ABSTRACT
The ubiquitous environmental bacterium Legionella pneumophila survives and replicates within amoebae and human macrophages by forming a Legionella-containing vacuole (LCV). In an intricate process governed by the bacterial Icm/Dot type IV secretion system and a plethora of effector proteins, the nascent LCV interferes with a number of intracellular trafficking pathways, including retrograde transport from endosomes to the Golgi apparatus. Conserved retrograde trafficking components, such as the retromer coat complex or the phosphoinositide (PI) 5-phosphatase D. discoideum 5-phosphatase 4 (Dd5P4)/oculocerebrorenal syndrome of Lowe (OCRL), restrict intracellular replication of L. pneumophila by an unknown mechanism. Here, we established an imaging flow cytometry (IFC) approach to assess in a rapid, unbiased, and large-scale quantitative manner the role of retrograde-linked PI metabolism and actin dynamics in the LCV composition. Exploiting Dictyostelium discoideum genetics, we found that Dd5P4 modulates the acquisition of fluorescently labeled LCV markers, such as calnexin, the small GTPase Rab1 (but not Rab7 and Rab8), and retrograde trafficking components (Vps5, Vps26, Vps35). The actin-nucleating protein and retromer interactor WASH (Wiskott-Aldrich syndrome protein [WASP] and suppressor of cAMP receptor [SCAR] homologue) promotes the accumulation of Rab1 and Rab8 on LCVs. Collectively, our findings validate IFC for the quantitative and unbiased analysis of the pathogen vacuole composition and reveal the impact of retrograde-linked PI metabolism and actin dynamics on the LCV composition. The IFC approach employed here can be adapted for a molecular analysis of the pathogen vacuole composition of other amoeba-resistant pathogens.
IMPORTANCE Legionella pneumophila is an amoeba-resistant environmental bacterium which can cause a life-threatening pneumonia termed Legionnaires' disease. In order to replicate intracellularly, the opportunistic pathogen forms a protective compartment, the Legionella-containing vacuole (LCV). An in-depth analysis of the LCV composition and the complex process of pathogen vacuole formation is crucial for understanding the virulence of L. pneumophila. Here, we established an imaging flow cytometry (IFC) approach to assess in a rapid, unbiased, and quantitative manner the accumulation of fluorescently labeled markers and probes on LCVs. Using IFC and L. pneumophila-infected Dictyostelium discoideum or defined mutant amoebae, a role for phosphoinositide (PI) metabolism, retrograde trafficking, and the actin cytoskeleton in the LCV composition was revealed. In principle, the powerful IFC approach can be used to analyze the molecular composition of any cellular compartment harboring bacterial pathogens.
KEYWORDS: amoeba, Dictyostelium discoideum, effector protein, endosome, GTPase, Golgi apparatus, host-pathogen interaction, Legionella pneumophila, pathogen vacuole, phosphoinositide lipid, OCRL, retrograde transport, retromer, sorting nexin, type IV secretion, vesicle trafficking
INTRODUCTION
Legionella pneumophila causes Legionnaires' disease, a severe pneumonia occurring in periodic outbreaks stemming from contaminated water sources (1, 2). The ubiquitous Gram-negative environmental bacterium resists and parasitizes free-living protozoa, with natural hosts including Acanthamoeba, Tetrahymena, and Vermamoeba species (3). Moreover, the bacterium thrives within Dictyostelium discoideum, a frequently employed protozoan experimental host (4). The bacterium avoids killing by protozoan phagocytes and human alveolar macrophages using an evolutionarily conserved mechanism. To this end, the bacteria establish a distinct replication-permissive intracellular compartment termed the Legionella-containing vacuole (LCV).
LCV formation is a complex process strictly dependent on the bacterial intracellular multiplication/defective organelle trafficking (Icm/Dot) type IV secretion system (T4SS) (5, 6). The LCV hijacks the secretory pathway, interacting intimately with the endoplasmic reticulum (ER) while concomitantly avoiding killing by the endocytic pathway. The hallmark of this compartment is its intimate interaction with the ER, resulting from the Rab1-dependent interception of secretory vesicles egressing from the ER (7–9). Thus, the LCV stains positively for several markers of the secretory pathway, including Rab1 and other Rab GTPases (10–12), and markers of the ER, including the ER-resident protein calnexin (13). Phosphatidylinositol 4-phosphate [PtdIns(4)P] is a major regulator of secretory vesicle trafficking through the Golgi apparatus (14, 15) and is also present on the LCV immediately upon infection (in a transient Icm/Dot-independent manner), and it later accumulates in an Icm/Dot-dependent manner (16).
The compartments of the endocytic and secretory pathways in eukaryotic cells communicate through vesicle trafficking. Mono- or polyphosphorylated phosphoinositide (PI) lipids and small GTPases of the Rab family define different intracellular membranes and govern the timely recruitment of vesicle trafficking transport machineries (17, 18). In addition to anterograde trafficking, functioning to transport newly synthesized proteins from the ER through the Golgi apparatus to their destination organelles and membranes, retrograde trafficking from endosomes back to the ER is also continually ongoing in the cell. Retrograde trafficking functions to maintain organelle integrity, prevents lysosomal degradation, and recycles cargo receptors of the secretory transport machinery (19, 20). Cargo of the retrograde pathway includes membrane proteins, such as cation-dependent or cation-independent mannose 6-phosphate receptor (CD- or CI-MPR) and sortilin/Vps10, as well as soluble proteins and lipids (21).
The budding of a retrograde trafficking vesicle from a donor membrane takes place in several steps, which have been elucidated only in part. First, the cargo receptors to be transported accumulate in clathrin-coated pits, and the Vps26-Vps29-Vps35 coat protein heterotrimer (termed retromer) is recruited by active, GTP-bound Rab7 (22, 23). Subsequently, sorting nexin (SNX) proteins in different combinations are recruited to the retromer complex by binding to resident phosphatidylinositol 3-phosphate [PtdIns(3)P], inducing membrane curvature and remodeling (24–26). The actin nucleation complex WASH (Wiskott-Aldrich syndrome protein [WASP] and suppressor of cAMP receptor [SCAR] homologue) binds the retromer (27) and triggers actin polymerization to push the emerging tubule (28). Simultaneously, the microtubule motor complex protein p150Glued/dynein bound to SNXs pulls the tubule (29). Then, scission factors, including EHD1 and dynamin, cut the elongating tubule and seclude the now cargo-loaded vesicle from the endosome (30). Finally, after uncoating, the vesicle is delivered to the target membrane (23).
D. discoideum 5-phosphatase 4 (Dd5P4) and its mammalian homologue, oculocerebrorenal syndrome of Lowe (OCRL), are PI 5-phosphatases that hydrolyze phosphatidylinositol 4,5-diphosphate [PtdIns(4,5)P2] and phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3] to yield PtdIns(4)P and PtdIns(3,4)P2, respectively (31, 32). Dd5P4/OCRL is crucially involved in retrograde trafficking from early endosomes to the trans-Golgi network as well as in receptor recycling between endosomes and the plasma membrane (33–36). The enzyme acts in the retrograde pathway by binding to clathrin (37, 38) and to several different Rab GTPases (39–41). Studies using D. discoideum as host cells revealed that upon infection with L. pneumophila, Dd5P4/OCRL is present and catalytically active on the LCV (42). Further, the intracellular growth of L. pneumophila is enhanced in the absence of Dd5P4/OCRL, as evidenced by deletion of the gene in D. discoideum (42) as well as depletion by small interfering RNA in human A549 lung epithelial cells (43), meaning that the phosphatase restricts intracellular replication of the pathogen. The mechanism by which this restriction is mediated is not known.
The L. pneumophila Icm/Dot effector RidL binds the retromer subunit Vps29, inhibits retrograde trafficking, and promotes intracellular bacterial replication, as demonstrated in D. discoideum and mouse macrophage experimental systems (43). By binding to Vps29, a hydrophobic β-hairpin in an N-terminal domain of RidL displaces the Rab7 GTPase-activating protein (GAP) and negative regulator of retrograde trafficking TBC1D5, possibly contributing to the effector function (44–46). In fact, RidL might block incoming retrograde traffic on the LCV, serving as an acceptor compartment of retrograde transport (47). While it is clear that retrograde trafficking is detrimental for and actively avoided by L. pneumophila (48), it is not known at which step in the retrograde pathway Dd5P4/OCRL has an effect or how exactly the bacterium interferes with the pathway.
In order to study the LCV composition and the hierarchical relationship of components of cellular trafficking pathways, objective and quantitative methods are pivotal. Flow cytometry has been established as an unbiased high-throughput method to investigate some Legionella-host cell interactions (49). Imaging flow cytometry (IFC) combines the advantages of flow cytometry with structural information from fluorescence microscopy and is a quantitative technique for rapidly analyzing the image content of cells in flow. In contrast to traditional microscopic imaging and manual scoring of pathogen vacuoles for the presence or absence of different markers, IFC provides a means for unbiased and rapid quantification (50). Here, we employed IFC to quantify different host factors on LCVs dependent on the D. discoideum genotype. Specifically, we assessed the impact of the PI 5-phosphatase Dd5P4 or the actin nucleator WASH on the acquisition of components implicated in the retrograde route or other trafficking pathways.
RESULTS AND DISCUSSION
Quantification of calnexin, PtdIns(4)P, and Rab1 on LCVs by IFC.
