ABSTRACT
In recent years, global warming has led to a growing number of Vibrio cholerae infections in bathing water users in regions formerly unaffected by this pathogen. It is therefore of high importance to monitor V. cholerae in aquatic environments and to elucidate the main factors governing its prevalence and abundance. For this purpose, rapid and standardizable methods that can be performed by routine water laboratories are prerequisite. In this study, we applied a recently developed multiplex quantitative PCR (qPCR) strategy (i) to monitor the spatiotemporal variability of V. cholerae abundance in two small soda pools and a large lake that is intensively used for recreation and (ii) to elucidate the main factors driving V. cholerae dynamics in these environments. V. cholerae was detected with qPCR at high concentrations of up to 970,000 genomic units 100 ml−1 during the warm season, up to 2 orders of magnitude higher than values obtained by cultivation. An independent cytometric approach led to results comparable to qPCR data but with significantly more positive samples due to problems with DNA recovery for qPCR. Not a single sample was positive for toxigenic V. cholerae, indicating that only nontoxigenic V. cholerae (NTVC) was present. Temperature was the main predictor of NTVC abundance, but the quality and quantity of dissolved organic matter were also important environmental correlates. Based on this study, we recommend using the developed qPCR strategy for quantification of toxigenic and nontoxigenic V. cholerae in bathing waters with the need for improvements in DNA recovery.
IMPORTANCE There is a definitive need for rapid and standardizable methods to quantify waterborne bacterial pathogens. Such methods have to be thoroughly tested for their applicability to environmental samples. In this study, we critically tested a recently developed multiplex qPCR strategy for its applicability to determine the spatiotemporal variability of V. cholerae abundance in lakes with a challenging water matrix. Several qPCR protocols for V. cholerae detection have been developed in the laboratory, but comprehensive studies on the application to environmental samples are extremely scarce. In our study, we demonstrate that our developed qPCR approach is a valuable tool but that there is a need for improvement in DNA recovery for complex water matrices. Furthermore, we found that nontoxigenic V. cholerae is present in very high numbers in the investigated ecosystems, while toxigenic V. cholerae is apparently absent. Such information is of importance for public health.
KEYWORDS: Vibrio cholerae non-O1/non-O139, bathing water, detection, qPCR, quantification
INTRODUCTION
Vibrio cholerae is one of the most important waterborne bacterial pathogens and the causative agent of epidemic cholera. Besides cholera, nontoxigenic V. cholerae (NTVC) strains can cause a variety of other infections, such as gastroenteritis and ear, wound, blood, or soft tissue infections (1–3). Attributable to climate change, the numbers of infections caused by nontoxigenic V. cholerae non-O1/non-139 have been increasing in Europe and the United States in the past 2 decades (1, 4–8). Most of the reported infections have been related to recreational activities (mainly bathing) in natural marine (2, 9, 10) and inland aquatic ecosystems (11, 12). It is thus of high importance that the prevalence and abundance of NTVC are monitored and linked to environmental parameters (i) to identify and monitor the hot spots of NTVC abundance in recreational waters and (ii) to elucidate the main factors governing their prevalence and abundance. For this purpose, reliable methods for the detection and quantification of NTVC are required. Cultivation by microbiological media is still the most commonly used approach (13); however, standard protocols rely on extremely tedious and elaborate most-probable-number procedures (14–16). Moreover, cultivation may underestimate the number of V. cholerae cells because of the presence of viable-but-nonculturable (VBNC) cells (17, 18), and more studies that compare cultivation results with results obtained by culture-independent methods are needed (19). Besides cultivation, quantitative PCR (qPCR) is a ubiquitously used method to quantify pathogenic bacteria in aquatic environments that has the potential to become standardized and used by routine laboratories (20). Several promising qPCR protocols for V. cholerae have been developed in past years (21–26), but they have not been widely applied in large environmental investigations (26). It is therefore of utmost importance to scrutinize existing qPCR protocols for their applicability and reliability to quantify NTVC over a wide range of environmental conditions.
The aim of the study was to comprehensively test a multiplex qPCR strategy for V. cholerae quantification (21) that was recently developed for Neusiedler See, a turbid shallow subsaline lake in eastern Austria with a water matrix that is extremely challenging for sample analysis. Neusiedler See has been associated with NTVC infections of bathers and other recreational visitors for approximately 15 years (3; Steliana Huhulescu, Austrian Agency for Health and Food Safety, personal communication), with a few cases per year. Most reported infections have been ear infections, but in 2005, a lethal case of septicemia occurred (3). Within the region of eastern Austria, two severe cases of necrotizing fasciitis, one with a fatal outcome, occurred in 2015 during a summer heat wave, related to two small bathing ponds approximately 40 km away from Neusiedler See (11). Neusiedler See was shown to harbor a community where NTVC is endemic (27, 28). In close vicinity to the lake, there are several small and very shallow soda pools that have recently been identified as hot spots of NTVC (19). V. cholerae abundance was monitored at several stations in the lake and in two selected soda pools over two annual cycles with the recently developed multiplex qPCR strategy, allowing the simultaneous detection of nontoxigenic and toxigenic V. cholerae. The applicability of the qPCR along a wide range of environmental gradients was critically investigated and compared to results obtained by cultivation and an independent cytometric method. In addition, the obtained qPCR abundance values were statistically related to a variety of important environmental factors to elucidate the main factors driving NTVC abundance in the lake and the soda pools.
RESULTS
Environmental conditions in the investigated ecosystems.
Neusiedler See (47°42′N, 16°46′E) is the largest shallow alkaline brown-water lake in central Europe and covers an area of approximately 320 km2 (29). About 55% of its area is covered with reed (Phragmites australis), and within this vegetation, extended brown-water areas are found. The water level of the lake (maximum depth, 1.8 m) is mainly controlled by precipitation (500 to 700 mm year−1) and evaporation. Due to the shallow water column, the water temperature changes rapidly in response to weather events (29). The intensive resuspension of the sediment due to winds and currents results in a high concentration of suspended solids in the water (Table 1). The lake is characterized by high pH values up to 8.9, elevated conductivity values up to 3,000 μS cm−1, and high inorganic and organic nutrient concentrations (Table 1). Due to its high turbidity (open water) and brown color (reed stand), chlorophyll a values are comparably low (Table 1). Altogether, the chemophysical conditions of the lake result in a water matrix that is extremely challenging for any kind of sample analysis. Four sites in the lake were selected for sampling to represent a wide range of ecological variability (see Fig. S1 in the supplemental material).
TABLE 1.
