading bacteria (12–14). This indicates that in Burkholderia sp. MAK1, 2HP is catabolized via the formation of 25DHP and through the maleamate pathway (Fig. 3). Also, the presence of the aforementioned genes supports the hypothesis that 2HP is being directly hydroxylated to form 25DHP. The maleamate pathway begins with the dioxygenolysis of 25DHP to N-formylmaleamic acid, catalyzed by 25DHP 5,6-dioxygenase (12). HpdF of Burkholderia sp. MAK1 shows a significant similarity to the well-characterized dioxygenases, such as NicX (55%) from Pseudomonas putida KT2440, Hpo (41%) from Pseudomonas putida S16, and VppE (41%) from Ochrobactrum sp. strain SJY1, suggesting that it likely is a 25DHP 5,6-dioxygenase. In addition, HpdF possesses the canonical 2-His-1-carboxylate (H265, H316, and D318) metal-binding motif found in the classical extradiol ring cleavage dioxygenases. The hydrolysis of N-formylmaleamic acid to maleamic and formic acids is catalyzed by N-formylmaleamate deformylase, a member of α/β-hydrolase-fold superfamily. HpdG from Burkholderia sp. MAK1 has homology to the enzymes from the α/β-hydrolase-fold superfamily and shows 30% similarity to the well-studied N-formylmaleamate deformylases Nfo (13) and NicD (12). The next step in the biodegradation pathway is the conversion of maleamate to maleic acid catalyzed by maleamate amidase, a member of the cysteine hydrolase superfamily. In the hpd gene cluster, the corresponding enzyme has been found to be encoded by hpdH. The degradation pathway of 2HP ends with the isomerization of maleic acid to fumaric acid, a Krebs cycle intermediate. In the case of Burkholderia sp. MAK1, the isomerization activity has been assigned to HpdI, as it shows a significant homology to the maleate cis/trans-isomerase, an enzyme belonging to the Asp/Glu racemase superfamily. Three genes (hpdJ, hpdL, and hpdK) from the hpd locus appear to be indirectly involved in the biodegradation of 2HP and have been associated with transcription regulation, protein folding, and transport. Based on the results of bioinformatics analysis, the hypothetical 2HP 5-monooxygenase of Burkholderia sp. MAK1 is a soluble diiron monooxygenase (SDIMO) encoded by a five-gene locus (hpdA, hpaB, hpdC, hpdD, and hpdE). The family of SDIMOs can be divided into the following five major groups: phenol hydroxylases, soluble methane monooxygenases, four-component alkene/aromatic monooxygenases, alkane/alkene monooxygenases, and tetrahydrofuran monooxygenases (26). The BLAST analysis revealed similarity between hpdA and toluene monooxygenase β-subunit. In the case of the gene product of hpdB, however, homology searches returned only a few low-similarity hits, one of which was a putative phenol hydroxylase. Based on a Pfam search, HpdB of Burkholderia sp. MAK1 has been predicted to belong to the Mmob/DmpM family, which mostly consists of methane monooxygenase regulatory proteins. The protein encoded by hpdC shows similarity to the toluene monooxygenase α-subunit, which harbors a carboxylate-bridged diiron center at the active site. According to the literature, SDIMOs form a dimeric (bi- or tri-component) hydroxylase complex, α2β2γ2 or α2β2 (27). Based on the results of bioinformatics analysis, hpdC and hpdA of Burkholderia sp. MAK1 code for the α- and β-subunits, respectively, whereas the hpdB-encoded protein possesses regulatory function. Most SDIMOs contain a sulfur-iron cluster and flavin adenine dinucleotide (FAD)-dependent oxidoreductase that shuttles electrons from NADH to a catalytic complex, as well as a small Rieske protein that mediates electron transfer between oxidoreductase and catalytic complex. In the case of the hpd gene cluster, these functions have been assigned to hpdE and hpdD, respectively. The multiple-amino-acid sequence alignment of HpdC with various well-studied α-subunits of SDIMOs revealed that these proteins share a set of conservative carboxylate-bridged diiron center-forming amino acids (E120, E150, H153, E211, E245, and H248), an observation that has been supported by the tertiary structure model of HpdC (see Fig. S1 in the supplemental material). Despite these similarities, the phylogenetic analysis revealed that HpdC likely differs from any known SDIMOs, as it appears to occupy a distinct branch in the phylogenetic tree (see Fig. 4A). Notably, homology searches with the HpdC amino acid sequence have returned only low-similarity hits. The closest matches (which all were putative proteins from Gram-positive bacteria) shared 41% amino acid (aa) sequence identity with HpdC, while the similarity between HpdC and other characterized homologous proteins was <35%. The remaining Burkholderia sp. MAK1 monooxygenase genes, hpdA, hpdB, hpdD, and hpdE, show even lower similarity to their characterized counterparts (Fig. 4B). Also, as seen in Fig. 4B, the arrangement of the Burkholderia sp. MAK1 hpdABCDE gene cluster differs profoundly from that of bacteria representing other SDIMO groups. Taken together, all these findings illustrate how unusual and unique is the hpdABCDE gene cluster in Burkholderia sp. MAK1.
TABLE 1.
Functional annotation of hypothetical proteins encoded in hpd gene cluster
| Protein | Size (amino acids) | Putative function | Superfamily designation information |
|
|---|---|---|---|---|
| Specific hit/conserved domain | Accession no. | |||
| HpdJ | 345 | Transcription regulator | HTH_AraC | cl26290 |
| HpdI | 233 | Maleate isomerase | Asp_Glu_race | cl00518 |
| HpdA | 348 | 2HP 5-monooxygenase subunit | Ferritin-like | cl00264 |
| HpdB | 105 | 2HP 5-monooxygenase regulatory subunit | MmoB/DmpM | PF02406 |
| HpdC | 491 | 2HP 5-monooxygenase catalytic subunit | Ferritin_like | cl00264 |
| HpdD | 118 | Ferredoxin | Thioredoxin_like | cl00388 |
| HpdE | 404 | NADH-ubiquinone oxidoreductase | NADH_4Fe-4S | cl26507 |
| HpdK | 555 | Permease | SLC5-6-like_sbd | cl00456 |
| HpdG | 285 | N-Formylmaleamate deformylase | Abhydrolase_1 | cl26327 |
| HpdL | 535 | Chaperone | Chaperonin_like | cl02777 |
| HpdF | 348 | 25DHP 5,6-dioxygenase | Peptidase_M29 | cl19596 |
| HpdH | 307 | Maleamate amidase | Cysteine_hydrolases | cl00220 |
FIG 4.