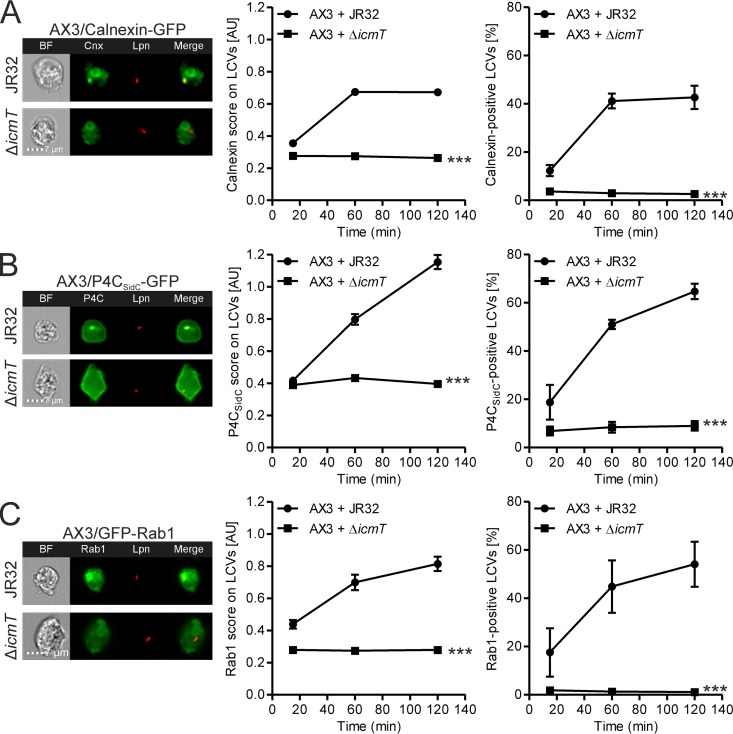
Upon infection, L. pneumophila creates a niche that favors its own survival and replication inside the cell, the LCV. In order to study the composition of the LCV, we developed two IFC-based methods, using either intact amoebae (Fig. 1) or LCVs from homogenized host cells (Fig. 2). We validated the IFC-based method for quantification of cellular components on the LCV in intact amoebae using well-studied markers, such as calnexin (13), the PtdIns(4)P probe P4CSidC (16), or the small GTPase Rab1 (10, 11). D. discoideum amoebae producing green fluorescent protein (GFP) fusions of one of the three proteins were infected with L. pneumophila producing mPlum and fixed, and 10,000 cells were acquired by IFC. The images were analyzed for colocalization between mPlum and GFP (Fig. 3), upon gating of cells containing only a single bacterium, as described in the legend to Fig. 1. As the infection progressed, the accumulation of calnexin, PtdIns(4)P, and Rab1 was evident for wild-type L. pneumophila, while a mutant lacking a functional Icm/Dot secretion system failed to recruit any of the markers to the vacuole (Fig. 3A to C). The data are presented as IFC scores, which is an efficient way of comparing different strains and allows for robust statistical analysis of the >1,000 cells per sample, but constitute arbitrary units with a certain degree of background signal. Alternatively, by placing a gate in the last step of the IFC analysis including only cells with a high degree of mPlum/GFP colocalization (Fig. 1), a percentage value for cells positive for the marker of interest is obtained (Fig. 3A to C). This representation corresponds more closely to how analogous microscopy data are usually presented (42, 43, 51, 52).
FIG 1.
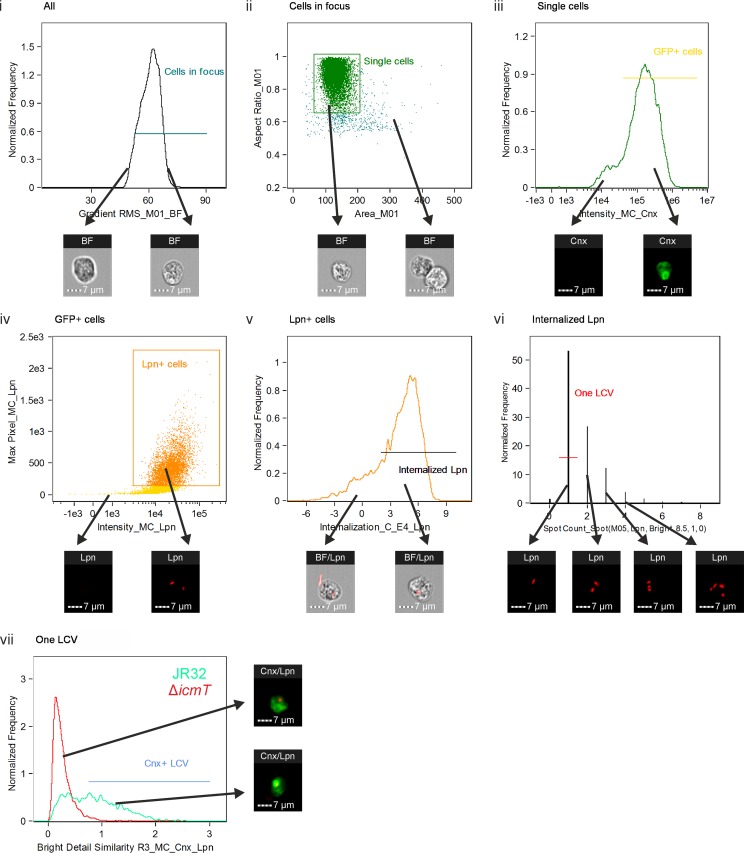
Imaging flow cytometry-based quantification of calnexin-GFP on LCVs in intact amoebae. D. discoideum Ax3 producing calnexin-GFP (pAW16) was infected (MOI, 5) for up to 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14). After fixation, 10,000 cells were acquired with an imaging flow cytometer. The analysis was performed in 7 steps, where the cells gated in one step were carried on to the subsequent step. Examples of included and excluded cells are shown for each step (arrows). (i) Cells in focus were gated using the feature Gradient RMS_M01_BF. (ii) Single cells were gated using Area_M01 versus Aspect Ratio_M01. (iii) Cells producing calnexin-GFP (Cnx) were gated using the feature Intensity_MC_Cnx. (iv) Cells positive for L. pneumophila (Lpn) were gated using Intensity_MC_Lpn versus Max Pixel_MC_Lpn. (v) Cells having internalized L. pneumophila rather than L. pneumophila merely attached to the surface were gated using Internalization_C_E4_Lpn. (vi) Cells containing exactly one bacterium were selected using Spot Count_Spot(M05, Lpn, Bright, 8.5, 1, 0). (vii) The included cells were finally analyzed for colocalization between an L. pneumophila bacterium and calnexin-GFP using the feature Bright Detail Similarity R3_MC_Cnx_Lpn, and the result was termed the IFC colocalization score. A gate was set at 0.75 to include only cells containing a calnexin-positive LCV. The histogram shows an overlay of JR32-infected (green) and ΔicmT mutant-infected (red) cells at 2 h postinfection.
FIG 2.
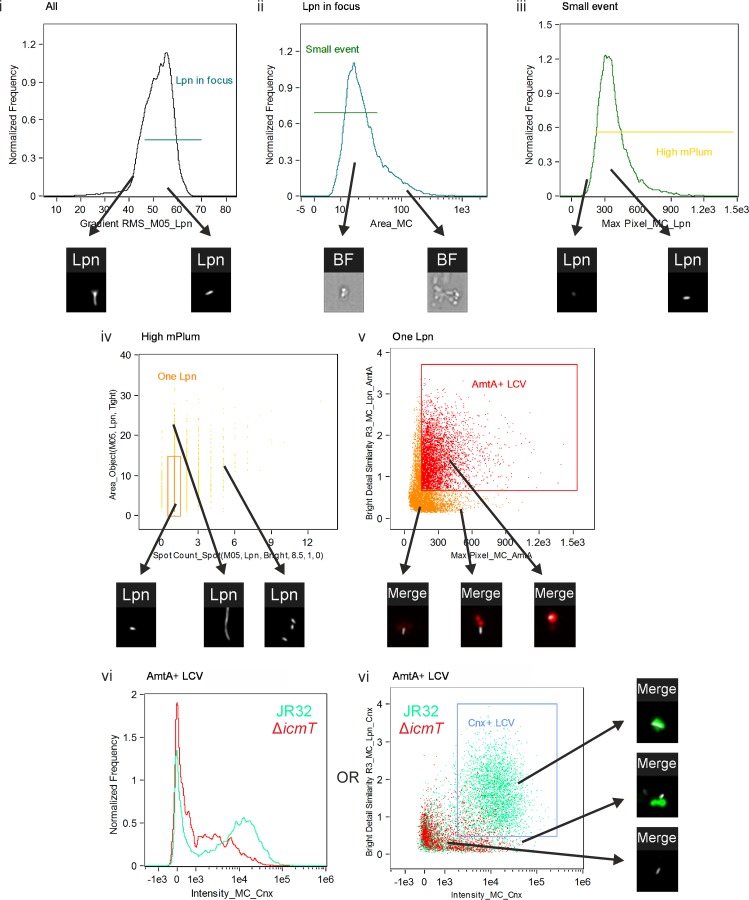
Imaging flow cytometry-based quantification of calnexin-GFP on LCVs in homogenized amoebae. D. discoideum Ax3 dually producing AmtA-mCherry and calnexin-GFP (pAW16) were infected (MOI, 50) for up to 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14). After homogenization and fixation, 20,000 events were acquired with an imaging flow cytometer. The analysis was performed in 6 steps, where the cells gated in one step were carried on to the subsequent step. Examples of included and excluded cells are shown for each step (arrows). (i) Events in focus were gated using the feature Gradient RMS_M05_Lpn. (ii) Intact cells and large membrane aggregates were excluded using the feature Area_MC. (iii) Events displaying a sufficiently high mPlum signal were gated using Max Pixel_MC_Lpn. (iv) LCVs containing one bacterium only were selected using the features Spot Count_Spot(M05, Lpn, Bright, 8.5, 1, 0) versus Area_Object(M05, Lpn, Tight). (v) AmtA-mCherry-positive events with the AmtA signal colocalizing with the mPlum signal were gated using the features Max Pixel_MC_AmtA versus Bright Detail Similarity R3_MC_Lpn_AmtA. (vi) Finally, in the gated AmtA-positive LCVs, the intensity of calnexin-GFP was quantified using Intensity_MC_Cnx. (Left) The histogram shows an overlay of JR32-containing (green) and ΔicmT mutant-containing (red) LCVs at 2 h postinfection. (Right) Alternatively, the gated AmtA-positive LCVs were analyzed using Intensity_MC_Cnx versus Bright Detail Similarity R3_MC_Lpn_Cnx, yielding a percentage of LCVs positive for calnexin-GFP and with the GFP colocalizing with the bacterium. The dot plot shows an overlay of JR32-containing (green) and ΔicmT mutant-containing (red) LCVs at 2 h postinfection.