Physicochemical variables of the different sampling sitesa
| Sampling site | pH | Conductivity (μS cm−1) | Carbonate alkalinity (mmol−1) | DOC (mg liter−1) | Ptot (μg liter−1) | O2 (mg liter−1) | TSS (mg liter−1) | Chl a (μg liter−1) | NH4-N (μg liter−1) | NO3-N (μg liter−1) | Secchi depth (cm) | Temp (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 8.6 (8.1–8.9) | 1,975 (1,500–3,000) | 11.5 (7.0–17.5) | 18.5 (12.0–26.0) | 36 (17–87) | 8.5 (5.2–11.6) | 68.9 (18.1–130) | 7.8 (1.8–22.8) | 39 (10–86) | 115 (47–154) | 65 (8–137) | 17.8 (3.4–26.9) |
| 24 | 8.8 (8.6–8.9) | 1,850 (1,500–2,200) | 11.9 (10.6–12.8) | 12.5 (11.5–14.8) | 30 (13–51) | 9.9 (7.9–12.6) | NA | 8.8 (1.1–20.2) | 13 (2–34) | 82 (14–176) | 31 (8–95) | 16.3 (3.8–24.9) |
| 29 | 8.3 (7.4–8.8) | 1,625 (800–2,600) | 10.2 (5.7–12.5) | 9.8 (5.8–15.2) | 62 (28–112) | 7.1 (1.3–11.9) | NA | 8.1 (1.7–17.7) | 56 (6–411) | 116 (37–340) | 44 (12–120) | 16.0 (4.6–25.0) |
| 36 | 8.4 (8.1–8.6) | 2,113 (1,600–3,000) | 12.7 (8.2–17.5) | 21.8 (15.6–26.8) | 31 (17–64) | 7.9 (5.2–10.5) | 24.1 (11.7–88.7) | 5.4 (1.8–12.2) | 12 (1–54) | 86 (10–146) | 90 (12–137) | 18.3 (3.4–26.9) |
| ZL | 9.4 (8.7–9.9) | 7,057 (1,100–29,600) | 38.0 (5.5–201) | 110.7 (17.5–431) | 627 (40–3,120) | 8.3 (1.9–16.8) | 8,117 (5–75,250) | 102.8 (0.2–476) | NA | NA | 14 (2–45) | 14.6 (0.7–24.3) |
| US | 9.1 (8.7–9.6) | 3,550 (2,100–6,500) | 31.6 (8.0–71.1) | 49.5 (32.0–79.8) | 89 (24–245) | 8.1 (5.2–12.5) | NA | 23.9 (1.4–186) | NA | NA | 16 (2–45) | 16.0 (2.3–25.6) |
Data are mean values, with minimum and maximum values in parentheses. DOC, dissolved organic carbon; Ptot, total phosphorus; TSS, total suspended solids; Chl a, chlorophyll a; NA, not analyzed.
Adjacent to the eastern shore of the lake, small, shallow (maximum depth, 80 cm) soda pools are situated. Two of them, Zicklacke (ZL) and Unterstinker (US), were selected as additional sampling sites. These soda pools are characterized by very high pH values, up to 9.9, very high inorganic and organic nutrient concentrations and chlorophyll a values (Table 1), an extremely active microbial community (30, 31), high turbidity, and thus an extremely challenging water matrix.
GR rate and sample limit of detection.
In order to determine the abundances of V. cholerae in the lake water samples, the global recovery (GR) rate was determined for each sample. GR describes the efficiency of the entire cell concentration and DNA extraction procedure to recover the target bacteria in a defined volume of lake water. GR values showed extremely high seasonal variability and ranged from 0.0% to 102% (Table 2). There was one outlier of 132% observed at site US in January 2014. At this time of the year, no V. cholerae cells were detectable by PCR (no positive signal in any of the unspiked triplicate samples). The lowest value was observed in September 2014 at site 5 (open water), when no positive qPCR signal was obtained even in the spiked sample. Median GR values ranged from 2.1% for site ZL in 2014 to 15.7% for site 24 in 2014, with a tendency to lower values at site 36 (reed stand) and ZL than at the other sites (Table 2). Several environmental parameters were significantly correlated with the GR values (Table S1). For the lake, negative correlations were found with water temperature (rho = −0.44, P < 0.001), ammonium (rho = −0.57, P < 0.001), and dissolved organic carbon (DOC; rho = −0.29, P < 0.01); positive correlations were found with oxygen (rho = 0.46, P < 0.001), Secchi depth (rho = 0.44, P < 0.001), pH (rho = 0.37, P < 0.001), and the 250 nm/365 nm absorbance ratio (rho = 0.34, P < 0.01). The 250 nm/365 nm absorbance ratio was calculated to determine possible shifts in the molecular size spectrum of DOC (32). A lower ratio indicates a higher percentage of high-molecular-weight matter than of low-molecular-weight substances and vice versa. Stepwise multiple linear regression was applied to predict the GR from environmental parameters. Of all parameters fed into the model, only water temperature and Secchi depth were significant predictors, together explaining 41.8% of the variability of the GR (F = 33.97, df = 92, P < 0.001). Despite their high correlation coefficients, ammonium and oxygen were not included in the model calculations. Both parameters were also correlated with temperature, and we assume that neither parameter has a mechanistic influence on the GR. We also examined if nonlinear relationships would better predict the GR values. From the visual inspection of the correlation scatterplots of the individual environmental variables, supported by mathematical calculations, we found that polynomial models were the only ones that exhibited a slightly higher goodness of fit. From the parameters fed into the model, a second-order polynomial function, including temperature and the 250 nm/365 nm absorbance ratio, gave the best fit and explained 49.5% of the variability of the GR (F = 23.58, df = 92, P < 0.001).
TABLE 2.
Global recovery rates and sample limit of detection at all sampling sitesa
| Sampling site | Yr 2011 |
Yr 2014 |
||
|---|---|---|---|---|
| GR rate (%) | SLOD (GU 100 ml−1) | GR rate (%) | SLOD (GU 100 ml−1) | |
| 5 | 7.8 (0.6–54.6) | 1,600 (230–20,800) | 8.0 (0.0–75.5) | 1,550 (165–NC)b |
| 24 | NA | NA | 15.7 (0.5–83.6) | 790 (150–25,000) |
| 29 | NA | NA | 4.1 (0.6–102) | 3,100 (125–20,800) |
| 36 | 2.8 (0.7–42.5) | 4,460 (295–17,800) | 2.5 (0.3–5.7) | 5,000 (1,750–41,700) |
| ZL | 4.0 (0.2–42.5) | 3,120 (295–62,400) | 2.1 (0.7–55.6) | 5,950 (225–17,800) |
| US | NA | NA | 6.1 (0.3–132) | 2,030 (125–41,700) |
Data are median values, with minimum and maximum values in parentheses. GR, global recovery; SLOD, sample limit of detection; NA, not analyzed.
NC, no calculation possible.
In the soda pools, GR values showed significant negative correlations with water temperature (rho = −0.60, P < 0.001), pH (rho = −0.30, P < 0.05), and the 250 nm/365 nm absorbance ratio (rho = −0.48, P < 0.001), and positive correlations were found with oxygen (rho = 0.51, P < 0.001) (Table S2). The stepwise multiple linear regression model revealed that temperature was the only significant predictor of GR, explaining 37.0% of its variability (F = 28.06, df = 46, P < 0.001). Similar to the lake results, a second-order polynomial function, including water temperature alone, showed a higher goodness of fit than the linear model and explained 52.9% of the variability of the GR (F = 26.84, df = 46, P < 0.001).
The sample limit of detection (SLOD) varied depending on the GR rate (21). Calculated SLOD values ranged between a minimum of 125 genomic units (GU) 100 ml−1, when GR was near 100%, and 62,400 GU 100 ml−1, when a GR rate of 0.2% was obtained. Median SLOD values were between 790 and 5,950 GU 100 ml−1 (Table 2).
Seasonal variation of V. cholerae abundances obtained by qPCR.