Phylogenetic analysis of hpdABCDE genes. (A) The phylogenetic tree of SDIMO α-subunits. The tree was generated using the maximum likelihood method with 500 bootstrap iterations. The scale bar represents 0.20 substitutions per site. The five closest BLAST hits of HpdC are given in bold. (B) Structural alignment between representatives of all five SDIMO groups and the hpdABCDE cluster. Identities (%) of amino acid sequence between hpdABCDE and homologous proteins are indicated under the corresponding ORFs. Blue arrows, α-subunits; green arrows, β-subunits; purple arrows, γ-subunits; dark red arrows, regulatory subunits; orange arrows, ferredoxins; brown arrows, NADH-ubiquinone oxidoreductases; gray arrows, putative or unknown-function genes.
The bioinformatics analysis of the hpd gene cluster revealed homologues of the characterized enzymes involved in the catabolism of 25DHP. In Burkholderia sp. MAK1, the hypothetical 2HP 5-monooxygenase is encoded by the hpdABCDE gene cluster and is only distantly related to SDIMOs from other bacteria. SDIMOs are usually found in xenobiotic-degrading microorganisms, where they usually catalyze the first steps of the degradation of xenobiotics by hydroxylating the substrate (26, 27). To date, a number of SDIMOs capable of hydroxylating pyridine derivatives (8, 9) have been described in the literature. However, to our knowledge, no SDIMO enzymes directly involved in the biodegradation of pyridine and its derivatives have been reported thus far.
qPCR analysis of the hpdABCDE gene cluster.
To confirm that the identified hpdABCDE genes were indeed expressed in response to 2HP, quantitative PCR (qPCR) experiments were performed using total RNA extracted from the 2HP-induced cells of MAK1, as described in Materials and Methods. The reverse transcription-PCR (RT-PCR) produced PCR products of the expected size (Fig. 5A). A comparison of hpdABCDE mRNA synthesis levels between 2HP-grown Burkholderia sp. MAK1 cells (induced conditions) and succinate-grown cells (no induction) was made. The results revealed that the transcription of hpdABCDE genes increased 100-fold in the presence of 2HP (Fig. 5B); hence, HpdABCDE synthesis was shown to be specifically induced by growth on 2HP.
FIG 5.
Qualitative (A) and quantitative (B) RT-PCR analysis of the expression of hpdABCDE genes. Strain MAK1 was cultivated in liquid EFA medium supplemented with either 1.0 g/liter 2 HP (induced conditions) or 1.0 g/liter succinate (Suc; no induction) as a single source of carbon. Primers were designed to amplify the regions in hpdC, hpdD, and hpdE genes. The data are presented as relative RNA amounts calculated from the threshold cycles using the threshold cycle of 16S RNA as a reference. Bars show the increase in the expression of the hpdC, hpdD, and hpdE genes under 2HP induction compared to growth without 2HP (on succinate). The averages of three independent runs are presented.
Cloning and expression of hpdABCDE and hpdF genes in E. coli.
Since the genes and enzymes implicated in the maleamate pathway are well described (12–14), we have concentrated our investigation on the apparent 2HP hydroxylation to 25DHP, presumably catalyzed by HpdABCDE, and the following ring opening of 25DHP potentially driven by HpdF. The overexpression of HpdF was successfully achieved by transforming E. coli BL21(DE3) or Rosetta(DE3)pLysS cells with the plasmid HpdF_pET. SDS-PAGE analysis revealed that the recombinant HpdF (theoretical mass, 38 kDa) was expressed as a soluble protein (Fig. S2A), and the cells harbored an active 25DHP dioxygenase (specific activity, 0.54 ± 0.1 nmol · min−1 · mg−1 of cell [wet weight]). To obtain a 2HP hydroxylase, the hpdABCDE cluster was cloned as a continuous DNA fragment into the pET-28b plasmid vector. The resulting recombinant plasmid was then used to transform E. coli BL21(DE3) and Rosetta(DE3)pLysS cells. The expression of the recombinant proteins was observed even though the proteins were found mainly in an insoluble fraction (Fig. S2B), and no enzyme activity was detected. By lowering the cultivation temperature to 16°C, the fraction of a soluble protein was increased, although the monooxygenase activity remained undetected.
Identification of catalytic properties of HpdABCDE and HpdF proteins.
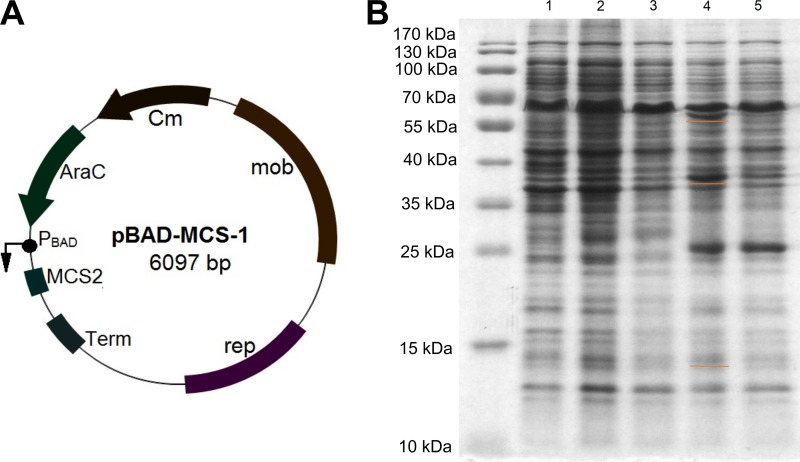
Since E. coli cells appeared to be an unsuitable host for the expression of HpdABCDE, Burkholderia sp. MAK1 ΔP5 cells were considered as an alternative. We tested pBBR1MCS plasmids (28) as suitable expression vectors and were satisfied with their transformation efficiency and stability in Burkholderia sp. MAK1 ΔP5 cells. However, the Plac promoter used in these vectors seemed to be inactive in this host, as no significant fluorescence was detected in bacteria harboring plasmid gfp_pBBR1MCS-1 containing the gfp gene downstream of the Plac promoter (data not shown). Hence, the arabinose-inducible PBAD promoter was inserted, since it was successfully used for protein expression in the Burkholderia genus (29). Plac promoter elements of pBBR1MCS-1 were cut out with the restriction endonucleases VspI and SacI and replaced with a PCR amplicon containing the PBAD promoter, araC regulator, and terminator sequences from the pBAD24 vector (30). Thus, a broad-host-range arabinose-inducible expression vector, pBAD-MCS-1, was generated (Fig. 6A). The activity of the PBAD promoter was successfully tested by cloning the gfp gene into the pBAD-MCS-1 (gfp_pBAD-MCS-1) vector and transforming the Burkholderia sp. MAK1 cells with this plasmid (data not shown).
