ABSTRACT
Fungi can decompose plant biomass into small oligo- and monosaccharides to be used as carbon sources. Some of these small molecules may induce metabolic pathways and the production of extracellular enzymes targeted for degradation of plant cell wall polymers. Despite extensive studies in ascomycete fungi, little is known about the nature of inducers for the lignocellulolytic systems of basidiomycetes. In this study, we analyzed six sugars known to induce the expression of lignocellulolytic genes in ascomycetes for their role as inducers in the basidiomycete white-rot fungus Dichomitus squalens using a transcriptomic approach. This identified cellobiose and l-rhamnose as the main inducers of cellulolytic and pectinolytic genes, respectively, of D. squalens. Our results also identified differences in gene expression patterns between dikaryotic and monokaryotic strains of D. squalens cultivated on plant biomass-derived monosaccharides and the disaccharide cellobiose. This suggests that despite conservation of the induction between these two genetic forms of D. squalens, the fine-tuning in the gene regulation of lignocellulose conversion is differently organized in these strains.
IMPORTANCE Wood-decomposing basidiomycete fungi have a major role in the global carbon cycle and are promising candidates for lignocellulosic biorefinery applications. However, information on which components trigger enzyme production is currently lacking, which is crucial for the efficient use of these fungi in biotechnology. In this study, transcriptomes of the white-rot fungus Dichomitus squalens from plant biomass-derived monosaccharide and cellobiose cultures were studied to identify compounds that induce the expression of genes involved in plant biomass degradation.
KEYWORDS: basidiomycetes, cellobiose, plant biomass degradation, regulation, rhamnose
INTRODUCTION
Plant biomass is an emerging source of fuels and chemicals, since it is the most abundant renewable organic material on earth (1, 2). However, its depolymerization is usually required before it can be used in biotechnological applications. White-rot fungi are a group of basidiomycetes whose natural substrate is wood (3); therefore, they produce a set of enzymes that are ideally suited for the depolymerization of components present in wood.
Wood cell walls consist mainly of four different polymers, cellulose, hemicellulose, pectin, and lignin, with cellulose being the major structural component (4). All of these polymers form a complex and compact lignocellulose network. White-rot fungi have the unique ability to efficiently degrade all the polymeric components of the plant cell walls, including the recalcitrant aromatic lignin. This makes them attractive candidates for various applications in the biobased economy (5). The polysaccharide fraction of plant biomass, which comprises cellulose, hemicellulose, and pectin, is composed of several different monomeric sugars. While cellulose consists solely of d-glucose, hemicelluloses (xylan, xyloglucan, or mannan) and pectin consist of various sugar monomers, such as d-glucose, d-galactose, d-mannose, d-xylose, l-arabinose, l-rhamnose, d-galacturonic acid, d-glucuronic acid, and l-fucose (6).
Fungi produce a broad range of extracellular enzymes to facilitate the depolymerization of plant biomass (7), most of which are classified in the Carbohydrate-Active enZYmes (CAZy) database (www.cazy.org) (8). The CAZy database divides the enzymes and associated modules into families and subfamilies and provides genomic, structural, and biochemical information on them. The action of carbohydrate-active enzymes (CAZymes) results in the formation of monomeric and small oligomeric components that are taken up into the fungal cells and catabolized for growth and reproduction.
The expression of the genes encoding plant biomass-degrading enzymes is tightly controlled to ensure efficient utilization of the carbon and energy sources available in fungal habitats (9). Production of the most suitable set of enzymes is achieved by inducing or repressing regulatory systems. Monosaccharides or oligosaccharides from plant biomass can trigger signaling pathways resulting in the activation of a transcriptional regulator. Several transcriptional activators and repressors related to lignocellulose degradation have been characterized from ascomycete fungi (10, 11); however, except for Ace3 and Cre1, none of these have orthologs in basidiomycete fungi (12). Interestingly, the sets of genes that are expressed in response to specific plant biomass substrates appear to be highly similar between ascomycetes and basidiomycetes, despite the absence of orthologous regulators (13, 14). This similarity in the expression profiles of the CAZyme-encoding genes together with the absence of orthologous regulators suggests parallel evolution of corresponding regulatory systems in these two fungal phyla (14).
Uncovering the regulatory systems driving wood degradation in the white-rot basidiomycetes is crucial to understanding the abilities of this group of fungi and to fully exploit them in biotechnological applications. A first step is the identification of the low-molecular-weight inducers of these regulators. In this study, the white-rot fungus Dichomitus squalens was cultivated on six monosaccharides derived from cellulose, hemicellulose, and pectin, as well as on the disaccharide cellobiose. Transcriptomics was used to identify which of these compounds induce the expression of genes encoding plant cell wall polymer-degrading enzymes. In nature, the predominant form of D. squalens is dikaryotic, but colonization of wood initiates from a spore that forms monokaryotic mycelium until it encounters a compatible mate. Previously, mono- and dikaryotic strains of D. squalens were suggested to have highly diverse abilities to degrade plant biomass (15). To evaluate if the diversity between these forms is due to regulatory-level differences, comparative transcriptomics of a monokaryotic progeny of the dikaryotic D. squalens strain was performed on the three carbon sources that were observed to induce plant biomass-degrading enzyme-encoding genes in the dikaryon.
RESULTS
Transcriptome analysis reveals cellobiose and l-rhamnose to be the main inducers of CAZyme-encoding genes in a D. squalens dikaryon.
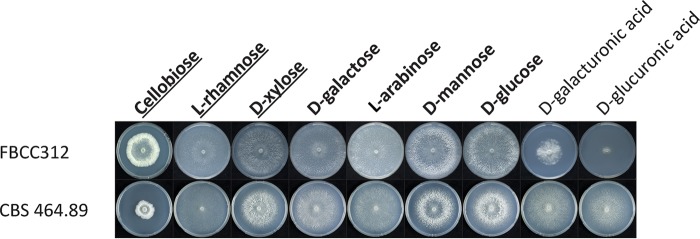
The dikaryotic and monokaryotic strains of D. squalens showed varied morphologies after 5 days of growth on the main plant polysaccharide-derived monomers and cellobiose (Fig. 1) (15). Both strains grew well on d-glucose, d-galactose, d-mannose, d-xylose, l-arabinose, and l-rhamnose. While the mycelium of the dikaryon FBCC312 fully covered those plates, the radial growth of the monokaryon CBS 464.89 was overall slower, except on l-rhamnose, and it formed a less dense mycelium. Compared to the dikaryon, the colony diameter of the monokaryon was smaller also on cellobiose, but the mycelium was denser, thus possibly indicating a better ability of CBS 464.89 to convert this carbon source. In contrast, compared to the monokaryon CBS 464.89, growth of the dikaryon FBCC312 was slow on d-galacturonic acid and especially poor on d-glucuronic acid. Most possibly due to the poor growth, extraction of high-quality RNA was not successful from these cultures, which were therefore omitted from the RNA sequencing (RNA-seq) analysis.