FIG 3.
Quantification of calnexin, PtdIns(4)P, and Rab1 on LCVs by IFC. D. discoideum Ax3 producing calnexin-GFP (pAW16) (A), the PtdIns(4)P probe P4CSidC-GFP (pWS34) (B), or GFP-Rab1 (pAW7) (C) were infected (MOI, 5) for 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14). (Left) Representative IFC images; (middle) quantifications of IFC colocalization scores between GFP and mPlum for >500 cells per sample at the time points postinfection indicated (means and 95% confidence intervals from one representative experiment out of three independent experiments are shown); (right) quantifications of the percentage of cells containing a GFP-positive LCV, based on the IFC colocalization scores (means and SEMs from three independent experiments are shown) (***, P < 0.001 at 2 h). AU, arbitrary units.
The data obtained closely match previously published values for LCV accumulation of calnexin (16, 53), PtdIns(4)P (16), and Rab1 (10, 54) during the first 2 h of infection. We have previously found that the majority of the trafficking events leading to the formation of a mature LCV, including PI conversion and calnexin accumulation, are completed within the first 2 h of infection (16, 42, 53). Furthermore, the IFC scores for wild-type L. pneumophila are clearly different from those for bacteria lacking a functional Icm/Dot T4SS, demonstrating that IFC is a reliable, robust, and high-throughput-compatible method to quantify cellular markers on the LCV inside amoebae. Taken together, we provide a comprehensive biological validation of the use of IFC for assessing the LCV composition.
The endosomal transporter AmtA is an LCV component.
The amount of an LCV marker present on the pathogen vacuole relative to that present on the remainder of the cell varies greatly depending on the protein. Small amounts of LCV marker proteins necessitate the homogenization of the infected cell before analysis, in order to increase the signal-to-noise ratio and to allow detection by confocal microscopy (42–44).
To analyze LCVs from homogenized host cells by IFC, a pathogen vacuole marker that is insensitive to the infection time course and the presence of the Icm/Dot T4SS is required. The ammonium transporter AmtA is present in all membranes of the endocytic pathway in D. discoideum (55) and has previously been used as a marker of the phagosomal membrane in amoebae infected with Mycobacterium marinum (56). Thus, we investigated whether AmtA localized to the LCV by using D. discoideum strains that produced both calnexin-GFP and AmtA-mCherry in parallel and were infected with mPlum-producing L. pneumophila wild-type or ΔicmT strains (Fig. 4A). We observed a typical gradual acquisition of calnexin-GFP on the LCV over time, while AmtA-mCherry was present at all time points tested (up to 2 h) and independently of the Icm/Dot secretion system (Fig. 4B). Thus, AmtA is a component of the LCV and is suitable for IFC analysis of vacuoles containing L. pneumophila wild-type or Icm/Dot-deficient strains.
FIG 4.
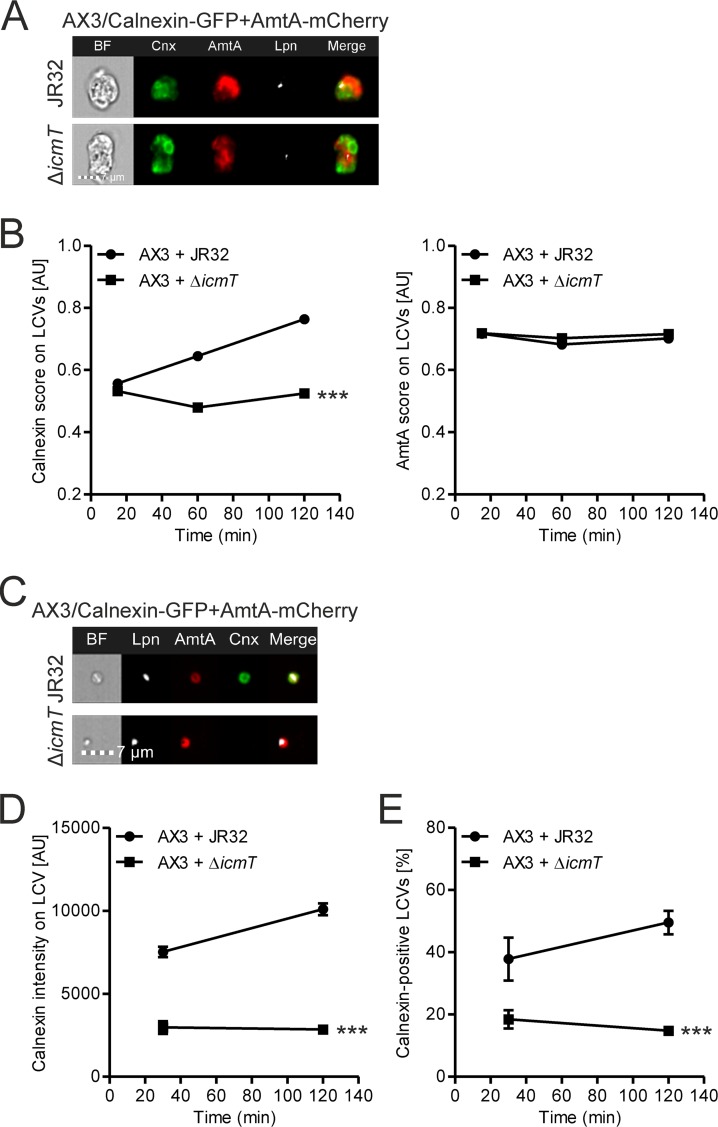
The endosomal transporter AmtA is a component of LCVs. (A) IFC images of intact D. discoideum Ax3 that dually produced AmtA-mCherry and calnexin-GFP (pAW16) and that was infected (MOI, 5) for 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14). (B) Simultaneous quantification of the IFC colocalization score between calnexin-GFP and mPlum (left) and AmtA-mCherry and mPlum (right) for >1,000 intact cells per sample at the time points postinfection indicated. (C) IFC images of LCVs from homogenized D. discoideum Ax3 that dually produced AmtA-mCherry and calnexin-GFP (pAW16) and that was infected (MOI, 50) for 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14). (D) Quantification of calnexin-GFP intensity on AmtA-positive LCVs by IFC for >1,000 LCVs per sample at the time points postinfection indicated. (E) Quantification of the percentage of LCVs positive for calnexin-GFP, based on the GFP intensity on the LCV together with the results of colocalization analysis of GFP and mPlum. The data show the means and 95% confidence intervals from one representative experiment out of three independent experiments (B, D) or the means and SEMs from three independent experiments (E) (***, P < 0.001 at 2 h).
Next, we adapted the mechanical host cell homogenization technique (57, 58) for IFC, using D. discoideum producing AmtA-mCherry and calnexin-GFP in parallel. After homogenization of amoebae infected with mPlum-producing L. pneumophila, fixation, and acquisition of 20,000 mPlum- and mCherry-positive events by IFC, the LCVs were analyzed as described in the legend to Fig. 2. The resulting images (Fig. 4C) and quantification of GFP fluorescence on the AmtA-positive LCVs containing a single bacterium show an Icm/Dot-dependent accumulation of calnexin on the LCV during the course of infection (Fig. 4D), as expected (16, 53). By setting a gate including only LCVs with a high GFP fluorescence intensity and comprising colocalizing signals of GFP and mPlum (Fig. 2), the percentage of LCVs that were positive for calnexin was obtained (Fig. 4E). This representation not only provides a more easily interpretable percentage value instead of an arbitrary fluorescence value but also ensures that only LCVs where the GFP signal localized exactly to the LCV and not to an adjacent structure in the same image were counted as positive. However, we opted to use arbitrary fluorescence values in the remainder of the study, since they were very similar to the percentage values and allowed robust statistical analysis.