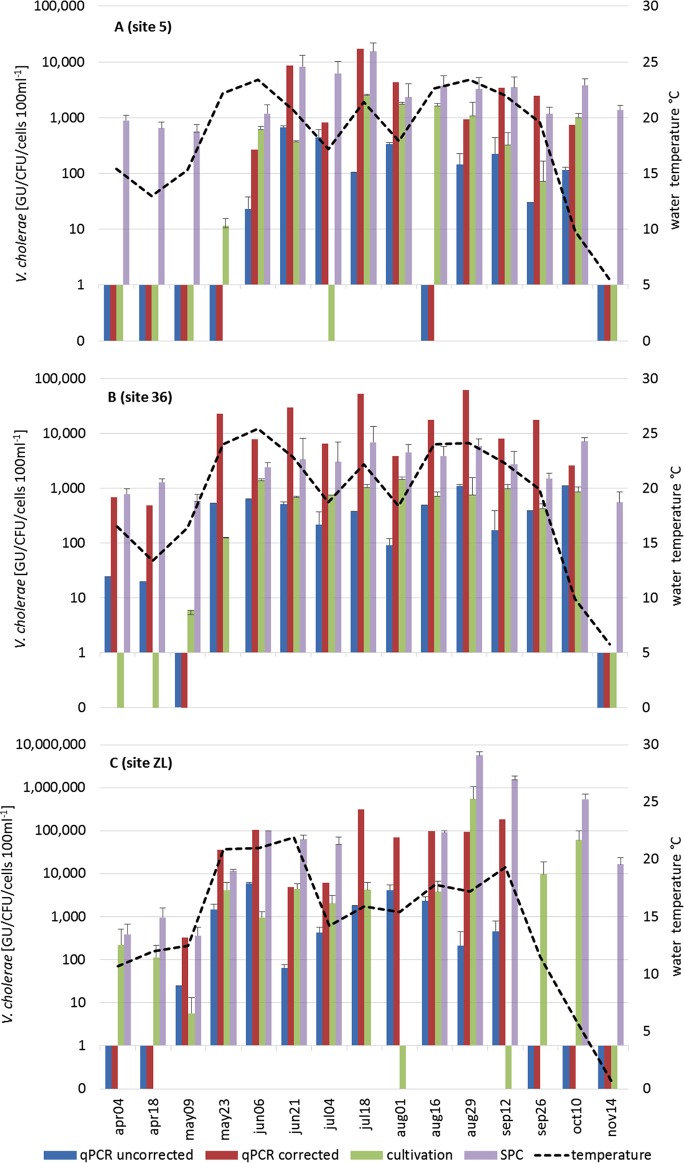
The qPCR used in this study allows the simultaneous detection of toxigenic V. cholerae (cholera toxin A gene target, ctxA) and nontoxigenic V. cholerae (outer membrane protein W gene target, ompW). At the three sampling sites investigated in 2011, V. cholerae was successfully quantified by qPCR in 9 (60%, site 5), 13 (87%, site 36), and 10 (67%, site ZL) out of 15 measurements (Fig. 1). qPCR results were considered quantitative only when all triplicate measurements gave a positive signal. Results from the exogenous inhibition control (IC; an egfp plasmid) amplification did not indicate inhibition of the PCRs for any of the samples. At all three sites, positive detection was linked to higher water temperatures (mid-May to mid-September); there was only one event during summer (site 5) when V. cholerae could not be detected by qPCR (16 August). Nondetections were otherwise observed only at the beginning and the end of the seasonal cycle. V. cholerae numbers, corrected with the GR values (see “Global recovery rate and calculation of V. cholerae abundances” in Materials and Methods), ranged from below the SLOD to 317,000 GU 100 ml−1. During the warm season, permanently higher numbers were found in soda pool ZL (median, 82,300 GU 100 ml−1) than in the lake sites (median, 2,500 and 17,600 GU 100 ml−1 for sites 5 and 36, respectively). Not a single sample was positive for ctxA (toxigenic V. cholerae).
FIG 1.
V. cholerae concentrations in 2011 as determined by qPCR, cultivation, and CARD-FISH/SPC. (A) Sampling site 5, Neusiedler See; (B) sampling site 36, Neusiedler See; (C) sampling site ZL (soda pool Zicklacke). qPCR-based concentrations are given without correction (blue bars) and with correction (corrected with global recovery rate; red bars). Cultivation-based (green bars) and CARD-FISH/SPC (purple bars) results are shown. A black dotted line indicates the water temperature. A missing bar means that an analysis with the respective method was not possible; bars between 0 and 1 indicate V. cholerae concentrations below the detection limit. Values and error bars represent the mean of three (cultivation, qPCR) and three to eight (SPC/CARD-FISH) replicates ± standard deviation. GU, genomic units.
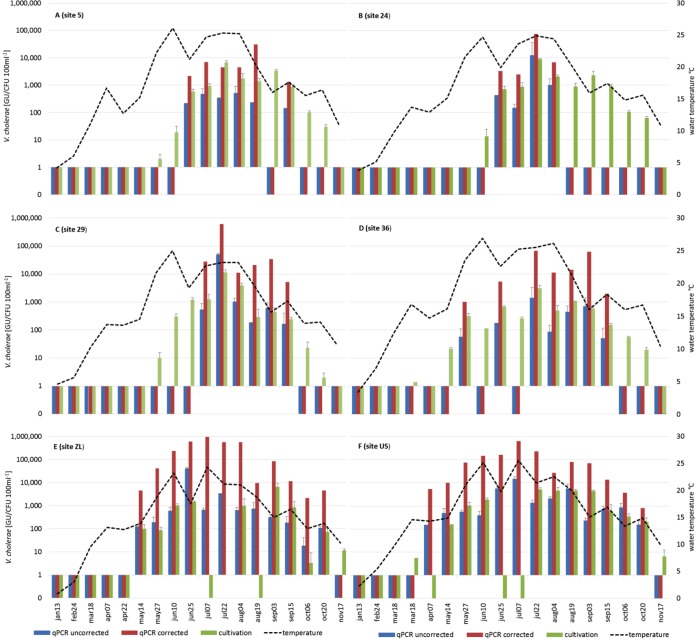
In 2014, V. cholerae abundance was determined by qPCR at four lake sites and two soda pools. As in 2011, no sample was positive for toxigenic V. cholerae. At the lake sites, positive qPCR results were obtained mostly during a short period of time during the warm season (end of June to mid-September), and a high number of nondetections was obtained (Fig. 2). In total, 23 out of 72 analyzed lake samples gave detectable results with qPCR (31.9%). As in 2011, no PCR inhibition was observed in any of the positive samples. V. cholerae numbers, corrected with the GR values, showed maximum values of 621,000 GU 100 ml−1 at sampling site 29 (median, 24,500 GU 100 ml−1). At the other three sites, maximum values ranged between 31,000 (site 5) and 74,000 (site 24) GU 100 ml−1. Median summer values at those sites were between 4,400 and 5,300 GU 100 ml−1. In the two soda pools, ZL and US, high maximum values of 966,000 and 644,000 GU 100 ml−1, respectively, were observed with GR-corrected qPCR (Fig. 2); median values amounted to 63,000 and 71,000 GU 100 ml−1, respectively.
FIG 2.
V. cholerae concentrations in 2014 as determined by qPCR and cultivation. (A) Sampling site 5, Neusiedler See; (B) sampling site 24, Neusiedler See; (C) sampling site 29, Neusiedler See; (D) sampling site 36, Neusiedler See; (E) sampling site ZL (soda pool Zicklacke); (F) sampling site US (soda pool Unterstinker). qPCR-based concentrations are given without correction (blue bars) and with correction (corrected with global recovery rate; red bars). Cultivation-based results (green bars) are shown. A black line indicates the water temperature. A missing bar means that an analysis with the respective method was not possible; bars between 0 and 1 indicate V. cholerae concentrations below the detection limit. Values and error bars represent the mean of three (cultivation, qPCR) replicates ± standard deviation. GU, genomic units.
When the data from both years were pooled, highly significant negative correlations between GR-corrected qPCR results and GR rates were observed (rho = −0.31, P < 0.001, n = 102 [for the lake]; rho = −0.68, P < 0.001, n = 51 [for the pools]). This was due to the strong contrasting effect of temperature on V. cholerae abundance (positive) and GR rates (negative). Thus, no bias of higher results caused by higher GR rates was observed.