FIG 1.
Growth profiles of D. squalens dikaryon FBCC312 and monokaryon CBS 464.89 on different sugars that were considered potential inducers of plant cell wall-degrading CAZymes, based on studies in ascomycetes (12). The plate cultures that were subsequently used for transcriptomics in FBCC312 are in bold, and those for CBS 464.89 are in bold and underlined.
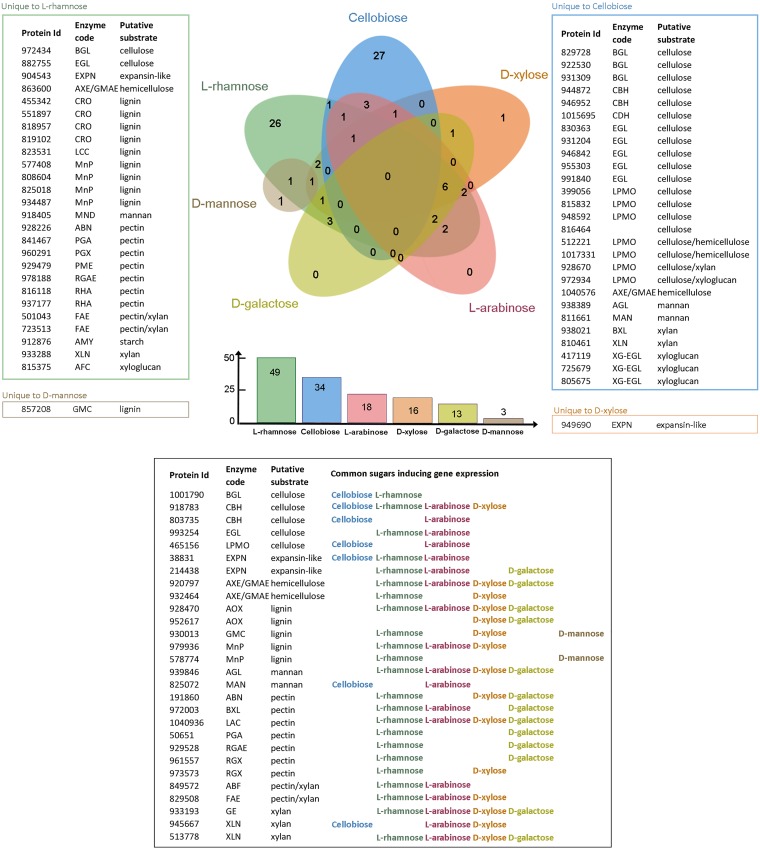
When the putative plant biomass-degrading CAZyme-encoding genes induced by the different sugars in D. squalens FBCC312 were compared to d-glucose (fold change, ≥2.5; P ≤ 0.01), 83 significantly upregulated genes were detected among all the sugars (Fig. 2; see also Table S1A to F in the supplemental material). Of these, 26 genes were unique to l-rhamnose and 27 genes were unique to cellobiose. In contrast, 28 genes were shared at least in two different sugars. On l-arabinose, d-xylose, l-rhamnose, d-galactose, and d-mannose, a smaller number of upregulated genes were present (Fig. 2). On one of the two pentoses studied, l-arabinose, 18 genes were upregulated, and all of them were also present in some of the other tested sugars. Similarly, none of the 13 genes upregulated in d-galactose were unique to this sugar. The growth of D. squalens on d-xylose, the other pentose studied, resulted in 16 upregulated CAZyme genes, with only one gene unique to this monosaccharide. On the hexose d-mannose, only three genes were upregulated, and two of them were also found on d-xylose or l-rhamnose. No upregulated CAZyme genes common to all the sugars were detected (Fig. 2). Downregulated genes were not analyzed.
FIG 2.
Comparison of the number of the plant cell wall-degrading CAZyme genes induced by the tested sugars in D. squalens FBCC312. The total number of induced CAZyme genes by each sugar is listed in the bar graph below the Venn diagram. CAZyme genes considered to be induced had a fold change value of ≥2.5 compared to d-glucose, a P value of ≤0.01, and an FPKM value of >10. See Fig. S1 for an explanation of the enzyme codes.
The proportion of the induced genes encoding CAZymes putatively acting on different polymeric components of plant biomass was determined from the transcriptome data (Fig. 2 and Tables 1 to 3). Cellobiose induced 34 CAZyme genes, of which most (68%) encode enzymes that putatively act on cellulose (Tables 1 and S1A and Fig. S1A). CAZyme family glycoside hydrolase 6 (GH6) and GH7 cellobiohydrolase-encoding genes were highly expressed in the presence of cellulose, with cel7c (JGI MycoCosm protein identification [ID] 944872, https://genome.jgi.doe.gov/Dicsqu464_1/Dicsqu464_1.home.html) showing 27-fold higher expression than d-glucose (Tables 1 and S1A). In contrast, the only highly expressed cellulolytic genes in the presence of other sugars were cellobiohydrolase genes cel6a (protein ID 803735) on l-arabinose (Fig. 2 and Table S1A and C) and cel7a (protein ID 918783) on l-rhamnose, d-xylose (Tables 1 to 3), and l-arabinose (Fig. 2 and Table S1). On l-rhamnose, 38% of the 49 induced genes encoded enzymes putatively involved in pectin degradation (Tables 2 and S1B and Fig. S1B). The number of genes with increased expression on d-xylose was lower (16), and thus, its role as an inducer was less evident than with cellobiose and l-rhamnose. However, 25% of the CAZyme genes induced on d-xylose were related to xylan degradation (Tables 3 and S1D and Fig. S1C). Lignin degradation-related CAZyme genes were induced on l-rhamnose (27%) and d-xylose (25%) cultures of D. squalens FBCC312 but not on cellobiose (Fig. 2 and Tables 1 to 3).