Of note, calnexin and AmtA do not colocalize on LCVs in intact D. discoideum (Fig. 4A) and form concentric rings on isolated pathogen vacuoles (Fig. 4C). This observation is in agreement with the finding that during LCV maturation calnexin is recruited to but does not fuse with the PtdIns(4)P-positive pathogen vacuole (16). Since AmtA is present on vacuoles harboring L. pneumophila JR32 or ΔicmT mutant bacteria, the Icm/Dot-dependent acquisition of PtdIns(4)P is clearly independent of AmtA. Collectively, our data show that AmtA is a suitable marker to quantify by IFC different GFP fusion proteins on LCVs obtained from homogenized amoebae, regardless of the infection time and bacterial strain used.
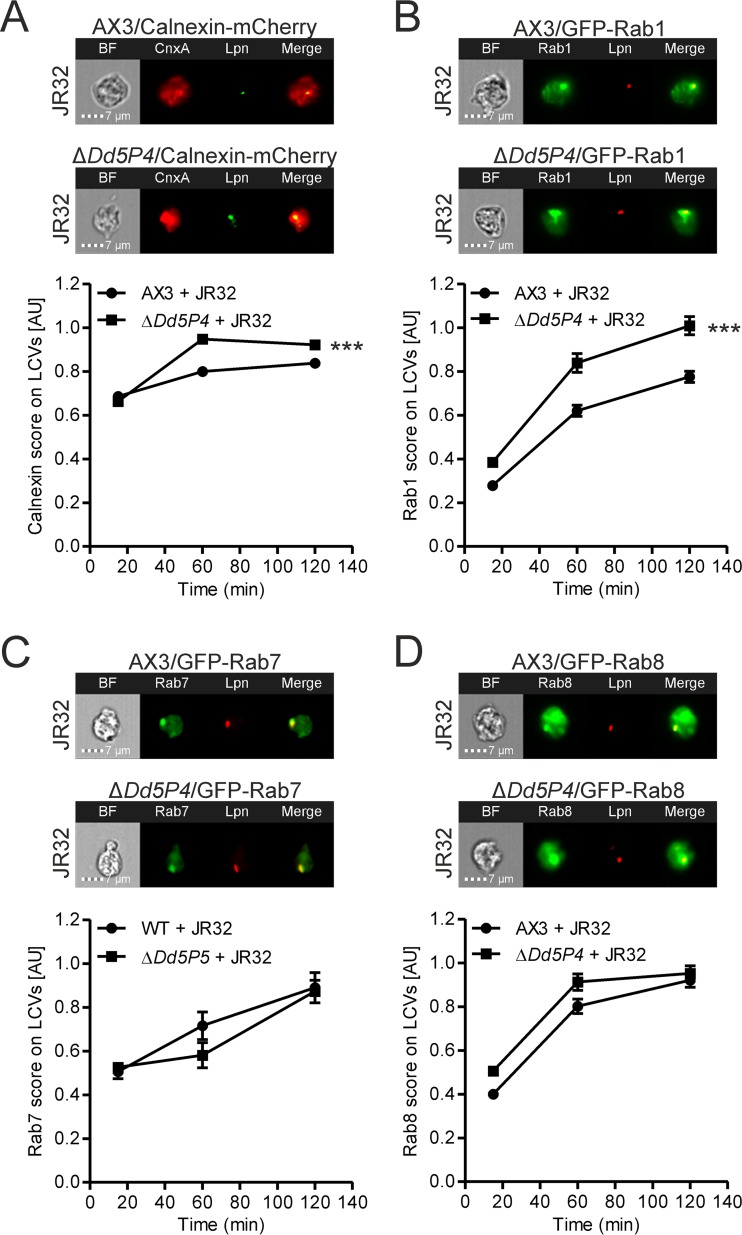
The PI 5-phosphatase Dd5P4 affects LCV acquisition of early secretory pathway components.
The endosomal PI 5-phosphatase Dd5P4 binds clathrin and several small GTPases, is crucially involved in retrograde trafficking, and restricts the intracellular replication of L. pneumophila (35, 36, 42). In order to study how Dd5P4 regulates intracellular trafficking to the LCV, we employed our newly developed IFC-based method and focused on the recruitment of four proteins known to accumulate on LCVs (10–12). These included the ER-resident protein calnexin and the small GTPases Rab1 (a regulator of the secretory pathway between the ER and Golgi apparatus), Rab7 (a late endosomal marker regulating phagolysosomal fusion and the initiation of retrograde trafficking), and Rab8 (a regulator in the secretory pathway beyond the Golgi apparatus) (59).
We infected the D. discoideum parental strain Ax3 or the isogenic ΔDd5P4 mutant producing mCherry or GFP fusions of calnexin, Rab1, Rab7, or Rab8 with the virulent L. pneumophila strain JR32 and quantified the colocalization of mCherry or GFP with the bacterium in intact amoebae (Fig. 5). In the absence of Dd5P4, the recruitment of calnexin and Rab1 was slightly but significantly enhanced (Fig. 5A and B). However, Dd5P4 did not affect the accumulation of Rab7 or Rab8 on LCVs (Fig. 5C and D). The effect on calnexin is consistent with previously published data obtained using conventional confocal microscopy (42). This earlier work validates the IFC approach documented here and shows that in the absence of Dd5P4, more calnexin is recruited to the LCV and the calnexin-positive LCVs formed are more spacious. These findings likely reflect a more efficient pathogen vacuole formation by L. pneumophila in the absence of a factor that restricts this process. The slightly enhanced acquisition of Rab1 may also reflect that the bacteria create a more favorable niche for replication in the absence of Dd5P4, as hijacking of Rab1 is a virulence mechanism used by L. pneumophila (54, 60, 61). The lack of an effect of Dd5P4 deletion on Rab7 and Rab8 indicates that the PI 5-phosphatase is not involved in their recruitment to the LCV. Collectively, the data are in agreement with the notion that Dd5P4 plays a role in the communication of LCVs with components of the early secretory pathway (ER-Golgi apparatus) but not with those of the late endosomal (Rab7) or late secretory (Rab8) pathway.
FIG 5.
Deletion of Dd5P4 affects LCV acquisition of early secretory pathway components. D. discoideum Ax3 or ΔDd5P4 producing calnexin-mCherry (pAW12) (A), GFP-Rab1 (pAW7) (B), GFP-Rab7 (pAW9) (C), or GFP-Rab8 (pAW10) (D) was infected (MOI, 5) for 2 h with GFP- or mPlum-producing virulent L. pneumophila JR32(pCR76) or JR32(pAW14). Shown are representative IFC images (top) and quantifications of the IFC colocalization scores (bottom) between mCherry and GFP (for calnexin) or GFP and mPlum (for Rab1, Rab7, and Rab8) for >350 cells per sample at the time points postinfection indicated. Data show the means and 95% confidence intervals from one representative experiment out of three independent experiments (***, P < 0.001).
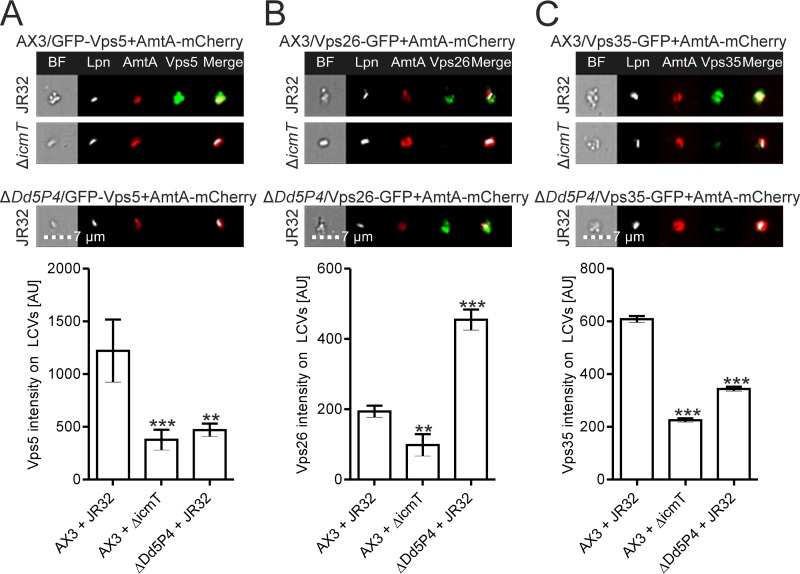
Dd5P4 modulates retrograde trafficking components on LCVs.
Dd5P4/OCRL is involved in retrograde trafficking from endosomes to the trans-Golgi network, as well as in the recycling of receptors between endosomes and the plasma membrane (35, 36). Since Dd5P4/OCRL and retrograde trafficking restrict intracellular L. pneumophila replication (42, 43), we sought to determine precisely how Dd5P4 controls the LCV composition with regard to retrograde trafficking components.
Retrograde trafficking components, including Dd5P4 and retromer subunits, localize in relatively small amounts on the LCV compared to the amounts at which they localize in the rest of the cell (43, 44). This issue, together with the fact that the resolution of IFC is not as high as that of confocal microscopy, implies that retrograde trafficking components are difficult to detect by IFC on the LCV in intact cells. For IFC quantification, we thus employed host cell lysates, using L. pneumophila-infected amoebae dually producing AmtA-mCherry as an LCV component together with a GFP fusion protein of interest.