Comparison of qPCR data with data obtained by cultivation and a cytometric approach.
For the lake sites in 2011, V. cholerae abundance was additionally determined by two independent methods: a cultivation-based approach and a cytometric approach (catalyzed reporter deposition-fluorescence in situ hybridization combined with solid-phase cytometry [CARD-FISH/SPC]) (33). Cultivation-based results were lower than corrected qPCR results (positive samples only; paired t test, P < 0.01). They ranged from below the detection limit (3 CFU 100 ml−1) to 2,500 CFU 100 ml−1 (median, 600 CFU 100 ml−1) for site 5 and up to 1,400 CFU 100 ml−1 (median, 750 CFU 100 ml−1) for site 36 (Fig. 1). In soda pool ZL, maximum concentrations (546,000 CFU 100 ml−1) were of magnitudes comparable to those obtained by qPCR. Similar to qPCR results, culture-negative results were mainly obtained in the beginning and at the end of the seasonal cycle; only at site ZL did a negative culture result occur in the beginning of August. In 2011, V. cholerae abundances were also determined via CARD-FISH/SPC (Fig. 1). At site 5, CARD-FISH/SPC results were comparable to corrected qPCR data (positive samples only; paired t test, P > 0.05), ranging from below the detection limit (200 cells 100 ml−1) to 15,500 cells 100 ml−1 (median, 3,500 cells 100 ml−1). However, CARD-FISH/SPC resulted in more positive results (14 out of 15) than did qPCR. At site 36, CARD-FISH/SPC results were significantly lower than corrected qPCR results (positive samples only; paired t test, P < 0.01), with maximum and median values of 7,200 and 3,300 cells 100 ml−1, respectively. Fourteen out of 15 samples gave positive results with CARD-FISH/SPC, in comparison to 13 with qPCR. For all lake samples, a significant correlation between log10-transformed corrected qPCR data and CARD-FISH/SPC data was observed (rho = 0.55, P < 0.01, n = 30). At site ZL, CARD-FISH/SPC led to higher numbers than those obtained by corrected qPCR, specifically during the period from the end of August until November 2011. Maximum numbers of 5,630,000 cells 100 ml−1 were observed at the end of August. In contrast, CARD-FISH/SPC failed to detect V. cholerae on 18 July and 1 August, when qPCR gave positive results. A significant correlation between log10-transformed corrected qPCR data and CARD-FISH/SPC data was observed (rho = 0.63, P < 0.05, n = 13). In contrast to the corrected qPCR results, uncorrected qPCR data in the lake and the soda pool were 1 to 3 log10 lower than the CARD-FISH/SPC results and up to 2 log10 lower than cultivation-based results (Fig. 1).
In 2014, V. cholerae abundance was additionally determined by cultivation. Results obtained by cultivation for the lake samples were on average 1 log lower than corrected qPCR results (positive samples only; paired t test, P < 0.001). Maximum concentrations of 11,500 CFU 100 ml−1 were observed at site 29; at the other sites, maximum values ranged from 3,100 (site 36) to 8,900 (site 24) CFU 100 ml−1. Concentrations of culturable V. cholerae in the soda pools were on average 2 orders of magnitude lower than the values obtained by corrected qPCR. In addition, there were several occasions during summer when no V. cholerae could be detected by cultivation. Despite the lower cultivation-based abundance values in the soda pools, V. cholerae was detected with cultivation at a rate (23 positive out of 36 samples, 63.8%) similar to that with qPCR (25 out of 36, 69.4%). As already observed in 2011, median uncorrected qPCR data in the lake were approximately 1 log10 lower than cultivation results; in the soda pools, they were of comparable magnitudes (Fig. 2).
When the data from both years were pooled, a highly significant correlation between corrected qPCR results and cultivation-based results was obtained for the lake data set (rho = 0.71, P < 0.001, n = 102) (Table S1). For the soda pool samples, a much weaker relationship was observed (rho = 0.29, P < 0.05, n = 51) (Table S2). Positive qPCR results were obtained for 80 out of 153 samples (52.3%); due to a lower SLOD, 102 positive results (66.7%) were obtained with cultivation.
Relationship of V. cholerae concentrations to environmental variables.
For the lake, three statistically significant correlations between V. cholerae concentrations obtained with qPCR and environmental parameters were observed (pooled data from 2011 and 2014). Slightly higher correlation coefficients were obtained for the GR-corrected data than for the uncorrected data (Table S1); therefore, in the following, only the corrected data set is described. Temperature and ammonium showed positive correlation coefficients (rho = 0.58, P < 0.001, n = 102, and rho = 0.25, P = 0.05, n = 64, respectively); oxygen was negatively correlated (rho = −0.45, P < 0.001, n = 96). All other parameters did not show any relationship to the V. cholerae concentrations. In multiple linear regression analysis, temperature and chlorophyll a were significant predictors of V. cholerae abundance, both explaining 36.8% of its variability (F = 23.84, df = 97, P < 0.001), while temperature alone already explained 30.1%. As for the GR rates, multiple nonlinear regression models were calculated. No nonlinear model was found that better explained the variability of V. cholerae abundance in the lake than the linear model.
For the soda pools, a variety of environmental parameters showed significant correlation with V. cholerae abundance (Table S2). Temperature exhibited the highest correlation coefficient (rho = 0.87, P < 0.001, n = 49), followed by the 250 nm/365 nm absorbance ratio (rho = 0.64, P < 0.001), oxygen (rho = −0.63, P < 0.001), DOC (rho = 0.44, P < 0.01), conductivity (rho = 0.36, P < 0.01), and pH (rho = 0.33, P < 0.05). As for the lake, slightly higher correlation coefficients were observed for the corrected than for the uncorrected values (Table S2). With multiple linear regression, temperature remained the only significant predictor, explaining 66.0% of the variability of V. cholerae abundance in the pools (F = 88.34, df = 45, P < 0.001). No nonlinear model was found that better explained the variability of V. cholerae abundance than the linear model in the soda pools.
DISCUSSION
Due to the increasing number of NTVC infections related to recreational activities in marine and inland aquatic ecosystems, reliable monitoring tools for quantification of these pathogenic bacteria in water samples are required. In this study, we used a recently developed triplex qPCR strategy (21) to critically examine its applicability to environmental samples along large spatiotemporal environmental gradients for (i) quantifying V. cholerae in a lake intensively used for recreation and two small soda pools nearby and (ii) elucidating the main environmental factors which drive V. cholerae abundance in the investigated ecosystems. This strategy comprises (i) a cell standard spike for each triplicate sample to obtain a global recovery (GR) rate, (ii) the correction of the measured qPCR results with the GR values to obtain the “true” V. cholerae concentrations in the water samples, and (iii) the search for environmental correlates to predict GR rates.