TABLE 1.
Expression of genes encoding putative plant biomass polysaccharide-degrading enzymes during the growth of D. squalens dikaryon FBCC312 in the presence of cellobiose compared to d-glucosea
| Protein ID | Enzyme code | Gene name | Fold change over d-glucose | P value over d-glucose | FPKM cellobiose | Putative substrate | CAZyme familyb |
|---|---|---|---|---|---|---|---|
| 829728 | BGL | 18.7 | 1.32E−75 | 229.2 | Cellulose | GH1 | |
| 1001790 | BGL | 7.2 | 5.90E−30 | 38.5 | Cellulose | CBM1-GH3 | |
| 922530 | BGL | 3.5 | 1.79E−15 | 44.3 | Cellulose | GH3 | |
| 931309 | BGL | 3.0 | 4.16E−20 | 96.5 | Cellulose | GH3 | |
| 944872 | CBH | cel7c | 27.3 | 3.19E−88 | 969.9 | Cellulose | GH7 |
| 918783 | CBH | cel7a | 14.6 | 1.83E−19 | 203.0 | Cellulose | GH7 |
| 803735 | CBH | cel6a | 9.6 | 7.73E−46 | 451.4 | Cellulose | CBM1-GH6 |
| 946952 | CBH | cel7b | 8.6 | 3.96E−11 | 252.4 | Cellulose | GH7 |
| 1015695 | CDH | 12.4 | 6.64E−47 | 107.7 | Cellulose | AA8-AA3_1 | |
| 931204 | EGL | 19.7 | 7.98E−44 | 182.3 | Cellulose | GH131-CBM1 | |
| 830363 | EGL | 18.1 | 6.75E−69 | 199.8 | Cellulose | CBM1-GH5_5 | |
| 955303 | EGL | 9.6 | 1.09E−63 | 426.1 | Cellulose | CBM1-GH5_5 | |
| 946842 | EGL | 8.2 | 5.82E−23 | 52.8 | Cellulose | CBM1-GH5_5 | |
| 991840 | EGL | 5.0 | 3.33E−12 | 140.6 | Cellulose | GH45 | |
| 399056 | LPMO | lpmo2 | 20.3 | 1.86E−56 | 417.2 | Cellulose | AA9 |
| 815832 | LPMO | 14.3 | 5.92E−34 | 99.1 | Cellulose | AA9-CBM1 | |
| 948592 | LPMO | 14.2 | 1.73E−45 | 269.7 | Cellulose | AA9-CBM1 | |
| 465156 | LPMO | 9.1 | 1.09E−33 | 340.1 | Cellulose | AA9 | |
| 816464 | 23.8 | 3.88E−40 | 160.6 | Cellulose | CBM1 | ||
| 1017331 | LPMO | 20.7 | 8.23E−52 | 280.7 | Cellulose/hemicellulose | AA9 | |
| 512221 | LPMO | 18.3 | 3.22E−54 | 417.8 | Cellulose/hemicellulose | AA9 | |
| 928670 | LPMO | 12.0 | 6.85E−26 | 195.7 | Cellulose/xylan | AA9 | |
| 972934 | LPMO | 2.8 | 0.003 | 17.6 | Cellulose/xyloglucan | AA9 | |
| 38831 | EXPN | 2.8 | 0.0001 | 24.5 | Expansin-like | NA | |
| 1040576 | AXE/GMAE | 16.2 | 1.08E−33 | 79.1 | Hemicellulose | CBM1-CE16 | |
| 938389 | AGL | 3.4 | 5.43E−13 | 109.6 | Mannan | GH27 | |
| 825072 | MAN | 3.5 | 8.65E−18 | 82.4 | Mannan | CBM1-GH5_7 | |
| 811661 | MAN | 2.8 | 1.99E−11 | 134.1 | Mannan | GH5_7 | |
| 938021 | BXL | 7.0 | 4.32E−22 | 101.1 | Xylan | GH5_22 | |
| 945667 | XLN | 9.7 | 9.97E−34 | 82.2 | Xylan | CBM1-GH10 | |
| 810461 | XLN | 6.4 | 7.48E−07 | 11.5 | Xylan | CBM1-GH10 | |
| 725679 | XG-EGL | 36.0 | 7.12E−53 | 200.8 | Xyloglucan | GH12 | |
| 805675 | XG-EGL | 13.8 | 5.31E−48 | 243.5 | Xyloglucan | GH74-CBM1 | |
| 417119 | XG-EGL | 3.8 | 8.38E−07 | 67.7 | Xyloglucan | GH12 |
Only genes for which expression was at least 2.5-fold higher than on d-glucose with a P value of ≤0.01 and an FPKM value of >10 are included.
NA, not applicable.
TABLE 2.