Infecting D. discoideum Ax3 or ΔDd5P4 with L. pneumophila, we investigated the levels on the LCV of GFP-Vps5, a D. discoideum SNX homologue (62) (Fig. 6A). The IFC quantification showed that Vps5 was recruited to the LCV in an Icm/Dot-dependent manner and that deletion of Dd5P4 significantly diminished its recruitment (Fig. 6A). These data show that Dd5P4 promotes sorting nexin localization to the LCV membrane. Moreover, the results suggest that Dd5P4 is upstream of Vps5 during LCV maturation and, in broader terms, in the cascade of events leading to the formation of a retrograde trafficking vesicle.
FIG 6.
Deletion of Dd5P4 modulates retrograde trafficking components on LCVs. Results for LCVs from homogenized D. discoideum Ax3 or ΔDd5P4 that dually produced AmtA-mCherry and GFP-Vps5 (pAW18) (A), Vps26-GFP (pAW1) (B), or Vps35-GFP (pAW3) (C) and that was infected (MOI, 50) for 2 h with mPlum-producing virulent L. pneumophila JR32 or ΔicmT(pAW14) are shown. Shown are representative IFC images (top) and IFC quantifications (bottom) of GFP intensities on AmtA-positive LCVs for >400 LCVs per sample. Data show the means and SEMs from one representative experiment out of three independent experiments (**, P < 0.01; ***, P < 0.001).
To further investigate the role of Dd5P4 in retrograde trafficking on the LCV, we turned to the subunits of the retromer complex. This protein complex consists of a Vps26-Vps29-Vps35 heterotrimer, which recruits sorting nexins and acts to bind cargo destined for retrograde trafficking (19, 20, 48). Although it is clear that Dd5P4 is needed for retrograde trafficking and that the enzyme has a number of interaction partners in the process (35, 36), its relationship to the retromer is not known. To address this question, we studied by IFC LCVs from homogenized D. discoideum (Ax3 or the ΔDd5P4 mutant) dually producing AmtA-mCherry and Vps26-GFP (Fig. 6B) or Vps35-GFP (Fig. 6C). Vps26-GFP and Vps35-GFP accumulated on LCVs in a manner dependent on the Icm/Dot secretion system, consistent with previously published data (43). While the level of Vps26-GFP on LCVs was increased in the absence of Dd5P4 (Fig. 6B), the level of Vps35-GFP was reduced (Fig. 6C).
It is currently not clear why the LCV localization of the Vps26 and Vps35 retromer subunits is apparently regulated differently by Dd5P4. However, at least in mammalian cells, we previously observed that the subunits do not contribute equally to the intracellular replication of L. pneumophila. While the depletion of Vps26A and Vps26B (as well as Vps29) reduced intracellular bacterial growth, depletion of Vps35 had no effect (43). This finding might be explained by the recent discovery that (at least in mammals) different distinct retrograde coat complexes which share some, but not all, cargo recognition subunits are present (63). It remains to be determined which retrograde coat complexes play a role in LCV formation by and intracellular replication of L. pneumophila. Taken together, the modulation of retromer components on LCVs by Dd5P4 indicates that the PI 5-phosphatase is active directly on the LCV membrane, and our findings support the hypothesis that the enzyme acts upstream of the retromer complex during LCV maturation and in the retrograde trafficking pathway.
Dd5P4 also seems to modulate the recruitment to LCVs of the retromer subunit Vps29 and sortilin/Vps10, a retrograde transport cargo molecule (64) (see Fig. S1 in the supplemental material). Compared to the levels in the parental D. discoideum strain, the levels of Vps29-GFP and Vps10-GFP were already increased in uninfected ΔDd5P4 mutant amoebae (Fig. S2), rendering the interpretation of the data more difficult. A potential pitfall is indeed that the overall production level of the fusion protein of interest might bias the result when comparing the fluorescence intensities on the LCV in different D. discoideum strains (the parental strain versus isogenic mutants). This is not the case for the colocalization analysis used here for intact cells, which is based on Pearson's correlation coefficient of the localized bright spots in two images, a coefficient independent of the total intensity (for technical details, see reference 65). To account for this potential bias, we quantified the overall fluorescence intensity of GFP fusion proteins of interest in each (uninfected) D. discoideum strain. This analysis revealed no major differences for all LCV markers tested, except Vps10 and Vps29 (Fig. S2).
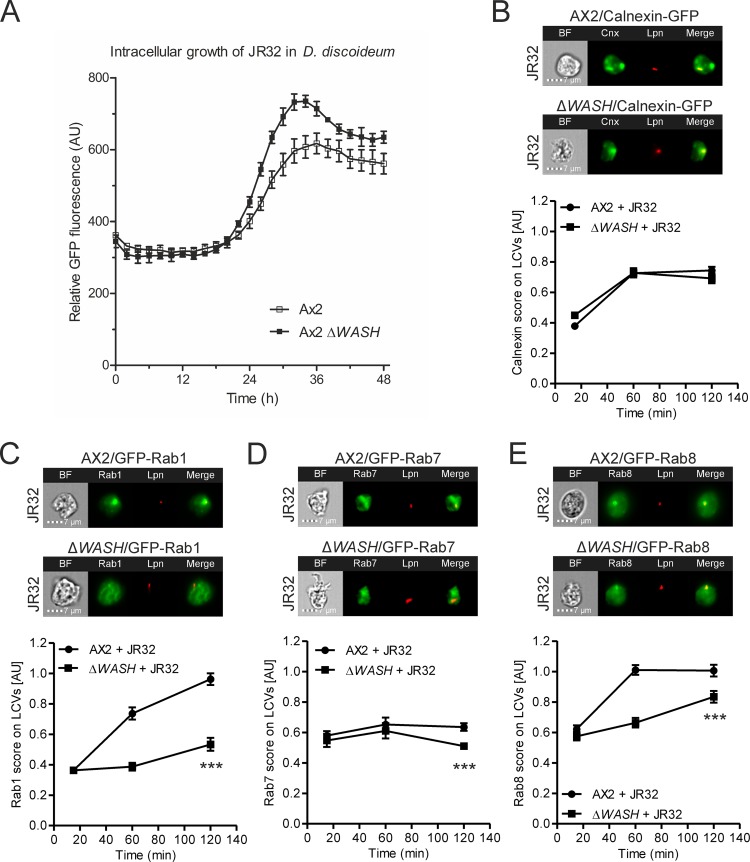
The actin nucleator WASH restricts the intracellular replication of L. pneumophila and affects the LCV composition.
The WASH actin nucleation complex binds the retromer (27), plays a role in driving endosomal actin polymerization, and mediates protein sorting and recycling from endosomes through retromer-dependent and -independent pathways (62). Specifically, the WASH complex plays a role in pushing the nascent retrograde trafficking vesicle away from the endosomal network (28). In order to expand our investigation from the PI phosphatase Dd5P4, implicated early during retrograde trafficking by interactions with Rab GTPases and clathrin (35, 36), to later steps involving retromer interactions and vesicle budding (27, 28), we investigated the effect of WASH deletion on the intracellular replication of L. pneumophila. D. discoideum Ax2 or ΔWASH was infected with GFP-producing L. pneumophila JR32, and intracellular replication was assessed by fluorescence. This experiment revealed that in the absence of WASH, L. pneumophila replicates more efficiently (Fig. 7A), and therefore, WASH restricts intracellular replication similarly to other components of the retrograde pathway (42, 43).
FIG 7.
The actin nucleator WASH restricts the intracellular replication of L. pneumophila and affects the LCV composition. (A) D. discoideum Ax2 or ΔWASH was infected (MOI, 1) with GFP-producing L. pneumophila JR32(pNT-28), and intracellular replication was assessed by fluorescence analysis. Data show the means and standard deviations for three biological replicates (means of technical replicates each). (B to E) D. discoideum Ax2 or ΔWASH producing calnexin-GFP (pAW16) (B), GFP-Rab1 (pAW7) (C), GFP-Rab7 (pAW9) (D), or GFP-Rab8 (pAW10) (E) was infected (MOI, 5) for 2 h with mPlum-producing virulent L. pneumophila JR32 (pAW14). Shown are representative IFC images (top) and quantifications of IFC colocalization scores (bottom) between GFP and mPlum for >230 cells per sample at the time points postinfection indicated. Data show the means and 95% confidence intervals from one representative experiment out of three independent experiments (***, P < 0.001).
Moreover, we directly compared the intracellular replication of L. pneumophila in the D. discoideum Ax2 and Ax3 parental strains, wherein the ΔWASH or ΔDd5P4 mutant strains were generated. The results indicate that during the first 15 h postinfection (p.i.), the growth of L. pneumophila was similar in the two amoeba strains, and at later time points, the bacteria grew more efficiently in Ax3 (Fig. S3). Hence, at the early time points (2 h p.i.) used for IFC measurements throughout this study, L. pneumophila infection appears to proceed similarly in D. discoideum Ax2 and Ax3.