In this study, 80 out of 153 samples (52.3%) were positive for NTVC as determined by qPCR (ompW target). Not a single sample was positive for ctxA, corroborating the findings of previous investigations in which no toxigenic isolates were found in the lake and the soda pools (19, 27). Potential false-negative ctxA results can be ruled out based on the results of the template competition assays (21) (see “Quantification of V. cholerae” in Materials and Methods). Due to the fact that a toxigenic V. cholerae strain was used as the cell spike standard and that all samples were negative for ctxA, the ctxA target could be used as a marker for calculation of the GR rates. In these challenging environments, the correction of the obtained “raw” qPCR results with the GR rates was obviously necessary to obtain more realistic NTVC numbers: corrected qPCR results were mostly of magnitudes similar to those of the CARD-FISH/SPC data and higher than those of cultivation-based data. In contrast, uncorrected values would have underestimated NTVC levels, as median concentrations were up to 3 log10 lower than results obtained by CARD-FISH/SPC and even up to 1 log10 lower than cultivation-based results. Thus, standard cell spikes have to be performed for each individual sample. PCR inhibition was obviously not a problem in any of the samples, as indicated by the used IC spike and dilution of DNA extracts. The GR rates displayed an extremely high spatiotemporal variability and were influenced by a combination of different environmental factors. With multiple linear regression, we tried to set up models to predict the GR rates from the environmental variables for the lake and the soda pools separately. With linear regression, temperature and water transparency (Secchi depth) and temperature alone were the only significant predictive variables, explaining 41.8% and 37.0% of the GR variability in the lake and the soda pool samples, respectively, which is insufficient to be used in a predictive model. With nonlinear (second-order polynomial) regression, the goodness of fit was slightly higher. For the lake, temperature and the 250 nm/365 nm absorbance ratio explained 49.5% of the variability of the GR rate, and in the soda pools, temperature alone explained 52.9% of the variability of the GR rate. It has to be mentioned again that the investigated ecosystems have an extremely challenging water matrix for any DNA-based analysis approach. Due to the shallowness of these water bodies, the resuspension of sediment results in a practically permanent presence of various amounts of different kinds of organic and inorganic particles in the water column (34) that may bind DNA to their surfaces and inhibit efficient DNA extraction (35). Obviously, lower GR rates were obtained when temperatures were higher. We speculate that this was due to an indirect effect of temperature on the presence and behavior (emulsion, DNA binding capacity) of specific (colloid) particles. More particles and/or particles with a higher DNA binding capacity may be present at higher temperatures than at lower temperatures. In addition to turbidity, the large amounts of refractory DOC (high abundance of high-molecular-weight substances like humic acids, reflected in a low 250 nm/365 nm absorbance ratio) (27, 32) may also have a negative influence on the DNA extraction efficiency (36) and GR rates. Indeed, in the lake, the GR rates were negatively correlated with DOC and a positive correlation with the 250 nm/365 nm absorbance ratio was found (a higher ratio means a lower percentage of refractory high-molecular-weight substances). In the soda pools, GR rates correlated negatively with the 250 nm/365 nm absorbance ratios. The annual course of the 250 nm/365 nm absorbance ratios in the pools was opposite to that of the ratios in the lake. While refractory material in the lake from the reed stand mainly fuels the DOC pool in summer (32), benthic and pelagic algae releasing low-molecular-weight substances mainly fuel the DOC pool in the soda pools (30). Thus, we propose that in the soda pools, the negative effect of the high-molecular-weight substances on the GR rates (36) (positive correlation with 250 nm/365 nm absorbance ratios) is masked by the specific ecological conditions in the soda pools (high percentage of low-molecular-weight substances present in summer, when GR rates are low due to temperature-induced effects on the presence of DNA binding particles).
The high number of negative results obtained by qPCR is caused by a combination of two factors. First, in the beginning of the warm season, when increasing numbers of NTVC can be expected, the GR rates are decreasing due to the indirect “temperature effect” mentioned above. False-negative results are then obtained because the DNA extraction efficiency is too low and the corresponding SLOD values are too high to enable successful quantification of NTVC by the qPCR. In summer, when NTVC concentrations are high enough, positive results are obtained despite lower GR rates. Such a scenario was especially evident in 2014, when positive NTVC detection in the lake was possible only in a short period, from mid-June to mid-September. In the soda pools, where NTVC concentrations are generally higher, such a seasonal delay did not occur. Second, when temperatures were low, low V. cholerae concentrations (or no NTVC at all) were also present, resulting in negative qPCR results. In general, 66.7% positive results were obtained with cultivation (all data from both years) and 88.9% positive results were obtained with CARD-FISH/SPC (data from 2011 only). Despite the large differences in positive detections between the two methods, qPCR results displayed a significant positive correlation with both cultivation and CARD-FISH/SPC results for the lake and the soda pools. Apart from small site-specific differences, qPCR delivered results comparable to those of CARD-FISH/SPC but which were 1 log10 (lake) to 2 log10 (soda pools in 2014) higher than results obtained by cultivation. A very similar finding was also obtained in a preliminary study (21), in which a much smaller number of samples was examined. The main reasons for these observations are that qPCR (as well as CARD-FISH/SPC) also detects VBNC and dead cells and that V. cholerae cells that are attached to particles end up in a single colony on agar medium but are detected separately by qPCR and CARD-FISH/SPC.
The maximum NTVC concentrations obtained in this study by qPCR were 6.2 × 105 100 ml−1 in the lake and 9.7 × 105 100 ml−1 in the soda pools. These results lie in the upper range of V. cholerae concentrations found in marine, brackish, and freshwater ecosystems (8, 26, 37–39). Higher values have been reported so far only for water samples in an area where cholera is endemic (2.9 × 106 CFU 100 ml−1) (40) and in an earlier study on the investigated soda pools (5.6 × 106 CFU 100 ml−1) (19).
In both the lake and the soda pools, temperature was the main predictor of NTVC abundance over the spatiotemporal scale, corroborating findings from an earlier study (19). In the soda pools, temperature alone explained 66.0% of the NTVC variability, but positive correlations were also found with the 250 nm/365 nm absorbance ratio, DOC, conductivity, and pH. It is well established that temperature is a main driver of V. cholerae growth in many ecosystems (41), and as already shown in earlier studies, increases in concentration of nutrients and salinity, but also in pH, occur in the soda pools during summer due to the evaporation of water (30), leading to higher NTVC abundances (19). Also, the positive correlation with the 250 nm/365 nm absorbance ratio corroborates this finding; a higher ratio means a higher percentage of low-molecular-weight substances, readily available for the bacterial metabolism (see above) (32). The negative correlation with oxygen can be explained simply by the lower solubility of this gas at higher temperatures in summer.
Also in the lake, temperature was the best predictor of NTVC abundance, alone explaining 36.8% of NTVC variability. Together with temperature, chlorophyll a was the only other significant predictor in the multiple linear regression model, but it explained only an additional 6.7% of NTVC variability. As mentioned above, eukaryotic and prokaryotic algae produce dissolved organic matter (DOM) that can be readily utilized by V. cholerae (42, 43). As observed previously for Neusiedler See (19), ammonium concentrations were positively correlated with NTVC abundance. No such positive relationship has been reported in the literature for other ecosystems so far. In contrast, a negative correlation was found by Blackwell and Oliver (44), and lake-specific characteristics may be responsible for this observation (19). In this context, it must be mentioned that crustacean zooplankton have been shown to be the dominant reservoir for environmental V. cholerae in many marine, estuarine, or brackish water habitats (38, 45, 46). However, the ecosystems studied here offer perfect conditions for V. cholerae (optimal pH, low salinity, and high DOM concentrations), leading to their effective growth in the water column (27). Zooplankton have been shown to be a major reservoir for NTVC only during relatively short periods in summer (19) and were therefore not specifically investigated in the present study.
Summary and conclusions.
With the used qPCR strategy (assessing GR rates for each single sample and correcting the obtained qPCR results with the GR rates), we were able to determine the spatiotemporal variability of V. cholerae abundance in lakes with a highly challenging matrix. In the investigated lakes, the large number of suspended particles and the concentrations of high-molecular-weight organic matter influenced the recovery of target DNA, resulting in a high variability in recovery rates. Such challenging conditions may not occur in other recreational water bodies, but the determination of recovery rates for each individual sample (standard cell spike) and strategies to improve DNA recovery (such as the addition of nontarget DNA saturating potential DNA binding sites on the surface of particles) should be eventually considered (47, 48). Despite the methodological challenges, highly significant correlations of the corrected qPCR data to the results of a culture-based method and a cytometric quantification method were found. While the corrected qPCR results were of a magnitude comparable to that of the cytometric approach, they were 1 to 2 orders of magnitude higher than the cultivation data.