Expression of genes encoding putative plant biomass polysaccharide-degrading enzymes during the growth of D. squalens dikaryon FBCC312 on l-rhamnose compared to d-glucosea
| Protein ID | Enzyme code | Gene name | Fold change over d-glucose | P value over d-glucose | FPKM l-rhamnose | Putative substrate | CAZyme family |
|---|---|---|---|---|---|---|---|
| 972434 | BGL | 2.7 | 5.64E−09 | 36.4 | Cellulose | GH3 | |
| 1001790 | BGL | 3.7 | 5.41E−13 | 18.1 | Cellulose | CBM1-GH3 | |
| 918783 | CBH | cel7a | 3.3 | 1.66E−04 | 33.5 | Cellulose | GH7 |
| 882755 | EGL | 7.5 | 3.31E−68 | 714.4 | Cellulose | GH45 | |
| 993254 | EGL | 3.4 | 7.38E−05 | 21.6 | Cellulose | GH45 | |
| 214438 | EXPN | 2.7 | 5.02E−13 | 543.1 | Expansin-like | ||
| 904543 | EXPN | 4.0 | 1.56E−06 | 130.5 | Expansin-like | ||
| 38831 | EXPN | 2.8 | 1.44E−04 | 22.4 | Expansin-like | ||
| 920797 | AXE/GMAE | 3.5 | 1.67E−11 | 93.9 | Hemicellulose | CE16 | |
| 932464 | AXE/GMAE | 2.6 | 1.64E−07 | 69.8 | Hemicellulose | CE16 | |
| 863600 | AXE/GMAE | 3.6 | 1.05E−04 | 15.5 | Hemicellulose | CE16 | |
| 818957 | CRO | 22.6 | 2.94E−65 | 567.6 | Lignin | AA5_1 | |
| 455342 | CRO | 11.0 | 1.85E−49 | 432.9 | Lignin | AA5_1 | |
| 819102 | CRO | 5.9 | 1.18E−19 | 186.2 | Lignin | AA5_1 | |
| 551897 | CRO | 40.8 | 5.43E−63 | 153.0 | Lignin | AA5_1 | |
| 930013 | GMC | 3.3 | 3.20E−06 | 20.1 | Lignin | AA3_2 | |
| 823531 | LCC | lcc8 | 2.6 | 2.79E−05 | 19.9 | Lignin | AA1_1 |
| 979936 | MnP | mnp7_short | 76.4 | 2.19E−153 | 3,594.2 | Lignin | AA2 |
| 808604 | MnP | mnp4_extralong | 7.2 | 4.16E−14 | 2,539.5 | Lignin | AA2 |
| 825018 | MnP | mnp5_short | 6.6 | 1.04E−10 | 2,001.9 | Lignin | AA2 |
| 578774 | MnP | mnp2_extralong | 2.8 | 6.53E−03 | 560.0 | Lignin | AA2 |
| 577408 | MnP | mnp3_extralong | 7.2 | 2.31E−26 | 153.4 | Lignin | AA2 |
| 934487 | MnP | mnp1_extralong | 2.7 | 1.19E−03 | 13.9 | Lignin | AA2 |
| 928470 | AOX | 6.2 | 1.09E−65 | 4,376.5 | Lignin | AA3_3 | |
| 939846 | AGL | 7.2 | 7.47E−40 | 448.6 | Mannan | GH27 | |
| 918405 | MND | 3.3 | 4.75E−08 | 14.2 | Mannan | GH2 | |
| 191860 | ABN | 6.0 | 6.28E−34 | 744.9 | Pectin | GH43 | |
| 972003 | BXL | 7.6 | 2.06E−41 | 142.4 | Pectin | GH43-CBM35 | |
| 928226 | ABN | 2.5 | 1.53E−03 | 16.5 | Pectin | GH43 | |
| 1040936 | LAC | 11.1 | 4.39E−104 | 1,101.3 | Pectin | GH35 | |
| 841467 | PGA | 4.0 | 2.30E−07 | 49.1 | Pectin | GH28 | |
| 960291 | PGX | 3.7 | 1.68E−09 | 59.3 | Pectin | GH28 | |
| 929479 | PME | 2.8 | 7.92E−08 | 50.6 | Pectin | CE8 | |
| 978188 | RGAE | 2.7 | 7.88E−04 | 26.2 | Pectin | CE12 | |
| 929528 | RGAE | 3.5 | 1.66E−04 | 21.7 | Pectin | CE12 | |
| 961557 | RGX | 3.7 | 5.27E−13 | 89.4 | Pectin | GH28 | |
| 973573 | RGX | 4.0 | 6.67E−10 | 27.6 | Pectin | GH28 | |
| 937177 | RHA | 2.9 | 5.64E−08 | 23.4 | Pectin | GH78 | |
| 816118 | RHA | 3.1 | 2.31E−08 | 21.5 | Pectin | GH78 | |
| 50651 | PGA | 2.9 | 2.11E−06 | 39.6 | Pectin | GH28 | |
| 849572 | ABF | 2.6 | 1.78E−10 | 75.9 | Pectin/xylan | GH51 | |
| 501043 | FAE | 3.1 | 1.59E−16 | 211.5 | Pectin/xylan | ||
| 723513 | FAE | 3.2 | 1.82E−18 | 208.0 | Pectin/xylan | ||
| 829508 | FAE | 4.5 | 2.20E−34 | 152.1 | Pectin/xylan | ||
| 912876 | AMY | 3.0 | 1.16E−23 | 296.9 | Starch | GH13_1 | |
| 933193 | GE | 20.4 | 8.94E−50 | 94.5 | Xylan | CBM1-CE15 | |
| 513778 | XLN | 31.9 | 2.07E−66 | 161.1 | Xylan | CBM1-GH10 | |
| 933288 | XLN | 14.4 | 2.35E−17 | 33.3 | Xylan | GH10 | |
| 815375 | AFC | 2.5 | 2.04E−17 | 275.5 | Xyloglucan | GH95 |
Only genes for which expression was at least 2.5-fold higher than on d-glucose with a P value of ≤0.01 and an FPKM value of >10 are included.
TABLE 3.
Expression of genes encoding putative plant biomass polysaccharide-degrading enzymes during growth of D. squalens dikaryon FBCC312 on d-xylose compared to d-glucosea
| Protein ID | Enzyme code | Gene name | Fold change over d-glucose | P value over d-glucose | FPKM l-rhamnose | Putative substrate | CAZyme family |
|---|---|---|---|---|---|---|---|
| 918783 | CBH | cel7a | 4.0 | 3.1E−05 | 44.555 | Cellulose | GH7 |
| 949690 | EXPN | 3.5 | 9.3E−04 | 21.47 | Expansin-like | EXPN | |
| 920797 | AXE/GMAE | 2.6 | 2.2E−06 | 72.855 | Hemicellulose | CE16 | |
| 932464 | AXE/GMAE | 2.5 | 2.5E−06 | 70.8 | Hemicellulose | CE16 | |
| 930013 | GMC | 4.2 | 5.6E−08 | 27.8 | Lignin | AA3_2 | |
| 979936 | MnP | mnp7_short | 4.0 | 5.5E−15 | 177.31 | Lignin | AA2 |
| 952617 | AOX | 2.8 | 2.1E−08 | 346.085 | Lignin | AA3_3 | |
| 928470 | AOX | 2.8 | 2.5E−20 | 2,079.17 | Lignin | AA3_3 | |
| 939846 | AGL | 3.4 | 7.3E−15 | 221.745 | Mannan | GH27 | |
| 1040936 | LAC | 3.2 | 5.7E−24 | 335.375 | Pectin | GH35 | |
| 973573 | RGX | 2.5 | 2.6E−04 | 17.97 | Pectin | GH28 | |
| 191860 | ABN | 3.1 | 1.8E−13 | 409.23 | Pectin | GH43 | |
| 829508 | FAE | 3.0 | 2.0E−17 | 107.605 | Pectin/xylan | ||
| 933193 | GE | 4.2 | 2.4E−10 | 19.25 | Xylan | CBM1-CE15 | |
| 513778 | XLN | 5.5 | 7.2E−15 | 27.48 | Xylan | CBM1-GH10 | |
| 945667 | XLN | 2.7 | 4.1E−06 | 22.06 | Xylan | CBM1-GH10 |
Only genes for which expression was at least 2.5-fold higher than on d-glucose with a P value of ≤0.01 and an FPKM value of >10 are included.