Next, we tested whether deletion of WASH affects the LCV composition. We focused on the ER marker calnexin and the small GTPases Rab1, Rab7, and Rab8. To this end, we infected the D. discoideum parental strain Ax2 or the ΔWASH mutant producing one of these GFP fusion proteins with the virulent L. pneumophila strain JR32 (Fig. 7B to E) and quantified the colocalization of GFP with the bacterium in intact amoebae. The deletion of WASH strongly impaired the recruitment of Rab1 and Rab8 to LCVs (Fig. 7C and E) and also reduced, to a smaller extent, the recruitment of Rab7 to LCVs (Fig. 7D), while the recruitment of the ER marker calnexin was not affected (Fig. 7B). These results indicate a major role for WASH and retrograde-linked actin polymerization during LCV maturation, consistent with the idea that endosomal sorting in general and retrograde trafficking in particular are important aspects of this process. Since WASH deletion had no effect on calnexin recruitment to the LCV, the LCV-ER interaction seems to be independent of actin polymerization or might require other factors affecting actin dynamics.
In summary, by providing comprehensive technical details, we have established and biologically validated two distinct IFC-based techniques for quantifying ectopically produced proteins on LCVs in intact D. discoideum or in amoeba homogenates, respectively. The IFC approach enables a quantitative, unbiased, and rapid analysis of both intact infected host cells and homogenized cells. Further advantages of the method include flexibility regarding the time points and the number of strains analyzed, as well as the ability to analyze proteins that are present at only low levels on the LCV compared to their levels in the remainder of the cell.
A limitation of the IFC approach is that the high abundance of ectopically produced proteins may affect their intracellular distribution, localization, and function. However, we did not observe any major differences between the intracellular replication kinetics of L. pneumophila infecting D. discoideum strains lacking calnexin or overproducing calnexin-GFP (53, 66) or strains overproducing small GTPases (10) and those of L. pneumophila infecting the corresponding parental D. discoideum strains. In contrast, overproduction of the large GTPase atlastin/Sey1 has been shown to affect the intracellular replication of L. pneumophila (53). Moreover, to allow direct comparison, the IFC technique requires equal levels of production of the proteins studied in different D. discoideum host strains, which is not always the case (Fig. S2). Finally, the relatively low resolution of IFC compared to that of confocal microscopy limits the spatial analysis of L. pneumophila infection and LCV formation.
We employed IFC to assess the impact of the PI 5-phosphatase Dd5P4 on the LCV composition in terms of canonical pathogen vacuole markers of the secretory pathway, as well as in terms of components of the retrograde trafficking pathway, such as the retromer coat complex. Our findings emphasize the role of retrograde trafficking in modulating the LCV composition and in restricting LCV formation. The IFC-based LCV analysis established here should prove useful to study the pathogen vacuole composition of other amoeba-resistant intracellular pathogens, in particular, those for which the D. discoideum model has already been established, including Mycobacterium spp. (67, 68) and Francisella spp. (69, 70).
MATERIALS AND METHODS
Molecular cloning.
All plasmids used in this study are listed in Table 1, and oligonucleotides are specified in Table 2. The plasmids pSW34, pAW1, pAW2, pAW3, and pAW5, which were used for overproduction of the PtdIns(4)P probe P4CSidC, the retromer components (Vps26, Vps29, Vps35), and sortilin/Vps10, respectively, and which had C-terminal GFP tags, were constructed by amplifying the respective D. discoideum gene from pWS22, pCR111, pCR112, pCR113, or pCR110 using the primers oWS17/oWS18 (pSW34), oAW1/oAW2 (Vps26), oAW3/oAW4 (Vps29), oAW5/oAW6 (Vps35), or oAW21/oAW22 (Vps10) and cloning the PCR fragments into the BglII and SpeI sites of pDM323. The plasmids pAW7, pAW10, and pAW18, which were used for overproduction of Rab1, Rab8, and Vps5, respectively, and which had N-terminal GFP tags, were constructed by amplifying the respective D. discoideum gene from pSU22, pSU12, or pFL1329 using the primers oAW9/oAW10 (Rab1), oAW15/oAW16 (Rab8), or oAW19/oAW20 (Vps5) and cloning the PCR fragments into the BglII and SpeI sites of pDM317. The plasmid pFL1329 was constructed by PCR amplification of the gene encoding Vps5 from a D. discoideum cDNA library and cloning the fragment into pBluescript and, after sequencing, into pDXA-GFP-Ct. All constructs were sequenced before transformation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| Strains | ||
| D. discoideum | ||
| Ax3 | Parental strain | 32 |
| Ax3 ΔDd5P4 | Dd5P4 deletion mutant, Blsr | 32 |
| Ax2 | Parental strain | 71 |
| Ax2 ΔWASH | WASH deletion mutant, Blsr | 71 |
| E. coli TOP10 | Invitrogen | |
| L. pneumophila | ||
| GS3011 | L. pneumophila JR32 icmT3011::Kanr (ΔicmT) | 74 |
| JR32 | Virulent L. pneumophila serogroup 1 strain, derivative of strain Philadelphia-1 | 75 |
| Plasmids | ||
| AmtA-mCherry | pDM1044-amtA-mCherry | 56 |
| pAW1 | pDM323-vps26-gfp | This work |
| pAW2 | pDM323-vps29-gfp | This work |
| pAW3 | pDM323-vps35-gfp | This work |
| pAW5 | pDM323-vps10-gfp | This work |
| pAW7 | pDM317-gfp-rab1A | This work |
| pAW9 | pDM317-gfp-rab7A | 44 |
| pAW10 | pDM317-gfp-rab8A | This work |
| pAW12 | pDM1044-cnxA-mCherry | 53 |
| pAW14 | pMMB207-C-Ptac-mplum, ΔlacIq (constitutive mPlum), Camr | 53 |
| pAW16 | pDM323-cnxA-gfp | 44 |
| pAW18 | pDM317-gfp-vps5 | This work |
| pCR76 | pMMB207-C-Ptac-RBS-gfp-RBS-MCS (constitutive gfp) Camr | 43 |
| pCR110 | pDXA-HC-vps10-gfp | 43 |
| pCR111 | pDXA-HC-vps26-gfp | 43 |
| pCR112 | pDXA-HC-vps29-gfp | 43 |
| pCR113 | pDXA-HC-vps35-gfp | 43 |
| pDM317 | Dictyostelium expression vector, extrachromosomal, N-terminal GFP, G418r | 76 |
| pDM323 | Dictyostelium expression vector, extrachromosomal, C-terminal GFP, G418r | 76 |
| pDM1044 | Dictyostelium expression vector, extrachromosomal, C-terminal mCherry, Hygr | 56 |
| pFL1329 | pDXA-vps5-gfp | This work |
| pNT-28 | pMMB207-C Ptac-gfp ΔlacIq (constitutive gfp) Camr | 77 |
| pSU12 | pDXA-gfp-rab8A | 11 |
| pSU22 | pDXA-gfp-rab1A | 11 |
| pWS22 | pBsrH-P4CSidC-mRFPmars | 16 |
| pWS34 | pDM323-P4CSidC-gfp | This work |
Abbreviations: Bls, blasticidin; Cam, chloramphenicol; Hyg, hygromycin; Kan, kanamycin; G418, Geneticin.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′)a | Comment |
|---|---|---|
| oAW1 | ATA TAT GGA TCC ATG AAC TTT TTT AGT AAT AAT | 5′ of vps26, BamHI |
| oAW2 | ATA TAT ACT AGT ATC ACT TTC ATC GGA AGA G | 3′ of vps26, SpeI |
| oAW3 | ATA TAT GGA TCC ATG TTT ATT ATT GCA ATT G | 5′ of vps29, BamHI |
| oAW4 | ATA TAT ACT AGT TTG TTG TTT GAC ATG ATC | 3′ of vps29, SpeI |
| oAW5 | ATA TAT GGA TCC ATG ATG AAA AAA AAT AAA CC | 5′ of vps35, BamHI |
| oAW6 | ATA TAT ACT AGT AAT TGA AAT ACC TTT ATA ATT TTG | 3′ of vps35, SpeI |
| oAW9 | ATA TAT GGA TCC ATG AAC CCA GAT TAT C | 5′ of rab1A, BamHI |
| oAW10 | ATA TAT ACT AGT ACA ACA ACC AGA TTT TG | 3′ of rab1A, SpeI |
| oAW15 | ATA TAT GGA TCC ATG ACT TCT CCA GC | 5′ of rab8A, BamHI |
| oAW16 | ATA TAT ACT AGT ACA ACA AGC TTT TTT CTT ATT G | 3′ of rab8A, SpeI |
| oAW19 | ATA TAT GGA TCC ATG TCA GAT TAT AAT AGT AAT C | 5′ of vps5, BamHI |
| oAW20 | ATA TAT ACT AGT AAT AGA AGT TGA ACC CCA AG | 3′ of vps5, SpeI |
| oAW21 | ATA TAT AGA TCT ATG AAG ATT TCA TC | 5′ of vps10, BglII |
| oAW22 | ATA TAT ACT AGT AAA ATC TTC GTT ATC | 3′ of vps10, SpeI |
| oWS17 | AAA AAG ATC TAT GTC AAA ATA TTC CTC C | 5′ of P4CSidC, BglII |
| oWS18 | AAA AACTAGTAA AGA ATT CAA TTG CTT CAT | 3′ of P4CSidC, SpeI |
Restriction sites are underlined.
Culture of L. pneumophila and D. discoideum.