In general, the detected V. cholerae concentrations were in the upper range of values reported in the literature; higher V. cholerae abundances have been reported only in areas where cholera is endemic. Thus, the lake used for recreation and the soda pools nearby can be regarded as hot spots of NTVC, which is of importance for public health. Moreover, global warming scenarios indicate a drastic further increase in average surface water temperatures by about 2°C for Neusiedler See and the Central European region over the next 30 years, after an already observed increase of about 2°C during the past 30 years (49, 50). As the spatiotemporal pattern in NTVC concentrations was mainly driven by temperature, increasing concentrations and potential threats can be expected in the future due to global warming. To be prepared for such scenarios, monitoring of recreational waters concerning the abundance of NTVC with suitable, standardizable methods that can be used in routine laboratories is a prerequisite. In addition to the determination of NTVC, toxigenic V. cholerae can also be quantified with this multiplex qPCR. Based on the current study, we can recommend using the developed qPCR strategy for such purposes, with the need for significant improvements in DNA recovery.
MATERIALS AND METHODS
Sample collection.
At the Neusiedler See, samples were taken from four sampling sites. Among them there were two open water sites, one in the southern area (sampling site 5) and one in the northern part of the lake (sampling site 24); one site was situated in a small open brown-water area inside the reed stand (sampling site 36), and one sampling site was close to the runoff from a municipal wastewater treatment plant (sampling site 29). In addition, samples were taken from the two soda pools, Zicklacke (ZL) and Unterer Stinker (US). For this study, sampling sites 5, 36, and ZL were monitored from April to November 2011, and in the year 2014 all described sampling sites were monitored from January to November. In the warm period (April to October), sampling took place in biweekly intervals, and during the rest of the year, sampling was done in monthly intervals. Lake water samples were taken with a motorboat, and soda pool samples were taken by foot with chest waders. From each site, triplicate water samples were collected from a water depth of 20 to 30 cm in autoclaved 500-ml glass flasks and transported to the laboratory at in situ temperature (±2°C) in a cooling box within 1.5 h after the last sample was taken.
Environmental parameters.
For each sampling site and time point, a variety of environmental variables were determined. Conductivity (LF330; WTW, Germany), water temperature, pH (GHM, Seibold Vienna, Austria), oxygen concentration (OXI 330i; WTW), and Secchi depth were measured in situ. Additionally, 1 liter of water was collected in a clean plastic bottle to analyze inorganic nutrients, chlorophyll a, total suspended solids (TSS), and DOC, and the spectrophotometric absorption ratio was measured at 250 nm and 365 nm according to the methods described by Eiler et al. (30) and Kirschner et al. (27).
Sample preparation.
The triplicate water samples were thoroughly mixed and subdivided into two 200-ml aliquots each. One aliquot was used for analysis of V. cholerae concentration by qPCR, and the second one was used for the standard addition to calculate the global recovery rate as described in detail by Bliem and colleagues (21) and for calculating the V. cholerae concentrations (see “Global recovery rate and calculation of V. cholerae abundances”).
Quantification of V. cholerae. (i) Multiplex qPCR.
The DNA extraction and multiplex qPCR protocol developed by Bliem et al. (21) was used to quantify V. cholerae in the water samples. The multiplex qPCR targets toxigenic (ctxA gene [cholera toxin A gene]) and nontoxigenic (ompW gene [outer membrane protein W gene]) V. cholerae and includes an exogenous inhibition control (egfp gene [enhanced green fluorescent protein gene]). Briefly, DNA was extracted from the pellet of centrifuged 200-ml samples (4,600 × g for 30 min) by use of the MoBio PowerSoil DNA extraction kit (MoBio, Carlsbad, CA, USA). Eluted DNA was stored at −80°C until qPCR analysis was performed. All multiplex qPCRs were carried out in a final reaction volume of 20 μl containing 10 μl 2× QuantiTect Multiplex PCR NoROX master mix (Qiagen), 2 μl of each primer-probe mixture, 2 μl of DNA template, and PCR-grade water. The exogenous inhibition control (IC) was added to the qPCR master mix at a final concentration of 1 × 103 per reaction mixture. Each qPCR analysis included a standard dilution series of ctxA, ompW, and egfp plasmid, a positive control containing all three targets, and a negative (no-template) control. Measurements were performed on a LightCycler 480 II, and analysis was done with LightCycler 480 II software release 1.5 (Roche). Quantification cycle (Cq) values were calculated based on the curve fit method, and a Cq cutoff level of 39 was defined for both ctxA and ompW based on the results of the standard dilution series. Color compensation was performed according to the manufacturer's guidelines. With the applied settings, a method detection limit of 5 copies per reaction was achieved for all three targets (21). To visualize problematic cases which might lead to a false-negative result, our qPCR was tested against a panel of various target concentrations (template matrix assay [21]). Delayed Cq values were recognized only for low ompW concentrations in the presence of high concentrations of ctxA and IC. Thus, false-negative results may only occur in the unrealistic or rare situation of a higher concentration of ctxA than of ompW (i.e., when the ctxA gene is present at high concentrations in a “host” other than V. cholerae) (21).
(ii) Global recovery rate and calculation of V. cholerae abundances.
To assess the global recovery rate for each individual sample, a known number of toxigenic V. cholerae O395 cells (5.4 × 105 cells, exact number determined by CARD-FISH/SPC; see below) was seeded into the replicate samples (see sample preparation described in reference 21), and the recovered number of V. cholerae cells was determined with multiplex qPCR. As a positive ctxA signal could not be detected in any sample (i.e., no toxigenic V. cholerae strains were present in the investigated lakes), the ctxA marker was used for calculating GR. The V. cholerae numbers (“corrected data”) were calculated from the measured qPCR results of the DNA extract from the nonspiked samples (“uncorrected” data) divided by the GR rate determined for each individual sample according to the following formula: corrected data = uncorrected data/GR rate.
V. cholerae abundance data were expressed in genomic units (GU) per 100 ml. Due to the fact that the target genes are single-copy genes, GU values are considered cell equivalents.
(iii) Cultivation.
The cultivation of V. cholerae from the water samples was done by membrane filtration as described by Schauer et al. (33). From each of the triplicate samples, subsamples of appropriate volumes (1, 10, and 100 ml for samples from Neusiedler See, and 0.01, 0.1, and 1 ml, filled up to 10 ml with sterile deionized water, for samples from the shallow soda pools) were taken and filtered, and the membrane was placed on thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates (Merck, Darmstadt, Germany). TCBS agar plates were incubated for 18 h at 37°C. Yellow, flat, 1- to 3-mm-diameter colonies were picked and streaked on nutrient agar without NaCl. Isolates showing growth on this agar were considered presumptive V. cholerae. Five representative presumptive V. cholerae isolates of each water sample were confirmed via species-specific ompW-based PCR (33). The SLOD of the cultivation procedure was 3 CFU 100 ml−1 (A. Kirschner, S. Hirk, S. Jakwerth, S. Rehak, A. H. Farnleitner, S. Huhulescu, and A. Indra, submitted for publication).
(iv) CARD-FISH/SPC.
For 2011, V. cholerae concentrations determined via a cytometric method (CARD-FISH/SPC) (33) were taken for comparison with the qPCR data. All CARD-FISH/SPC data used here were retrieved from the report of Schauer et al. (19).