l-Rhamnose resulted in significant differences in gene expression of mono- and dikaryotic strains of D. squalens.
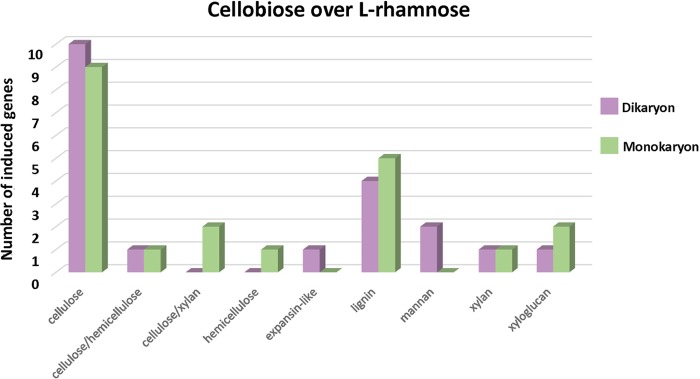
To compare possible regulatory differences in plant cell wall degradation between mono- and dikaryotic strains of D. squalens, the D. squalens monokaryon CBS 464.89 was cultivated on cellobiose, l-rhamnose, and d-xylose. When the transcriptomes from the three sugars were analyzed, a considerably smaller amount of highly expressed (fold change, ≥2.5; P ≤ 0.01; fragments per kilobase per million [FPKM], ≥10) CAZyme genes was detected in d-xylose (7–11) (Table S2) than in cellobiose (21) and l-rhamnose (45) (Tables S3B and S4B); therefore, the d-xylose transcriptome was excluded from further analyses. When the putative plant biomass-degrading CAZyme-encoding genes induced by cellobiose (FPKM, >10) were compared to those induced by l-rhamnose (fold change, ≥2.5; P ≤ 0.01) in the two D. squalens strains, 20 genes were highly expressed in the dikaryon, 11 (55%) of which are related to cellulose degradation (Table S3A). In the monokaryon, the number of genes (17) induced by cellobiose and the proportion of the genes putatively targeted for cellulose degradation were similar (12 [57%]), but 1/3 of these were different genes in the two strains (Fig. 3 and Table S3B). Overall, when the CAZyme gene expression of the mono- and dikaryotic strains in the presence of cellobiose was compared to that in the presence of l-rhamnose, no large differences were detected in the number of highly expressed genes targeting different putative plant cell wall compounds (Fig. 3).
FIG 3.
Plant biomass degradation-related CAZyme genes induced by cellobiose in comparison to l-rhamnose in the D. squalens dikaryon FBCC312 and monokaryon CBS 464.89. CAZyme genes considered to be induced by cellobiose were expressed at a 2.5-fold higher level (P ≤ 0.01) than on l-rhamnose with an FPKM value of >10.
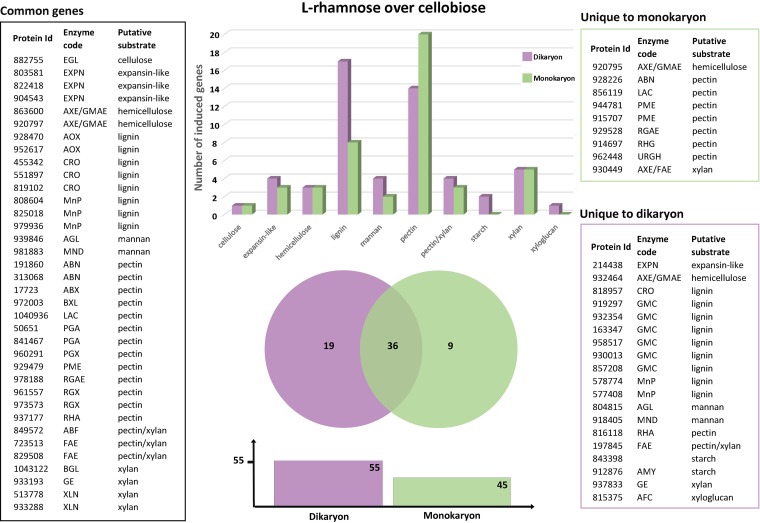
When the putative plant biomass-degrading CAZyme-encoding genes of D. squalens induced by l-rhamnose (FPKM, >10) were compared to those induced by cellobiose (fold change, ≤2.5; P ≤ 0.01), the differences between the mono- and dikaryotic strains were evident. The number of expressed genes was higher in the dikaryon (55) than in the monokaryon (45) (Fig. 4 and Table S4). Also, the proportion of the induced pectinolytic genes was higher in the monokaryon (44%) than in the dikaryon (25%) (Fig. 4 and Table S4). Nearly all pectinolytic genes upregulated in the dikaryon were also upregulated in the monokaryon, indicating that in the monokaryon, an additional set of genes responds to l-rhamnose. Interestingly, the proportion of the lignin degradation-related genes varied notably between the strains, as 31% and 18% of the induced genes in the dikaryon and the monokaryon, respectively, were putatively involved in lignin modification (Fig. 4). Here, the opposite pattern was observed from that for the pectinolytic genes, as all induced genes in the monokaryon were also induced in the dikaryon, suggesting that in the dikaryon, an additional set of ligninolytic genes responds to l-rhamnose.
FIG 4.