The strains used in this study are listed in Table 1. L. pneumophila strain JR32 or the L. pneumophila ΔicmT mutant containing a plasmid for constitutive production of a fluorescent protein (mPlum or GFP) was cultured for 3 days at 37°C on charcoal yeast extract (CYE) agar plates buffered with N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) and supplemented with 5 μg/ml chloramphenicol (Cam). On the day before infection, an early-stationary-phase broth culture (optical density at 600 nm [OD600] = 5) was prepared by inoculating 3 ml ACES yeast extract (AYE) broth (containing 5 μg/ml Cam) with the bacteria at an OD600 of 0.1 and incubating for 21 h on a rotating wheel at 37°C, as described previously (13).
D. discoideum parental strain Ax3 or Ax2 or isogenic mutants thereof lacking Dd5P4 (32) or WASH (71), respectively, were cultured axenically by standard procedures in HL5 medium (11 g glucose, 5 g yeast extract, 5 g proteose peptone, 5 g thiotone E peptone, 2.5 mM Na2HPO4, 2.5 mM KH2PO4 in 1 liter H2O; pH 6.5) at 23°C. Transformation with plasmids for the production of pathogen vacuole markers fused with GFP or mCherry was performed by electroporation as previously described (16, 72), and appropriate antibiotics were used for selection of amoebae harboring plasmids (5 μg/ml G418 and/or 50 μg/ml hygromycin). Transformation was repeated to obtain tandem fluorescent strains. On the day prior to infection, subconfluent amoebae were seeded in 12-well tissue culture plates (5 × 105 per well, for analysis of intact cells) or 25-cm2 tissue culture flasks (3.5 × 106 per flask, for analysis of cell homogenates).
Experimental infection of D. discoideum for IFC.
Infection and processing of D. discoideum amoebae were performed essentially as described previously (43, 53, 57). Briefly, the HL5 medium was replaced with early-stationary-phase L. pneumophila diluted in medium without antibiotics to give a multiplicity of infection (MOI) of 5 (for analysis of intact cells) or 50 (for analysis of cell homogenates). The low MOI for intact cells was used to optimize the number of amoeba infected with low numbers of bacteria, while the high MOI for homogenates was used to obtain as many LCVs as possible. For tissue culture plates, uptake of the bacteria was synchronized by centrifugation (450 × g, 10 min, room temperature), before incubation at 23°C, while the flasks were incubated right away. Infected cells to be analyzed in intact form were removed from the plate by pipetting at the indicated time, pelleted by centrifugation (500 × g, 5 min, 4°C), and fixed with 4% paraformaldehyde for 30 min at 4°C. After one wash in phosphate-buffered saline (PBS), the cells were resuspended in 20 μl PBS for IFC. Cells to be homogenized before analysis were detached from the flask surface by tapping, pelleted by centrifugation (500 × g, 5 min, 4°C), washed once in Sörensen phosphate buffer (SorC buffer; 2 mM Na2HPO4, 15 mM KH2PO4, 50 μM CaCl2, pH 6.0), and resuspended in homogenization buffer (20 mM HEPES, 250 mM sucrose, 0.5 mM EGTA, pH 7.2) (73), before passage 11 times through a ball homogenizer (Isobiotec) with an exclusion size of 8 μm. The homogenates were pelleted (2,700 × g, 15 min, 4°C), fixed, washed, and resuspended in the same way as the intact cells.
Intracellular replication assay.
To determine the intracellular growth of L. pneumophila in D. discoideum Ax3, Ax2, or Ax2 ΔWASH, the amoebae were washed with LoFlo medium (ForMedium) and then seeded in 200 μl LoFlo medium at 5 × 104 cells/well in a 96-well plate 2 h prior to experimentation. GFP-producing L. pneumophila JR32(pNT-28), grown to early stationary phase, was diluted in LoFlo medium and added to the amoebae at an MOI of 1 in a volume of 20 μl. The fluorescence intensity of GFP was measured by a plate reader (BioTek) over the course of 48 h.
Imaging flow cytometry.
For IFC of intact amoebae, D. discoideum Ax3 or Ax2 (parental strains or mutants, as indicated) producing calnexin-GFP (pAW16), calnexin-mCherry (pAW12), the PtdIns(4)P probe P4CSidC-GFP (pWS34), GFP-Rab1 (pAW7), GFP-Rab7 (pAW9), or GFP-Rab8 (pAW10) or, alternatively, producing in tandem calnexin-GFP (pAW16) and AmtA-mCherry was used. The amoebae were infected with L. pneumophila JR32 or ΔicmT producing mPlum (pAW14) or GFP (pCR76) and processed as described above. With an imaging flow cytometer (ImageStreamX MkII; Amnis), 10,000 events were acquired, and analysis was carried out using IDEAS (v.6.2) software (Amnis), as detailed in the legend to Fig. 1. Amoebae having internalized exactly one L. pneumophila bacterium (generally, >1,000 cells per sample; the lowest number of events in the data set is indicated in the figure legends) were analyzed for bright detail colocalization of mPlum and GFP and/or mCherry (host factors), when the bacteria were producing mPlum, or of GFP and mCherry (host factors), when the bacteria were producing GFP. The resulting value was termed the IFC colocalization score and is the log-transformed Pearson's correlation coefficient of the localized bright spots with a radius of 3 pixels or less in two images.
For IFC of LCVs from homogenized amoebae, D. discoideum Ax3 or an isogenic ΔDd5P4 mutant strain producing calnexin-GFP (pAW16), GFP-Vps5 (pAW18), Vps10-GFP (pAW5), Vps26-GFP (pAW1), Vps29-GFP (pAW2), or Vps35-GFP (pAW3) and AmtA-mCherry in tandem was employed. The amoebae were infected and processed as described above with the indicated L. pneumophila JR32 or ΔicmT strain producing mPlum (pAW14). With an imaging flow cytometer, 20,000 mPlum- and AmtA-positive events were acquired, and after color compensation, analysis was carried out using IDEAS (v.6.2) software, as detailed in the legend to Fig. 2. AmtA-positive LCVs containing only one L. pneumophila bacterium (generally, >1,000 events per sample; the lowest number of events in the data set is indicated in the figure legends) were gated and analyzed for the intensity of GFP.
Statistical analysis.
GraphPad Prism (v.7.0) software was used for statistical analysis. All IFC data were analyzed by regular two-way analysis of variance followed by Bonferroni post hoc tests, comparing each sample to the corresponding wild type.
Supplementary Material
ACKNOWLEDGMENTS
We thank François Letourneur (CNRS/University of Montpellier) for constructing pFL1329.
Research in the laboratory of H.H. was supported by the Swiss National Science Foundation (SNF; 31003A_153200), the Novartis Foundation for Medical-Biological Research, the OPO Foundation, and the German Ministry of Education and Research (BMBF) in the context of the EU Infect-ERA initiative (project EUGENPATH). Imaging flow cytometry was carried out at the Flow Cytometry Facility, University of Zürich. A.W. was supported by a grant from the Swedish Research Council (2014-396).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00158-18.