FIG 6.
(A) Map of the plasmid vector pBAD-MCS-1. (B) SDS-PAGE analysis of HpdABCDE overexpression in Burkholderia sp. MAK1 ΔP5 cells. Lanes 1 to 3, free-cell extracts of Burkholderia sp. MAK1 grown on succinate (lane 1), and on 2HP (lane 2, total fraction; lane 3, soluble fraction). Lanes 4 and 5, extracts of Burkholderia sp. MAK1 ΔP5/HpdABCDE_pBAD-MCS-1 and supplemented with 0.2% arabinose (lane 4, total fraction; lane 5, soluble fraction). Orange underlines indicate recombinant proteins.
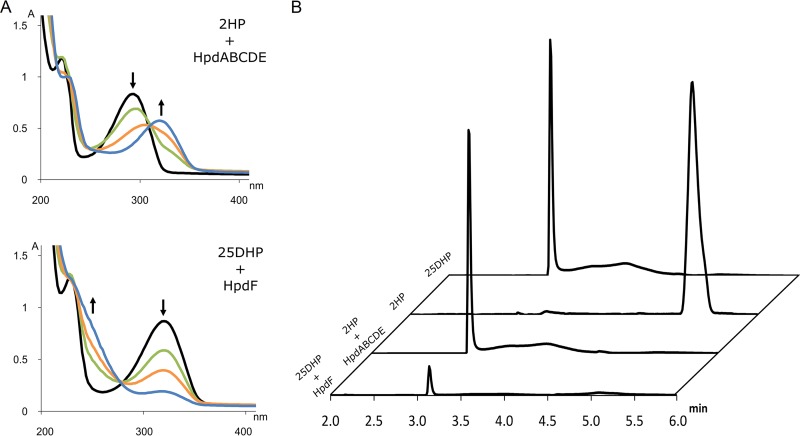
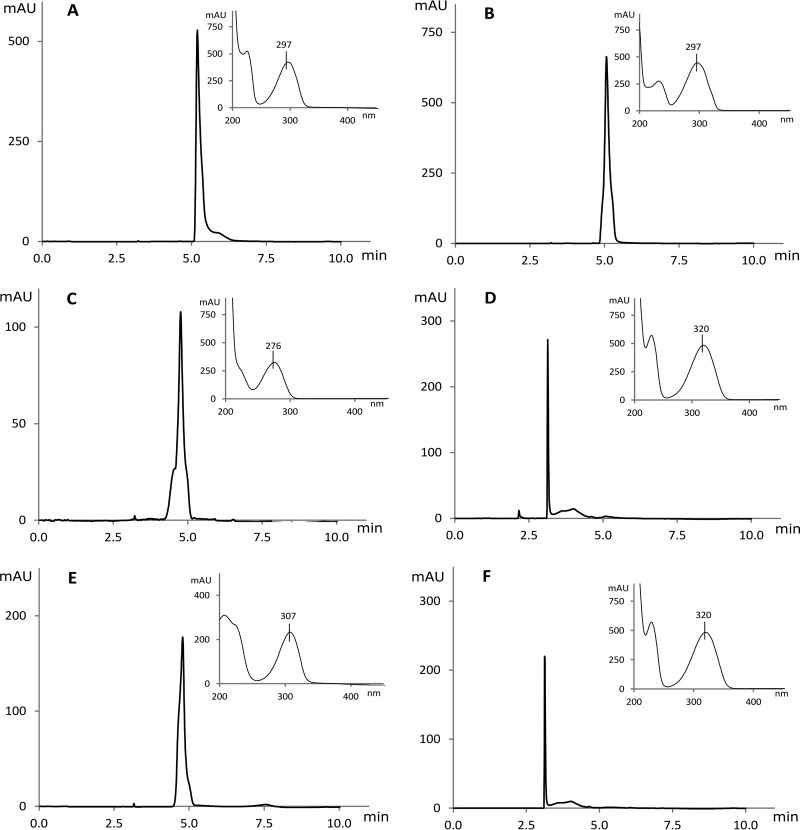
Encouraged by these results, we cloned the hpdABCDE cluster as a continuous DNA fragment into the pBAD-MCS1 vector (plasmid HpdABCDE_pBAD-MCS-1). Burkholderia sp. MAK1 ΔP5 cells transformed with this plasmid were grown in liquid EFA medium, and the expression of the recombinant proteins was induced with arabinose (final concentration, 0.2%). The SDS-PAGE analysis revealed the presence of the recombinant proteins in cell extracts (Fig. 6B), though a major portion of these proteins were synthesized as inclusion bodies. However, the most noticeable effect of the expression of HpdABCDE was observed on agar plates. When plated on medium containing succinate, 2HP, and arabinose, both the transformed cells and the medium turned bright green at first. Later, both turned deep brown and, finally, to black. In the case of Burkholderia sp. MAK1 ΔP5 cells plated on the same medium, as expected, no such change in color was observed. Since a freshly prepared aqueous solution of 25DHP exhibits the same color change during incubation under aerobic conditions, it was a strong indication that recombinant HpdABCDE catalyzes 2HP turnover to 25DHP. To confirm this, a more detailed analysis was carried out. First, the biomass of Burkholderia sp. MAK1 ΔP5 cells carrying the plasmid HpdABCDE_pBAD-MCS-1 was prepared, as described in Materials and Methods. Then, the intact cells were transferred to a reaction mixture containing 2HP, and the UV spectra were recorded at fixed time intervals. The time-dependent decline of 2HP absorbance at 290 nm and the formation of a new compound with an absorbance peak at 320 nm were observed (Fig. 7). Then, this conversion mixture was analyzed by high-performance liquid chromatography mass spectrometry (HPLC-MS), where a 2HP-specific peak (retention time, 5 min 12 s; m/z 96) disappeared, and a new one (retention time, 3 min 8 s; m/z 112) was detected. An increase in mass by 16 Da indicated a typical monooxygenase activity, i.e., the incorporation of one oxygen atom. To evaluate the regioselectivity of the hydroxylation, a chromatogram of the conversion product was compared with those of dihydroxypyridine standards (Fig. 8). The properties (retention time and absorbance spectrum) of the conversion product were found to exactly match those of 25DHP. These results confirmed that when Burkholderia sp. MAK1 ΔP5 cells were carrying a plasmid with hpdABCDE monooxygenase genes, they produced an active enzyme and efficiently hydroxylated 2HP to 25DHP. Although the enzyme activity of Burkholderia sp. MAK1 ΔP5 carrying HpdABCDE_pBAD-MCS-1 does not surpass that of the wild-type bacterium (0.75 ± 0.12 nmol · min−1 · mg−1 compared to 1.1 ± 0.15 nmol · min−1 · mg−1 of cell [wet weight], respectively), such a system offers a desirable advantage, the production of detectable hydroxylated intermediates. It should be noted that the formation of 25DHP has never been tracked in the wild-type Burkholderia sp. MAK1 bacterium. As 25DHP was readily used up by heterologously expressed HpdF (Fig. 7), we speculated that under 2HP-induced growth conditions, Burkholderia sp. MAK1 cells expressed genome-encoded HpdF, which reduced 25DHP to an imperceptible concentration. The HpdF-catalyzed dioxygenolysis would also explain why, during our previous work, a big portion of possible conversion products remained undetected. Notably, no such obstacle was encountered when HpdABCDE was expressed in Burkholderia sp. MAK1 ΔP5. Here, bacteria were grown without 2HP and, as a result, the expression of the genome-encoded HpdF was disabled. This not only led to the detection of 25DHP but also allowed us to perform HpdABCDE-catalyzed conversions (such as hydroxylation of 3-fluoropyridin-2-amine, 6-fluoropyridin-2-amine, 3-fluoropyridin-2-ol, 3-chloropyridin-2-ol, 3-bromopyridin-2-ol, 3-methylpyridin-2-ol, 6-methylpyridin-2-ol, and 2-hydroxy-6-methylpyridine-3-carbonitrile [Fig. S3]), all of which had been unsuccessfully attempted previously using 2HP-induced wild-type Burkholderia sp. MAK1. Therefore, the Burkholderia sp. MAK1 ΔP5/HpdABCDE_pBAD-MCS-1 system not only provided evidence of 2HP catabolism but also proved to be a superior biocatalyst, as opposed to the wild-type Burkholderia sp. MAK1.