Common and strain-specific plant cell wall degradation-related CAZyme genes of D. squalens dikaryon FBCC312 (purple) and monokaryon CBS 464.89 (green) induced by l-rhamnose in comparison to cellobiose. The genes considered to be induced by l-rhamnose were expressed at a 2.5-fold higher level (P ≤ 0.01) than on cellobiose and an FPKM value of >10. See Table S2 for an explanation of the enzyme codes.
The largest difference in gene expression between the mono- and dikaryon was observed on l-rhamnose, since despite the similar number of genes induced by l-rhamnose and by cellobiose, only 36 of those genes were common in the two strains. From the nine genes unique to the monokaryon, seven (78%) genes were related to pectin degradation, whereas almost half (47%) of the 19 unique dikaryotic genes were related to lignin degradation (Fig. 4 and Table S4).
DISCUSSION
Gene regulation has been extensively studied in fungi, especially in the representatives from the phylum Ascomycota (10, 11). In contrast, the understanding of gene regulation in basidiomycetes is less extensive, largely due to the lack of characterized regulators and complications with gene transformation systems in these fungi. With the recent development of a transformation system (18), the white-rot fungus D. squalens has become an interesting species to study the regulation of plant biomass degradation in basidiomycetes. This species can degrade both hardwood and softwood (3), and several dikaryotic and related monokaryotic strains are available (15), as well as genome sequences for four monokaryotic strains (19).
In response to different conditions, fungi have the ability to up- or downregulate the expression of specific genes in order to adapt to their environment. Since the two major fungal phyla, Ascomycota and Basidiomycota, separated 500 million years ago (20), we may expect different responses in their gene expression. In addition, that dichotomy is possibly a reason for the scarcity of orthologous transcriptional regulators between ascomycetes and basidiomycetes (12, 14). The present study reveals detailed insights into the induction system of plant cell wall polymer degradation in D. squalens.
Similar to what has been observed in ascomycetes (21), genes encoding enzymes acting on a variety of plant polymers were upregulated in D. squalens when cultivated on cellobiose, l-rhamnose, and d-xylose. However, the distribution of these genes revealed a clear pattern for two of these compounds. On cellobiose, more than half of the upregulated genes encode enzymes acting on cellulose, while on l-rhamnose, more than 1/3 of the upregulated genes encoded enzymes acting on pectin. In contrast, on d-xylose, a more randomly distributed set of genes was upregulated, including a very small number of cellulolytic genes. At this point, it is not clear whether a specific inducer for (subsets of) hemicellulolytic genes exists in D. squalens.
Therefore, our results demonstrate that the dimer cellobiose is the primary inducer for cellulose degradation in D. squalens, while l-rhamnose is an inducer for pectin degradation. This role for cellobiose was previously suggested for the basidiomycete brown-rot fungus Postia placenta, where cellulases were induced by soluble sugars, especially cellobiose, after the repression of oxidoreductases (22).
In the well-studied white-rot fungus Phanerochaete chrysosporium, cellotriose and cellotetraose, but not cellobiose, were suggested to result in the strongest induction of cellulases (23). In light of the results of our study, we had a closer look at the data from Suzuki et al. (23) and do not think those results exclude a role for cellobiose as inducer. In that study, cellobiose is accumulating after 1 h cultivation of P. chrysosporium on cellotriose and cellotetraose. This is also the time point at which high induction of the cellobiohydrolase-encoding genes cel7C, cel7D, cel7F, celG, and cel6A is observed on cellotriose and cellotetraose cultures (23), suggesting that, in fact, the accumulated cellobiose may have caused induction of these genes. Microcrystalline cellulose (Avicel) has been found to upregulate the expression of cellobiohydrolase-encoding genes in D. squalens FBCC312, with the strongest effect on the gene cel7c (JGI MycoCosm protein ID 944872) (24, 25). This observation is in agreement with our results determined for this particular gene in the cellobiose cultures of D. squalens FBCC312, indicating that the results from the Avicel cultures may in fact also reflect the induction by cellobiose released from this substrate. Cellobiose has also been proposed as an inducer for several, but not all, ascomycete species (12), suggesting that there possibly are some similarities in the signaling mechanism of ascomycete and basidiomycete fungi, even if no orthologs for ascomycete cellobiose-responsive regulators were identified in basidiomycete genomes.
Considering the broad range of pectinolytic genes that were upregulated in the presence of l-rhamnose, this sugar appears to be a main inducer for pectinolytic gene expression in D. squalens. An l-rhamnose-responsive pectinolytic regulator (RhaR) has been identified in the ascomycete fungus Aspergillus niger (26), but this regulator seems to have a narrower role in activating pectinolytic genes than our data suggest for D. squalens (27). Furthermore, D. squalens does not have an ortholog of RhaR, nor of the other two A. niger pectinolytic regulators GaaR and AraR (12), indicating significant differences in pectinolytic regulation in D. squalens. In A. niger, genes encoding pectinolytic enzymes have recently been shown to be induced mainly by d-galacturonic acid (27), while l-rhamnose and l-arabinose affect smaller sets of pectinolytic genes (28). Although we were not able to assess the role of d-galacturonic acid as a pectinolytic inducer in D. squalens due to the low quality of RNA extracted from those cultures, our results demonstrated l-rhamnose induction for a wide set of pectinolytic genes, especially from the glycoside hydrolase (GH) families. Considering the low percentage of pectin in wood (29), the development of an efficient regulatory system for pectin degradation in D. squalens may seem less important than that for cellulose degradation. However, it has been previously reported that pectin in wood is particularly present at high levels in bordered pits, which connect adjacent xylem parenchyma cells, and are often the point of entry for basidiomycete hyphae to the wood cell walls (30). Therefore, this polysaccharide may still be a significant target for white-rot fungi during the onset of wood colonization.
Differences in gene expression between the D. squalens mono- and dikaryon were observed especially on l-rhamnose. This effect was not limited to pectinolytic genes but was observed across all CAZyme genes. When both strains were exposed to l-rhamnose, the dikaryon showed higher upregulation for genes encoding lignin-modifying enzymes than did the monokaryon. The monokaryon instead showed higher upregulation for pectinolytic genes than the dikaryon. The proportion of induced ligninolytic genes during growth on l-rhamnose correlated with a previously reported comparison of enzymatic activities in mono- and dikaryotic strains of D. squalens (15). In addition, the dikaryon had a broader set of upregulated genes encoding CAZymes, like those active on starch and xyloglucan, which were not expressed in the monokaryon, as well as a larger number of genes involved in mannan degradation.