REFERENCES
- 1.Bangsborg JM. 1997. Antigenic and genetic characterization of Legionella proteins: contributions to taxonomy, diagnosis and pathogenesis. APMIS Suppl 70:1–53. [PubMed] [Google Scholar]
- 2.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilbi H, Hoffmann C, Harrison CF. 2011. Legionella spp. outdoors: colonization, communication and persistence. Environ Microbiol Rep 3:286–296. doi: 10.1111/j.1758-2229.2011.00247.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann C, Harrison CF, Hilbi H. 2014. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16:15–26. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 5.Isaac DT, Isberg R. 2014. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol 9:343–359. doi: 10.2217/fmb.13.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finsel I, Hilbi H. 2015. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 7.Swanson MS, Isberg RR. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H, Clarke M. 2005. Dynamic properties of Legionella-containing phagosomes in Dictyostelium amoebae. Cell Microbiol 7:995–1007. doi: 10.1111/j.1462-5822.2005.00528.x. [DOI] [PubMed] [Google Scholar]
- 9.Robinson CG, Roy CR. 2006. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol 8:793–805. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann C, Finsel I, Otto A, Pfaffinger G, Rothmeier E, Hecker M, Becher D, Hilbi H. 2014. Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol 16:1034–1052. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 11.Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. 2009. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10:76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- 12.Schmölders J, Manske C, Otto A, Hoffmann C, Steiner B, Welin A, Becher D, Hilbi H. 2017. Comparative proteomics of purified pathogen vacuoles correlates intracellular replication of Legionella pneumophila with the small GTPase Ras-related protein 1 (Rap1). Mol Cell Proteomics 16:622–641. doi: 10.1074/mcp.M116.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. 2008. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 14.Jean S, Kiger AA. 2012. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol 13:463–470. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- 15.Haneburger I, Hilbi H. 2013. Phosphoinositide lipids and the Legionella pathogen vacuole. Curr Top Microbiol Immunol 376:155–173. doi: 10.1007/82_2013_341. [DOI] [PubMed] [Google Scholar]
- 16.Weber S, Wagner M, Hilbi H. 2014. Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. mBio 5:e00839-13. doi: 10.1128/mBio.00839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 18.Wandinger-Ness A, Zerial M. 2014. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol 6:a022616. doi: 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannes L, Popoff V. 2008. Tracing the retrograde route in protein trafficking. Cell 135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Lu L, Hong W. 2014. From endosomes to the trans-Golgi network. Semin Cell Dev Biol 31:30–39. doi: 10.1016/j.semcdb.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Chi RJ, Harrison MS, Burd CG. 2015. Biogenesis of endosome-derived transport carriers. Cell Mol Life Sci 72:3441–3455. doi: 10.1007/s00018-015-1935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. 2008. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol 183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. 2016. Structural mechanism for cargo recognition by the retromer complex. Cell 167:1623–1635. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. 2004. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol 14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 25.van Weering JR, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, Carlsson SR, Hurley JH, Cullen PJ. 2012. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J 31:4466–4480. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen PJ, Korswagen HC. 2011. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol 14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seaman MN, Gautreau A, Billadeau DD. 2013. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol 23:522–528. doi: 10.1016/j.tcb.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour ME, Breusegem SY, Antrobus R, Freeman C, Reid E, Seaman MN. 2010. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci 123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wassmer T, Attar N, Harterink M, van Weering JR, Traer CJ, Oakley J, Goud B, Stephens DJ, Verkade P, Korswagen HC, Cullen PJ. 2009. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell 17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gokool S, Tattersall D, Seaman MN. 2007. EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic 8:1873–1886. doi: 10.1111/j.1600-0854.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Jefferson AB, Auethavekiat V, Majerus PW. 1995. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci U S A 92:4853–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loovers HM, Kortholt A, de Groote H, Whitty L, Nussbaum RL, van Haastert PJ. 2007. Regulation of phagocytosis in Dictyostelium by the inositol 5-phosphatase OCRL homolog Dd5P4. Traffic 8:618–628. doi: 10.1111/j.1600-0854.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- 33.Noakes CJ, Lee G, Lowe M. 2011. The PH domain proteins IPIP27A and B link OCRL1 to receptor recycling in the endocytic pathway. Mol Biol Cell 22:606–623. doi: 10.1091/mbc.E10-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicinanza M, Di Campli A, Polishchuk E, Santoro M, Di Tullio G, Godi A, Levtchenko E, De Leo MG, Polishchuk R, Sandoval L, Marzolo MP, De Matteis MA. 2011. OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. EMBO J 30:4970–4985. doi: 10.1038/emboj.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta ZB, Pietka G, Lowe M. 2014. The cellular and physiological functions of the Lowe syndrome protein OCRL1. Traffic 15:471–487. doi: 10.1111/tra.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S, Skowronek A, Erdmann KS. 2015. The role of the Lowe syndrome protein OCRL in the endocytic pathway. Biol Chem 396:1293–1300. doi: 10.1515/hsz-2015-0180. [DOI] [PubMed] [Google Scholar]
- 37.Mao Y, Balkin DM, Zoncu R, Erdmann KS, Tomasini L, Hu F, Jin MM, Hodsdon ME, De Camilli P. 2009. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J 28:1831–1842. doi: 10.1038/emboj.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ungewickell A, Ward ME, Ungewickell E, Majerus PW. 2004. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci U S A 101:13501–13506. doi: 10.1073/pnas.0405664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda M, Kanno E, Ishibashi K, Itoh T. 2008. Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 7:1031–1042. doi: 10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. 2006. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by Rab GTPases. EMBO J 25:3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. 2011. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J 30:1659–1670. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber SS, Ragaz C, Hilbi H. 2009. The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol 11:442–460. doi: 10.1111/j.1462-5822.2008.01266.x. [DOI] [PubMed] [Google Scholar]
- 43.Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. 2013. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Bärlocher K, Hutter CAJ, Swart AL, Steiner B, Welin A, Hohl M, Letourneur F, Seeger MA, Hilbi H. 2017. Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5. Nat Commun 8:1543. doi: 10.1038/s41467-017-01512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romano-Moreno M, Rojas AL, Williamson CD, Gershlick DC, Lucas M, Isupov MN, Bonifacino JS, Machner MP, Hierro A. 2017. Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL. Proc Natl Acad Sci U S A 114:E11151–E11160. doi: 10.1073/pnas.1715361115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao J, Yang F, Sun X, Wang S, Gan N, Liu Q, Liu D, Zhang X, Niu D, Wei Y, Ma C, Luo ZQ, Sun Q, Jia D. 2018. Mechanism of inhibition of retromer transport by the bacterial effector RidL. Proc Natl Acad Sci U S A 115:E1446–E1454. doi: 10.1073/pnas.1717383115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bärlocher K, Welin A, Hilbi H. 2017. Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Front Cell Infect Microbiol 7:482. doi: 10.3389/fcimb.2017.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Personnic N, Bärlocher K, Finsel I, Hilbi H. 2016. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 24:450–462. doi: 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Tiaden AN, Kessler A, Hilbi H. 2013. Analysis of Legionella infection by flow cytometry. Methods Mol Biol 954:233–249. doi: 10.1007/978-1-62703-161-5_14. [DOI] [PubMed] [Google Scholar]
- 50.Johansson J, Karlsson A, Bylund J, Welin A. 2015. Phagocyte interactions with Mycobacterium tuberculosis—simultaneous analysis of phagocytosis, phagosome maturation and intracellular replication by imaging flow cytometry. J Immunol Methods 427:73–84. doi: 10.1016/j.jim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Rothmeier E, Pfaffinger G, Hoffmann C, Harrison CF, Grabmayr H, Repnik U, Hannemann M, Wölke S, Bausch A, Griffiths G, Müller-Taubenberger A, Itzen A, Hilbi H. 2013. Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog 9:e1003598. doi: 10.1371/journal.ppat.1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon S, Wagner MA, Rothmeier E, Müller-Taubenberger A, Hilbi H. 2014. Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell Microbiol 16:977–992. doi: 10.1111/cmi.12258. [DOI] [PubMed] [Google Scholar]
- 53.Steiner B, Swart AL, Welin A, Weber S, Personnic N, Kaech A, Freyre C, Ziegler U, Klemm RW, Hilbi H. 2017. ER remodeling by the large GTPase atlastin promotes vacuolar growth of Legionella pneumophila. EMBO Rep 18:1817–1836. doi: 10.15252/embr.201743903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 55.Kirsten JH, Xiong Y, Davis CT, Singleton CK. 2008. Subcellular localization of ammonium transporters in Dictyostelium discoideum. BMC Cell Biol 9:71. doi: 10.1186/1471-2121-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barisch C, Paschke P, Hagedorn M, Maniak M, Soldati T. 2015. Lipid droplet dynamics at early stages of Mycobacterium marinum infection in Dictyostelium. Cell Microbiol 17:1332–1349. doi: 10.1111/cmi.12437. [DOI] [PubMed] [Google Scholar]
- 57.Finsel I, Hoffmann C, Hilbi H. 2013. Immunomagnetic purification of fluorescent Legionella-containing vacuoles. Methods Mol Biol 983:431–443. doi: 10.1007/978-1-62703-302-2_24. [DOI] [PubMed] [Google Scholar]
- 58.Hoffmann C, Finsel I, Hilbi H. 2013. Pathogen vacuole purification from Legionella-infected amoeba and macrophages. Methods Mol Biol 954:309–321. doi: 10.1007/978-1-62703-161-5_18. [DOI] [PubMed] [Google Scholar]
- 59.Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 60.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. 2009. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherwood RK, Roy CR. 2013. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buckley CM, Gopaldass N, Bosmani C, Johnston SA, Soldati T, Insall RH, King JS. 2016. WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. Proc Natl Acad Sci U S A 113:E5906–E5915. doi: 10.1073/pnas.1524532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nagele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ. 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol 19:1214–1225. doi: 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seaman MN, Marcusson EG, Cereghino JL, Emr SD. 1997. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol 137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beum PV, Lindorfer MA, Hall BE, George TC, Frost K, Morrissey PJ, Taylor RP. 2006. Quantitative analysis of protein co-localization on B cells opsonized with rituximab and complement using the ImageStream multispectral imaging flow cytometer. J Immunol Methods 317:90–99. doi: 10.1016/j.jim.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 66.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. 2006. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagedorn M, Soldati T. 2007. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol 9:2716–2733. doi: 10.1111/j.1462-5822.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 68.Hagedorn M, Rohde KH, Russell DG, Soldati T. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lampe EO, Brenz Y, Herrmann L, Repnik U, Griffiths G, Zingmark C, Sjostedt A, Winther-Larsen HC, Hagedorn M. 2015. Dissection of Francisella-host cell interactions in Dictyostelium discoideum. Appl Environ Microbiol 82:1586–1598. doi: 10.1128/AEM.02950-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brenz Y, Ohnezeit D, Winther-Larsen HC, Hagedorn M. 2017. Nramp1 and NrampB contribute to resistance against Francisella in Dictyostelium. Front Cell Infect Microbiol 7:282. doi: 10.3389/fcimb.2017.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH. 2013. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev Cell 24:169–181. doi: 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 72.Weber S, Hilbi H. 2014. Live cell imaging of phosphoinositide dynamics during Legionella infection. Methods Mol Biol 1197:153–167. doi: 10.1007/978-1-4939-1261-2_9. [DOI] [PubMed] [Google Scholar]
- 73.Derre I, Isberg RR. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect Immun 72:3048–3053. doi: 10.1128/IAI.72.5.3048-3053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Segal G, Shuman HA. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol 30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 75.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veltman DM, Akar G, Bosgraaf L, Van Haastert PJM. 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid 61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 77.Tiaden A, Spirig T, Weber SS, Brüggemann H, Bosshard R, Buchrieser C, Hilbi H. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.