FIG 7.
2HP and 25DHP biotransformation in vivo with recombinant cells [Burkholderia sp. MAK1 ΔP5/HpdABCDE_pBAD-MCS-1 and E. coli Rosetta(DE3)pLysS/HpdF_pET]. (A) The progress of the reaction was monitored using a UV-Vis spectrophotometer; initial spectra showed by black curves, after 1 h of incubation (green), after 2 h (orange), and after 3 h (blue). (B) The end products of biotransformation reaction were analyzed by HPLC and are presented as stacked HPLC chromatograms (310 nm).
FIG 8.
HPLC chromatograms (295 nm) and UV spectra of dihydroxypyridine standards (2-hydroxypyridine N-oxide [A], 2,3-dihydroxypyridine [B], 2,4-dihydroxypyridine [C], 2,5-dihydroxypyridine [D], 2,6-dihydroxypyridine [E]) and conversion product of 2HP (F) using Burkholderia sp. MAK1 ΔP5 whole cells carrying HpdABCDE_pBAD-MCS-1 plasmid. mAU, milli-absorbance units.
Diversity of pyridine derivative-attacking oxygenases.
Aromatic and aliphatic compounds (hydrocarbons) are devoid of functional groups and therefore exhibit a low chemical reactivity (31). Aerobic utilization of various hydrocarbons as organic growth substrates by microorganisms generally involves oxidizing enzymes (32). Pyridine derivatives, despite their heterocyclic nature, appear to follow this rule as well. Although the proposed pathways for the catabolism of pyridines predict the participation of oxygenases, only a limited number of the putative biocatalysts responsible for these conversions have been identified to date. These include the following nicotine and nicotinic acid degradation enzymes: 6-hydroxynicotinate 3-monooxygenase NicC from Pseudomonas putida KT2440 (12), 6-hydroxy-3-succinoylpyridine 3-monooxygenase HspB from Pseudomonas putida S16 (13), and another 6-hydroxy-3-succinoylpyridine 3-monooxygenase VppD from Ochrobactrum sp. SJY1 (14). The catabolism of 2HP or closely related compounds in different bacteria is started by the following structurally different enzymes: the four-component 2HP dioxygenase HpoBCDF from Rhodococcus sp. PY11 (22), the 26DHP 3-hydroxylase from Arthrobacter nicotinovorans (20), the two-component 2HP monooxygenase HpyB from Arthrobacter sp. PY22 (21), or the 2-hydroxypyridine 5-monooxygenase encoded by hpdABCDE from Burkholderia sp. MAK1, characterized in this work. Although only a small fraction of possible biocatalysts has been identified, it is clear that bacteria code for a variety of different oxygenases that initialize the degradation of pyridine derivatives. The structural diversity of oxygenases with the same specificity is not new to the microbial world. A good example is the aerobic degradation of toluene that can be initiated by at least five different pathways. Monooxygenase-driven hydroxylation may occur at all three positions on the benzene ring (ortho, meta, and para) as well as at the methyl group, while dioxygenases catalyze incorporation of both oxygen atoms releasing toluene-cis-1,2-dihydrodiol (33). All of these conversions are catalyzed by structurally and functionally different enzymes. The reason behind this phenomenon may be the abundance of the toluene sources. Toluene is found naturally in petroleum and coal. It is a major component of gasoline and is also produced by the burning of organic materials (31). Toluene is also produced industrially for use as a solvent and in the production of various chemicals (32). In addition, toluene is produced and emitted by plants (33). All of this makes toluene obtainable in different ecological niches that are occupied by diverse bacteria, which employ different enzymes to initiate the assimilation of toluene. The pyridine derivatives are also abundant in nature. Both natural and anthropogenic sources contribute to the presence of pyridines in the environment.