Statistical analysis.
Statistical analysis was performed with SPSS 22.0. For correlation analysis, Spearman rank correlation was used. Multiple linear stepwise regression was used for predicting global recovery rates and V. cholerae abundance from environmental parameters. For evaluating whether nonlinear models are better suited for prediction, correlation scatterplots of the GR rates and V. cholerae abundance with all individual environmental variables were visually inspected and supported by mathematical calculations. Polynomial models were the only ones that partly exhibited a higher goodness of fit than the linear models and were used in multiple nonlinear regression analysis. For testing differences between sites and methods, analysis of variance (ANOVA) and Student's t test were applied. Microbiological data were log10 transformed to obtain normal distribution.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Haider and Rudolf Schalli (Biological Research Institute, Burgenland) for sampling and on-board analysis of environmental parameters. We give special thanks to Stefan Jakwerth (Medical University of Vienna) for cultivation-based enumeration of V. cholerae as well as to Franz Rauchwarter, Peter Gisch, Jutta Prückler, Alois Herzig, and Thomas Zechmeister (Biological Research Institute, Burgenland) for providing chlorophyll a, DOC, total phosphorus, NH4, NO3, and total suspended solids data. We also thank István Hatvani (Institute for Geological and Geochemical Research, Hungarian Academy of Sciences) for his help in multivariate nonlinear regression analysis.
This study was financed by Austrian Science Fund (FWF) project no. P21625-B20.
Footnotes
For this virtual institution, see http://www.waterandhealth.at.
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00317-18.
REFERENCES
- 1.Crowe SJ, Newton AE, Gould LH, Parsons MB, Stroika S, Bopp CA, Freeman M, Greene K, Mahon BE. 2016. Vibriosis, not cholera: toxigenic Vibrio cholerae non-O1, non-O139 infections in the United States, 1984-2014. Epidemiol Infect 144:3335–3341. doi: 10.1017/S0950268816001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engel MF, Muijsken MA, Mooi-Kokenberg E, Kuijper EJ, van Westerloo DJ. 2016. Vibrio cholerae non-O1 bacteraemia: description of three cases in the Netherlands and a literature review. Euro Surveill 21:pii=30197. doi: 10.2807/1560-7917.ES.2016.21.15.30197. [DOI] [PubMed] [Google Scholar]
- 3.Huhulescu S, Indra A, Feierl G, Stoeger A, Ruppitsch W, Sarkar B, Allerberger F. 2007. Occurrence of Vibrio cholerae serogroups other than O1 and O139 in Austria. Wien Klin Wochenschr 119:235–241. doi: 10.1007/s00508-006-0747-2. [DOI] [PubMed] [Google Scholar]
- 4.Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez-Urtaza J. 2013. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Chang 3:73–77. doi: 10.1038/nclimate1628. [DOI] [Google Scholar]
- 5.Maraki S, Christidou A, Anastasaki M, Scoulica E. 2016. Non-O1, non-O139 Vibrio cholerae bacteremic skin and soft tissue infections. Infect Dis (Lond) 48:171–176. doi: 10.3109/23744235.2015.1104720. [DOI] [PubMed] [Google Scholar]
- 6.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. 2012. Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin Infect Dis 54(Suppl 5):S391–S395. doi: 10.1093/cid/cis243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vezzulli L, Grande C, Reid PC, Helaouet P, Edwards M, Hofle MG, Brettar I, Colwell RR, Pruzzo C. 2016. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc Natl Acad Sci U S A 113:E5062–E5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sterk A, Schets FM, de Roda Husman AM, de Nijs T, Schijven JF. 2015. Effect of climate change on the concentration and associated risks of Vibrio spp. in Dutch recreational waters. Risk Anal 35:1717–1729. doi: 10.1111/risa.12365. [DOI] [PubMed] [Google Scholar]
- 9.Baker-Austin C, Trinanes JA, Salmenlinna S, Lofdahl M, Siitonen A, Taylor NG, Martinez-Urtaza J. 2016. Heat wave-associated vibriosis, Sweden and Finland, 2014. Emerg Infect Dis 22:1216–1220. doi: 10.3201/eid2207.151996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stypulkowska-Misiurewicz H, Pancer K, Roszkowiak A. 2006. Two unrelated cases of septicaemia due to Vibrio cholerae non-O1, non-O139 in Poland, July and August 2006. Euro Surveill 11:E061130.2. [DOI] [PubMed] [Google Scholar]
- 11.Hirk S, Huhulescu S, Allerberger F, Lepuschitz S, Rehak S, Weil S, Gschwandtner E, Hermann M, Neuhold S, Zoufaly A, Indra A. 2016. Necrotizing fasciitis due to Vibrio cholerae non-O1/non-O139 after exposure to Austrian bathing sites. Wien Klin Wochenschr 128:141–145. doi: 10.1007/s00508-015-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrovic K, Rudman F, Ottaviani D, Sestan Crnek S, Leoni F, Skrlin J. 2016. A rare case of necrotizing fasciitis caused by Vibrio cholerae O8 in an immunocompetent patient. Wien Klin Wochenschr 128:728–730. doi: 10.1007/s00508-016-1060-3. [DOI] [PubMed] [Google Scholar]
- 13.Huq A, Haley BJ, Taviani E, Chen A, Hasan NA, Colwell RR. 2012. Detection, isolation, and identification of Vibrio cholerae from the environment. Curr Protoc Microbiol Chapter 6:Unit6A.5. doi: 10.1002/9780471729259.mc06a05s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDC. 2014. Laboratory methods for the diagnosis of Vibrio cholerae, p 27–37 Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 15.ISO. 2007. Microbiology of food and animal feeding stuffs—horizontal method for the detection of potentially enteropathogenic Vibrio spp., vol ISO/TS 21872-1, p 1–19 International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 16.Public Health England. 2005. Identification of Vibrio species, vol BSOP ID 19, p 1–11 Public Health England, London, United Kingdom. [Google Scholar]
- 17.Wu B, Liang W, Kan B. 2016. Growth phase, oxygen, temperature, and starvation affect the development of viable but non-culturable state of Vibrio cholerae. Front Microbiol 7:404. doi: 10.3389/fmicb.2016.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B, Liang W, Kan B. 2015. Enumeration of viable non-culturable Vibrio cholerae using propidium monoazide combined with quantitative PCR. J Microbiol Methods 115:147–152. doi: 10.1016/j.mimet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Schauer S, Jakwerth S, Bliem R, Baudart J, Lebaron P, Huhulescu S, Kundi M, Herzig A, Farnleitner AH, Sommer R, Kirschner A. 2015. Dynamics of Vibrio cholerae abundance in Austrian saline lakes, assessed with quantitative solid-phase cytometry. Environ Microbiol 17:4366–4378. doi: 10.1111/1462-2920.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ISO. 2012. ISO/TS 12869: water quality—detection and quantification of Legionella spp. and/or Legionella pneumophila by concentration and genic amplification by quantitative polymerase chain reaction (qPCR). International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 21.Bliem R, Schauer S, Plicka H, Obwaller A, Sommer R, Steinrigl A, Alam M, Reischer GH, Farnleitner AH, Kirschner A. 2015. A novel triplex quantitative PCR strategy for quantification of toxigenic and nontoxigenic Vibrio cholerae in aquatic environments. Appl Environ Microbiol 81:3077–3085. doi: 10.1128/AEM.03516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid RB, Ferdous J, Tulsiani S, Jensen PKM, Begum A. 2017. Development and validation of a novel real-time assay for the detection and quantification of Vibrio cholerae. Front Public Health 5:109. doi: 10.3389/fpubh.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vezzulli L, Stauder M, Grande C, Pezzati E, Verheye HM, Owens NJ, Pruzzo C. 2015. gbpA as a novel qPCR target for the species-specific detection of Vibrio cholerae O1, O139, non-O1/non-O139 in environmental, stool, and historical continuous plankton recorder samples. PLoS One 10:e0123983. doi: 10.1371/journal.pone.0123983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubala AJ. 2006. Multiplex real-time PCR detection of Vibrio cholerae. J Microbiol Methods 65:278–293. doi: 10.1016/j.mimet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Ferdous J, Hossain ZZ, Tulsiani S, Rashid RB, Jensen PKM, Begum A. 2016. Optimization and validation of real time PCR assays for absolute quantification of toxigenic Vibrio cholerae and Escherichia coli. Trop Biomed 33:641–651. [PubMed] [Google Scholar]