On cellobiose, the strains showed a similar pattern of CAZyme gene expression, both having a large proportion of cellulolytic genes upregulated but showing differences between individual genes. However, genes encoding mannanases and expansin-like proteins were induced only in the dikaryon. The broader induction of CAZyme-encoding genes in the dikaryon indicates that this form of the fungus has better ability to grow or utilize plant biomass in nature than the monokaryon. This was not only observed during growth on plant biomass substrates for D. squalens (15) but also for another white-rot species (Pleurotus ostreatus), a litter-degrading species (Agaricus bisporus), and a brown-rot species (Serpula lacrymans) (www.fung-growth.org).
The results of this study are an important step toward dissecting the regulatory network driving the expression of genes encoding plant biomass-degrading enzymes in the white-rot fungus D. squalens and, possibly, other basidiomycetes. Together with the recent development of a transformation system for D. squalens (18), this species is a good candidate as a model system for the molecular biological analysis of wood decay by white-rot fungi.
MATERIALS AND METHODS
Fungal strains and cultivation.
The dikaryotic D. squalens strain FBCC312 (the Fungal Biotechnology Culture Collection [FBCC], Department of Microbiology, University of Helsinki) and monokaryotic D. squalens strain CBS 464.89 (the CBS collection, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands), which is a direct offspring of the dikaryon FBCC312, were maintained on 2% (wt/vol) malt extract–1.5% (wt/vol) agar (MEA) plates. The strains were grown on low-nitrogen asparagine-succinate (LN-AS) medium (16) with 1.5% (wt/vol) agar plates supplemented with main monosaccharides derived from plant biomass polysaccharides, i.e., 25 mM d-glucose, d-glucuronic acid, d-galacturonic acid, l-rhamnose, d-galactose, d-xylose, d-mannose, or l-arabinose or 25 mM disaccharide cellobiose. For growth profiling, an agar plug (diameter, 0.5 cm) from an MEA plate that was covered with fresh fungal mycelium was placed at the center of the LN-AS plate and incubated at 28°C. As the mycelium of the dikaryon FBCC312 reached the edge of the d-glucose-supplemented plate after 5 days, this was chosen as the incubation time for all the plates.
RNA isolation.
Based on the growth profiling, six plant cell wall polysaccharide-derived monosaccharides and cellobiose were selected to be used as carbon sources to test the fungal response at the gene expression level. The dikaryon FBCC312 was cultivated as mentioned before on 25 mM d-glucose, l-rhamnose, d-galactose, d-xylose, d-mannose, l-arabinose, or cellobiose, but this time using a polycarbonate membrane (Maine Manufacturing, LLC) on the top of the agar plate to facilitate harvesting of the mycelium. The monokaryon CBS 464.89 was cultivated similarly, with l-rhamnose, d-xylose, and cellobiose as carbon sources. After 5 days of growth at 28°C, the most external mycelium from the edge of the colony (1.5 cm) of each plate was harvested and transferred to 2-ml Eppendorf tubes containing two carbon steel balls (diameter, 3/16”) and frozen in liquid nitrogen. The tubes were placed in precooled adapters (Qiagen) and ground for 1 min at a frequency of 25 s−1 using a TissueLyser II (Qiagen). TRIzol (Ambion) and the Nucleospin RNA extraction kit (Macherey-Nagel) were used for RNA isolation according to the instructions of the manufacturers. RNA was eluted using RNase-free H2O and stored at −45°C.
RNA sequencing analysis.
The quantity and quality of RNA were checked with a RNA6000 Nano assay using the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples were single-end sequenced using the Illumina HiSeq 2000 platform (Illumina). Purification of mRNA, synthesis of the cDNA library, and sequencing were conducted at BGI Tech Solutions Co., Ltd. (Hong Kong, China).
Raw reads were produced from the original image data by base calling. After data filtering, the adaptor sequences, reads with high “N” content (>10% of unknown bases), and low-quality reads (more than 50% bases with a quality value of <5%) were removed. After data filtering, on average, 99.9% clean reads remained in each sample. The clean reads were then mapped to the genome of D. squalens CBS 464.89 (http://genome.jgi.doe.gov/Dicsqu464_1) using Bowtie2 (31) and the BWA software (32). In average, 64.9% total mapped reads to the genome were achieved. The gene expression level was measured in FPKM (33) using the RSEM tool (34). Differential expression was identified by the DESeq2 (17), with a cutoff value of ≥2.5-fold change, FPKM value of ≥10, and adjusted P value of ≤0.01. For the comparison of the plant cell wall CAZyme genes considered highly expressed with previous parameters, a Venn diagram was constructed using the online tool (35). For CAZyme annotations, the JGI MycoCosm website was used (https://genome.jgi.doe.gov/mycocosm/proteins-browser/browse;qLeIA4?p=Dicsqu464_1).
Accession number(s).
The RNA-seq data have been submitted to the Gene Expression Omnibus (GEO) (36) with accession number GSE105076.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthieu Hainaut and Bernard Henrissat from Aix-Marseille University, France, for analysis of the CAZyme genes of D. squalens.
P.D. was supported by a grant from the ALW division of NWO (824.15.023) to R.P.D.V. The Academy of Finland grant no. 308284 to M.R.M. is acknowledged.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00403-18.