In the presence of a new growth substrate, bacteria may not necessarily adapt their own enzymatic repertoire via the duplications of corresponding genes and their subsequent mutations. In addition, bacteria constantly borrow genetic material by horizontal gene transfer (31). The hpd gene cluster is likely to have been recently horizontally transferred into Burkholderia MAK1 for the following reasons. First, all of the closest homologues of hpd cluster genes are not from the Burkholderia genus. Second, the first enzyme of the pathway, HpdABCDE monooxygenase, is very unusual among SDIMOs. It exhibits only a very low sequence identity (∼30%) with other known proteins and features a unique genetic architecture. Last, there is no experimental evidence of the direct involvement of SDIMOs in the biodegradation of pyridine derivatives. Most likely, the monooxygenase genes have been acquired by horizontal transfer and have been recruited by the appropriate lower pathway (maleamate pathway) for cleavage of hydroxylated products. We hypothesize that Burkholderia sp. MAK1 may have acquired either the entire hpd cluster or only the hpdABCDE genes from other bacteria, and the novel enzymes may have been recruited to serve in a preexisting maleamate pathway. Over time, these new catabolic genes may have been modified, allowing utilization of an alternative carbon source, such as 2HP. The results of HpdABCDE overexpression in Burkholderia sp. MAK1 ΔP5 may also be attributed to the “external origin” of the hpd cluster. It is possible that in the presence of 2HP, the wild-type bacteria produced a sufficient, albeit small, amount of enzymes needed to assimilate 2HP. When the expression of HpdABCDE was induced from a plasmid, the protein synthesis machinery of Burkholderia sp. MAK1 ΔP5 cells was not able to overcome the overproduction of “foreign” proteins, which became misfolded and ended up insoluble.
Overall, the discovery of HpdABCDE monooxygenase is a major contribution to our understanding of the initial steps of degradation of pyridine derivatives in bacteria and the diversity of enzymes that govern this process. This study has identified the genetic locus corresponding to a long-elusive 2HP 5-monooxygenase and thus has added an important piece to the puzzle of the biodegradation of pyridine derivatives in nature.
Conclusions.
A gene cluster, hpd, from Burkholderia sp. MAK1 containing all the necessary genes for the degradation of 2HP has been identified. We show that the 2HP biodegradation pathway starts with the hydroxylation of 2HP to 25DHP catalyzed by the 2-hydroxypyridine 5-monooxygenase encoded by hpdABCDE, a gene encoding an enzyme (HpdABCDE) that shows similarity to soluble diiron monooxygenases, which participate in the biodegradation of various aromatic and aliphatic pollutants but have never been implicated in the biodegradation of pyridines. The next step in the catabolic pathway is the ring opening of 25DHP. This reaction is catalyzed by 25DHP 5,6-dioxygenase HpdF, which is very similar to enzymes found in the catabolic pathways of nicotine and nicotinate. Notably, no 25DHP 5,6-dioxygenase has been implicated in the 2HP degradation pathway to date. After the ring opening, the degradation process possibly proceeds via the so-called maleamate pathway, since all genes associated with this pathway have homologues in the hpd cluster. Our study not only provides evidence of the new degradation pathway of 2HP but also deepens our understanding of the microbial biodegradation of pyridine and its derivatives in nature. A broad-range substrate specificity allows the recommendation of HpdABCDE to be used as a regioselective biocatalyst for the synthesis of the hydroxylated pyridine derivatives.
MATERIALS AND METHODS
Bacterial strains, chemicals, and standard techniques.
The bacterial strains used in this study are listed in Table 2. Burkholderia cells were cultivated at 30°C in liquid EFA medium [10.0 g/liter K2HPO4, 4.0 g/liter KH2PO4, 0.5 g/liter yeast extract, 1.0 g/liter (NH4)2SO4, 0.2 g/liter MgSO4·7H2O (pH 7.0)] supplemented with salt solution (2.0 g/liter CaCl2·2H2O, 1.0 g/liter MnSO4·4H2O, 0.5 g/liter FeSO4·7H2O; all components were dissolved in 0.1 M HCl) and with an appropriate carbon source (1.0 g/liter succinate or 1.0 g/liter 2HP). Burkholderia cells transformed with plasmids were cultivated on agar plates containing EFA medium. The medium that was used to cultivate transformed Burkholderia cells was also supplemented with the appropriate antibiotics (50 μg/ml kanamycin and 20 μg/ml chloramphenicol). E. coli cells carrying recombinant plasmids were cultivated in LB (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) or 37 g/liter brain heart infusion (BHI) medium supplemented with the appropriate antibiotics (50 μg/ml kanamycin, 20 μg/ml chloramphenicol, or 100 μg/ml ampicillin). Electroporation was used to introduce plasmid DNA.
TABLE 2.
Bacterial strains used in this study
| Strain | Genotype or characteristics of the straina | Source or reference |
|---|---|---|
| Burkholderia sp. MAK1 (DSM 102049) | 2HP-degrading bacterium | 39 |
| Burkholderia sp. MAK1Δ P5 | Mutant of Burkholderia sp. MAK1, which is unable to consume 2HP | This study |
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 ϕ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK− mK+), λ− | Thermo Fisher Scientific, Lithuania |
| E. coli BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λs) | Novagen, Germany |
| E. coli Rosetta(DE3)pLysS | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λs) pLysSRARE[T7p20 ileX argU thrU tyrU glyT thrT argW metT leuW proL orip15A] (Cmr) | Novagen, Germany |
Cmr, chloramphenicol resistance.