- 26.Siboni N, Balaraju V, Carney R, Labbate M, Seymour JR. 2016. Spatiotemporal dynamics of Vibrio spp. within the Sydney Harbour estuary. Front Microbiol 7:460. doi: 10.3389/fmicb.2016.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirschner AKT, Schlesinger J, Farnleitner AH, Hornek R, Süß B, Golda B, Herzig A, Reitner B. 2008. Rapid growth of planktonic Vibrio cholerae non-O1/non-O139 strains in a large alkaline lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl Environ Microbiol 74:2004–2015. doi: 10.1128/AEM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pretzer C, Druzhinina IS, Amaro C, Benediktsdottir E, Hedenstrom I, Hervio-Heath D, Huhulescu S, Schets FM, Farnleitner AH, Kirschner AK. 2017. High genetic diversity of Vibrio cholerae in the European lake Neusiedler See is associated with intensive recombination in the reed habitat and the long-distance transfer of strains. Environ Microbiol 19:328–344. doi: 10.1111/1462-2920.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herzig A, Dokulil M. 2001. Neusiedler See—a steppe lake in Europe, p 401–415. In Ecology and protection of lakes. Facultas UTB, Vienna, Austria: (In German.) [Google Scholar]
- 30.Eiler A, Farnleitner AH, Zechmeister TC, Herzig A, Hurban C, Wesner W, Krachler R, Velimirov B, Kirschner AK. 2003. Factors controlling extremely productive heterotrophic bacterial communities in shallow soda pools. Microb Ecol 46:43–54. doi: 10.1007/s00248-002-2041-9. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner AK, Eiler A, Zechmeister TC, Velimirov B, Herzig A, Mach R, Farnleitner AH. 2002. Extremely productive microbial communities in shallow saline pools respond immediately to changing meteorological conditions. Environ Microbiol 4:546–555. doi: 10.1046/j.1462-2920.2002.00334.x. [DOI] [PubMed] [Google Scholar]
- 32.Reitner B, Herzig A, Herndl GJ. 1999. Dynamics in bacterioplankton production in a shallow, temperate lake (Neusiedl, Austria): evidence for dependence on macrophyte production rather than on phytoplankton. Aquat Microb Ecol 19:245–254. doi: 10.3354/ame019245. [DOI] [Google Scholar]
- 33.Schauer S, Sommer R, Farnleitner AH, Kirschner AKT. 2012. Rapid and sensitive quantification of Vibrio cholerae and Vibrio mimicus cells in water samples by use of catalyzed reporter deposition fluorescence in situ hybridization combined with solid-phase cytometry. Appl Environ Microbiol 78:7369–7375. doi: 10.1128/AEM.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stojanovic A, Kogelnig D, Mitteregger B, Mader D, Jirsa F, Krachler R, Krachler R. 2009. Major and trace element geochemistry of superficial sediments and suspended particulate matter of shallow saline lakes in Eastern Austria. Chem Erde 69:223–234. doi: 10.1016/j.chemer.2009.03.001. [DOI] [Google Scholar]
- 35.Yu WH, Li N, Tong DS, Zhou CH, Lin CXC, Xu CY. 2013. Adsorption of proteins and nucleic acids on clay minerals and their interactions: a review. Appl Clay Sci 80-81:443–452. [Google Scholar]
- 36.Saeki K, Ihyo Y, Sakai M, Kunito T. 2011. Strong adsorption of DNA molecules on humic acids. Environ Chem Lett 9:505–509. doi: 10.1007/s10311-011-0310-x. [DOI] [Google Scholar]
- 37.Jiang SC, Fu W. 2001. Seasonal abundance and distribution of Vibrio cholerae in coastal waters quantified by a 16S-23S intergenic spacer probe. Microb Ecol 42:540–548. doi: 10.1007/s00248-001-0029-5. [DOI] [PubMed] [Google Scholar]
- 38.Batabyal P, Mookerjee S, Einsporn MH, Lara RJ, Palit A. 2016. Environmental drivers on seasonal abundance of riverine-estuarine V. cholerae in the Indian Sundarban mangrove. Ecol Indic 69:59–65. doi: 10.1016/j.ecolind.2016.04.004. [DOI] [Google Scholar]
- 39.Machado A, Bordalo AA. 2016. Detection and quantification of Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus in coastal waters of Guinea-Bissau (West Africa). Ecohealth 13:339–349. doi: 10.1007/s10393-016-1104-1. [DOI] [PubMed] [Google Scholar]
- 40.Neogi SB, Islam MS, Nair GB, Yamasaki S, Lara RJ. 2012. Occurrence and distribution of plankton-associated and free-living toxigenic Vibrio cholerae in a tropical estuary of a cholera endemic zone. Wetl Ecol Manag 20:271–285. doi: 10.1007/s11273-012-9247-5. [DOI] [Google Scholar]
- 41.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eiler A, Gonzalez-Rey C, Allen S, Bertilsson S. 2007. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol Ecol 60:411–418. doi: 10.1111/j.1574-6941.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 43.Worden AZ, Seidel M, Smriga S, Wick A, Malfatti F, Bartlett D, Azam F. 2006. Trophic regulation of Vibrio cholerae in coastal marine waters. Environ Microbiol 8:21–29. doi: 10.1111/j.1462-2920.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- 44.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. doi: 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 45.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vezzulli L, Pruzzo C, Huq A, Colwell RR. 2010. Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ Microbiol Rep 2:27–33. doi: 10.1111/j.1758-2229.2009.00128.x. [DOI] [PubMed] [Google Scholar]
- 47.Lever MA, Torti A, Eickenbusch P, Michaud AB, Šantl-Temkiv T, Jørgensen BB. 2015. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front Microbiol 6:476. doi: 10.3389/fmicb.2015.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart JR, Gast RJ, Fujioka RS, Solo-Gabriele HM, Meschke JS, Amaral-Zettler LA, del Castillo E, Polz MF, Collier TK, Strom MS, Sinigalliano CD, Moeller PD, Holland AF. 2008. The coastal environment and human health: microbial indicators, pathogens, sentinels and reservoirs. Environ Health 7(Suppl 2):S3. doi: 10.1186/1476-069X-7-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dokulil MT. 2014. Predicting summer surface water temperatures for large Austrian lakes in 2050 under climate change scenarios. Hydrobiologia 731:19–29. doi: 10.1007/s10750-013-1550-5. [DOI] [Google Scholar]
- 50.Woolway RI, Dokulil MT, Marszelewski W, Schmid M, Bouffard D, Merchant CJ. 2017. Warming of Central European lakes and their response to the 1980s climate regime shift. Clim Change 142:505–520. doi: 10.1007/s10584-017-1966-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.