REFERENCES
- 1.Casado López S, Sietiö O-M, Hildén K, de Vries RP, Mäkelä MR. 2016. Homologous and heterologous expression of basidiomycete genes related to plant biomass degradation, p 119–160. In Schmoll M, Dattenböck C (ed), Gene expression systems in fungi: advancements and applications. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 2.Rytioja J, Hildén KS, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. 2014. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol Mol Biol Rev 78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rytioja J, Hildén K, Di Falco M, Zhou M, Aguilar-Pontes MV, Sietiö O-M, Tsang A, de Vries RP, Mäkelä MR. 2017. The molecular response of the white-rot fungus Dichomitus squalens to wood and non-woody biomass as examined by transcriptome and exoproteome analyses. Environ Microbiol 19:1237–1250. doi: 10.1111/1462-2920.13652. [DOI] [PubMed] [Google Scholar]
- 4.Sorieul M, Dickson A, Hill SJ, Pearson H. 2016. Plant fibre: molecular structure and biomechanical properties, of a complex living material, influencing its deconstruction towards a biobased composite. Materials 9:618. doi: 10.3390/ma9080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mäkelä MR, Bredeweg EL, Magnuson JK, Baker SE, de Vries RP, Hildén K. 2016. Fungal ligninolytic enzymes and their applications. Microbiol Spectr 4(6):FUNK-0017-2016. doi: 10.1128/microbiolspec.FUNK-0017-2016. [DOI] [PubMed] [Google Scholar]
- 6.Voragen AGJ, Coenen G, Verhoef RP, Schols HA. 2009. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem 20:263–275. doi: 10.1007/s11224-009-9442-z. [DOI] [Google Scholar]
- 7.van den Brink J, de Vries RP. 2011. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol 91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng M, Aguilar-Pontes MV, Hainaut M, Henrissat B, Hildén K, Mäkelä MR, de Vries RP. 2018. Comparative analysis of basidiomycete transcriptomes reveals a core set of expressed genes encoding plant biomass degrading enzymes. Fungal Genet Biol 112:40–46. doi: 10.1016/j.fgb.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Kowalczyk JE, Benoit I, de Vries RP. 2014. Regulation of plant biomass utilization in Aspergillus. Adv Appl Microbiol 88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 11.Daly P, van Munster JM, Archer DB, Raulo R. 2015. Transcriptional regulation and responses in filamentous fungi exposed to lignocellulose, p 82–127. In Silva RN. (ed), Mycology: current and future developments: fungal biotechnology for biofuel production. Bentham Science, University of Nottingham, Nottingham, United Kingdom. [Google Scholar]
- 12.Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, de Vries RP. 2017. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol Biofuels 10:152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rytioja J, Hildén K, Mäkinen S, Vehmaanperä J, Hatakka A, Mäkelä MR. 2015. Saccharification of lignocelluloses by carbohydrate active enzymes of the white rot fungus Dichomitus squalens. PLoS One 10:e0145166. doi: 10.1371/journal.pone.0145166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd RB, Zhou M, Ohm RA, Leeggangers HA, Visser L, de Vries RP. 2014. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics 15:214. doi: 10.1186/1471-2164-15-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casado López S, Theelen B, Manserra S, Issak TY, Rytioja J, Mäkelä MR, de Vries RP. 2017. Functional diversity in Dichomitus squalens monokaryons. IMA Fungus 8:17–25. doi: 10.5598/imafungus.2017.08.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatakka AI, Uusi-Rauva AK. 1983. Degradation of 14C-labelled poplar wood lignin by selected white-rot fungi. Eur J Appl Microbiol Biotechnol 17:235–242. doi: 10.1007/BF00510422. [DOI] [Google Scholar]
- 17.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daly P, Slaghek GG, Casado López S, Wiebenga A, Hilden KS, de Vries RP, Mäkelä MR. 2017. Genetic transformation of the white-rot fungus Dichomitus squalens using a new commercial protoplasting cocktail. J Microbiol Meth 143:38–43. doi: 10.1016/j.mimet.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 20.Berbee ML, Taylor JW. 2001. Fungal molecular evolution: gene trees and geologic time, p 229–245. In McLaughlin DJ, McLaughlin EG, Lemke PA (ed), Systematics and evolution. Springer Berlin Heidelberg, Berlin, Heidelberg, Germany. [Google Scholar]
- 21.Gruben BS, Mäkelä MR, Kowalczyk JE, Zhou M, Benoit-Gelber I, de Vries RP. 2017. Expression-based clustering of CAZyme-encoding genes of Aspergillus niger. BMC Genomics 18:900. doi: 10.1186/s12864-017-4164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Schilling JS. 2017. Role of carbon source in the shift from oxidative to hydrolytic wood decomposition by Postia placenta. Fungal Genet Biol 106:1–8. doi: 10.1016/j.fgb.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Igarashi K, Samejima M. 2010. Cellotriose and cellotetraose as inducers of the genes encoding cellobiohydrolases in the basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 76:6164–6170. doi: 10.1128/AEM.00724-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanden Wymelenberg A, Gaskell J, Mozuch M, Kersten P, Sabat G, Martinez D, Cullen D. 2009. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol 75:4058–4068. doi: 10.1128/AEM.00314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rytioja J, Hildén K, Hatakka A, Mäkelä MR. 2014. Transcriptional analysis of selected cellulose-acting enzymes encoding genes of the white-rot fungus Dichomitus squalens on spruce wood and microcrystalline cellulose. Fungal Genet Biol 72:91–98. doi: 10.1016/j.fgb.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Gruben BS, Zhou M, Wiebenga A, Ballering J, Overkamp KM, Punt PJ, de Vries RP. 2014. Aspergillus niger RhaR, a regulator involved in l-rhamnose release and catabolism. Appl Microbiol Biotechnol 98:5531–5540. [DOI] [PubMed] [Google Scholar]
- 27.Alazi E, Niu J, Kowalczyk JE, Peng M, Aguilar Pontes MV, van Kan JAL, Visser J, de Vries RP, Ram AFJ. 2016. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of d-galacturonic acid from pectin. FEBS Lett 590:1804–1815. doi: 10.1002/1873-3468.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczyk JE, Lubbers RJM, Peng M, Battaglia E, Visser J, de Vries RP. 2017. Combinatorial control of gene expression in Aspergillus niger grown on sugar beet pectin. Sci Rep 7:12356. doi: 10.1038/s41598-017-12362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjöström E. 1993. Wood chemistry: fundamentals and applications, 2nd ed Academic Press, San Diego, CA. [Google Scholar]
- 30.Ohm RA, de Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, de Vries RP, Record E, Levasseur A, Baker SE, Bartholomew KA, Coutinho PM, Erdmann S, Fowler TJ, Gathman AC, Lombard V, Henrissat B, Knabe N, Kües U, Lilly WW, Lindquist E, Lucas S, Magnuson JK, Piumi F, Raudaskoski M, Salamov A, Schmutz J, Schwarze FW, vanKuyk PA, Horton JS, Grigoriev IV, Wösten HA. 2010. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 28:957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinform 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.