Qualitative and quantitative RT-PCR.
Burkholderia sp. MAK1 cells were cultivated in EFA medium containing 1.0 g/liter succinate or 1.0 g/liter 2HP as the sole source or carbon until the culture reached an optical density at 600 nm (OD600) of 0.3. Total RNA was isolated using a ZR fungal/bacterial RNA MicroPrep kit (Zymo Research Corporation). Copy DNA synthesis was carried out using the Maxima H Minus first-strand cDNA synthesis kit (Thermo Fisher Scientific, Lithuania). qPCR was conducted in a 25-μl reaction mixture containing 12.5 μl of Maxima SYBR green/ROX qPCR master mix (2×) (Fermentas, Lithuania), 200 nM each primer (see Table 3), ∼50 ng of cDNA, and water up to 25 μl. qPCR amplification was performed using a 7500 Fast real-time PCR system (Applied Biosystems, USA). The first step in qPCR was initial denaturation at 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, 62°C for 30 s, and 72°C for 30 s. For qualitative evaluation, endpoint PCR products were analyzed using electrophoresis. For quantitative analysis, fluorescence data were recorded after the annealing step. All experiments were carried out three times. To verify the absence of DNA in the RNA samples, the procedure was performed without the reverse transcriptase step. The threshold cycle (CT) (threshold value, 0.05) values were obtained using 7500 Software version 2.0.6. Relative target RNA analysis was performed using the ΔΔCT algorithm and 16S RNA as a reference for normalization.
TABLE 3.
Primers used in this study
| Primer name | Primer sequence (5′→3′)a | Use |
|---|---|---|
| Vsp_F_BAD | AACATTAATGTCTGATTCGTTACC | Designing pBAD-MCS-1 |
| Sac_R_BAD | ATGAGCTCGAGTTTGTAGAAACG | Designing pBAD-MCS-1 |
| P5_Hind_R | TTAAGCTTTCAAGCGTCAGCCCGCAAGGGTTCTC | Cloning |
| P1_PciI_F | TAACATGTCAGCACGGTCAAGCTGGTCC | Cloning |
| HpdF_F_Nco | ATCCATGGGCTCCTTAAGCGATGCAG | Cloning |
| HpdF_R_Hind | ACTAAGCTTTCATGCCATCACCTCATCG | Cloning |
| TnModSekv_F | GCAACACCTTCTTCACGAGG | Sequencing |
| TnModSekv_R | GTTCCTGGCCTTTTGCTGGC | Sequencing |
| P4M_R | GCCGACCACTGGCAAGGCGG | Sequencing |
| CB1F2 | GCGAACACAACCTTGCCATT | Sequencing |
| CB2F2 | TCATTCCTTCGCCATCGTTCAG | Sequencing |
| Cont84R2 | CTTCCAGCCTGACGATCACA | Sequencing |
| diox1_R | TTGCTCAACGATTATATGGACG | Sequencing |
| Hinc_R | ATCGACGTGGCGACGCGCAGG | Sequencing |
| Groel_F | ATACATATGAGCGTTAAGCGGATGG | Sequencing |
| MAK_Tat_F | GTATCACGGGGGCAAGTTCATCTGC | Sequencing |
| MAK_Rsa_F | ACATCGCACTGATGCTCCTCGG | Sequencing |
| P3Mut_F | TCTATCGCGAGTATGTGCGC | RT-PCR |
| P3Mut_R | AACGGGTCGCCGGAGAACAG | RT-PCR |
| P4Mut_F | GCCGACCACTGGCAAGGCGG | RT-PCR |
| P4Mut_R | ACGCGGAGAAGCACAGATAG | RT-PCR |
| P5Mut_F | GGGGGCGGAGGCTTACCGGG | RT-PCR |
| P5Mut_R | AGCCCGCGGAGAAGAAACCG | RT-PCR |
| Frrs | GATTAGATACCCTGGTAGTCC | RT-PCR, 16S RNA |
| Rrrs | GTTGCGGGACTTAACCCAAC | RT-PCR, 16S RNA |
Restriction sites are underlined.
Plasmids and primers.
The plasmids and primers used in this study are listed in Tables 3 and 4. Primers were ordered from Metabion, Germany. All the targeted genes were amplified by PCR using either Taq or Phusion (Thermo Fisher Scientific, Lithuania) DNA polymerase, according to the manufacturer's recommendations. The blunt-ended amplicons were cloned into the pJET1.2/blunt vector, while sticky-ended amplicons were cloned into pTZ57R/T. Before recloning into expression vectors, the gene sequences were confirmed by DNA sequencing (Macrogen, The Netherlands).
TABLE 4.
Plasmids used in this study
| Plasmid | Plasmid characteristicsa | Source or reference |
|---|---|---|
| pJET1.2/blunt | Apr ori ColE1 eco47IR, high copy-no. cloning vector | Thermo Fisher Scientific, Lithuania |
| pTZ57R/T | Apr ori ColE1 lacZα, high copy-no. cloning vector | Thermo Fisher Scientific, Lithuania |
| pET-28b | pBR322-derived ColE1, T7 lac promoter, Kmr, protein expression vector | Novagen, Germany |
| pRSFDuet-1 | RSF1030-replicon, T7 lac promoter, two MCSs, Kmr, protein expression vector | Novagen, Germany |
| pCDFDuet-1 | CloDF13 replicon, T7 lac promoter, two MCSs, Smr, protein expression vector | Novagen, Germany |
| pBAD24 | ColE1 ori, Apr, araC promoter, protein expression vector | Invitrogen |
| pBBR1MCS-1 | pBBR1-derived, Cmr, lacZα, broad-host-range vector | 28 |
| pBAD-MCS-1 | Amplicon containing regulatory elements of pBAD24 was amplified by PCR using primers Vsp_F_BAD and Sac_R_BAD, digested with VspI and SacI, and cloned into pBBR1MCS vector | This study |
| HpdF_pET | hpdF gene was amplified by PCR using HpdF_F_Nco and HpdF_R_Hind primers, digested with NcoI and HindIII, and cloned into pET-28b vector | This study |
| HpdABCDE_pET | Monooxygenase gene cluster was amplified by PCR using P1_PciI_F and P5_Hind_R primers, digested with PciI and HindIII, and cloned into pET-28b vector | This study |
| HpdABCDE_pBAD-MCS-1 | pET_ HpdABCDE plasmid was digested with XbaI and HindIII, monooxygenase gene containing fragment was isolated, and cloned into pBAD-MCS-1 vector | This study |
| pART3-gfp | Escherichia coli-Arthrobacter shuttle vector | 40 |
| gfp_pBAD-MCS-1 | pART3-gfp plasmid was digested with XbaI and HindIII, gfp gene containing fragment was isolated and cloned into pBAD-MCS-1 vector | This study |
| gfp_pBBR1MCS-1 | pART3-gfp plasmid was digested with KpnI and HindIII, gfp gene containing fragment was isolated and cloned into pBBR1MCS-1 vector | This study |
| pSal_4 | 2HP degradation genes containing plasmid obtained during transposon mutagenesis | This study |
| pDra_1 | 2HP degradation genes containing plasmid obtained during transposon mutagenesis | This study |
Apr, apramycin resistance; Kmr, kanamycin resistance; MCS, membrane contact sites; Smr, streptomycin resistance.
Induction of recombinant protein synthesis.
E. coli Rosetta(DE3)pLysS cells transformed with plasmid HpdF_pET were grown in BHI or LB medium containing kanamycin (final concentration, 40 μg/ml) at 30°C until the OD600 reached 0.5 to 0.6. Then, the medium was supplemented with isopropyl-thio-β-d-galactopyranoside (IPTG; final concentration, 1 mM) and FeSO4·7H2O (final Fe2+ concentration, 100 μM), and the cultivation continued for another 4 h.
Burkholderia sp. MAK1 ΔP5 cells carrying the HpdABCDE_pBAD-MCS-1 plasmid were grown for about 40 h at 30°C in EFA medium containing 0.1% succinate, 0.2% arabinose, and 20 μg/ml chloramphenicol. Also, FeSO4·7H2O was added twice, at the beginning of bacterial growth and after 20 h of cultivation, to a final concentration of 100 μM.
Biotransformation of pyridine derivatives.
After protein synthesis induction, the cells were harvested by centrifugation and then washed twice with 50 mM potassium phosphate buffer (pH 7.0). In total, 0.005 g of wet biomass was resuspended in 1 ml of potassium phosphate buffer, and the resulting suspension was supplemented with the appropriate substrate at 0.25 to 1.0 mM and incubated at 30°C. The conversion process was monitored periodically by recording the UV-Vis spectrum of the substrate, as well as by performing HPLC-MS analysis.
HPLC-MS analysis.
The parameters and conditions used in HPLC-MS analysis of biotransformation mixtures were described in our previous work (23).
Whole-genome sequencing.
The sequencing and subsequent assembly of the Burkholderia sp. MAK1 genome was performed by BaseClear (Leiden, The Netherlands). The quality-filtered Illumina FASTQ sequence 3,339,832 reads were assembled into a number of contig sequences. The analysis was performed using ABySS version 1.5.1. The contigs were linked and placed into scaffolds based on the alignment of the 200,298 PacBio reads. Alignment was performed with BLASR. The de novo assembly statistics are the following: total number of scaffolds, 355 (maximum scaffold size, 2.9 Mb; minimum, 306 bp; average, 35.2 kb); number of gaps, 530; and total coverage, 12.5 Mb.
Gene sequence analysis and protein tertiary structure modeling.
The deduced amino acid sequences of the proteins encoded by the hpd locus were searched against the NCBI database using BLAST (34). Protein functions were assigned based on a sequence similarity search against the NCBI Conserved Domain Database (35). Phylogenetic analyses and the multiple-sequence alignment were conducted using MEGA version 7 (36). The model of the HpdC subunit was generated using the i-TASSER server (37), while visualizations were made using the PyMOL 1.7.1.3 software (38).
Accession number(s).
The sequence of the 2HP degradation cluster hpd of Burkholderia sp. MAK1 was deposited in GenBank under the accession number MF957200.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the European Social Fund according to the activity “Improvement of researchers' qualification by implementing world-class R&D projects” of measure no. 09.3.3-LMT-K-712 [grant no. DOTSUT-34(09.3.3-LMT-K712-01-0058/LSS-600000-58)].
We thank L. Kalinienė for assistance with the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00387-18.
REFERENCES
- 1.Kaiser JP, Feng Y, Bollag JM. 1996. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol Rev 60:483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fetzner S. 1998. Bacterial degradation of pyridine, indole, quinoline, and their derivatives under different redox conditions. Appl Microbiol Biotechnol 49:237–250. doi: 10.1007/s002530051164. [DOI] [Google Scholar]
- 3.Padoley KV, Mudliar SN, Pandey RA. 2008. Heterocyclic nitrogenous pollutants in the environment and their treatment options–an overview. Bioresour Technol 99:4029–4043. doi: 10.1016/j.biortech.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 4.Sims GK, O'Loughlin EJ, Crawford RL. 1989. Degradation of pyridines in the environment. Crit Rev Environ Contr 19:309–340. doi: 10.1080/10643388909388372. [DOI] [Google Scholar]
- 5.Kuhn EP, Suflita JM. 1989. Microbial degradation of nitrogen, oxygen and sulfur heterocyclic compounds under anaerobic conditions: studies with aquifer samples. Environ Toxicol Chem 8:1149–1158. doi: 10.1002/etc.5620081207. [DOI] [Google Scholar]
- 6.Richards DJ, Shieh WK. 1986. Biological fate of organic priority pollutants in the aquatic environment. Water Res 20:1077–1090. doi: 10.1016/0043-1354(86)90054-0. [DOI] [Google Scholar]
- 7.Khasaeva F, Vasilyuk N, Terentyev P, Troshina M, Lebedev A. 2011. A novel soil bacterial strain degrading pyridines. Environ Chem Lett 9:439–445. doi: 10.1007/s10311-010-0299-6. [DOI] [Google Scholar]
- 8.Sun JQ, Xu L, Tang YQ, Chen FM, Liu WQ, Wu XL. 2011. Degradation of pyridine by one Rhodococcus strain in the presence of chromium (VI) or phenol. J Hazard Mater 191:62–68. doi: 10.1016/j.jhazmat.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 9.Sun JQ, Xu L, Tang YQ, Chen FM, Zhao JJ, Wu XL. 2014. Bacterial pyridine hydroxylation is ubiquitous in environment. Appl Microbiol Biotechnol 98:455–464. doi: 10.1007/s00253-013-4818-9. [DOI] [PubMed] [Google Scholar]
- 10.Shukla OP, Kaul SM. 1986. Microbiological transformation of pyridine N-oxide and pyridine by Nocardia sp. Can J Microbiol 32:330–341. doi: 10.1139/m86-065. [DOI] [Google Scholar]
- 11.Damani LA, Crooks PA, Shaker MS, Caldwell J, D'Souza J, Smith RL. 1982. Species differences in the metabolic C- and N-oxidation, and N-methylation of [14C]pyridine in vivo. Xenobiotica 12:527–534. doi: 10.3109/00498258209038931. [DOI] [PubMed] [Google Scholar]
- 12.Jiménez JI, Canales A, Jiménez-Barbero J, Ginalski K, Rychlewski L, García JL, Díaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377–385. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Tang H, Zhu X, Li Y, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1 Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cain RB, Houghton C, Wright KA. 1974. Microbial metabolism of the pyridine ring. Metabolism of 2- and 3-hydroxypyridines by the maleamate pathway in Achromobacter sp. Biochem J 140:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, Tang H, Ren H, Yu H, Wang L, Zhang W, Behrman JE, Xu P. 2013. Iron(II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci Rep 3:3235. doi: 10.1038/srep03235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semėnaitė R, Gasparavičiūtė R, Duran R, Precigou S, Marcinkevičienė L, Bachmatova I, Meškys R. 2003. Genetic diversity of 2-hydroxypyridine-degrading soil bacteria. Biologija 2:27–29. [Google Scholar]
- 18.Kolenbrander PE, Weinberger M. 1977. 2-Hydroxypyridine metabolism and pigment formation in three Arthrobacter species. J Bacteriol 132:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RC, Shukla OP. 1975. Microbial metabolism of 2-hydroxypyridine. Indian J Biochem Biophys 12:296–298. [PubMed] [Google Scholar]
- 20.Baitsch D, Sandu C, Brandsch R, Igloi GL. 2001. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J Bacteriol 183:5262–5267. doi: 10.1128/JB.183.18.5262-5267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanislauskiene R, Gasparaviciute R, Vaitekunas J, Meskiene R, Rutkiene R, Casaite V, Meskys R. 2012. Construction of Escherichia coli-Arthrobacter-Rhodococcus shuttle vectors based on a cryptic plasmid from Arthrobacter rhombi and investigation of their application for functional screening. FEMS Microbiol Lett 327:78–86. doi: 10.1111/j.1574-6968.2011.02462.x. [DOI] [PubMed] [Google Scholar]
- 22.Vaitekūnas J, Gasparaviciute R, Rutkiene R, Tauraite D, Meskys R. 2015. A 2-hydroxypyridine catabolism pathway in Rhodococcus rhodochrous strain PY11. Appl Environ Microbiol 82:1264–1273. doi: 10.1128/AEM.02975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stankevičiūtė J, Vaitekūnas J, Petkevičius V, Gasparavičiūtė R, Tauraitė D, Meškys R. 2016. Oxyfunctionalization of pyridine derivatives using whole cells of Burkholderia sp. MAK1. Sci Rep 6:39129. doi: 10.1038/srep39129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han XH, Wang W, Xiao XG. 2008. Microbial biosynthesis and biotransformation of indigo and indigo-like pigments. Sheng Wu Gong Cheng Xue Bao 24:921–926. (In Chinese.) doi: 10.1016/S1872-2075(08)60043-6. [DOI] [PubMed] [Google Scholar]
- 25.Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl Environ Microbiol 64:2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leahy JG, Batchelor PJ, Morcomb SM. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev 27:449–479. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 27.Notomista E, Lahm A, Di Donato A, Tramontano A. 2003. Evolution of bacterial and archaeal multicomponent monooxygenases. J Mol Evol 56:435–445. doi: 10.1007/s00239-002-2414-1. [DOI] [PubMed] [Google Scholar]
- 28.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM Jr, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 29.Lefebre MD, Valvano MA. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl Environ Microbiol 68:5956–5964. doi: 10.1128/AEM.68.12.5956-5964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widdel F, Musat F. 2010. Diversity and common principles in enzymatic activation of hydrocarbons, p 984–1004. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Heidelberg, Germany. [Google Scholar]
- 32.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds–from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 33.Parales RE, Parales JV, Pelletier DA, Ditty JL. 2008. Chapter 1 diversity of microbial toluene degradation pathways. Adv Appl Microbiol 64:2–43. doi: 10.1016/S0065-2164(08)00401-2. [DOI] [PubMed] [Google Scholar]
- 34.Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A 85:2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y. 2008. I-TASSER: server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLano WL. 2002. The PyMOL molecular graphics system on World Wide Web. http://www.pymol.org.
- 39.Gasparavičiūtė R, Kropa A, Meškys R. 2006. A new Arthrobacter strain utilizing 4-hydroxypyridine. Biologija 4:41–45. [Google Scholar]
- 40.Kutanovas S, Stankeviciute J, Urbelis G, Tauraite D, Rutkiene R, Meskys R. 2013. Identification and characterization of a tetramethylpyrazine catabolic pathway in Rhodococcus jostii TMP1. Appl Environ Microbiol 79:3649–3657. doi: 10.1128/AEM.00011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.